ABSTRACT
Endothelial-mesenchymal transition (EndoMT) is associated with damage to blood-brain barrier (BBB) integrity. Circular RNAs (circRNAs) are highly expressed in the brain and are involved in brain diseases; however, whether circRNAs regulate the EndoMT in the brain remains unknown. Our study demonstrated that circHECW2 regulated the EndoMT by directly binding to MIR30D, a significantly downregulated miRNA from miRNA profiling, which subsequently caused an increased expression of ATG5. These findings shed new light on the understanding of the noncanonical role of ATG5 in the EndoMT induced by methamphetamine (Meth) or lipopolysaccharide (LPS). The in vivo relevance was confirmed as microinjection of circHecw2 siRNA lentivirus into the mouse hippocampus suppressed the EndoMT induced by LPS. These findings provide novel insights regarding the contribution of circHECW2 to the nonautophagic role of ATG5 in the EndoMT process in the context of drug abuse and the broad range of neuroinflammatory disorders.
KEYWORDS: ATG5, circular RNA HECW2, EndoMT, endothelial cell, LPS, methamphetamine, MIR30D, nonautophagy
Abbreviations
- ACTA2
actin, alpha 2, smooth muscle, aorta
- ACTB
actin beta
- ATG5
autophagy-related 5
- BBB
blood-brain barrier
- BECN1
beclin 1
- circRNA
circular RNA
- circHECW2
circular HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2
- CLDN5
claudin 5
- COL1A2
collagen type I alpha 2 chain
- EC
endothelial cell
- EndoMT
endothelial-mesenchymal transition
- FISH
fluorescence in situ hybridization
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HBMECs
human brain microvascular endothelial cells
- LPS
lipopolysaccharide
- MAP1LC3B
microtubule associated protein 1 light chain 3 beta
- Meth
methamphetamine
- MIR30D
microRNA30d-5p
- miRs
microRNAs
- OCLN
occludin
- PCR
polymerase chain reaction
- SQSTM1
sequestosome 1
- TJP
tight junction protein
- TJP1
tight junction protein 1
- UTR
untranslated region
- WT
wild type
Introduction
It has been established that the disruption of the blood-brain barrier (BBB) integrity is involved in the context of drug abuse and other brain disorders [1–6]; however, the mechanisms that underlie the damage of the BBB integrity remain unclear. Under this pathological condition, cerebral microvascular endothelial cells lose their protective function, which leads to BBB damage. Endothelial to mesenchymal transition (EndoMT) is a dynamic process by which endothelial cells (ECs) dedifferentiate into mesenchymal cells and, as a result, lose their specific phenotype and function [7]. Dedifferentiation of endothelial cells to mesenchymal cells has also been identified in brain disorders, particularly multiple sclerosis (MS), thus lending credence to the hypothesis that the EndoMT may contribute to BBB damage [8]. However, few studies have investigated the potential role of the EndoMT in BBB damage during drug abuse and the broad neuroinflammation context.
MicroRNAs (miRNAs) are involved in numerous central nervous system pathologies, including brain tumors, neurodegeneration and MS [9–12]. Moreover, miRNAs are envisioned as therapeutic strategies to treat neurovascular-dependent brain diseases because a number of miRNAs have been implicated in the regulation of EC function under normal and inflammatory conditions [13]. A previous study demonstrates that Mir31 acts as a positive modulator of the EndoMT in mouse pancreatic microvascular endothelial cells [14]. However, the specific roles of miRNAs in the EndoMT in the brain are largely unknown. Therefore, miRNA profiling was performed to identify the specific miRNA in the cell function regulation of human brain microvascular endothelial cells (HBMECs) treated with methamphetamine (Meth). MIR30D was characterized because its downregulation was validated in HBMECs treated with Meth or lipopolysaccharide (LPS). Thus, we deemed it necessary to identify the downstream target of MIR30D and gain further insights regarding whether MIR30D plays a role in the EndoMT.
A genome-wide bioinformatic analysis indicated that the circular RNA HECW2 (circHECW2), derived from exon 12 and exon 13 of the HECW2 gene, acts as a sponge for MIR30D. Circular RNAs (circRNAs), generated from back-spliced exons, have recently been identified as a naturally occurring family of noncoding RNAs that are highly represented in the eukaryotic transcriptome [15]. circRNAs are highly homologous and, in general, more stable than their linear counterparts because they lack accessible ends and are thus resistant to exonucleases [16]. Several classes of noncoding RNAs are involved in the regulation of physiological and pathophysiological processes, including neural development and plasticity, Alzheimer disease, heart senescence, hypertrophy and failure, and cell growth [17–22]. However, whether circHECW2 is involved in the EndoMT remains unknown, and more extensive studies are required.
Therefore, our current study demonstrated that the circHECW2-MIR30D axis was involved in the nonautophagic role of ATG5 in the EndoMT process, which provides a potential novel therapeutic strategy for cerebrovascular damage in the context of drug abuse and broad neuroinflammation-related diseases.
Results
MIR30D is downregulated in vitro and in vivo
As miRNAs are signaling factors that affect cerebrovascular permeability by regulating structural components [23,24], we aimed to identify the potential role of specific miRNAs in the cerebrovascular dysfunction induced by Meth using a microRNA array. Variations in the miRNA expression between the controls and samples were shown with a heat map and scatter plot, MIR30D was significantly downregulated (up to 53.56-fold) (Figure 1A and B). The reduction in MIR30D was further confirmed via real-time PCR in HBMECs treated with Meth (Figure 1C). Interestingly, this finding was also validated in HBMECs stimulated with LPS (Figure 1D). Consistent with this in vitro finding, LPS administration in mice significantly decreased Mir30d expression in the hippocampus (Figure 1E). A TargetScan algorithm was subsequently applied to predict the target genes of MIR30D. As shown in Figure S1, the ATG5 (autophagy-related 5) gene that has a conserved MIR30D binding site within its 3'-UTR in most species, was identified as the putative target of MIR30D. Intriguingly, cotransfection of a MIR30D-overexpressing vector and pmiR-GLO plasmid with the ATG5 WT 3'-UTR resulted in the downregulation of luciferase activity, and this effect was reversed in HEK293T cells transfected with a mutated ATG5 3′-UTR (ATG5 3'-UTRΔ) (Figure 1F). Consistent with this finding, MIR30D decreased ATG5 expression, whereas anti-MIR30D increased its expression in HBMECs at the protein level (Figure 1G).
Figure 1.
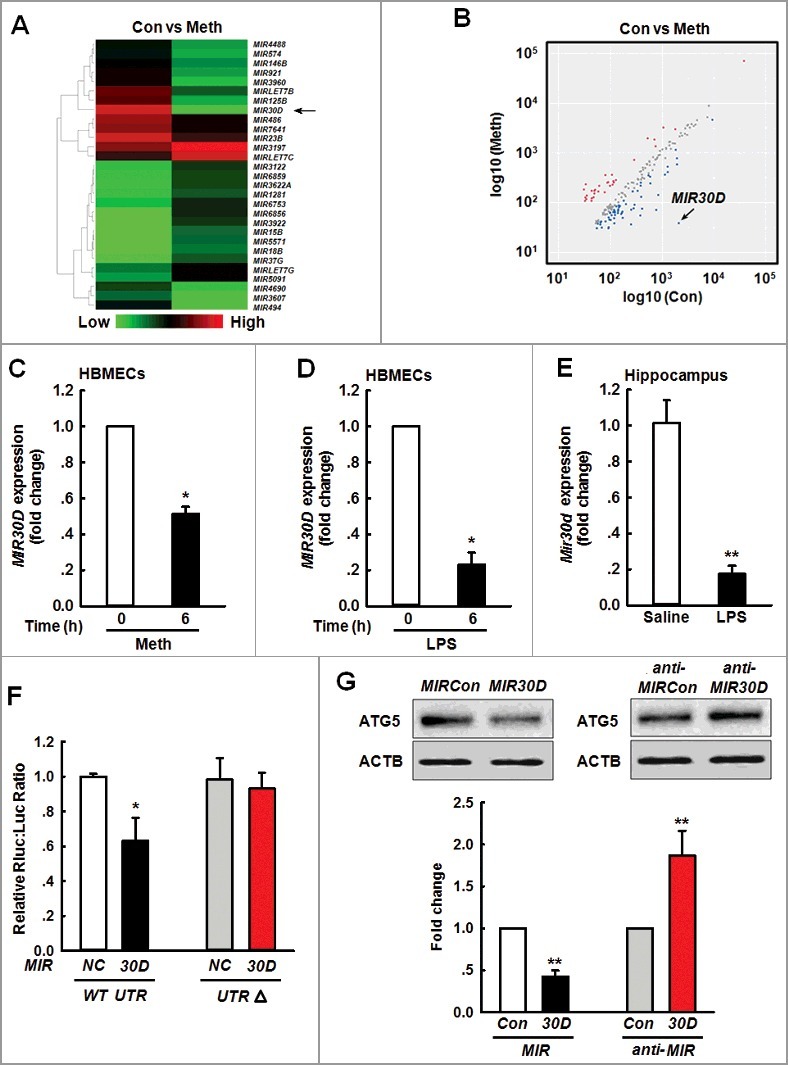
MIR30D is downregulated in vitro and in vivo. (A) Heat map indicates the differential expressions of microRNAs in HBMECs between control and Meth-treated groups. MIR30D expression is downregulated in HBMECs. Down- and upregulated genes are indicated by green and red, respectively. (B) Scatter plot represents the visualization method used to assess variations in microRNA expression between control and Meth-treated groups. The values for the X and Y axes represent normalized signal values (log10 scale). 6 h after the treatment of HBMECs with Meth (100 μM), RNA was extracted and pooled from triplicate samples in each control and Meth-treated group. (C and D) Effect of Meth or LPS on MIR30D expression in HBMECs. HBMECs were treated with Meth (100 μM) or LPS (10 ng/ml) for 6 h, followed by RNA extraction. All data are presented as the mean ± SD of 4 independent experiments. *P<0.05 vs. vehicle control using the Mann-Whitney test. (E) Effect of LPS on Mir30D expression in the hippocampus of mice. Mice were treated with saline or LPS (0.83 mg/kg) once per day for 7 d. n = 6 animals/group. **P<0.01 vs. saline group using the Mann-Whitney test. (F) Relative luciferase activity of wild-type and 3'-UTRΔ mutant constructs of ATG5 cotransfected with a MIR30D overexpression vector and pmiR-GLO plasmid. All data are presented as the mean ± SD of 3 individual experiments. *P<0.05 vs. the MIRCon cotransfected with the WT construct via one-way ANOVA followed by the Holm-Sidak test. (G) MIR30D regulated the expression of ATG5 in HBMECs. Representative western blots indicate the ATG5 expression in HBMECs exposed to MIR30D or anti-MIR30D lentivirus. All data are presented as the mean ± SD of 5 independent experiments. **P<0.01 vs. MIRCon group or anti-MIRCon group using the Mann-Whitney test. HBMECs: human brain microvascular endothelial cells; Meth: methamphetamine.
