ABSTRACT
Stromal/cytoplasmic collagen XIαI (COL11A1) has been highlighted in the process of neoplastic transformation, including epithelial mesenchymal transition (EMT), metastasis and invasiveness. In this study, we aim to illuminate the clinical significance and biological role of COL11A1 in esophageal squamous cell carcinoma (ESCC). Herein, we investigated COL11A1 expression in 16 pairs of ESCC and adjacent normal tissues by RT-PCR and western blotting analysis. Correlations of COL11A1 expression with clinicopathologic parameters and survival status were then determined by immunohistochemistry in 116 ESCC and 50 normal specimens. Furthermore, bioinformatics was used for mechanisms exploration. And in vitro knockdown experiments were also performed. We found that COL11A1 expression was significantly higher in ESCC than in paired normal tissues at both mRNA and protein level. Immunohistochemistry showed that COL11A1 was predominantly localized to the cytoplasm rather than tumor stroma, patients with high COL11A1 expression had a poorer overall survival (OS) rate than those with low COL11A1 expression. Besides, increased COL11A1 expression was dramatically correlated with advanced clinical stage, invasion depth and lymph node metastases and served as an independent prognostic marker for ESCC. Likewise, COL11A1 dependent nomogram predicted a more precise survival outcome than traditional staging system. Moreover, COL11A1 silencing resulted in impaired cell proliferation and EMT, and subdued EMT inhibited cells aggressiveness. These biological processes (BPs) might be modulated by COL11A1 via the intracellular AKT/ERK/c-Myc cascades.
KEYWORDS: AKT, COL11A1, cytoplasm, EMT, ESCC, nomogram, prognosis
Introduction
ESCC is broadly pervaded in east Asia and serves as the most common variant of esophageal cancer in China.1,2 Regardless of highly evolutive therapies comprising endoscopic resection and ablation, surgery and neoadjuvant chemoradiotherapy, ESCC remains a devastating 5-year survival rate.2 Advanced stage at initial diagnosis due to dormant clinical manifestation is the primary cause in contribution to the grim outcome. Besides, dysregulation of oncogenes and tumor suppressor genes reinforces lethality of ESCC. Digging of somatic genomic alternations and underlying mechanisms is urgently required.
Collagens, a family of 28 members, is the main structural protein of the extracellular matrix consisting of three subtypes, namely fibrillar collagens, non-fibril forming collagens, and fibril-associated collagens.3 Collagen XI, a minor fibrillar collagen found in many tissues, is mostly detected in cartilage. It is a heterotrimeric protein composed of three α chains. Both αI and αII are genetically diverse, and αIII is a hyperglycosylated version of collagen IIαI.4 Fischer et al firstly advocated the oncogenic property of stromal COL11A1 in colorectal carcinoma.5 Another study clarified the tumorigenic character of COL11A1 in ovarian cancer and its role as a downstream molecular of TGFβ1 signaling pathway.6 With rare foci of epithelial cells expressing COL11A1, Cheon et al also found the criticality of COL11A1 confined to intra/peritumoral stroma in accelerating ovarian cancer progression.7 Additionally, COL11A1 protein expression was found in Golgi apparatus and cytoplasm of normal and malignant colon specimens.8 In situ hybridization signals of COL11A1 were acquired in stromal cells, which may be a potential marker for malignant and premalignant stomach lesion discrimination.9 However, cytoplasmic COL11A1 was also observed in breast cancer tissues.10 Known that the role of COL11A1 in cancer progression is complex and its localization varies. We thought to investigate the clinical significance and biological role of COL11A1 in ESCC with previous conclusion that COL11A1 was frequently amplified in ESCC.11
In this study, we identified that COL11A1 was remarkably overexpressed in ESCC tissues and primarily localized to the cytoplasm. Overexpression of COL11A1 was significantly correlated with advanced stage, lymph nodes involvement, and poor prognosis. Furthermore, nomogram established upon COL11A1 expression level showed a more precise and reliable prognostic prediction. In vitro study showed that downregulated COL11A1 inhibited ESCC growth and aggressiveness, corresponding underlying mechanism was also investigated.
Materials and methods
Patients and specimens
A total of 116 paraffin-embedded ESCC specimens and 50 stochastically selected normal ones were collected, which were histopathologically and clinically diagnosed at The First Affiliated Hospital of Chongqing Medical University (Chongqing, China) from 2009 to 2013. All patients received no chemotherapy or radiotherapy before surgery. Pathological TNM classification and corresponding staging were defined in accordance with the criteria proposed by 7th American Joint Committee on Cancer (AJCC). All patients signed informed consent for sample collection and Institutional Research Ethics Committee approval was obtained. Another 16 paired ESCC samples were snap-frozen in liquid nitrogen after resection and stored until required.
Immunohistochemistry (IHC)
Briefly, 4-μm formalin-fixed and paraffin-embedded ESCC specimens were baked at 60°C for 30 min, deparaffinized and rehydrated in xylenes and graded ethanol respectively. Sections were submerged into citrate buffer (pH 6.0) and microwaved for antigenic retrieval. Next, 3% hydrogen peroxide in methanol was utilized to quench endogenous peroxidase activity, followed by interdiction for nonspecific binding. Rabbit anti-COL11A1 (1:50, Cat. No. PA5-36227, Invitrogen) was incubated with the slides overnight in a moist chamber at 4°C. After washing in phosphate-buffered saline (PBS), slides were treated with biotinylated goat anti-rabbit secondary antibody, stained by 3,3′-diaminobenzidine (DAB) system and counterstained with hematoxylin. The expression status of immunostaining was reviewed and scored independently by two pathologists based on positive cells proportion and staining intensity. Scoring for percentage was as follows: 0% (0), <10% (1), 10–35% (2), 35–75% (3), and >75% (4). Scoring for intensity: no staining (1), yellowish (2), claybank (3), and tawny (4). Staining index (SI), the product of percentage score and intensity score comprising 0, 1, 2, 3, 4, 6, 8, 9, 12 and 16, was the final criterion for the evaluation. Samples were categorized as either high expression (SI ≥ 8) or low expression (SI<8).
