ABSTRACT
Objectives: Due to the absence of reliable and accurate biomarkers for the early detection of liver malignancy, circulating microRNAs have recently emerged as great candidates for prompt cancer identification. Therefore, the aim of this study was to investigate the potential of liver-specific circulating microRNAs as an accurate non-invasive diagnostic tool for early diagnosis of hepatitis C virus (HCV)-induced hepatocellular carcinoma (HCC).
Methodology: A total of 165 patients were enrolled in this study and categorized into four main groups: 42 chronic hepatitis C (CHC) without cirrhosis, 45 CHC with cirrhosis (LC), 38 HCC with HCV patients, and 40 healthy controls. The expression profiles of seven miRNAs (miR-16, miR-34a, miR-125a, miR-139, miR-145, miR-199a, and miR-221) were analyzed using real-time PCR.
Results: Serum levels of miRNA-125a, miRNA-139, miRNA-145, and miRNA199a were significantly lower (p < 0.01) in HCC than in both CHC and LC groups. On the other hand, no significant difference was shown in the expression of miR-16, miR-34a, and miR-221 between the CHC, LC, and HCC groups. MiR-16, miR-34a, and miR-221 were significantly elevated in the HCC group compared to the control group. MiR-34a showed the highest specificity and sensitivity.
Conclusions: The results indicated that the measurement of serum levels of miR-125a, miR-139, miR-145, and miR-199a can help to differentiate HCC from CHC and LC. Also, miR-16, miR-34a, and miR-221 serum levels would have a prognostic value. MiR-34a had the highest specificity and sensitivity, indicating that it might serve as a novel and potential non-invasive biomarker for HCV-induced HCC.
KEYWORDS: HCV, Hepatocellular Carcinoma, miRNAs
Introduction
The significance of reliable biomarkers is evaluated based on their ability to be easily detected at the early onset of the disease and their sensitivity and specificity for disease diagnosis and prognosis. Therefore, a reliable and accurate biomarker must lack limitations such as low sensitivity, low specificity, low predictive power, and low robustness, in addition to exerting noninvasive characteristics.1
In 1993, Lee et al.2 were the first to discover the small noncoding RNA lin-4 in Caenorhabditis elegans. Around 20% of all annotated miRNAs are highly conservative between organisms like C. elegans and Homo sapiens. MiRNAs are assumed to target more than 60% of the human genome, hence their high abundance in mammalian cells. Several research groups have illustrated the deregulation of miRNAs in various pathological processes, which was evident in the development of many illnesses as well as malignancy.1
Measuring the levels of the circulating miRNAs to be used as biomarkers for malignancies has recently been investigated in the hope of identifying unique miRNA signatures in human tumors that will help in better understanding cancer pathways and carcinogenesis in general, thus facilitating the proper design of effective diagnostic, therapeutic, as well as prognostic tools. This diagnostic approach shows a lot of promise due to the high stability of miRNAs in human formalin-fixed tissue, serum, and plasma, in addition to their tissue-specific expression pattern.3 MiRNAs that were observed to be upregulated in tumors probably function as oncogenes, while other miRNAs have been described as tumor suppressors due to their loss or down-regulation. Moreover, some miRNAs have been recognized to participate in tumor invasion, metastasis, and angiogenesis.4
Hepatocellular carcinoma (HCC) is categorized as the 3rd most common cause of cancer-related death as well as the major cause of deaths among patients suffering from liver cirrhosis.5 Additionally, it is the 5th most frequent cancer type in men and the 7th for women worldwide. HCC is usually presented around the age of 40 years, where it reaches its peak at the age of 70 years.6
HCC resulting from hepatitis C virus (HCV) is becoming a fast-rising cause of cancer-related deaths in the United States, in which the incidence has seen to be tripled throughout the past few decades.7 Further HCC risk factors are excessive alcohol intake, non-alcoholic fatty liver disease, α1-antitrypsin deficiency, Wilson's disease, hemochromatosis, and autoimmune hepatitis.
Symptoms caused by HCC are typically absent, patients usually show symptoms related to cirrhosis, which is a condition found in 80–90% of HCC patients. As a result, HCC morbidity and mortality are almost the same due to the fact that the majority of HCC patients are diagnosed at late stages of the disease where the cancer is significantly advanced for good therapeutic options. The absence of accurate diagnostic tools for early HCC detection as well as curative therapy resulted in very poor patient prognosis.5
Therefore, the aims of this study were to 1) investigate the expression profiles of 7 specific miRNAs: miR-16, miR-34a, miR-125a, miR-139, miR-145, miR-199a, and miR-221 in the sera of Egyptian patients with HCV-induced HCC; 2) evaluate the potential of using these miRNAs as promising non-invasive biomarkers to differentiate between normal, fibrotic, cirrhotic, and HCC patients; 3) pick the miRNAs with the highest sensitivity and specificity as potential biomarkers to diagnose HCV-induced HCC during the early disease stages.