Meth or LPS increases ATG5 expression independent of autophagy
Because ATG5 is the target of MIR30D, we subsequently aimed to explore the effects of Meth or LPS on the autophagy of HBMECs. As shown in Figure 2A, LPS (10 ng/ml) treatment significantly increased the expression of BECN1/Beclin 1 and ATG5/ATG5; however, the same treatment failed to induce the conversion from cytosolic MAP1LC3B/LC3B-I to membrane-bound MAP1LC3B /LC3B-II (Figure 2B). Consistent with this unexpected finding, LPS (10 ng/ml) failed to decrease the expression of SQSTM1/p62, a marker protein that functions as a link between MAP1LC3B and the ubiquitinated substrate [25]; instead, treatment increased its expression (Figure 2B). In addition to LPS treatment, exposure of cells to Meth (100 μM) increased the expression of BECN1 and ATG5 (Figure S2A), but failed to increase the level of MAP1LC3B-II as well as decrease the expression of SQSTM1 (Figure S2B). To rule out false-negative findings, HBMECs were exposed to a higher concentration of Meth or LPS for different time points. As shown in Figure 2C, LPS (100 ng/ml) significantly increased the conversion of MAP1LC3B-II and decreased the expression of SQSTM1, which was further confirmed in the cells treated with Meth (1 mM) (Figure S2C).
Figure 2.
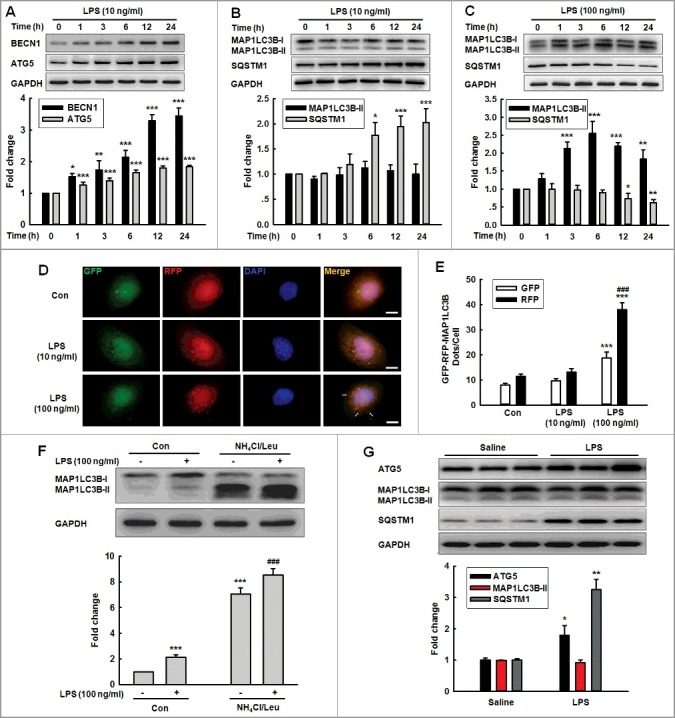
Effects of different concentrations of LPS on the expression of autophagy-associated proteins. (A and B) Effects of LPS (10 ng/ml) on the expression of BECN1 and ATG5 (A), MAP1LC3B-II and SQSTM1 (B). Cells were treated with LPS (10 ng/ml) for different time points. (C) Effects of LPS (100 ng/ml) on the expression of MAP1LC3B-II and SQSTM1. Cells were treated with LPS (100 ng/ml) for different time points. All data are presented as the mean ± SD of 4 independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. vehicle control using one-way ANOVA. (D and E) Effects of LPS on RFP- and GFP-MAP1LC3B puncta (D). HBMECs transduced with the RFP-GFP-MAP1LC3B adenovirus and subsequently treated with different concentrations of LPS for 24 h. The RFP- and GFP-MAP1LC3B puncta per cell were counted, and the quantification is shown in (E). Scale bar: 5 μm. Quantification of cell numbers is presented as the mean ± SD of 3 independent experiments. ***P<0.001 vs. vehicle control. ###P<0.001 vs. the LPS (10 ng/ml)-treated group using one-way ANOVA followed by the Holm-Sidak test. (F) Effect of LPS on the MAP1LC3B-II expression. HBMECs were treated with LPS (100 ng/ml) for 24 h. A lysosomal inhibitor mix (NH4Cl [20 mM], Leu [200 μM]) was added 1 h prior to cell harvesting, and the cell lysate was collected and analyzed via immunoblotting for MAP1LC3B-II accumulation. ***P<0.001 vs. vehicle control. ###P<0.001 vs. the LPS (100 ng/ml)-treated group using one-way ANOVA followed by the Holm-Sidak test. (G) Expression of ATG5, MAP1LC3B-II and SQSTM1 in the hippocampus isolated from mice treated with LPS. C57BL/6J mice were treated with LPS (0.83 mg/kg) or saline once per day for 7 d followed by determination of the expression of autophagic proteins. n = 6 animals/group. *P<0.05, **P<0.01 vs. saline group using the Mann-Whitney test. Leu, leupeptin.
We then directly investigated the macroautophagic/autophagic flux by visualizing the HBMECs transfected with a tandem fluorescent-mRFP-GFP-MAP1LC3B-adenovirus. The colocalization of GFP and RFP (yellow puncta) indicates the lack of fusion phagophores or autophagosomes with lysosomes, whereas the RFP-only signals (red puncta) indicate the presence of autolysosomes. As shown in Figure 2D and E, LPS (10 ng/ml) exposure failed to induce an autophagic influx, whereas there are significantly increased yellow dots per cell accompanied by an increase in RFP-only dots after the LPS (100 ng/ml) treatment; we identified similar evidence following Meth treatment (Figure S2D). Consistent with this finding, treatment with LPS (100 ng/ml) alone caused accumulation of MAP1LC3B-II, and treatment with LPS (100 ng/ml), and NH4Cl/Leu further enhanced the accumulation of MAP1LC3B-II (Figure 2F). This finding was further confirmed in HBMECs treated with Meth (1 mM) (Figure S2E). These findings indicated that the lower concentrations of Meth or LPS increased ATG5 expression without autophagy activation; however, higher concentrations of Meth or LPS increased ATG5 expression and autophagy activation. Consistent with these in vitro findings, the administration of LPS in mice increased ATG5 expression and concomitantly increased SQSTM1 expression without affecting the MAP1LC3B-II level (Figure 2G).
ATG5 is involved in the EndoMT
Following the detection of the surprising phenomenon that ATG5 expression was increased in the HBMECs treated with Meth (100 μM) or LPS (10 ng/ml) without autophagy activation, we aimed to examine the role of ATG5 in endothelial cell dysfunction because Meth or LPS treatment have been determined to cause BBB injury [26,27]. As shown in Figure 3A, the LPS administration significantly decreased the expression of all 3 tight junction proteins (TJPs). The EndoMT is a dynamic process by which ECs dedifferentiate into mesenchymal cells and thus lose their specific phenotype and function; thus, we aimed to examine whether the EndoMT is responsible for EC damage after LPS treatment. As shown in Figure 3B, LPS administration significantly increased the expression of COL1A2/Collagen I and ACTA2/α-SMA (actin, alpha 2, smooth muscle, aorta), which are markers of mesenchymal cells. To verify this finding, the BBB integrity and EndoMT were further confirmed using a TEK/Tie2-GFP transgenic mouse model combined with a 2-photon analysis. As shown in Figure 3C, the Dil-labeled monocytes out of blood vessels in the LPS mice were significantly increased compared with those in the saline mice as accessed via 2-photon laser scanning microscopy (TPLSM). In this same set of experiments, the LPS group exhibited a significantly increased colocalization rate, which was 2.21-fold (Pearson correlation) and 3.07-fold (Manders overlap) higher than that in the saline group (Figure 3D and E).
Figure 3.