Cell culture
The ESCC cell lines TE-10, EC109, KYSE150, KYSE410, KYSE510 and EC9706, obtained from Chongqing Key Laboratory of Molecular Oncology and Epigenetics (Chongqing, China), were maintained in RPMI-1640 medium (HyClone) supplemented with 10% fetal bovine serum (FBS) (PAN Biotech), 100Unist/mL penicillin and 100μg/mL streptomycin (Gibco), at 37°C in a humidified chamber with 5% CO2.
RNAi construction
The short hairpin RNA (shRNA) targeting human COL11A1 (5′-ACCGGTGAGACTGGTCCAATA-3′) and the negative control shRNA (5′-TTCTCCGAACGTGTCACGT-3′) were designed, synthesized and cloned into the GV248 vector by GeneChem. Compound vector, pHelper1.0 and pHelper2.0 were co-transfected into HEK293T cells, followed by virus supernatants collection, concentration and purification. KYSE510 was infected with acquired lentivirus particles using polybrene (5μg/mL) and enhanced infection solution (ENI.S.) according to the manufacturer's protocol. Application of 1μg/mL puromycin (BioVision) selected stable cells expressing COL11A1 RNA interference (COL11A1-RNAi) and negative control (COL11A1-vector).
RNA extraction, reverse transcription, and real-time PCR (RT-PCR)
Total RNA from ESCC tissues was extracted using the RNAiso Plus (Cat. No. 9108, Takara) according to the manufacturer's instruction. Subsequently, cDNA was synthesized for quantitative RT-PCR (qPCR) and semi-quantitative RT-PCR (SqPCR) with employment of PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (Cat. No. RR047A, Takara). qPCR was performed on the 7500 Real-Time PCR System (Applied Biosystems) with SYBR Premix Ex Taq™ II (Tli RNaseH Plus) (Cat. No. RR820A) according to the standard procedure. For SqPCR, the amplification was performed on an ABI Prism 7000 detection system (Applied Biosystems) under the following conditions: 1. 95°C for 2 min; 2. 95°C for 30s, 55°C for 30s, and 72°C for 30s (COL11A1 – 36cycle, GAPDH – 23cycle); 3. 72°C for 3 min. After electrophoresis, the PCR products were visualized by Quantity One 4.6.2 (Bio-Rad) via ultraviolet ray. Primers designed by Primer Premier 5 and Oligo 6.0 were as follows: (1) COL11A1 F: 5′-TTGATGTTACCGTTCCGTTATG-3′ and R: 5′-AATCCGAGCCTGCTGAAGAAT-3′ (qPCR); F: 5′-TGGTGATCAGAATCAGAAGTTCG-3′ and R: 5′-AGGAGAGTTGAGAATTGGGAATC-3′ (SqPCR). (2) GAPDH F: 5′-GGAGTCAACGGATTTGGT-3′ and R: 5′-GTGATGGGATTTCCATTGAT-3′ (qPCR); F: 5′-ATCTCTGCCCCCTCTGCTGA-3′ and R: 5′-GATGACCTTGCCCACAGCCT-3′ (SqPCR).
Western blotting
Western blotting was performed according to standard procedures using primary antibodies, anti-COL11A1 (1:500, AP21844a, ABGENT), anti-β-catenin (1:500, sc-7963), anti-E-cadherin (1:500, sc-8426), anti-Vimentin (1:300, sc-6260), anti-FAK (1:500, sc-271126), anti-p-FAK (1:500, sc-81493), anti-p-Akt (1:300, sc-514032), anti-ERK1/2 (1:500, sc-514302), anti-p-ERK1/2 (1:500, sc-7383), anti-c-Myc (1:300, sc-40) (Santa Cruz Biotechnology), anti-N-cadherin (1:300, WL01047), anti-MMP2 (1:500, WL1579), anti-MMP9 (1:500, WL01580), anti-Slug (1:750, WL01508) (Wanleibio), anti-Akt (1:1000, #4691), anti-p-Akt (1:1000, #2965) (Cell Signaling Technology), anti-CyclinD1 (1:1000, bs-0623R), and anti-GAPDH (1:1000, bs-2188R) (Bioss). Band was visualized by Immobilon Western Chemiluminescent HRP Substrate (Cat. No. WBKLS0100, Merck Millipore).
Cell proliferation and foci formation assays
Log-phase cells were seeded in 96-well plate at a density of 2000/well. Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies) was used to detect cell proliferation rate. The optical density (OD) at 450 nm was acquired using a microplate reader (Infinite 200 PRO, Tecan). To perform foci formation assay, log-phase cells were seeded in 6-well plate at a density of 500/well. After incubated for 12 d, viable colonies were counted after 4% paraformaldehyde fixation and crystal violet staining. Triplicate independent experiments were performed.
Cell migration and invasion assays
Transwell cell migration assay was performed using 8-μm pore size transwell insert chambers (Cat. No. 3422, Corning). Matrigel-coated (Cat. No. 356234, BD Biosciences) chambers were used for invasion assay. Cells (5 × 104) were suspended in serum-free RPMI-1640 onto the upper chambers, with the lower ones containing complete medium (10% FBS), and incubated for 24 hours. Penetrated cells were fixed in 4% paraformaldehyde, stained with crystal violet, and counted under a × 100 magnification (six random fields) after gentle removal of inside cells with cotton swabs. Implementation of wound healing assay via artificial wound scraping in highly confluent cells was also for cells migration ability examination, percentage of wound closure was measured by relative closure distance (24 h to 0 h) under identical magnification. Triplicate independent experiments were performed.