Results
Demographics, laboratory investigations, and clinical features of the patients:
Clinically and biochemically healthy, roughly age- and gender-matched individuals (n = 40) served as a control population for patients with HCV-related liver fibrosis, cirrhosis, and primary HCC, respectively. Table I summarizes the demographic and biochemical profiles of the 42 patients with chronic hepatitis C (CHC), the 45 patients with liver cirrhosis (LC), the 48 patients with HCC, and the 40 healthy controls. The biochemical parameters, i.e. AST, ALT, albumin, ALP, prothrombin concentration, AFP, and DCP, were as expected, within the reference range for control subjects but significantly elevated in the three patient groups (CHC, LC, and HCC).
Serum HCV-RNA revealed that all patients were genotype 4a. All the studied groups' patients had increased ALT, AST, albumin, ALP, prothrombin concentration, AFP, and DCP (P < 0.001) compared to the control group. AFP and DCP serum levels were increased significantly in HCC patients compared to HCV and cirrhosis patients (P<0.001) (Table 1).
Table 1.
Demographic and laboratory data of all patients and controls.
| Variables | Controls (n = 40) | CHC (n = 42) | LC (n = 45) | HCC (n = 38) |
|---|---|---|---|---|
| Age | 47.0 ± 3.5 | 46.4 ± 7.3 | 51.3 ± 4.7 | 48.1 ± 4.1 |
| Gender (M:F) | 3:1 | 3:2 | 7:5 | 6:4 |
| Liver Function Tests (mean ± SD) | ||||
| AST (U/L) | 21.32 ± 1.07 | 46.14 ± 4.77 a | 50.84 ± 5.00 a | 76.40 ± 7.34 b |
| ALT (U/L) | 23.16 ± 1.95 | 44.12 ± 5.50 a | 50.84 ± 5.00 a | 76.40 ± 7.34 b |
| ALP (U/L) | 189 ± 41 | 331 ± 36 a | 336 ± 48 a | 420 ± 33 b |
| Albumin (g/dL) | 4.4 ± 0.5 | 3.60 ± 0.74 | 3.8 ± 0.72 | 3.08 ± 0.48 |
| Prothrombin Concentration | 95.6 ± 3.4 | 89.6 ± 4.8 | 41.5 ± 11.1 b | 69.4 ± 3.7 a |
| AFP (IU/mL) | 3.12 ± 0.08 | 8.86 ± 0.11 a | 10.11 ± 0.11 a | 55.18 ± 0.44 b |
| DCP (mAU/ ml) | 30.42 ± 0.70 | 121.49 ± 0.59 a | 123.62 ± 0.38 a | 456.52 ± 0.66 b |
Data are expressed as mean ± standard deviations (SD).
Chronic hepatitis C (CHC); liver cirrhosis (LC); hepatocellular carcinoma (HCC).
Normal range for alanine aminotransferase (ALT) and aspartate aminotransferase (AST) is up to 40 IU/L.
Normal range for alkaline phosphatase (ALP) is up to 250 U/L.
Normal range for albumin is 3.5–5 g/dl.
Normal range for prothrombin concentration is 80–1 00%.
Normal range for alpha-fetoprotein (AFP) is 0.1–9.6 IU/mL.
mAU/ml = milli-absorbance unit/ml.
Normal range for des-γ-carboxy prothrombin (DCP) ≤40 mAU/ml.
p < 0.001 significant increase than control group.
p < 0.001 significant increase than CHC and LC groups.
Expression levels of miRNAs:
All miRNAs showed a significant increase than control in both CHC and LC groups (p < 0.01). Serum levels of miRNA-125a, miRNA-139, miRNA-145, and miRNA-199a were significantly lower (p < 0.01) in HCC group than in both CHC and LC groups. MiR-34a showed a significant decrease (p < 0.01) in LC group compared to CHC and HCC groups. On the other hand, miR-16 was significantly decreased (p < 0.01) in HCC group compared to the CHC group. MiR-139 was also significantly decreased (p < 0.01) in LC group compared to the CHC group (Table 2, Fig. 1).
Table 2.