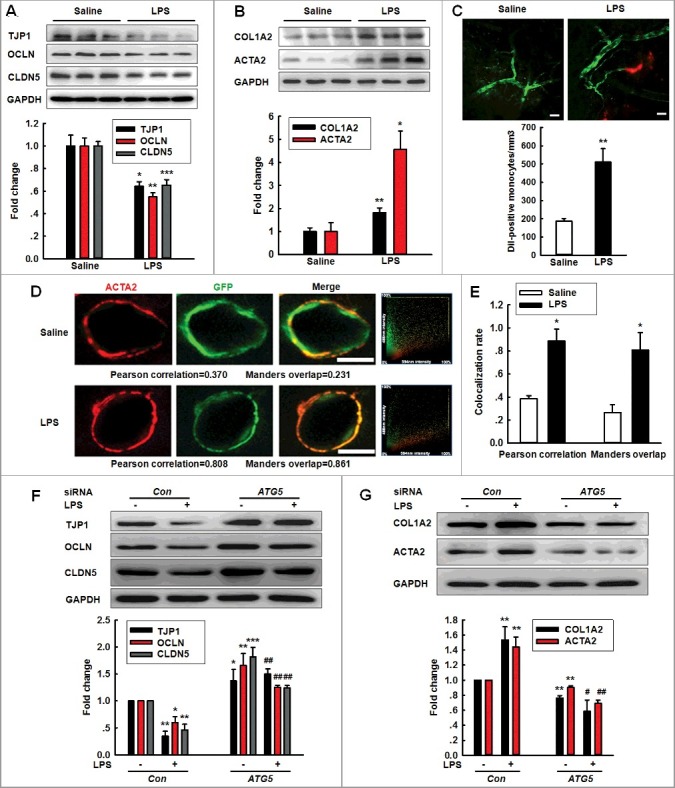
Role of ATG5 in LPS-induced EndoMT. (A and B) Effects of LPS treatment on the expression of TJP (TJP1, OCLN and CLDN5) (A) and mesenchymal cell markers (COL1A2 and ACTA2) (B) in vivo. (C) The number of Dil-labeled monocytes increased out of the blood vessels of LPS-treated TEK-GFP transgenic mice compared with that in the saline treatment. Mice were treated with LPS (0.83 mg/kg) once per day for 7 d and were subsequently accessed via TPLSM. Scale bar: 50 μm. (D and E) Effect of LPS on the GFP+ and ACTA2 colocalization in vivo. Immunostaining for ACTA2 in the hippocampus from TEK-GFP transgenic mice. Z-tack images were acquired on a Carl Zeiss confocal laser scanning microscope (D). The Pearson correlation or Manders overlap of the GFP+ and ACTA2 immunofluorescence was calculated, respectively (E). Scale bar: 5 μm. Mice were administered LPS (0.83 mg/kg) once per day for 7 d. n = 6 animals/group. *P<0.05, **P<0.01, ***P<0.001 vs. saline group using the Mann-Whitney test. (F and G) Transfection of HBMECs with siRNA ATG5 significantly inhibited the decreases in the TJP expression levels (F) and the increases in the mesenchymal cell marker expression (G). HBMECs were cultured for 24 h after the transfection of ATG5 siRNA and were subsequently exposed to LPS (10 ng/ml) for 24 h. All data are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. the siRNA-control group. #P<0.05, ##P<0.01 vs. the LPS-incubated siRNA-control via one-way ANOVA followed by the Holm-Sidak test.
Consistent with the in vivo findings, the exposure of HBMECs to Meth or LPS significantly decreased the expression of TJPs with a concomitantly increased expression of mesenchymal cell markers (Figure S3). To examine the role of ATG5 in the EndoMT, HBMECs were transfected with ATG5 siRNA. As expected, the ATG5 siRNA successfully decreased ATG5 expression (Figure S4A). Knockdown of ATG5 expression significantly inhibited the decreased expression of TJPs induced by Meth or LPS (Figure S4B and Figure 3F). Consistently, Meth or LPS treatment displayed increased mesenchymal cell marker expression, which was significantly ameliorated by the ATG5 siRNA transfection (Figure S4C and Figure 3G).
Engagement of the MIR30D-ATG5 axis in the EndoMT
Following the determination that MIR30D regulates ATG5 expression, we aimed to examine whether MIR30D was involved in the EndoMT induced by Meth or LPS. As shown in Figure S5A and Figure 4A, the transduction of cells with MIR30D lentivirus significantly ameliorated the decreases in the TJP expression induced by Meth or LPS. In contrast, the increases in the mesenchymal cell marker expression were significantly inhibited by the transduction of HBMECs with MIR30D lentivirus (Figure S5B and Figure 4B). We subsequently aimed to examine whether ATG5 is the functional target of MIR30D. The transduction of HBMECs with anti-MIR30D lentivirus decreased the expression of TJPs, and this effect was significantly inhibited in cells cotransduced with the lentiviral vectors that expressed anti-MIR30D and ATG5 siRNA (Figure 4C). Moreover, the transduction of HBMECs with anti-MIR30D lentivirus resulted in enhanced mesenchymal cell marker expression, and this effect was inhibited in cells cotransduced with the lentiviral vectors that expressed anti-MIR30D and ATG5 siRNA (Figure 4D). These results indicate that ATG5 acts as a functional target of MIR30D to regulate the EndoMT.
Figure 4.
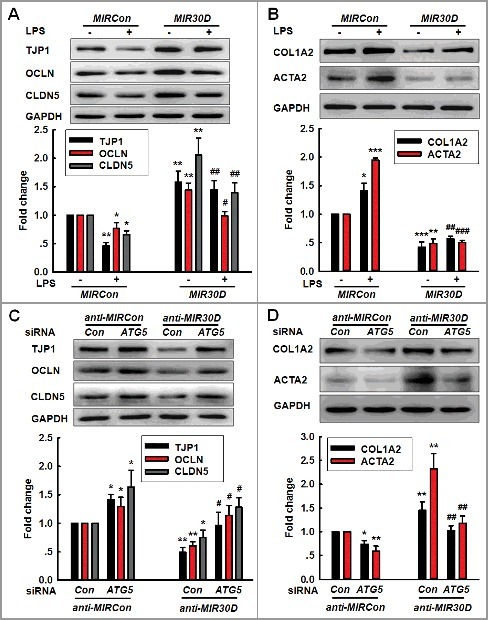
The MIR30D-ATG5 axis plays an important role in the EndoMT. (A and B) Transduction of HBMECs with MIR30D lentivirus significantly inhibited the LPS-induced decreases in the TJP expression levels (A) and increases in the mesenchymal cell marker expression (B). HBMECs were transduced with the MIR30D lentivirus for 24 h and were subsequently incubated with LPS (10 ng/ml) for 24 h. All data are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. the MIRCon group. #P<0.05, ##P<0.01, ###P<0.001 vs. the LPS-treated MIRCon group via one-way ANOVA followed by the Holm-Sidak test. (C and D) Transduction of HBMECs with the ATG5 siRNA significantly inhibited the decreases in the TJP expression levels (C) and the increases in the mesenchymal cell marker expression (D) induced by anti-MIR30D. HBMECs were cotransduced with anti-MIRCon or anti-MIR30D and either control siRNA or ATG5 siRNA for 24 h. All data are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01 vs. the anti-MIRCon group with the control siRNA. #P<0.05, ##P<0.01 vs. the anti-MIR30D cotransduced with the control siRNA via one-way ANOVA followed by the Holm-Sidak test.
circHECW2 functions to bind MIR30D
After determining the role of the MIR30D-ATG5 axis in the EndoMT, we subsequently obtained insight into the mechanisms of MIR30D regulation. Since circRNAs act as competing endogenous RNA (ceRNA) sponges to interact with miRNAs and influence their activity, we aimed to examine which circRNA binds MIR30D. Using a bioinformatic program, RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid), we determined that circHECW2 contains one MIR30D target site (Figure 5A). Consistent with the prediction, we subsequently accessed whether circHECW2 is involved in the EndoMT induced by different stimuli via real-time PCR using divergent primers (Figure 5B). The treatment of HBMECs with Meth or LPS significantly increased the circHECW2 expression in HBMECs (Figure 5C and D). This finding was further confirmed in LPS-treated mice as there was an increased expression of circHecw2 in the hippocampus (Figure 5E). Furthermore, we applied a biotin-coupled MIR30D mimic to determine whether MIR30D was able to pull down circHECW2. We identified an enrichment of circHECW2 in the MIR30D-captured fraction compared with that observed following the introduction of mutations that disrupt base paring between circHECW2 and MIR30D (Figure 5F). In contrast, by applying a biotin-labeled specific probe, MIR30D was pulled down by circHECW2 (Figure 5G). Furthermore, the double in situ hybridization indicated that there was a colocalization of circHECW2 and MIR30D in HBMECs (Figure 5H).
Figure 5.
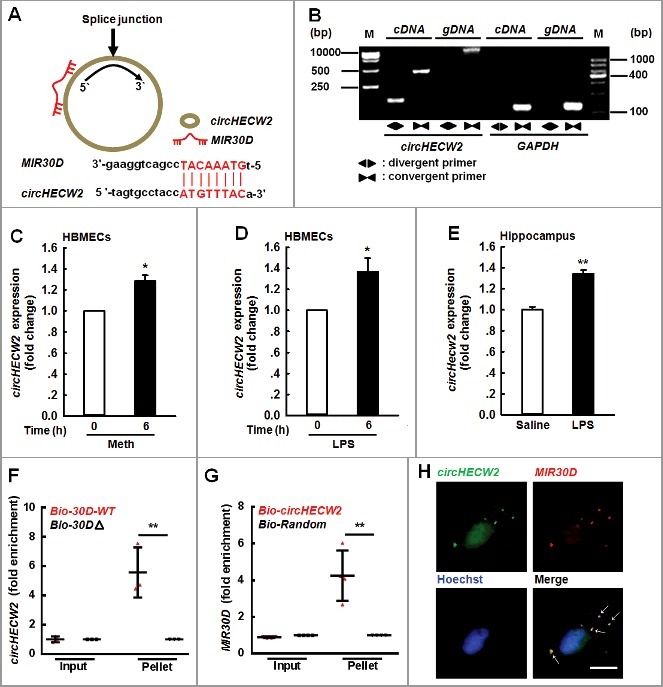
circHECW2 sponges MIR30D. (A) circHECW2 contained one site complementary to MIR30D. The potential complementary residues are presented in red, as determined using the bioinformatic program RNAhybrid. (B) Divergent primers amplified circHECW2 in cDNA but not genomic DNA (gDNA). GAPDH, linear control. (C and D) Effects of Meth or LPS on circHECW2 expression in HBMECs. Real-time PCR was performed to measure the circHECW2 expression levels in HBMECs treated with Meth (100 μM) (C) or LPS (10 ng/ml) (D) for 6 h. All data are presented as the mean ± SD of 4 independent experiments. *P<0.05 vs. vehicle control using the Mann-Whitney test. (E) Effect of LPS on circHecw2 expression in the hippocampus of mice. Mice were administered LPS (0.83 mg/kg) once per day for 7 d. n = 6 animals/group. **P<0.01 vs. saline group using the Mann-Whitney test. (F) Application of a biotin-coupled MIR30D mimic to determine whether MIR30D was able to pull down circHECW2. The biotinylated wild-type MIR30D (Bio-MIR30D-WT) or its biotinylated mutant (Bio-MIR30DΔ) was transfected into HEK293T cells. After streptavidin capture, the circHECW2 and GAPDH mRNA levels were quantified via real-time PCR, and the relative immunoprecipitate/input ratios were plotted. All data are presented as the mean ± SD of 3 independent experiments. **P<0.01 vs. the Bio-MIR30D-WT group via one-way ANOVA followed by the Holm-Sidak test. (G) Following the application of a biotin-labeled specific probe, MIR30D was pulled down by circHECW2. The biotinylated circHECW2 (Bio-circHECW2) or circRNA negative control (Bio-Random) was transfected into HEK293T cells. After streptavidin capture, the MIR30D and RNU6-6P microRNA levels were quantified via real-time PCR, and the relative immunoprecipitate/input ratios were plotted. All data are presented as the mean ± SD of 4 independent experiments. **P<0.01 vs. the Bio-Random group via one-way ANOVA followed by the Holm-Sidak test. (H) Double-label in situ hybridization assays showed that circHECW2 colocalized with MIR30D. Scale bar: 10 μm. bp, base pair; M, size markers; Meth, methamphetamine.