Statistical analysis
Accomplishment of all statistical analyses were based upon R version 3.4.0 (www.r-project.org), GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla California USA) and SPSS 20.0 (IBM, Armonk, NY, USA). Data were primarily tested for normal distribution with the One-Sample Kolmogorov Smirnov test. If so, paired t test or independent t test was adopted for two-group data comparisons. Otherwise the Wilcoxon matched-pairs signed rank test or nonparametric Mann-Whitney test were used. Enumeration data were examined by χ2 test. Estimation of linearity degree was built on Spearman's rho due to outliers (data not shown). Survival curves were constructed using Kaplan-Meier estimates, and variable values were compared using the Mantel-Cox test, followed by multivariate analyses for significant variables via Cox regression model (backward: LR). Data were presented as means ± SD or means ± SEM, p<0.05 was considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
Integrative analysis identified COL11A1 oncogenic property in ESCC
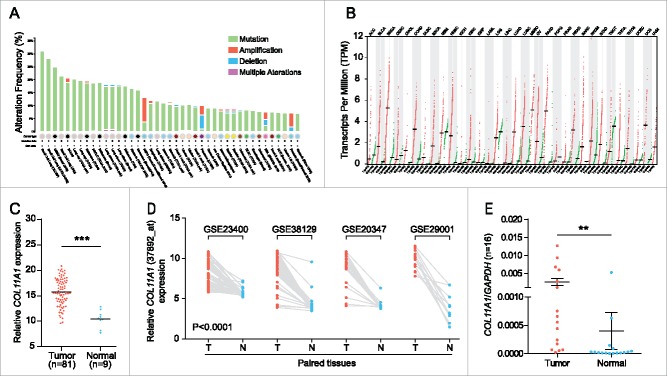
To understand the tumorigenic profiles of COL11A1, we initially mined the genome alterations of COL11A1 in numerous cancer types via The Cancer Genome Atlas (TCGA) using cBioPortal12 (www.cbioportal.org). A high somatic mutation rate with a moderate amplification rate was observed (Fig. 1A). Besides, higher COL11A1 mRNA level was found in several malignancies under RNAseq expression profiling data in TCGA (Fig. 1B); and ESCC samples sorted from TCGA esophageal carcinoma emphasized the distinct rise of COL11A1 in neoplasms (Fig. 1C). Subsequently, four Gene Expression Omnibus (GEO) microarrays were used to detect COL11A1 expression in paired ESCC and normal samples, and the latter exhibited lower transcription level of COL11A1 with valid reliability (Fig. 1D). We then examined COL11A1 mRNA expression level in 16 paired ESCC and matched normal tissues via qPCR. As expected, COL11A1 was significantly upregulated in tumor tissues (Fig. 1E). Furthermore, SqPCR and western blotting analysis in 8 selected pairs confirmed that COL11A1 indeed overexpressed in ESCC samples at mRNA/protein level (Fig. 2A and B). A large cohort of 116 ESCC with 50 randomly selected normal tissues were obtained for IHC detection. Intriguingly, COL11A1 was found located in the cytoplasm of all positive cases, with only 40.57% (43/106) malignant cases showed faint stroma staining (Fig. 2C). This indicated that COL11A1 was primarily produced by epithelial compartment instead of stromal fibroblasts. Immunoreactive score of COL11A1 not only exhibited a higher level in ESCC, but also showed a stage dependent manner (Fig. 2C). These results revealed the vital role of COL11A1 in ESCC.
Figure 1.
Pan-cancer genomic patterns and expression status of COL11A1. (A) High frequency of COL11A1 somatic mutation was observed across multiple cancers. (B) COL11A1 was overexpressed in multiple types of cancer. (C) Comparison of COL11A1 mRNA level using TCGA dataset. (D) Before-after graph of COL11A1 mRNA expressive discrepancy between ESCC (T) and non-cancerous esophagus tissues (N) in 4 microarrays. (E) qPCR results showed high COL11A1 expression in 16 paired ESCC tissues (p = 0.0092, Wilcoxon matched-pairs signed rank test). Data were presented by means ± SEM and 2−ΔCt. **p < 0.01, ***p < 0.001.
Figure 2.
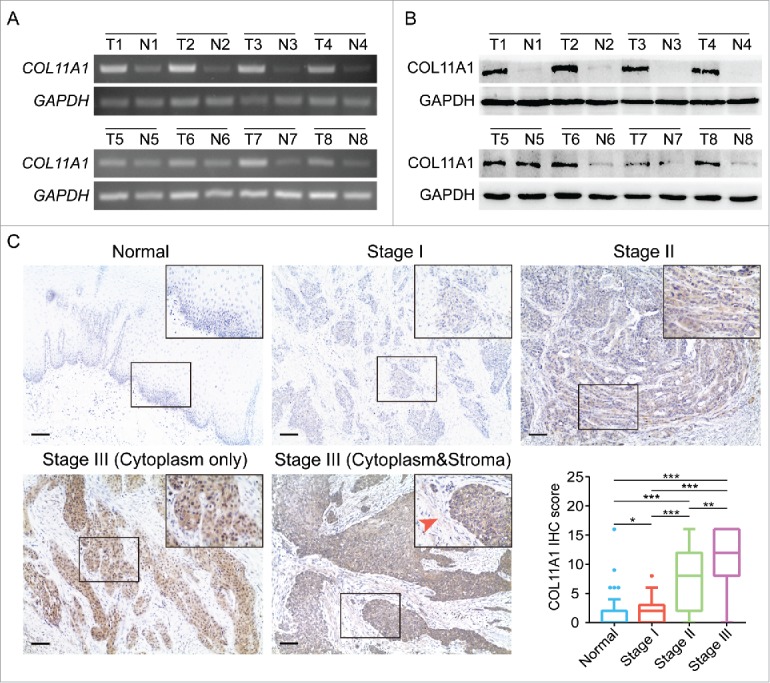
Expression of COL11A1 in ESCC samples. (A) and (B) SqPCR and western blotting analysis of COL11A1 expression in 8 paired ESCC tissues and the corresponding normal epithelial tissues. (C) Representative images of COL11A1 staining in normal and tumor samples. Immunoreactive intensity increased as cancer progressed (arrow: faint stroma expression). Scale bar, 100μm. * p<0.05, ** p<0.01, *** p<0.001.