The qPCR expression levels of the serum microRNAs in the studied groups.
| miRNAs | Normal | CHC | LC | HCC |
|---|---|---|---|---|
| miRNA-16 | 14.26 ± 0.69 | 24.09 ± 0.44** | 23.29 ± 0.46** | 22.35 ± 0.54**a |
| miRNA-34a | 27.32 ± 0.19 | 32.69 ± 0.34** | 30.01 ± 0.54**b | 32.50 ± 0.94** |
| miRNA-125a | 20.57 ± 0.54 | 96.01 ± 4.36** | 100.54 ± 0.81** | 29.96 ± 0.57**d |
| miRNA-139 | 29.96 ± 0.57 | 94.63 ± 0.38** | 86.02 ± 0.40**e | 30.03 ± 0.43d |
| miRNA-145 | 20.65 ± 0.52 | 85.31 ± 0.53** | 80.74 ± 0.59** | 20.64 ± 0.57d |
| miRNA-199a | 80.23 ± 0.72 | 330.38 ± 0.74** | 311.98 ± 0.72** | 66.16 ± 0.44c d |
| miRNA-221 | 22.82 ± 0.38 | 27.17 ± 1.44** | 28.22 ± 0.41** | 28.51 ± 0.46** |
p < 0.01 significant increase than control; ap < 0.01 significant decrease than CHC; bp < 0.01 significant decrease than CHC and HCC; cp < 0.01 significant decrease than control; dp < 0.01 significant decrease than CHC and LC; ep < 0.01 significant decrease than CHC
Figure 1.
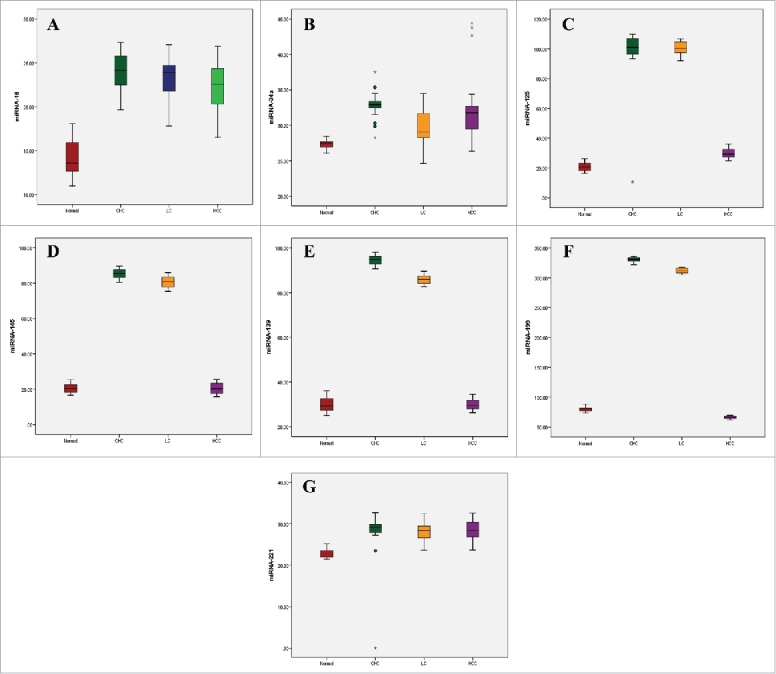
Box plot diagrams showing the expression of (A) miR-16, (B) miR-34a, (C) miR-125a, (D) miR-139, (E) miR-145, (F) miR-199a, and (G) miR-221 in Hepatitis C virus (HCV)-induced hepatocellular carcinoma (HCC) patients. The box indicates the 25th and 75th percentile of the data and the middle line indicates the median. A line extends from the minimum to the maximum value, excluding outliers that are displayed as separate points.
Diagnostic performance of circulating microRNAs in predicting HCC:
To assess the efficacy of the investigated serum miRNAs for predicting HCC, the AUC values (Table 3) were analyzed to calculate the sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) for each microRNAs (Table 4). The sensitivities and specificities of the 7 miRNAs were calculated from the ROC curves in order to evaluate the diagnostic potential of the different miRNAs. Out of the 7 miRNAs, miR-34a had the highest sensitivity and specificity, indicating that it is a promising biomarker for the early detection of liver cancer. Additionally, miR-125a showed the second highest specificity (83.33), therefore indicating its significance in diagnosis, but not in screening due to the low sensitivity (86.67).
Table 3.
Area Under the Curve (AUC), Confidence Interval (CI), and p-values for the circulating miRNAs.
| MicroRNAs | AUC | CI 95% | p-value |
|---|---|---|---|
| miRNA-16 | 0.716 ± 0.05 | 0.604 – 0.829 | <0.01 |
| miRNA-34a | 0.791 ± 0.04 | 0.695–0.888 | <0.001 |
| miRNA-125a | 0.806 ± 0.04 | 0.729–0.884 | <0.001 |
| miRNA-139 | 0.863 ± 0.05 | 0.763–0.962 | <0.001 |
| miRNA-145 | 0.941 ± 0.02 | 0.902–0.980 | <0.001 |
| miRNA-199a | 0.805 ± 0.03 | 0.704–0.906 | <0.001 |
| miRNA-221 | 0.633 ± 0.06 | 0.510–0.757 | <0.05 |
Table 4.