Knockdown of circHECW2 expression inhibits the EndoMT by targeting MIR30D in vitro
After determining that circHECW2 binds with MIR30D, we subsequently aimed to examine the role of circHECW2 in the EndoMT. As shown in Figure S6, the knockdown of circHECW2 successfully decreased the expression of circHECW2 in HBMECs. We then examined the effect of circHECW2 on the EndoMT. As shown in Figure 6A, the transduction of cells with circHECW2 siRNA significantly ameliorated the decreases in the TJP expression induced by LPS. In contrast, the increased mesenchymal cell marker expressions induced by LPS were significantly inhibited by the transduction of HBMECs with circHECW2 siRNA (Figure 6B). Consistent with this finding, the knockdown of the expression of circHECW2 also inhibited the EndoMT induced by Meth (Figure S7A and B). To verify whether MIR30D is a mediator of circHECW2, the transduction of HBMECs with anti-MIR30D significantly inhibited the decrease in the ATG5 expression induced by circHECW2 siRNA lentivirus (Figure S8). Expectedly, the transduction of HBMECs with anti-MIR30D significantly inhibited the increases in the TJP expression induced by circHECW2 siRNA lentivirus (Figure 6C). Moreover, the transduction of HBMECs with circHECW2 siRNA lentivirus resulted in decreased mesenchymal cell marker expression, and this effect was inhibited in cells cotransduced with the lentiviral vectors that expressed circHECW2 siRNA and anti-MIR30D (Figure 6D). These results indicate that circHECW2 acts as an endogenous MIR30D sponge to regulate the EndoMT.
Figure 6.
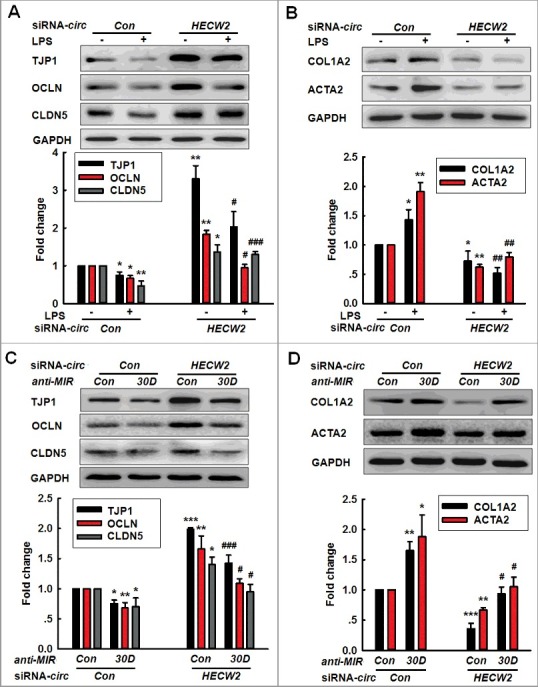
Knockdown of circHECW2 expression inhibits the EndoMT by targeting MIR30D in vitro. (A and B) Transduction of HBMECs with circHECW2 siRNA lentivirus significantly inhibited the LPS-induced decreases in the TJP expression levels (A) and the increases in the mesenchymal cell marker expression (B). HBMECs were transduced with the circHECW2 lentivirus for 24 h and were subsequently incubated with LPS (10 ng/ml) for 24 h. All data are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01 vs. the circCon siRNA group. #P<0.05, ##P<0.01, ###P<0.001 vs. the LPS-treated circCon siRNA via one-way ANOVA followed by the Holm-Sidak test. (C and D) Transduction of HBMECs with the anti-MIR30D lentivirus significantly inhibited the increases in the TJP expression levels (C) and the decreases in the mesenchymal cell marker expression (D) induced by circHECW2 siRNA. HBMECs were cotransduced with anti-MIR30D and circHECW2 siRNA for 24 h. All data are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. the circCon siRNA cotransduced with the anti-MIRCon group. #P<0.05, ###P<0.001 vs. the circHECW2 siRNA cotransduced with the anti-MIRCon group via a one-way ANOVA followed by the Holm-Sidak test.
Knockdown of circHecw2 expression inhibits the EndoMT in vivo
After determining that circHECW2 has an important influence on the EndoMT in vitro, we subsequently aimed to examine the in vivo relevance of circHECW2 by microinjecting a circHecw2 siRNA lentivirus into the hippocampus of C57BL/6J mice as illustrated in Figure 7A; the distribution of GFP+ lentiviruses was tracked in the hippocampus as expected (Figure 7B). We then examined the efficacy of circHecw2-GFP siRNA lentivirus transfection in vivo. As shown in Figure 7C, circHecw2 expression was decreased in the circHecw2 siRNA-microinjected side compared with that in the circCon siRNA-microinjected side. circHecw2 siRNA significantly ameliorated the decreases in the TJP expression induced by LPS (Figure 7D and E). In contrast, the increased expression of mesenchymal cell markers induced by LPS treatment was significantly inhibited with circHecw2 siRNA-microinjected (Figure 7F and G).
Figure 7.
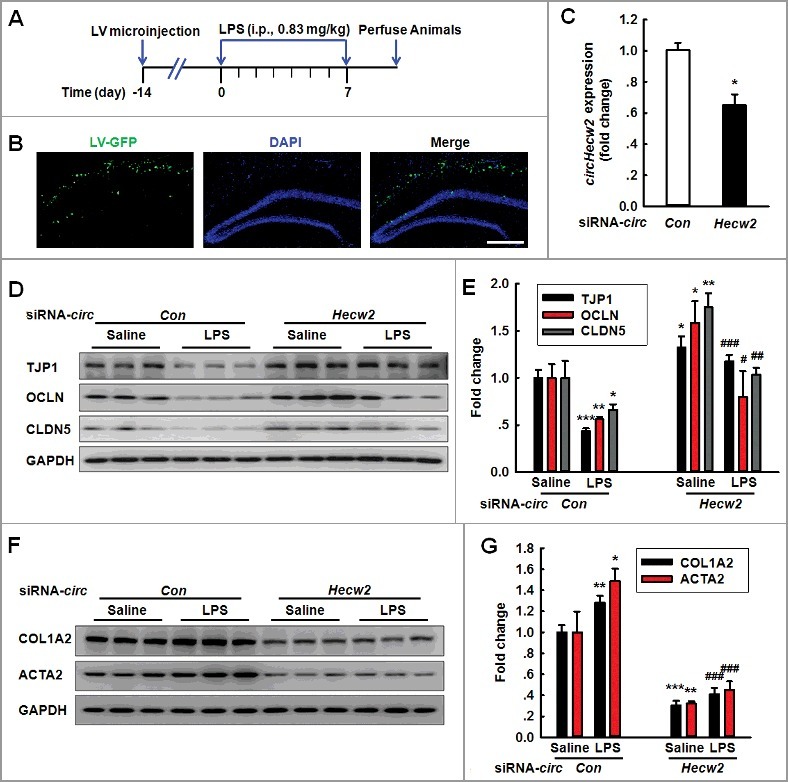
Knockdown of circHecw2 inhibits the EndoMT in vivo. (A) Schematic diagram presents the procedure for microinjection of circHecw2 siRNA lentivirus prior to LPS treatment in mice. (B) Representative images of C57BL/6J mice bilaterally microinjected with siRNA-GFP lentivirus (1 μl of 109 viral genomes μl−1) in the hippocampus. Scale bar: 200 μm. (C) circHecw2 siRNA lentivirus injection successfully decreased circHecw2 expression. Two wk after the microinjection, mice were sacrificed and assessed regarding the circHecw2 level as determined via real-time PCR. n = 6 animals/group. *P<0.05 vs. circCon using the Mann-Whitney test. (D to G) Microinjection of C57BL/6J mice with circHecw2 siRNA lentivirus significantly inhibited the LPS-induced decreases in the TJP expression levels (D and E) and the increases in the mesenchymal cell marker expression (F and G). Mice were microinjected with the circHecw2 siRNA or circCon siRNA lentivirus for 2 wk and were subsequently treated with LPS (0.83 mg/kg) once per day for 7 d. n = 6 animals/group. *P<0.05, **P<0.01, ***P<0.001 vs. the saline-treated circCon siRNA group. #P<0.05, ##P<0.01,###P<0.001 vs. the LPS-treated circCon siRNA group via one-way ANOVA followed by the Holm-Sidak test.
Involvement of the NOTCH1/Notch1 pathway in the circHECW2-MIR30D-ATG5-mediated EndoMT
NOTCH1/Notch1 signaling is involved in the EndoMT in cardiac ECs and kidney fibrosis [28–31]; thus, we subsequently examined whether this pathway was involved in the EndoMT regulated by the circHECW2-MIR30D-ATG5 axis. As shown in Figure 8A, LPS treatment significantly increased the NOTCH1/NICD (notch intracellular domain) level. ATG5 siRNA inhibited the enhanced NOTCH1 level induced by anti-MIR30D (Figure 8B). We subsequently examined the effect of circHECW2 on the NOTCH1 signaling pathway. As shown in Figure 8C, the transduction of HBMECs with circHECW2 siRNA lentivirus significantly ameliorated the increased NOTCH1 level induced by LPS. The transduction of HBMECs with circHECW2 siRNA lentivirus resulted in a decreased NOTCH1 level, and this effect was inhibited in cells cotransfected with the lentiviral vectors that expressed circHECW2 siRNA and anti-MIR30D (Figure 8D). These results indicate that circHECW2 acts as an endogenous MIR30D sponge to regulate NOTCH1 signaling.