High COL11A1 was associated with diminished overall survival time
Statistical data revealed that overexpression of COL11A1 was positively relevant to pT (p = 0.0007), pN (p = 0.0015), and clinical stage (p = 0.0070) (Table 1 and Fig. 2C). This signifies the invasiveness, lymph node metastases, and advanced stage signature in patients with high COL11A1 expression. In the cohort, 73 deaths were recorded. The median survival was 72 and 15 months in low and high COL11A1 expression group respectively. Kaplan-Meier survival curve showed that elevated COL11A1 expression predicted worse overall survival (p<0.0001; Fig. 3A).
Table 1.
Relationship between COL11A1 protein overexpression and clinicopathologic parameters in ESCC tissues.
| Parameter | COL11A1 high expression | p value |
|---|---|---|
| Sex | ||
| Female | 55.0%(11/20) | 0.9184 |
| Male | 56.3%(54/96) | |
| Location | ||
| Lower/Middle | 55.4%(56/101) | 0.7402 |
| Upper | 60.0%(9/15) | |
| Age, y | ||
| <61.5 | 56.9%(33/58) | 0.8516 |
| ≥61.5 | 55.2%(32/58) | |
| pT | ||
| T1/T2 | 37.5%(18/48) | 0.0007 |
| T3/T4 | 69.1%(47/68) | |
| pN | ||
| N0 | 43.1%(28/65) | 0.0015 |
| N1/N2/N3 | 72.5%(37/51) | |
| AJCC7 stage | ||
| I/IIA | 37.1%(13/35) | 0.0070 |
| IIB/III | 64.2%(52/81) | |
| Grade | ||
| G1 | 63.0%(17/27) | 0.4076 |
| G2/G3 | 53.9%(48/89) |
Abbreviations: AJCC7, American Joint Committee on Cancer; pN, lymph node metastases; pT, pathologic T stage.
Figure 3.
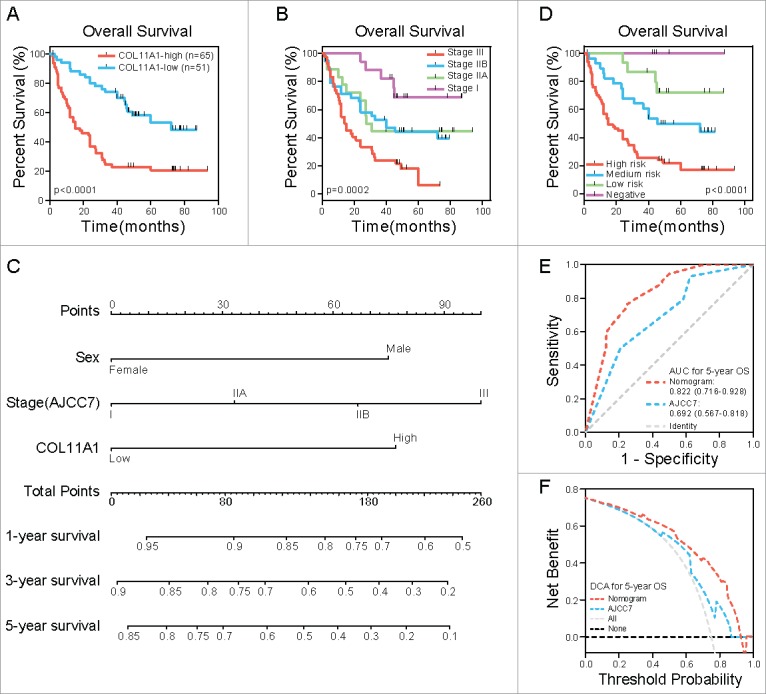
COL11A1 and nomogram in ESCC outcome prediction. (A) Kaplan-Meier analysis revealed that high expresion of COL11A1 is closely corrlated with dimished OS time. (B) Role of tranditional staging system in OS status. No significant difference was found between stage IIA and IIB. (C) Nomogram built on COL11A1, sex and stage concerning 1, 3, 5-year suvival probability of ESCC patients. (D) Patients grouped by nomogram derived scores showed distinct survival outcome. (E) and (F) ROC curve and DCA indicated that nomogram possessed a more precise prediction for 5-year OS.
Univariate analysis results were listed in Table 2. Female patients were associated with more pleasant outcome (p = 0.0176). Pathological T (p = 0.0067) and N status (p = 0.0184) both had influence on survival. Clinical stage was also linked to patients' terminal status (p = 0.0001), nevertheless, no statistically significance was found between IIA and IIB stage (Fig. 3B). Factors with significance were put together for multivariate analysis grounded on the Cox regression model. Sex (p = 0.0284), stage (p = 0.0376) and COL11A1 (p = 0.0012) were independent prognostic factors for OS (Table 2).