The sensitivity and specificity of the microRNAs.
| MicroRNAs | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| miR-16 | 80.95 | 70.59 | 87.18 | 60.00 |
| miR-34a | 96.77 | 81.82 | 93.75 | 90.0 |
| miR-125a | 86.67 | 83.33 | 92.86 | 71.43 |
| miR-139 | 85.71 | 64.29 | 82.76 | 69.23 |
| miR-145 | 90.91 | 77.78 | 93.75 | 70.00 |
| miR-199a | 87.10 | 72.73 | 90.0 | 66.67 |
| miR-221 | 90.32 | 81.82 | 93.33 | 75.0 |
PPV: Positive Predictive value
NPV: Negative Predictive Value
Correlations
Certain pairs of miRNAs within the same patient group or between two different groups, yielded the Pearson correlation coefficient, r, which is a value ranging from +1 to -1 indicating whether the correlation between the two certain miRNAs is a positive or negative correlation.
MiR-16 in the HCC group was shown to be negatively correlated with miR-34a in CHC group. A positive correlation has been shown in CHC group between miR-16 and miR-34a, in LC group between miR-16 and miR-34a, and in HCC group for miR-34a and CHC group for miR-16 (supplementary data Table II).
Negative correlations were shown between miR-221 and miR-125a in LC group. miR-221 in LC and CHC groups, and miR-125a in HCC and CHC groups, were positively correlated (supplementary data Table III). MiR-199a in LC group and miR-139 in HCC showed a negative correlation. miR-145 in HCC and miR-139 in CHC, and miR-199a in CHC and miR-145 in LC were positively correlated (supplementary data Table IV).
Discussion
Hepatitis C virus (HCV) infection is considered as a serious global health concern. It has been estimated that more than 80 million individuals suffer from HCV chronic infection worldwide and that 3–4 million new cases are infected annually, in addition to approximately 350,000 HCV-related deceases. Egypt is known to be the country with the highest prevalence of HCV infection worldwide, with genotype 4 being the most common.8,9 HCV infection usually results in fibrosis, cirrhosis, eventually leading to the development of hepatocellular carcinoma (HCC).10
HCC represents a major health issue worldwide, which is characterized by varied prognosis as well as its biological and clinical heterogeneity, due to different management approaches.11 HCC is still an extremely poor prognostic cancer that represents one of the most prevalent and most aggressive human malignancies worldwide. The early diagnosis of HCC is of great clinical desirability since it promises good prognosis if the patient could get early surgical treatment. Currently, AFP is one of the main biomarkers used clinically for diagnosing primary HCC; however, its sensitivity and specificity are not satisfying12; therefore, novel non-invasive biomarkers for early HCC detection are exceedingly required.
Chen et al.13 demonstrated that by extracting RNA from the serum or by utilizing serum directly, it is possible to identify unique miRNA expression profiles for lung cancer, colorectal cancer, and diabetic patients compared with healthy subjects. Circulating miRNAs have also been proposed as novel biomarkers for ovarian,14 pancreatic,15 and colorectal cancers.16 Although the clinical significance of these data has not been clarified in detail, they demonstrated that circulating miRNAs could be used as non-invasive diagnostic or prognostic biomarkers for cancer.
Several reports have revealed the significant association of miR-16 with hepatocarcinogenesis, Qu et al.17 showed that when combining miR-16 expression with the traditional liver biomarkers, the diagnostic accuracy is highly improved. As was shown in our results, miR-16 was significantly upregulated in HCC patient group. However, according to Qu et al.17 miR-16 is downregulated in HCC patients, in which HCV infection was the underlying etiology in the study subjects. In 2014, Ge et al.18 also showed that miR-16 is downregulated in the sera of HCC patients which in combination with let-7f and miR-21 they can be used biomarkers for estimating the tumor size as well as recurrence. As a matter of fact, in 2009, Huang et al.19 have revealed through microarray analysis that miR-16 was upregulated in HCC patients with mixed etiologies compared to normal subjects.