Figure 8.
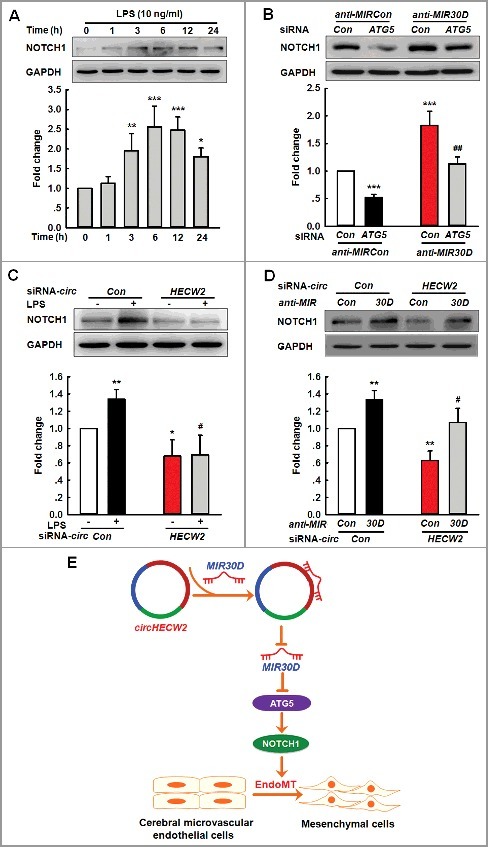
Involvement of the NOTCH1 pathway in the circHECW2-MIR30D-ATG5-mediated EndoMT. (A) LPS induced an increased cleavage of NOTCH1/NICD from NOTCH1. Cells were treated with LPS (10 ng/ml) for different time points. All data are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. the vehicle control via one-way ANOVA. (B) Transduction of HBMECs with ATG5 siRNA significantly inhibited the increases in the NOTCH1 level induced by anti-MIR30D lentivirus. HBMECs were cotransduced with anti-MIR30D and ATG5 siRNA for 24 h. All data are presented as the mean ± SD of 3 independent experiments. ***P<0.001 vs. the anti-MIRCon cotransduced with the control siRNA group. ##P<0.01 vs. the anti-MIR30D cotransduced with the ATG5 siRNA via one-way ANOVA followed by the Holm-Sidak test. (C) Transduction of HBMECs with circHECW2 siRNA lentivirus significantly inhibited the level of NOTCH1 induced by LPS. HBMECs were transduced with the circHECW2 siRNA or circCon siRNA for 24 h and were subsequently incubated with LPS (10 ng/ml). All data are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01 vs. the circCon siRNA group via one-way ANOVA. #P<0.05 vs. the LPS-treated circCon siRNA group via one-way ANOVA followed by the Holm-Sidak test. (D) Transduction of HBMECs with the anti-MIR30D lentivirus significantly inhibited the decreases in the NOTCH1 level induced by circHECW2 siRNA. HBMECs were cotransduced with anti-MIR30D and circHECW2 siRNA. All data are presented as the mean ± SD of 3 independent experiments. **P<0.01 vs. the circCon siRNA with anti-MIRCon-cotransduced group. #P<0.05 vs. the circHECW2 siRNA with anti-MIRCon-cotransduced group via one-way ANOVA followed by the Holm-Sidak test. (E) circHECW2, identified as ceRNA, functioned as an endogenous MIR30D sponge to sequester MIR30D and inhibited its activity, which resulted in increased ATG5 expression and consequent EndoMT via the NOTCH1 signaling pathway.
Discussion
Our study provides novel insights into the function of circHECW2, which was shown to be involved in the EndoMT induced by drug abuse and another inflammatory stimulus, LPS by binding with MIR30D and subsequently targeting ATG5 (Figure 8E). Another novel finding is the nonautophagic role of ATG5 in the EndoMT that contributes to cerebrovascular damage. Specific blockade of circHECW2 may be envisioned as a potential therapeutic target for the treatment of BBB damage.
Although miRNAs are envisioned as therapeutic strategies for the treatment of neurovascular-dependent brain diseases [13], the specific roles of miRNAs in the EndoMT in the brain are largely unknown. In addition to Mir31 involved in the EndoMT, other miRNAs, including MIR21, MIR23, MIR20A and MIR302C are also involved in the EndoMT in peripheral cells, such as cardiac endothelial cells and hepatocellular carcinoma cells [32–36]. The rationale for our selection of MIR30D was as follows: 1) MIR30D screened from a miRNA array was significantly decreased in the Meth-treated group compared with that in the control group; 2) the decreased expression of MIR30D was validated in HBMECs treated with Meth, which was also expanded in LPS-treated HBMECs. MIR30D plays a critical role in tumor cell proliferation and motility, cardiomyocyte apoptosis and the epithelial-mesenchymal transition [37–39]; however, whether MIR30D is involved in the regulation of the EndoMT has remained largely unknown. To the best of our knowledge, this study is the first investigation to demonstrate that MIR30D is involved in the EndoMT in drug abuse and LPS-induced neuroinflammation. In this study, MIR30D was initially identified using a bioinformatic approach; definitive proof of whether MIR30D is beneficial and its detailed mechanisms may be determined only with gain- or loss-of-function experiments in animal models with Meth or LPS injection in future studies.
Consistent with the TargetScan prediction and a previous study [40], our current study further confirmed that ATG5 was the target of MIR30D. ATG5, an essential component for canonical autophagy, plays nonautophagic roles in innate antiviral immune responses, T cell survival and proliferation [41]., [42] In this study, to our surprise, treatment with Meth (100 μM) or LPS (10 ng/ml) enhanced the ATG5 level but did not increase the level of autophagy as accessed via MAP1LC3B-II and SQSTM1 immunoblotting. Therefore, the function of ATG5 in this context remains questionable. Whether ATG5 is involved in the BBB integrity also remains elusive. To the best of our knowledge, this study is the first demonstration that ATG5, as the target of MIR30D, functions in the EndoMT induced by Meth or LPS, which lends credence to our speculation that MIR30D and ATG5 contributed to the EndoMT during BBB damage. To rule out false-negative findings, we also performed additional experiments with higher concentrations of Meth (1 mM) or LPS (100 ng/ml) as these treatments induced the activation of autophagy in HBMECs. These results indicated that in contrast to the lower concentrations of Meth or LPS, canonical autophagy was induced by the higher concentrations. Our findings are consistent with a previous study which indicates that for reprogramming of induced pluripotent stem cells (iPSCs), different reprogramming induction strengths with different concentrations of doxycycline determine the reprogramming pathway in ATG5-dependent canonical autophagy [43]. Our study also shares features with the noncanonical role of ATG5 proposed in a previous study in which the upregulation of ATG5 mediated by DNA-damaging agents promotes mitotic catastrophe independent of autophagy [44]. The detailed mechanisms of the noncanonical role of ATG5 remain largely unknown, and specific tools are lacking; thus, it is technically challenging to thoroughly characterize the noncanonical role of ATG5 in the EndoMT.
Although the functions of circRNAs are largely unknown, accumulating evidence suggests that they are key regulators of numerous biological functions in neural development and plasticity, as well as Alzheimer disease [21,22]; however, whether they are involved in the pathogenesis of drug abuse or in the broad neuroinflammation context has not currently been determined. Consistent with the findings of previous studies [45], [46] the current study indicated that circHECW2 acts as a MIR30D sponge to regulate the EndoMT and downregulates the expression of circHecw2, attenuating cerebrovascular damage in mice treated with LPS. The interaction between MIR30D and circHECW2 was confirmed by miRNA affinity isolation and double fluorescence in situ hybridization (FISH) assays. In addition to acting as sponges to interact with miRNAs, circRNAs exert their functions via another mechanism as illustrated in a recent study which indicates that circFox3 regulates cell cycle progression by conjugating with CDKN1A/p21 and CDK2 [47]. Although MIR30D is regulated by circHECW2, we could not rule out the other mechanisms that contribute to the functions of circHECW2.
circRNAs have been identified to play a specific role in the epithelial-mesenchymal transition [48]; however, none of these studies have investigated the role of circRNAs in the EndoMT. Increasing evidence indicates the role of the EndoMT in cardiac development and various pathological conditions [49–52]; however, most studies regarding the EndoMT have been performed in embryonic development or nonbrain-derived endothelial cells. To the best of our knowledge, this study is the first investigation to demonstrate that the EndoMT contributed to cerebrovascular damage in drug abuse or a broader neuroinflammation induced by LPS. Moreover, a TEK-GFP genetic mouse model was applied for a colocalization analysis as the mesenchymal cell marker ACTA2 colocalized with TEK-GFP. The molecular mechanisms involved in the switch from an endothelial phenotype to a mesenchymal status are complicated and controlled by various signaling pathways, such as the WNT, NOTCH and TGFB pathways [53–55]. Consistent with a previous study [54], NOTCH1 signaling lies downstream of circHECW2-MIR30D-ATG5 as shown in our current study. Given that ATG5 contains substantial numbers of acidic residues, a potential glycosylation site, protein kinase C and tyrosine kinase sites [56], further investigation was required to identify the precise mechanism that underlies the activation of the NOTCH1 pathway induced by circHECW2-MIR30D-ATG5.
In summary, circHECW2 was involved in the EndoMT that results in the damage of BBB integrity via the suppression of MIR30D activity as an endogenous MIR30D sponge and subsequent downstream activation of ATG5. The circHECW2-MIR30D axis thus lies upstream of the nonautophagic role of ATG5 in the EndoMT. Specific blockage of circHECW2 may be envisioned as a potential therapeutic target for the treatment of BBB integrity damage.