Table 2.
Univariate and Coxph regression analysis for overall survival time of patients with ESCC.
| Univariate | Multivariate analysis and point assignment |
||||
|---|---|---|---|---|---|
| Parameter | analysis p | HR | 95% CI | p value | Score |
| Sex | 0.0176 | 0.0284 | |||
| Female | reference | 0 | |||
| Male | 2.4168 | 1.0981 – 5.3195 | 0.0284 | 75 | |
| Location | 0.6969 | ||||
| Lower/Middle | |||||
| Upper | |||||
| Age, y | 0.4988 | ||||
| <61.5 | |||||
| ≥61.5 | |||||
| pT | 0.0067 | ||||
| T1 | |||||
| T2 | |||||
| T3 | |||||
| T4 | |||||
| pN | 0.0184 | ||||
| N0 | |||||
| N1 | |||||
| N2 | |||||
| N3 | |||||
| AJCC7 stage | 0.0001 | 0.0376 | |||
| I | reference | 0 | |||
| IIA | 1.1655 | 0.3712 – 3.6594 | 0.7930 | 33 | |
| IIB | 1.8573 | 0.6835 – 5.0473 | 0.2248 | 67 | |
| III | 2.7659 | 1.0047 – 7.6144 | 0.0490 | 100 | |
| Grade | 0.8074 | ||||
| G1 | |||||
| G2 | |||||
| G3 | |||||
| COL11A1 | 0.0000 | 0.0012 | |||
| Low | reference | 0 | |||
| High | 2.5059 | 1.4366 – 4.3711 | 0.0012 | 77 | |
Abbreviations: Coxph, cox proportional hazard; pT, pathologic T stage; pN, lymph node metastases; AJCC7, the 7th American Joint Committee on Cancer; COL11A1, collagen XIαI; HR, hazard ratio; CI, confidence interval.
Construction of prognostic nomogram for OS
With ambiguous outcome prediction among stage IIA & IIB patients, nomogram incorporating sex, stage and COL11A1 was established according to previously reported approach,13 aiming for rigorous outcome prediction (Fig. 3C). Scores were estimated from Fig. 3C and presented in Table 2. Total score and estimated survival probability at a specific time point were acquired as previously described.13 We then grouped the cohort evenly into four subgroups after total score systematization (0: negative, 33–100: low risk, 108–152: medium risk, 175–252: high risk); each group stood for distinct prognosis (Fig. 3D). To better understand the prognostic value of the nomogram, receiver operating characteristic (ROC) curve was applied to compare it with 7th AJCC staging system on 5-y OS. As illustrated in Fig. 3E, the nomogram displayed higher specificity and sensitivity. Besides, in the decision curve analysis (DCA), nomogram showed better net benefits basically throughout the whole spectrum of threshold probabilities (Fig. 3F).
COL11A1-knockdown attenuated cell proliferation and impaired migration/invasion ability by dissipating EMT
We measured the protein levels of COL11A1 in six ESCC cell lines, and KYSE510 exhibited strongest intensity. Afterwards, shRNA was stably infected into KYSE510, knockdown efficacy was confirmed by western blotting analysis (Fig. 4A). CCK8 assay found that COL11A1 depletion dramatically reduced the cell viabilities (Fig. 4B). Foci formation showed that decreased expression of COL11A1 curtailed the ESCC cells colony formation ability (Fig. 4C).
Figure 4.
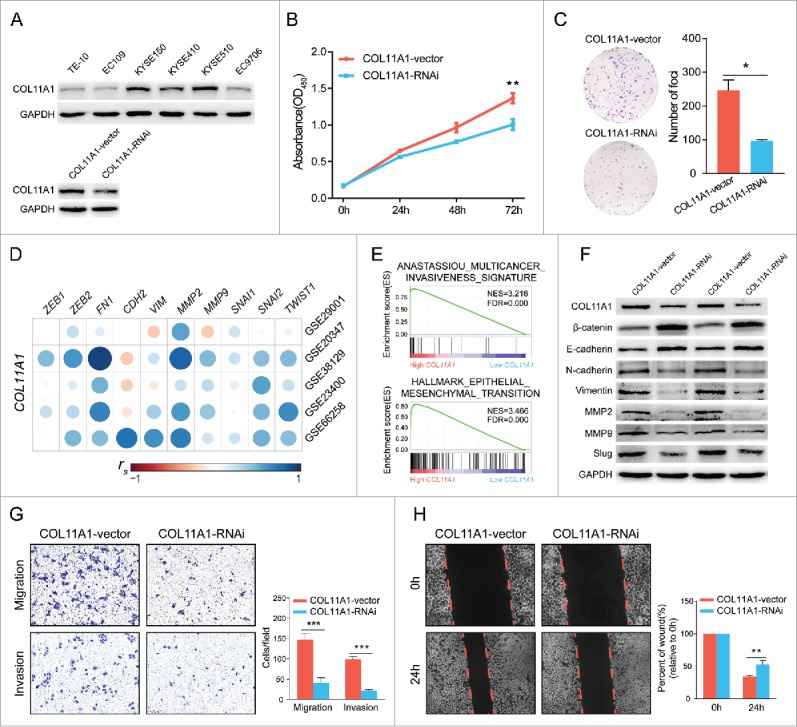
Bioinformatics analysis and in vitro experiments with silenced COL11A1 and negative control. (A) Expression levels of COL11A1 in 6 ESCC cell lines were measured through western blotting assay. Cells infected with shCOL11A1 showed remarkable silencing efficiency. (B) and (C) ESCC cells with inhibition of COL11A1 expression resulted in decreased growth rate (p = 0.0035) and impaired colony formation ability (p = 0.0126). (D) Correlation analysis revealed strong connections between COL11A1 and mesenchymal markers (spearman's correlation). (E) GSEA results revealed an enhancive effect of COL11A1 in cancer invasion and EMT. (F) Increased epithelial markers and decreased mesenchymal markers were found in COL11A1-knockdown cells. (G) Transwell assay showed that cells with down-regulated COL11A1 could weaken migration (p = 0.0008) and invasion (p<0.0001) capacity. (H) A delay of wound closure was observed in COL11A1-knockdown cells (p = 0.0062). Triplicate independent experiments were performed. * p<0.05, ** p<0.01, *** p<0.001.