Also, unlike what has been described in the literature, in the current study, miR-34a was upregulated in HCC patients compared to the healthy subjects. Both Miao et al.20 and Yu et al.21 have shown that miR-34a expression was declined in HCC. The conflicting results between the current study and what has been published in the literature might be attributed to a number of factors that impact the expression pattern of miRNAs in different studies. Such factors include the heterogeneity of the cancer patients, such as the tumor stage, treatment, and etiology. The type of specimen is also a major factor since different samples are continuously studied, like serum, plasma, paraffin-embedded tissue, or formalin-fixed tissue. Additionally, the differences in sample collection, processing, and preservation are all factors that might impact the outcome of the expression analysis. Different RNA isolation techniques, the quality and concentration of the isolated RNA, and the detection methods are all additional factors that might impact the miRNAs expression patterns. Even if the study was meticulously designed, using various methods and housekeeping transcripts for miRNA expression normalization might result in experimental bias and therefore yielding diverse miRNA expression profiles. Thus, normalization is essential in eliminating most of the non-biological variations in order to ensure accurate miRNA expression profiles.22
MiR-221 is known to be upregulated in HCC and it plays important roles in hepatocarcinogenesis, responsible for proliferation, migration, invasion, apoptosis, clonogenicity, and G1 arrest.23 In 2011, Li et al.24 demonstrated that miR-221 was highly expressed in the sera of HCC patients; however, it was concluded that it was not statistically significant and might not serve as a reliable diagnostic biomarker for HCC. On the other hand, Turchinovich et al.25 showed that the augmented expression of miR-221 in the sera of HCC patients was associated with decreased survival rate. These data were consistent with our results as miR-221 was upregulated in HCC group.
The majority of studies that focused on the expression profiles of miRNA in HCC have shown that in most cases the progression of the malignancy is correlated with miRNAs downregulation. However, it needs to be taken into consideration that at the posttranscriptional level, miRNAs regulate hundreds of targets that are part of many signal transduction pathways, which makes the role of miRNAs in the process of hepatocarcinogenesis very complicated. Bi et al.26 showed that the expression of miR-125a is lower in HCC malignant tissue compared to non-tumor adjacent liver tissue and that the low expression level of miR-125a was associated with cancer advancement as well as poor prognosis. In addition to the lower expression observed in tissues, also a low expression of miR-125a was seen in HCC cell lines. In fact, it has been exhibited that in vitro and in vivo ectopic expression of miR-125a can inhibit the proliferation, the migration, and the invasion of the HCC cells.
Unlike what the literature and other studies have shown, the expression level of miR-125a in our study was significantly higher in HCC patient samples compared to control group, which might be due to the fact that HCC was a result of a genotype 4 HCV infection. In addition, the ancestral background and the gene pool of the current study are different than that of the other studies that demonstrated the downregulation of miR-125a. Therefore, factors such as the genotype of HCV as well as the ancestral background of the study group might have had an impact on the expression profile of miR-125a. On the other hand, very high expression levels of miR-125a were observed in CHC and LC patient groups. However, very low expression of miR-125a was reported in HCC group compared to that of CHC and LC groups. miR-125a showed a high specificity suggesting its diagnostic value. Therefore, miR-125a can be used for the early detection of HCC-induced HCV.
Same as miR-125a, according to previous studies the expression level of miR-139 has been shown to be significantly lower in HCC patient samples compared to control individuals. MiR-139 was found to have a major role in the hepatocarcinogenesis. In fact, high expression levels of miR-139 were shown to inhibit cellular proliferation and invasion in malignant liver cells. In addition, in miR-139 transfected cells, apoptosis was shown to be highly induced, which suggests that miR-139 can act as a tumor suppressor through the inhibition of cell proliferation and invasion in HCC.27 As shown in our results, the expression levels of miR-139 have increased in HCC patient group compared to control group, which contradicts what has been reported in other studies, while CHC and LC patient groups showed a significant increase in miR-139 expression. As a matter of fact, a possible explanation for the elevated expression levels for miR-125a and miR-139 in HCC patient group can be due to the fact that HCV infection is the underlying cause for HCC patients in the study, which might have an effect on the expression level of the two miRNAs. Although according to the literature, the HCV infection should not affect the expression levels after the patient develops HCC; however, no study has examined that with HCV genotype 4. All patients in our study have developed HCC as a result of a genotype 4 HCV infection, which may have resulted in the high expression level of miR-125a and miR-139 contradicting other studies.
MiR-145, another miRNA tested among the miRNAs panel investigated, in which its expression level showed no difference between normal controls and HCC patient group. Same as the rest of the miRNAs investigated, miR-145 in CHC and LC groups showed a significant increase compared to control and HCC groups. Among several studies, Wang et al.28 displayed that miR-145 expression was significantly diminished in HCC patient samples as well as in HCC cell lines. As a matter of fact, the down-regulation of miR-145 was greatly linked to intrahepatic metastasis, tumor size, vascular invasion, and tumor grade, all suggesting that miR-145 acts as tumor suppressor and its decreased expression results in the progression of the hepatocarcinogenesis. Therefore, determining the tumor size and grade in the HCC patients enrolled in the current study will be a key factor in understanding such contradicting expression profiles between what has been reported in the literature versus the current study.