Materials and methods
Reagents
The lentiviral vectors that carried MIR30D overexpression, anti-MIR30D, circHECW2 siRNA and an adenoviral vector that carried mRFP-GFP-MAP1LC3B (HB-AP2100001) were obtained from HANBIO (Shanghai, China). The control siRNA (sc-37007) and human ATG5 siRNA (sc-41445) were acquired from Santa Cruz Biotechnology. Leupeptin (L2884), NH4Cl (A9434) and LPS (L2630) were obtained from Sigma-Aldrich. Methamphetamine was purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Lot, 171212–201104; ID, 26CYK7B6).
Animals
C57BL/6J mice (male, 5 to 6 wk) and STOCK TEK-GFP 287Sato/JNju mice were obtained from the Laboratory Animal Center of Nanjing Medical University (Nanjing, China); the GFP expresses only in endothelial cells. All animals were housed at a constant temperature and humidity with a 12 h light: 12 h dark cycle, with lights on at 08:00 am. Food and water were available ad libitum. The animals were deeply anesthetized with Nembutal, followed by perfusion. All animal procedures were conducted according to the protocols approved by the Institutional Animal Care and Use Committee of the Medical School, Southeast University.
Cell cultures
Primary human brain microvascular endothelial cells (HBMECs) were purchased from ScienCell, cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Life Technologies, 22400089) and used at passages 4 to 14. The culture was incubated to confluence in T25 flasks in a humid atmosphere at 37°C with 5% CO2. The cultured cells were negative for HIV-1, HBV, HCV, mycoplasma, bacteria, yeast, and fungi.
MicroRNA microarray assay
The microarray assay was performed by a service provider (LC-Bio, Hangzhou, China) as follow. HBMECs were treated with Meth at a concentration of 100 μM. After 6 h, RNA was extracted and pooled from triplicate samples in each control and Meth-treated group. The assay was performed with 4 to 5 μg of the total RNA samples, which were 3’-extended with a poly (A) tail using a poly (A) polymerase (NEB, M0276L). An oligonucleotide tag was subsequently ligated to the poly (A) tail for fluorescent dye staining. The hybridization was performed overnight on a μParaflo microfluidic chip using a microcirculation pump (Atactic Technologies, Houston, USA). The detection probes were created via in situ synthesis using photogenerated reagent chemistry. After the RNA hybridization, a tag-conjugating Cy3 dye was circulated through the microfluidic chip for the dye staining. Fluorescence images were collected using a laser scanner (GenePix 4000B, Molecular Device, USA) and digitized using Array-Pro image analysis software (Media Cybernetics). The data were initially analyzed by subtracting the background and then normalizing the signals using a LOWESS filter (locally weighted regression).
The Gene Expression Omnibus (GEO) accession code for the data in this manuscript is GSE96063.
Western blot (WB) analysis
According to our previous studies [57,58] proteins were extracted in a RIPA lysis buffer (Beyotime, P0013B). The proteins were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. After blocking with 5% nonfat dry milk in Tris-buffered saline (20 mM Tris-HCl, 500 mM NaCl, pH7.4) with 0.2% Tween-20 (Aladdin, T104863), the membranes were probed with antibodies overnight at 4°C, followed by incubation with a horseradish peroxidase-conjugated goat anti-mouse (ZSGB-BIO, ZB5305) or goat anti-rabbit (ZSGB-BIO, ZB5301) IgG secondary antibody (1:2000). The antibodies were as follows: anti-ATG5 (sc33210) obtained from Santa Cruz Biotechnology; anti-GAPDH (60004), anti-SQSTM1 (18420), anti-MAP1LC3B/LC3B (14600), anti-NOTCH1 (10062), anti-BECN1 (11306), anti-TJP1 (21773) and anti-ACTA2 (14395) were obtained from Proteintech; anti-COL1A2 (BS1530) was obtained from Bioworld Technology; anti-CLDN5 (AF0130) was obtained from Affinity; and anti-ACTB (3700) and anti-OCLN (5506) were obtained from Cell Signaling Technology. The detection was performed using a MicroChemi 4.2® (DNR, Jerusalem, Israel) digital image scanner. The band intensity was quantified using ImageJ software (NIH).
Real-time PCR
According to our previous studies [59,60] total RNA was extracted and subjected to reverse transcription using a HiScript RT SuperMix Kit (Vazyme, R133-01 for miRNAs, R123-01 for mRNAs). Real-time PCR was performed by StepOne™ Real-Time PCR System (Life Technologies, 4376357, Singapore) using the following primers: human circHECW2 (forward primer: 5’-CCCACCACTTTGAACGCTAC-3’; reverse primer: 5’-GGCTGTCAATGCGTGCCT-3’), human GAPDH (forward primer: 5’-ACCATCTTCCAGGAGCGAGAT-3’; reverse primer: 5’-GGGCAGAGATGATGACCCTTT-3’), mouse circHecw2 (forward primer: 5’-AACAGGGACCTCGTGGGATT-3’; reverse primer: 5’-GGCTGTCAATCCGTGCCTC-3’) and mouse Gapdh (forward primer: 5’-AGGTCGGTGTGAACGGATTTG-3’; reverse primer: 5’-GGGGTCGTTGATGGCAACA-3’). The relative quantification was performed using AceQ qPCR SYBR Green Master Mix (Vazyme, Q141-02). The specific primers for the mature MIR30D and RNU6-6P/RNU6B snRNA were purchased from RiboBio. All reactions were performed in triplicate.
Luciferase activity assays
The 3'-UTR of the 706-base pair human ATG5 gene that contained the putative MIR30D target site was PCR amplified from human genomic DNA using forward (5’-GCGGCTCGAGGAAAATGGAAGTATGTGCC-3’) and reverse (5’-AATGCGGCCGCACTTCAAAAGACTGGCTGC-3’) primers; the DNA fragment was cloned into the XhoI and NotI sites on the 3’ end of the luc2 gene in a pmiR-RB-GLO vector (RiboBio, GUR100000). For the pmiR-RB-ATG5-3'-UTRΔ-MIR30D-target-mutant vector, the MIR30D target site (TGTTTACA) within the ATG5 3'-UTR was changed to ACAAATGT via PCR mutagenesis with the primers ATG5-MIR30DT-F (5’-ATTTCCTAACAAATGTGTCTGTCTATTGGTATGC-3’) and ATG5 -MIR30DT-R (5’-AGACAGACACATTTGTTAGGAAATTTTCGTTAAG-3’). Briefly, HEK293T cells were transfected with a MIR30D overexpression pLV-[Hsa-MIR30D] vector (RiboBio) and a target plasmid, pmiR-RB-Bbc3-3’-UTR or pmiR-RB-ATG5-3’-UTRΔ-MIR30D-target-mutant (RiboBio), at a molar ratio of 50:1. A MIRCon pLV-[MIRCon] vector was used as a negative control. The luciferase activity was determined at 24 h post-transfection, and a reporter assay was performed following the manufacturer's protocol (Promega, E2920). The Renilla luciferase activity was normalized to the firefly luciferase activity and expressed as a percentage of the control.
Affinity isolation assay with biotinylated MIR30D
A total of 2 × 106 HEK293T cells were seeded the day before the transfection, and the cells were transfected with 3'-end biotinylated MIR30D at a final concentration of 50 nM for 36 h. The cells were subsequently washed with phosphate-buffered saline (PBS; 2.96 mM Na2HPO4.7H2O, 1.05 mM KH2PO4, 155.17 mM NaCl, pH 7.4) and incubated in a lysis buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.05% Igepal [Sigma-Aldrich, I3021], 60 U/ml SUPERase-In [Ambion, AM2694], 1 mM DTT [Sigma-Aldrich, 43816], and protease inhibitors [Biotool, B14001) on ice for 10 min. After being precleared, 50-μl aliquots of the samples were used as the input. The remaining lysates were incubated in M-280 Streptavidin magnetic beads (Life Technologies, 11205D), which were previously coated with yeast tRNA (Invitrogen, 15401011) to prevent the nonspecific binding of the RNA and protein complexes. The beads were incubated at 4°C for 2.5 h, followed by being washed twice with ice-cold lysis buffer, twice with a low-salt buffer (0.1% SDS [Invitrogen, 15553035], 1% Triton X-100 [Aladdin, T109027], 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl), and once with a high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl). The bound RNAs were isolated using Trizol (TaKaRa, 9109) to measure the circHECW2 level. The 3'-end biotinylated MIR30D was synthesized by GenePharma (Shanghai, China), and its sequence was 5'-UGUAAACAUCCCCGACUGGAAGUU-3'.
Affinity isolation assay with the biotinylated DNA probe
The biotinylated DNA probes, complementary to circHECW2, were dissolved in 500 ml of the wash/binding buffer (0.5 M NaCl, 20 mM Tris-HCl, pH 7.5, 1 mM EDTA). The probes were incubated in M-280 Streptavidin magnetic beads at 4°C for 3 h to generate probe-coated magnetic beads. The HEK293T cell lysates were incubated with the probe-coated beads at 25°C for 1.5 h, and the RNA complexes bound to the beads were eluted and extracted for real-time PCR analysis after being washed with the wash/binding buffer. The following primer sequence was used: circHECW2 affinity-isolation probe (Invitrogen), 5'-aaaATGAGTGCCTCCCAGTCTTGCCCTGGTGATCA-3'; and random affinity-isolation probe (Invitrogen), 5'-aaaCAGTACTGGTGTGTAGTACGAGTCTAAGCTAC-3'.
Microinjection of circHecw2 siRNA lentivirus
Six-wk-old C57BL/6J mice were bilaterally microinjected with control-GFP or circHecw2-GFP siRNA lentivirus (1 μl of 109 viral genomes µl−1, HANBIO) in the hippocampus using the following microinjection coordinates: 2.06 mm posterior to bregma, ±1.5 mm lateral from the sagittal midline, and 1.7 mm ventral to the skull surface. To evaluate the effect of circHECW2 on the LPS-induced EndoMT, twelve mice were divided into the following 2 groups (n = 6 animals/group, male) 2 weeks after lentivirus microinjection: the saline-treated mouse group and the LPS-treated mouse group. The rationale for the selection of this dosage of LPS (0.83 mg/kg, injected once per day for 7 d, intraperitoneal) was based on previous studies [6,61].