Given that COL11A1 was associated with EMT,7,14 we then detected correlations between COL11A1 and representative mesenchymal markers in five GEO datasets and obtained a positive conclusion (Fig. 4D). GSE66258, an expression array with high quality and relative vast ESCC samples, was then performed for gene set enrichment analysis (GSEA) on GSEA v3.0 (http://www.broad.mit.edu/gsea/) via the Molecular Signature Database (MSigDB).15 Prominent enrichment among high-COL11A1 cluster genes for multiple cancer invasiveness signature16 and EMT were obtained (Fig. 4E). Moreover, COL11A1 deficiency significantly induced the expression of epithelial markers (β-catenin and E-cadherin), inversely reduced mesenchymal markers (N-cadherin, vimentin, MMP2, MMP9, and slug) expression at protein level (Fig. 4F). Furthermore, transwell assay indicated that cells with silenced COL11A1 showed impaired migratory and invasive ability; wound healing assay reached a similar result on migration (Fig. 4G and H). These data conclusively suggested that downregulation of COL11A1 may affect ESCC migration-invasion cascades via deregulation of EMT.
COL11A1 depletion influenced intracellular signaling transduction
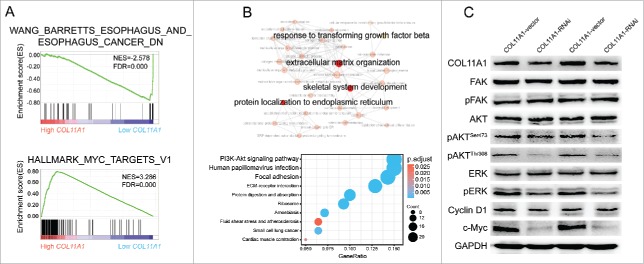
We have identified that impaired cell proliferation, migration, and invasion were generated by COL11A1 depletion. GSEA results also indicated that low-COL11A1 cluster genes overlapped prominently with minus esophagus caner probability, while high-COL11A1 cluster genes enriched significantly with target genes of MYC (c-Myc) signaling pathway (Fig. 5A). Moreover, we applied clusterProfiler17 to achieve Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. For GO annotation, highlighted hub BPs were presented; KEGG pathway analysis revealed that overexpression of COL11A1 may participate in PI3K/Akt and focal adhesion signaling pathway (Fig. 5B). Since ERK signaling pathway has been implicated in regulation of cell proliferation and EMT in various cancers, we therefore utilized western blotting for hypothesis verification that COL11A1 may facilitate ESCC growth and EMT via AKT/ERK/c-Myc signaling pathway. As expected, protein levels of phosphorylated-ERK (p-ERK), phosphorylated-Akt (p-Akt) at both serine 473 (Ser473) and threonine 308 (Thr308) sites, and c-Myc in cells with stable COL11A1-knockdown were conspicuously attenuated. Focal adhesion kinase (FAK), a protein involved in cellular adhesion, was also tested, yet the phosphorylation level of FAK at tyrosine 397 (Tyr397) appeared no change. Cyclin D1, a downstream molecular of AKT and ERK, showed monotonous expression after detection (Fig. 5C).
Figure 5.
Cytoplasmic COL11A1 might involved in Akt/ERK/c-Myc cascades. (A) GSEA enrichment results indicated that low COL11A1 was negatively correlated with esophagus cancer; and high COL11A1 may participate in c-Myc signaling pathway. (B) GO and KEGG annotation using R package clusterProfiler. COL11A1 might involved in PI3K-Akt cascades. (C) Protein levels of FAK, pFAK, AKT, pAKT, ERK, pERK, cyclin D1, c-Myc were assessed by western blotting assay. Triplicate independent experiments were performed.
Discussion
The gene mutations of structural protein collagen XIαI has been well investigated in musculoskeletal disorders, such as type II Stickler syndrome and Marshall syndrome.18,19 It is noteworthy that COL11A1 may serve as a promising putative tumor marker due to its barely-existing character in most tissues.18 In this study, we found that COL11A1 was not only amplified, but also prone to mutating in many types of cancer. Significant mRNA and protein expression differences among ESCC and bordering normal tissues were recorded. COL11A1 upregulation conferred poor outcome. Besides, nomogram built on COL11A1 showed a more precise and rigorous outcome prediction than AJCC7 staging system. Furthermore, in vitro study revealed attenuated cell proliferation and colony formation ability modulated by COL11A1 knockdown. COL11A1 silencing contributed to impaired migratory and invasive ability through EMT reversion. These effects might be regulated through AKT/ERK/c-Myc cascades.