The last miRNA investigated was miR-199a, which was downregulated in HCC patient group compared to the control group, while it was significantly upregulated in the CHC and the LC groups. The downregulation of miR-199a has been reported in several studies. Moreover, miR-199a has been one of the most consistently reported miRNAs to be involved in HCC. MiR199a is the third highly expressed miRNA in the liver, and in the case of HCC, it has been shown to be downregulated especially in patients with HCV, HBV infections, and alcohol abuse.29 The downregulation of miR-199a in HCC patients has been associated with poor prognosis. The identification of miRNAs associated with HCC is crucial to developing new diagnostic and therapeutic tools with high specificity and sensitivity in order to combat this vicious human cancer.
The correlation between miR-16 and miR-34a was investigated among the three patient groups, in which miR-34a and miR-16 in CHC and LC were positively correlated as well as miR-34a in HCC and miR-16 in CHC. On the other hand, miR-34a in CHC and miR-16 in HCC showed a highly significant negative correlation.
More positive correlations were shown among the following groups: miR-221 in LC and CHC, miR-145 in HCC and miR-139 in CHC, miR-125a in HCC and CHC, as well as miR-199a in CHC and miR-145 in LC. Finally, miR-199a in LC and miR-139 in HCC were negatively correlated. Establishing the Pearson correlation between different miRNAs among the patient groups has a significant diagnostic value because it indicates if two miRNAs were for example positively correlated in one or more patient group, then only one of these miRNAs is needed to be investigated. For example, in the CHC group, the r value for the correlation between miR-34a and miR-16 was shown to have a positive correlation, which means that if miR-34a is upregulated, then miR-16 will be upregulated as well, and vice versa. Therefore, from a diagnostic perspective, if for instance miR-34a was found to be elevated, then it can be inferred that miR-16 will be elevated as well.
Finally, circulating miRNAs in liver cancer patients represent promising biomarkers that possess great stability and reproducibility in peripheral blood. MiRNAs have the potential to be used in several clinical aspects of cancer management, such as cancer screening and early diagnosis, evaluating the malignancy in order to choose a surgical or a non-surgical approach, and to check for recurrence and cancer dynamics.30 Additionally, circulating miRNAs can be versatile, as it is anticipated that they will be used as efficient therapeutic agents in human tumors.
In conclusion, the measurement of serum levels of miR-125a, miR139, miR-145, and miR-199a can help to differentiate HCC from CHC and LC. The serum levels of miRNA-34a, miRNA-125a, miRNA-139, miRNA-145, and miRNA-199a showed a high accuracy for the early detection of HCC. On the other hand, measurement of serum levels of miR-16 and miR-221 was shown to have a prognostic rather than a diagnostic value since they did not significantly differentiate HCC group from CHC and LC groups. Moreover, miR-16, miR-34a, and miR-221 can be used to detect liver injuries, such as fibrosis and cirrhosis. Additionally, miR-34a was shown to be a potential HCC biomarker since it showed the highest specificity and sensitivity.
Methodology
Subjects and methods
Hundreds of patients are regularly admitted at the Hepato-Gastroenterology Department, Theodor Bilharz Research Institute (TBRI), Giza, Egypt, for evaluation of their HCV-related chronic liver disease, out of which 165 patients were selected to be enrolled in this study based on specific screening criteria. All participants signed informed consent before participating in the study, according to the guidelines of TBRI Institute's Human Research Ethics Committee and AUC's human research ethics committee and in compliance with the guidelines of the 1975 Declaration of Helsinki.
After acquiring the patients' full medical history, they were subjected to thorough clinical examination and were assessed by: (a) laboratory testing; including urine and stool analysis, liver function tests, prothrombin concentration, AFP, des-γ carboxy prothrombin (DCP), serological diagnosis of schistosomiasis and viral hepatitis and HCV-RNA RT-PCR. (b) ultrasonography.
Patients were enrolled in the study if they had clinical and laboratory evidence of chronic HCV infection, circulating anti-HCV (genotype-4) antibodies, HCV-RNA viremia, and hepatic lesion indicative of malignancy detected by abdominal ultrasonography and was confirmed to be HCC by triphasic abdominal CT.
Patients were disqualified from participating in this study if they had findings suggestive of other etiologies of chronic liver disease, such as Schistosoma infection, HBV infection, biliary disorders, other malignancies or HCV-infected patients receiving immunomodulatory interferon-α therapy.
Patients were divided into three groups. (i) chronic hepatitis C (CHC) virus infection group (n = 42), (ii) liver cirrhosis (LC) group (n = 45), and (iii) patients with HCC (n = 38). Age- and sex-matched individuals (n = 40) who had undergone laparoscopic cholecystectomy were included in this study as controls.