Immunostaining and image analysis
According to our previous studies [58,59] the sections that encompassed the entire hippocampus were cut into 30-μm slices with a cryostat. The sections were subsequently incubated with 0.3% Triton X-100 in PBS for 15 min and blocked with 10% normal goat serum (ZSGB-BIO, ZLI-0956) in 0.3% Triton X-100 for 1 h at room temperature. Next, the sections were incubated with a rabbit anti-ACTA2 antibody overnight at 4°C. On the following day, the sections were washed and incubated with Alexa Fluor 594 goat anti-rabbit IgG (Invitrogen, A-11037) for 1 h. After a final washing step with PBS, the sections were mounted onto glass slides. The immunofluorescence images were captured via microscopy (Carl Zeiss LSM700, Zeiss, Oberkhorn, Germany). The intensities of ACTA2 and GFP were calculated using PSC colocalization and Manders coefficients [62].
Tandem fluorescent-mRFP-GFP-MAP1LC3B-adenovirus transduction of HBMECs
According to our previous study [59], HBMECs were transfected with a fluorescent-mRFP-GFP-MAP1LC3B-adenovirus that expresses a specific marker of autophagosome formation to detect autophagy according to the manufacturer's instructions. Five fields were selected from 3 different cell preparations. GFP- and mRFP-expressing spots, which were indicated by fluorescent puncta, and 4′,6-diamidino-2-phenylindole (DAPI, SouthernBiotech, 010020) -stained nuclei were manually counted. The number of puncta per cell was determined by dividing the total number of spots by the number of nuclei in each field.
Bone marrow-derived monocyte (BMM) isolation and Dil staining
Cultured BMMs were 98% ITGAM+/CD11b+ as demonstrated via flow cytometry in our previous study [63]; to evaluate the monocyte migration, monocytes were washed with PBS and fluorescently labeled with 10 μM Dil (Beyotime, C1036) for 10 min at room temperature according to the manufacturer's instructions.
In vivo 2-photon laser scanning microscopy (TPLSM) for monocyte migration
According to our previous and other studies [58,64,65], for the monocyte migration studies, animals were injected with Dil-labeled BMM at a concentration at 107 cells/100 μl through the tail vein and subsequently subjected to TPLSM to detect the monocyte migration 24 h after the cell infusion. Cortical brain vessels 20 to 40 μm in diameter and 100 to 150 μm in depth below the cortical surface were selected for the 2-photon imaging. Z-stack images of 50-μm-thick tissue sections were collected at 2-μm steps, and 3-dimensional pictures were constructed using ZEN2011 Imaging Software (Zeiss) to calculate the number of monocytes that migrated to the parenchyma.
Fluorescence in situ hybridization (FISH)
According to our previous study [60], HBMECs cultured in a coverslip were fixed with 4% paraformaldehyde for 20 min and incubated in PBS overnight at 4°C. The cells were permeabilized with 0.25% Triton X-100 in PBS for 15 min and prehybridized in a hybridization buffer (50% formamide, 10 mM Tris-HCl [pH 8.0], 200 μg ml−1 yeast tRNA, 1X Denhardt's solution [Sigma-Aldrich, 30915], 600 mM NaCl, 0.25% SDS, 1 mM EDTA and 10% dextran sulfate [Sigma-Aldrich, D8906]) for 1 h at 37°C. The hybridization buffer, which contained 25 nM of a commercially available digoxigenin-labeled MIR30D probe (synthesized by Invitrogen) and 50 nM of a commercially available biotin-labeled circHECW2 probe (synthesized by Invitrogen), was subsequently heated to 65°C for 5 min; the hybridization was allowed to occur at 37°C overnight. On the following day, the coverslips were washed 3 times with 2XSSC (10% v/v 20X SSC [Invitrogen, 15557] in DEPC-treated water [Vetec, V900882]) and twice with 0.2X SSC at 42°C and then blocked with 1.5% BSA (Biosharp, BS043D) and 3% normal goat serum in PBS for 1 h at room temperature. The coverslips were subsequently incubated with a horseradish peroxidase-conjugated anti-digoxigenin antibody (1:200; Roche Diagnostics GmbH, 11207733910) and FITC-Streptavidin (1:200, Life Technologies, 434311) overnight at 4°C. After the coverslips were washed 3 times with TBS, they were incubated using a TSA Cy5 kit (PerkinElmer, NEL766B001KT) for 10 min at room temperature. The coverslips were subsequently washed twice with PBS and mounted with ProLong Gold anti-fade reagent, which contained Hoechst (Life Technologies, H3570). The digoxigenin-labeled MIR30D probe was CTTCCAGTCGGGGATGTTTACA (Invitrogen); biotin-labeled circHECW2 probe was 5'-aaaATGAGTGCCTCCCAGTCTTGCCCTGGTGATCA-3' (Invitrogen).
Statistical analyses
Statistical analyses were performed using the Mann-Whitney test or one-way analysis of variance (ANOVA) followed by the Holm-Sidak test (SPSS 16.0). The appropriate test is indicated in the figure legends. Results were judged statistically significant when P<0.05 via analysis of variance.
Supplementary Material
Funding Statement
This work was supported by the National Natural Science Foundation of China (NSFC) [grant number 81473190, 81673410 and 81603090]; Major State Basic Research Development Program of China [grant number 2013CB733803 and 2013CB733800].
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81473190, No. 81673410 and No. 81603090) and grants from the Jiangsu Specially Appointed Professor and the Major State Basic Research Development Program of China (973 Program) (2013CB733800 and 2013CB733803).
Disclosure of potential conflicts of interest
The authors declare that there are no competing financial interests.
References
- [1].Sajja RK, Rahman S, Cucullo L. Drugs of abuse and blood-brain barrier endothelial dysfunction: a focus on the role of oxidative stress. J Cereb Blood Flow Metab: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 2016;36(3):539–554. doi: 10.1177/0271678X15616978. PMID:26661236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chapy H, Smirnova M, Andre P, et al. Carrier-mediated cocaine transport at the blood-brain barrier as a putative mechanism in addiction liability. Int J Neuropsychopharmacol. 2014;18(1):1–10. doi: 10.1093/ijnp/pyu001. PMID:25539501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].O'Shea E, Urrutia A, Green AR, et al. Current preclinical studies on neuroinflammation and changes in blood-brain barrier integrity by MDMA and methamphetamine. Neuropharmacology. 2014;87:125–134. doi: 10.1016/j.neuropharm.2014.02.015. PMID:24594477 [DOI] [PubMed] [Google Scholar]
- [4].Ding G, Zhang Z, Chopp M, et al. MRI evaluation of BBB disruption after adjuvant AcSDKP treatment of stroke with tPA in rat. Neuroscience. 2014;271:1–8. doi: 10.1016/j.neuroscience.2014.04.025. PMID:24769225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu WY, Wang ZB, Wang Y, et al. Increasing the permeability of the blood-brain barrier in three different models in vivo. CNS Neurosci Ther. 2015;21(7):568–574. doi: 10.1111/cns.12405. PMID:25982054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Morita-Takemura S, Nakahara K, Tatsumi K, et al. Changes in endothelial cell proliferation and vascular permeability after systemic lipopolysaccharide administration in the subfornical organ. J Neuroimmunol. 2016;298:132–137. doi: 10.1016/j.jneuroim.2016.06.011. PMID:27609286 [DOI] [PubMed] [Google Scholar]
- [7].Potenta S, Zeisberg E, Kalluri R. The role of endothelial-to-mesenchymal transition in cancer progression. Br J Cancer. 2008;99(9):1375–1379. doi: 10.1038/sj.bjc.6604662. PMID:18797460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Troletti CD, de Goede P, Kamermans A, et al. Molecular alterations of the blood-brain barrier under inflammatory conditions: the role of endothelial to mesenchymal transition. Biochim Biophys Acta. 2016;1862(3):452–460. doi: 10.1016/j.bbadis.2015.10.010. PMID:26493443 [DOI] [PubMed] [Google Scholar]
- [9].Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurotoxins. 2009;32(4):199–206. doi: 10.1016/j.tins.2008.12.003. PMID:19268374 [DOI] [PubMed] [Google Scholar]
- [10].O'Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33(4):607–619. doi: 10.1016/j.immuni.2010.09.009. PMID:20888269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Junker A, Hohlfeld R, Meinl E. The emerging role of microRNAs in multiple sclerosis. Nat Rev Neurol. 2011;7(1):56–59. doi: 10.1038/nrneurol.2010.179. PMID:21151203 [DOI] [PubMed] [Google Scholar]
- [12].Smits M, Wurdinger T, van het Hof B, et al. Myc-associated zinc finger protein (MAZ) is regulated by miR-125b and mediates VEGF-induced angiogenesis in glioblastoma. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2012;26(6):2639–2647. doi: 10.1096/fj.11-202820. PMID:22415301 [DOI] [PubMed] [Google Scholar]
- [13].Reijerkerk A, Lopez-Ramirez MA, van Het Hof B, et al. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: implications for multiple sclerosis. The Journal of Neuroscience: The Official Journal of The Society for Neuroscience. 2013;33(16):6857–6863. doi: 10.1523/JNEUROSCI.3965-12.2013. PMID:23595744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Katsura A, Suzuki HI, Ueno T, et al. MicroRNA-31 is a positive modulator of endothelial-mesenchymal transition and associated secretory phenotype induced by TGF-beta. Genes to Cells : Devoted to Molecular & Cellular Mechanisms. 2016;21(1):99–116. doi: 10.1111/gtc.12323. PMID:26663584 [DOI] [PubMed] [Google Scholar]
- [15].Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. PMID:24811520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. PMID:23446348 [DOI] [PubMed] [Google Scholar]
- [17].Boeckel JN, Jae N, Heumuller AW, et al. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res. 2015;117(10):884–890. doi: 10.1161/CIRCRESAHA.115.306319. PMID:26377962 [DOI] [PubMed] [Google Scholar]
- [18].Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. PMID:26802132 [DOI] [PubMed] [Google Scholar]