We observed that COL11A1 is aberrantly mutated in various cancers. Besides, mutation of COL11A1 exon 54 was associated with colorectal cancer.18 Further studies focusing on COL11A1 mutation and cancer progression are required. In our cohort, COL11A1 was an independent prognostic marker for OS of ESCC patients (HR = 2.5059, p = 0.0012). COL11A1-positve patients had a lower median survival than COL11A1-negative patients (15 vs 72 months). Equivalently, GSEA revealed that high COL11A1 expression had a tight connection with liver cancer poor survival (data not shown). Collectively, COL11A1 appears to be an ideal outcome predictor for ESCC patients. A pluralistic outcome prediction system — nomogram, was then constructed, which demonstrated a first-class AUC and superb net benefits for 5-year OS prediction. However, by lacking of external validation and uncertainty of sex in ESCC, bias could exist. A multicenter study with larger sample size is needed.
Anastassiou el al conducted a meta-analysis to shed light on biological mechanisms of tumor invasiveness.16 They revealed the peak role of COL11A1 in a metastasis-associated gene expressing signature. To confirm the data, they carried out a xenograft experiment concerning neuroblastoma, and proved that mRNA levels of COL11A1 and other EMT-related genes were highly and merely expressed in xenografted human cells, not the ambient mouse stroma.14 Consistently, immunofluorescence confirmed that COL11A1 was localized to normal and neoplastic epithelium and stroma, whereas epithelial derived COL11A1 mRNA levels were statistically higher than those that derived from stroma.20 These are in agreement with our findings that COL11A1 was localized to cytoplasm of all positive tumor tissues and corresponding normal epithelial tissues, with only a small proportion of tumor samples showing faint expression in stroma. Paralleled results demonstrated that type VII collagen and COL6A1 were localized to the cytoplasm of ESCC.21,22 Difference as they may have in ways of signal transmission, oncogenic certainty of stromal and cytoplasmic COL11A1 do exist in some cancer types. The interplay and underlying mechanisms need further investigation.
EMT, a process that epithelial cells loosen adhesion ability and transdifferentiate into isolated mesenchymal cells, has a crucial role in modulation of tumor cell migration and invasion, including ESCC. Notably, in patients with advanced pathological T (T3-T4) stage and lymph node metastases, high COL11A1 expression accounted for 69.1% and 72.5% respectively. Suggesting that up-regulated COL11A1 might take part in the process of invasiveness and regional migration. Besides, in vitro assay indicated that COL11A1-knockdown cells had a lower probability of migration and invasion. Alternatively, we found that ESCC cells with inhibition of COL11A1 expressed higher levels of E-cadherin and β-catenin and lower levels of N-cadherin, vimentin, MMP2, MMP9 and slug. Together with correlation analysis and GSEA results, we conclude that COL11A1 contribute to the aggressiveness of ESCC via the EMT property. To our knowledge, being an important transcription factor, slug regulates EMT via suppressing E-cadherin expression. And slug, together with MMP2, MMP9 and c-Myc, have been well characterized as downstream targets of AKT and ERK.23-26 Ewald et al said that COL11A1 may promote bladder cancer invasiveness through PI3K/Akt and Ras/ERK signaling pathway.27 Wu et al highlighted the magnitude of COL11A1 in chemoresistance of ovarian cancer patients through increasing p-AKT expression; comparing to chemo-naive cells, chemoresistant cells exhibited stronger intensity and higher proportion of COL11A1 staining among positive cancer cells. Intriguingly, COL11A1 was also confined to cytoplasm.28 In our study, eliminating endogenous COL11A1 expression led to decreased expression of p-AKT, p-ERK and c-Myc. Above all, we infer that impacts made by cytoplasmic COL11A1 on ESCC might be mediated via EMT and AKT/ERK/c-Myc cascades. And it is reasonable to deduce that cytoplasmic COL11A1 initiate tumor progression and protection at least in part, through AKT signaling pathway.
GO analysis annotated “endoplasmic reticulum”, GSEA and KEGG enriched in “translation” and “ribosome”, these results gave evidence for COL11A1 cytoplasm localization and its role in intracellular signal transduction, protein synthesis and processing. KEGG analysis also revealed the connection between COL11A1 and human papillomavirus infection — a controversial risk factor of ESCC.2
In summary, our results illuminate that COL11A1 helps regulate malignant biological behavior and shows potential as a promising outcome predictor and therapeutic target in ESCC.