Histopathologic study
Assessment of the grade of inflammation and stage of fibrosis was carried out in 5 µm thick serial sections of formalin-fixed, paraffin-embedded blocks, stained with hematoxylin/eosin and Masson trichrome stains. The stage of hepatic fibrosis was determined according to the Metavir scoring system (F0, F1, F2, F3, and F4). The Metavir scoring system is used to evaluate the extent of liver inflammation and fibrosis through the histopathological examination of liver tissue of HCV patients. The stage indicates the extent of the fibrosis of the scarring. The fibrosis stages are as follows: F0: no fibrosis; F1: portal fibrosis with no septa; F2: portal fibrosis with few septa; F3: several septa with no cirrhosis; F4: cirrhosis. We collected the F0, F1, F2, and F3 in one group, the chronic hepatitis C (CHC) with cirrhosis (LC) in another group and the HCV-induced HCC group.
RNA extraction
For RNA isolation from serum, 250 µl of serum was homogenized in 750 µl of Trizol LS (Invitrogen, Carlsbad, CA, USA). Then 200 µl of chloroform was added to the sample followed by centrifugation. After an additional chloroform extraction and precipitation with isopropanol, the pellet was washed twice by centrifugation with 70% ethanol. The RNA pellet was dried for 10 min at room temperature and dissolved in 30 µl of diethylpyrocarbonate (DEPC)-treated water. DNase treatment (Qiagen, Valencia, CA) was carried out to remove any contaminating DNA. The concentration and quality of RNA were measured by A260/280 ratio and checked by gel electrophoresis individually.
Reverse Transcription (RT) and Quantitative PCR (qPCR)
RT reactions were performed using a TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Each 15 µL RT reaction included 7 µL of the RT master mix, 3 µL of the specific miRNA RT primer, and 5 µL of the total extracted RNA. All RT reactions were incubated for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C, and then maintained at 4°C. For qPCR, 1.33 µL of RT product was mixed with 10 µl of TaqMan 2X Universal PCR master mixture (Applied Biosystems, Foster City, CA, USA), 1 µl of the specific TaqMan MicroRNA assay (supplementary data table I), and 7.67 µl of nuclease-free water to bring the final reaction volume of 20 µl according to manufacturer protocol. All reactions were run on the Step One Plus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the following conditions: 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. qPCR was done in duplicates, including no-template controls. Relative expression of miRNA was calculated using the comparative cycle threshold (Ct) (2−ΔΔCT) method, with miR-39 as the endogenous control to normalize the data.
Statistical analysis
Data were analyzed using Statistical Package for the Social Science (SPSS) version 24 (IBM SPSS, Chicago, IL, USA). Continuous variables are expressed as the mean ± standard deviation. The analysis of variance (ANOVA) test and the Mann-Whitney (U-test) were used for comparisons of continuous (non-discrete) variables. In the current study, Mann-Whitney was used because the data is considered nonparametric since the data does not have a normal distribution. ANOVA was used to evaluate expression differences of the chosen miRNAs between patients and controls, while the Mann-Whitney was used for the analysis and comparison of the patient demographic and biochemical data. Finally, Pearson correlation was used to determine the correlation between the miRNA expression pattern and the patient group. A significant level of p < 0.05 was used in this test. To evaluate the diagnostic accuracy of the 7 miRNAs, a receiver operating characteristic (ROC) curve analysis was carried out. The area under the ROC curve (AUC), as well as the 95% confidence interval (CI), were calculated for each miRNA in order to determine the specificities and sensitivities. For the probability (p) value: p < 0.05 was considered significant, while p < 0.001 was considered highly significant.