- [19].Fu D, Yu JY, Yang S, et al. Survival or death: a dual role for autophagy in stress-induced pericyte loss in diabetic retinopathy. Diabetologia. 2016;59(10):2251–2261. doi: 10.1007/s00125-016-4058-5. PMID:27475954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. PMID:27050392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. PMID:24427167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–610. doi: 10.1038/nn.3975. PMID:25714049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Almutairi MM, Gong C, Xu YG, et al. Factors controlling permeability of the blood-brain barrier. Cell Mol Life Sci: CMLS. 2016;73(1):57–77. doi: 10.1007/s00018-015-2050-8. PMID:26403789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Miao YS, Zhao YY, Zhao LN, et al. MiR-18a increased the permeability of BTB via RUNX1 mediated down-regulation of ZO-1, occludin and claudin-5. Cell Signal. 2015;27(1):156–167. doi: 10.1016/j.cellsig.2014.10.008. PMID:25452107 [DOI] [PubMed] [Google Scholar]
- [25].Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. PMID:22966490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mayerhofer R, Frohlich EE, Reichmann F, et al. Diverse action of lipoteichoic acid and lipopolysaccharide on neuroinflammation, blood-brain barrier disruption, and anxiety in mice. Brain Behav Immun. 2017;60:174–187. doi: 10.1016/j.bbi.2016.10.011. PMID:27751870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Donelli MG, Colombo S, Garattini S. Experiments aiming at demonstrating microsomal drug metabolism in the tumor tissue. Eur J Cancer. 1972;8(2):181–183. doi: 10.1016/0014-2964(72)90042-4. PMID:5073292 [DOI] [PubMed] [Google Scholar]
- [28].Zhao Y, Qiao X, Tan TK, et al. Matrix metalloproteinase 9-dependent Notch signaling contributes to kidney fibrosis through peritubular endothelial-mesenchymal transition. Nephrology, Dialysis, Transplantation: Official Publication of the European Dialysis and Transplant Association – European Renal Association. 2017;32(5):781–791. doi: 10.1093/ndt/gfw308. PMID:27566305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13(6):427–437. doi: 10.1038/nri3445. PMID:23665520 [DOI] [PubMed] [Google Scholar]
- [30].Niessen K, Fu Y, Chang L, et al. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J Cell Biol. 2008;182(2):315–325. doi: 10.1083/jcb.200710067. PMID:18663143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Timmerman LA, Grego-Bessa J, Raya A, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. PMID:14701881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kumarswamy R, Volkmann I, Jazbutyte V, et al. Transforming growth factor-beta-induced endothelial-to-mesenchymal transition is partly mediated by microRNA-21. Arterioscler Thromb Vasc Biol. 2012;32(2):361–369. doi: 10.1161/ATVBAHA.111.234286. PMID:22095988 [DOI] [PubMed] [Google Scholar]
- [33].Lagendijk AK, Goumans MJ, Burkhard SB, et al. MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ Res. 2011;109(6):649–657. doi: 10.1161/CIRCRESAHA.111.247635. PMID:21778427 [DOI] [PubMed] [Google Scholar]
- [34].Zhu K, Pan Q, Jia LQ, et al. MiR-302c inhibits tumor growth of hepatocellular carcinoma by suppressing the endothelial-mesenchymal transition of endothelial cells. Sci Rep. 2014;4:5524. doi: 10.1038/srep05524. PMID:25027009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ghosh AK, Nagpal V, Covington JW, et al. Molecular basis of cardiac endothelial-to-mesenchymal transition (EndMT): differential expression of microRNAs during EndMT. Cell Signal. 2012;24(5):1031–1036. doi: 10.1016/j.cellsig.2011.12.024. PMID:22245495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Laurila EM, Kallioniemi A. The diverse role of miR-31 in regulating cancer associated phenotypes. Genes Chromosom Cancer. 2013;52(12):1103–1113. doi: 10.1002/gcc.22107. PMID:23999990 [DOI] [PubMed] [Google Scholar]
- [37].Chen D, Guo W, Qiu Z, et al. MicroRNA-30d-5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non-small cell lung cancer. Cancer Lett. 2015;362(2):208–217. doi: 10.1016/j.canlet.2015.03.041. PMID:25843294 [DOI] [PubMed] [Google Scholar]
- [38].Melman YF, Shah R, Danielson K, et al. Circulating MicroRNA-30d is associated with response to cardiac resynchronization therapy in heart failure and regulates cardiomyocyte apoptosis: a translational pilot study. Circulation. 2015;131(25):2202–2216. doi: 10.1161/CIRCULATIONAHA.114.013220. PMID:25995320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ye Z, Zhao L, Li J, et al. miR-30d blocked transforming growth factor beta1-induced epithelial-mesenchymal transition by targeting snail in ovarian cancer cells. International Journal of Gynecological Cancer: Official Journal of the International Gynecological Cancer Society. 2015;25(9):1574–1581. doi: 10.1097/IGC.0000000000000546. PMID:26501435 [DOI] [PubMed] [Google Scholar]
- [40].Yang X, Zhong X, Tanyi JL, et al. mir-30d Regulates multiple genes in the autophagy pathway and impairs autophagy process in human cancer cells. Biochem Biophys Res Commun. 2013;431(3):617–622. doi: 10.1016/j.bbrc.2012.12.083. PMID:23274497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kimmey JM, Huynh JP, Weiss LA, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528(7583):565–569. doi: 10.1038/nature16451. PMID:26649827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pua HH, Dzhagalov I, Chuck M, et al. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204(1):25–31. doi: 10.1084/jem.20061303. PMID:17190837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ma T, Li J, Xu Y, et al. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat Cell Biol. 2015;17(11):1379–1387. doi: 10.1038/ncb3256. PMID:26502054 [DOI] [PubMed] [Google Scholar]
- [44].Maskey D, Yousefi S, Schmid I, et al. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun. 2013;4:2130. doi: 10.1038/ncomms3130. PMID:23945651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. PMID:23446346 [DOI] [PubMed] [Google Scholar]
- [46].Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. PMID:26802132 [DOI] [PubMed] [Google Scholar]
- [47].Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. PMID:26861625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. PMID:25768908 [DOI] [PubMed] [Google Scholar]
- [49].Zeisberg EM, Potenta S, Xie L, et al. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67(21):10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. PMID:17974953 [DOI] [PubMed] [Google Scholar]
- [50].LeBleu VS, Taduri G, O'Connell J, et al. Origin and function of myofibroblasts in kidney fibrosis. Nat Med. 2013;19(8):1047–1053. doi: 10.1038/nm.3218. PMID:23817022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Maddaluno L, Rudini N, Cuttano R, et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498(7455):492–496. doi: 10.1038/nature12207. PMID:23748444 [DOI] [PubMed] [Google Scholar]
- [52].Cooley BC, Nevado J, Mellad J, et al. TGF-beta signaling mediates endothelial-to-mesenchymal transition (EndMT) during vein graft remodeling. Sci Transl Med. 2014;6:227ra34. doi: 10.1126/scitranslmed.3006927. PMID:24622514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wang SH, Chang JS, Hsiao JR, et al. Tumour cell-derived WNT5B modulates in vitro lymphangiogenesis via induction of partial endothelial-mesenchymal transition of lymphatic endothelial cells. Oncogene. 2017;36(11):1503–1515. doi: 10.1038/onc.2016.317. PMID:27593938 [DOI] [PubMed] [Google Scholar]
- [54].Liu J, Dong F, Jeong J, et al. Constitutively active Notch1 signaling promotes endothelialmesenchymal transition in a conditional transgenic mouse model. Int J Mol Med. 2014;34(3):669–676. doi: 10.3892/ijmm.2014.1818. PMID:24969754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Xavier S, Vasko R, Matsumoto K, et al. Curtailing endothelial TGF-beta signaling is sufficient to reduce endothelial-mesenchymal transition and fibrosis in CKD. Journal of the American Society of Nephrology: JASN. 2015;26(4):817–829. doi: 10.1681/ASN.2013101137. PMID:25535303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jounai N, Takeshita F, Kobiyama K, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U.S.A. 2007;104(35):14050–14055. doi: 10.1073/pnas.0704014104. PMID:17709747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yao H, Duan M, Buch S. Cocaine-mediated induction of platelet-derived growth factor: implication for increased vascular permeability. Blood. 2011;117(8):2538–2547. doi: 10.1182/blood-2010-10-313593. PMID:21148086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bai Y, Zhang Y, Hua J, et al. Silencing microRNA-143 protects the integrity of the blood-brain barrier: implications for methamphetamine abuse. Sci Rep. 2016;6:35642. doi: 10.1038/srep35642. PMID:27767041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang Y, Shen K, Bai Y, et al. Mir143-BBC3 cascade reduces microglial survival via interplay between apoptosis and autophagy: implications for methamphetamine-mediated neurotoxicity. Autophagy. 2016;12(9):1538–59. doi: 10.1080/15548627.2016.1191723. PMID:27464000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yao H, Ma R, Yang L, et al. MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun. 2014;5:4386. doi: 10.1038/ncomms5386. PMID:25019481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].O'Connor JC, Lawson MA, Andre C, et al. Lipopolysaccharide-induced depressive-like behavior is mediated by indoleamine 2,3-dioxygenase activation in mice. Mol Psychiatry. 2009;14(5):511–522. doi: 10.1038/sj.mp.4002148. PMID:18195714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].French AP, Mills S, Swarup R, et al. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc. 2008;3(4):619–628. doi: 10.1038/nprot.2008.31. PMID:18388944 [DOI] [PubMed] [Google Scholar]
- [63].Yao H, Yang Y, Kim KJ, et al. Molecular mechanisms involving sigma receptor-mediated induction of MCP-1: implication for increased monocyte transmigration. Blood. 2010;115(23):4951–4962. doi: 10.1182/blood-2010-01-266221. PMID:20354174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang H, Hong LJ, Huang JY, et al. P2RX7 sensitizes Mac-1/ICAM-1-dependent leukocyte-endothelial adhesion and promotes neurovascular injury during septic encephalopathy. Cell Res. 2015;25(6):674–690. doi: 10.1038/cr.2015.61. PMID:25998681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hill RA, Patel KD, Goncalves CM, et al. Modulation of oligodendrocyte generation during a critical temporal window after NG2 cell division. Nat Neurosci. 2014;17(11):1518–1527. doi: 10.1038/nn.3815. PMID:25262495 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