Funding Statement
This work was supported by the Key Scientific Research Project of Chongqing Municipal Bureau of Health (2012-1-015).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Rustgi AK, El-Serag HB. Esophageal carcinoma. The New England journal of medicine. 2014;371:2499–509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- 2.Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2015;149:1700–15. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 3.Ricard-Blum S. The collagen family. Cold Spring Harbor perspectives in biology. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris NP BH. Type XI collagen is a heterotrimer with the composition (1 alpha, 2 alpha, 3 alpha) retaining non-triple-helical domains. J Biol Chem. 1987;262:11345–50. [PubMed] [Google Scholar]
- 5.Fischer H. Colorectal carcinogenesis is associated with stromal expression of COL11A1 and COL5A2. Carcinogenesis. 2001;22:875–8. doi: 10.1093/carcin/22.6.875. [DOI] [PubMed] [Google Scholar]
- 6.Wu YH, Chang TH, Huang YF, Huang HD, Chou CY. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene. 2014;33:3432–40. doi: 10.1038/onc.2013.307. [DOI] [PubMed] [Google Scholar]
- 7.Cheon DJ, Tong Y, Sim MS, Dering J, Berel D, Cui X, Lester J, Beach JA, Tighiouart M, Walts AE, et al.. A collagen-remodeling gene signature regulated by TGF-beta signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin Cancer Res. 2014;20:711–23. doi: 10.1158/1078-0432.CCR-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowen KB, Reimers AP, Luman S, Kronz JD, Fyffe WE, Oxford JT. Immunohistochemical localization of collagen type XI alpha1 and alpha2 chains in human colon tissue. J Histochem Cytochem. 2008;56:275–83. doi: 10.1369/jhc.7A7310.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, Zhou T, Li A, Yao H, He F, Wang L, Si J. A potential role of collagens expression in distinguishing between premalignant and malignant lesions in stomach. Anatomical record. 2009;292:692–700. doi: 10.1002/ar.20874. [DOI] [PubMed] [Google Scholar]
- 10.Halsted KC, Bowen KB, Bond L, Luman SE, Jorcyk CL, Fyffe WE, Kronz JD, Oxford JT. Collagen α1(XI) in normal and malignant breast tissue. Modern Pathology. 2008;21:1246–54. doi: 10.1038/modpathol.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chattopadhyay I, Singh A, Phukan R, Purkayastha J, Kataki A, Mahanta J, Saxena S, Kapur S. Genome-wide analysis of chromosomal alterations in patients with esophageal squamous cell carcinoma exposed to tobacco and betel quid from high-risk area in India. Mutation research. 2010;696:130–8. doi: 10.1016/j.mrgentox.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al.. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang W, Zhang L, Jiang G, Wang Q, Liu L, Liu D, Wang Z, Zhu Z, Deng Q, Xiong X, et al.. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol. 2015;33:861–9. doi: 10.1200/JCO.2014.56.6661. [DOI] [PubMed] [Google Scholar]
- 14.Anastassiou D, Rumjantseva V, Cheng W, Huang J, Canoll PD, Yamashiro DJ, Kandel JJ. Human cancer cells express Slug-based epithelial-mesenchymal transition gene expression signature obtained in vivo. BMC cancer. 2011;11:529. doi: 10.1186/1471-2407-11-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al.. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H, Watkinson J, Varadan V, Anastassiou D. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC medical genomics. 2010;3:51. doi: 10.1186/1755-8794-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raglow Z, Thomas SM. Tumor matrix protein collagen XIalpha1 in cancer. Cancer letters. 2015;357:448–53. doi: 10.1016/j.canlet.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith AJ, Sprunger LK, Sirko-Osadsa DA, Tiller GE, Meisler MH, Warman ML. Marshall syndrome associated with a splicing defect at the COL11A1 locus. American journal of human genetics. 1998;62:816–23. doi: 10.1086/301789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vargas AC, McCart Reed AE, Waddell N, Lane A, Reid LE, Smart CE, Cocciardi S, da Silva L, Song S, Chenevix-Trench G, et al.. Gene expression profiling of tumour epithelial and stromal compartments during breast cancer progression. Breast Cancer Res Treat. 2012;135:153–65. doi: 10.1007/s10549-012-2123-4. [DOI] [PubMed] [Google Scholar]
- 21.Baba Y, Iyama K, Honda S, Ishikawa S, Miyanari N, Baba H. Cytoplasmic expression of type VII collagen is related to prognosis in patients with esophageal squamous cell carcinoma. Oncology. 2006;71:221–8. doi: 10.1159/000106426. [DOI] [PubMed] [Google Scholar]
- 22.Fan NJ, Gao CF, Wang CS, Zhao G, Lv JJ, Wang XL, Chu GH, Yin J, Li DH, Chen X, et al.. Identification of the up-regulation of TP-alpha, collagen alpha-1(VI) chain, and S100A9 in esophageal squamous cell carcinoma by a proteomic method. Journal of proteomics. 2012;75:3977–86. doi: 10.1016/j.jprot.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Cheng X, Wei L, Huang X, Zheng J, Shao M, Feng T, Li J, Han Y, Tan W, Tan W, et al.. Solute Carrier Family 39 Member 6 Gene Promotes Aggressiveness of Esophageal Carcinoma Cells by Increasing Intracellular Levels of Zinc, Activating Phosphatidylinositol 3-Kinase Signaling, and Up-regulating Genes That Regulate Metastasis. Gastroenterology. 2017;152:1985-97 e12. doi: 10.1053/j.gastro.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Luo Y, Liang F, Zhang ZY. PRL1 promotes cell migration and invasion by increasing MMP2 and MMP9 expression through Src and ERK1/2 pathways. Biochemistry. 2009;48:1838–46. doi: 10.1021/bi8020789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sze KM, Wong KL, Chu GK, Lee JM, Yau TO, Ng IO. Loss of phosphatase and tensin homolog enhances cell invasion and migration through AKT/Sp-1 transcription factor/matrix metalloproteinase 2 activation in hepatocellular carcinoma and has clinicopathologic significance. Hepatology. 2011;53:1558–69. doi: 10.1002/hep.24232. [DOI] [PubMed] [Google Scholar]
- 26.Wang RY, Chen L, Chen HY, Hu L, Li L, Sun HY, Jiang F, Zhao J, Liu GM, Tang J, et al.. MUC15 inhibits dimerization of EGFR and PI3K-AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology. 2013;145:1436-48 e1-12. doi: 10.1053/j.gastro.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Ewald JA, Downs TM, Cetnar JP, Ricke WA. Expression microarray meta-analysis identifies genes associated with Ras/MAPK and related pathways in progression of muscle-invasive bladder transition cell carcinoma. PloS one. 2013;8:e55414. doi: 10.1371/journal.pone.0055414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu YH, Chang TH, Huang YF, Chen CC, Chou CY. COL11A1 confers chemoresistance on ovarian cancer cells through the activation of Akt/c/EBPbeta pathway and PDK1 stabilization. Oncotarget. 2015;6:23748–63. doi: 10.18632/oncotarget.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]