Supplementary Material
Funding Statement
This work was funded by Theodor Bilharz Research Institute (internal project NO: 86 diagnosis) and by American University in Cairo.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Schöler N, Langer C, Döhner H, Buske C, Kuchenbauer F. Serum microRNAs as a novel class of biomarkers: a comprehensive review of literature. Exp Hematol. 2010;38(12):1126–130. doi: 10.1016/j.exphem.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans Heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-4. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 3.Barger JF, Rahman MA, Jackson D, Acunzo M, Nana-Sinkam SP. Extracellular miRNAs as biomarkers in cancer. Food Chem Toxicol. 2016;98:66–72. doi: 10.1016/j.fct.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function, and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 5.Tinkle CL, Haas-Kogan D. Hepatocellular carcinoma: natural history, current management, and emerging tools. Biologics. 2012;6:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding J, Wang H. Multiple interactive factors in hepatocarcinogenesis. Cancer Lett. 2014;346(1):17–23. doi: 10.1016/j.canlet.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 7.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 8.Kandeel A, Genedy M, El-Refai S, Funk AL, Fontanet A, Talaat M. The prevalence of hepatitis C virus infection in Egypt 2015: implications for future policy on prevention and treatment. Liver Int. 2017;37(1):45–53. doi: 10.1111/liv.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalifa RH, Labib DA, Kamel MA, Shahin RMH, Bahgat DMR, Riad NM, El Khateeb E, El-Deeb AM, Hassan M. Role of ApoB-516C/T promoter gene polymorphism in the risk of Hepatitis C virus infection in Egyptian patients and in gender susceptibility. J Med Virol. 2017;89(9):1584–1589. doi: 10.1002/jmv.24815. [DOI] [PubMed] [Google Scholar]
- 10.Elgharably A, Gomaa AI, Crossey MM, Norsworthy PJ, Waked I, Taylor-Robinson SD. Hepatitis C in Egypt- past, present, and future. Int J of Gen Med. 2017;10:1–6. doi: 10.2147/IJGM.S119301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabibbo G, Reig M, Gadaleta-Caldarola G, Galati G, Lombardi G, Mazza G, Marzi L, Saitta C, Nault JC, Sacco R. The calm before the storm: a report from the International Liver Cancer Association congress 2015- part 1. Future Oncol. 2016;12(3):281–284. doi: 10.2217/fon.15.323. [DOI] [PubMed] [Google Scholar]
- 12.Aubé C, Oberti F, Lonjon J, Pageaux G, Seror O, N'Kontchou G, Rode A, Radenne S, Cassinotto C, Vergniol J, et al.. EASL and AASLD recommendations for the diagnosis of HCC to the test of daily practice. Liver Int. 2017;37(10):1515–1525. doi: 10.1111/liv.13429. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al.. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 14.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112(1):55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 15.Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, Koong AC. Circulating miR-210 as a novel hypoxia marker in pancreatic Cancer. Transl Oncol. 2010;3(2):109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 17.Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol. 2011;45(4):355–360. doi: 10.1097/MCG.0b013e3181f18ac2. [DOI] [PubMed] [Google Scholar]
- 18.Ge W, Yu DC, Li QG, Chen X, Zhang CY, Ding YT. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin Lab. 2014;60(3):427–434. [DOI] [PubMed] [Google Scholar]
- 19.Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ, Li W, Bi J, Zhang LJ, Fu Q, et al.. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39(8):786–794. doi: 10.1111/j.1872-034X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- 20.Miao HL, Lei CJ, Qiu ZD, Liu ZK, Li R, Bao ST, Li MY. MicroRNA-520c-3p inhibits hepatocellular carcinoma cell proliferation and invasion through induction of cell apoptosis by targeting glypican-3. Hepatol Res. 2014;44(3):338–348. doi: 10.1111/hepr.12121. [DOI] [PubMed] [Google Scholar]
- 21.Yu L, Ding GF, He C, Sun L, Jiang Y, Zhu L. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PLoS One 2014;9(3):e91661. doi: 10.1371/journal.pone.0091661. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Shen J, Wang Q, Gurvich I, Remotti H, Santella RM. Evaluating normalization approaches for the better identification of aberrant microRNAs associated with hepatocellular carcinoma. Hepatoma Res. 2016;2:305–315. doi: 10.20517/2394-5079.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Q, Cicinnati VR, Zhang X, Iacob S, Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G, et al.. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma. Mol Cancer. 2010;9:227. doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406 (1):70–73. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 25.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bi Q, Tang S, Xia L, Du R, Fan R, Gao L, Jin J, Liang S, Chen Z, Xu G, et al.. Ectopic expression of miR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One 2012;7(6):e40169. doi: 10.1371/journal.pone.0040169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu W, Li X, Wang J. miR-139 regulates the proliferation and invasion of hepatocellular carcinoma through the WNT/TCF-4 pathway. Oncol Rep. 2014;31(1):397–404. doi: 10.3892/or.2013.2831. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Hu C, Cheng J, Chen B, Ke Q, Lv Z, Wu J, Zhou Y. MicroRNA-145 suppresses hepatocellular carcinoma by targeting IRS1 and its downstream Akt signaling. Biochem Biophys Res Commun. 2014;446(4):1255–1260. doi: 10.1016/j.bbrc.2014.03.107. [DOI] [PubMed] [Google Scholar]
- 29.Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, Farci P. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133(4):816–824. doi: 10.1002/ijc.28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawaguchi T, Komatsu S, Ichikawa D, Tsujiura M, Takeshita H, Hirajima S, Miyamae M, Okajima W, Ohashi T, Imamura T, et al.. Circulating microRNAs: a next-generation clinical biomarker for digestive system cancers. Int J Mol Sci. 2016;17(9): pii: E1459. doi: 10.3390/ijms17091459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


