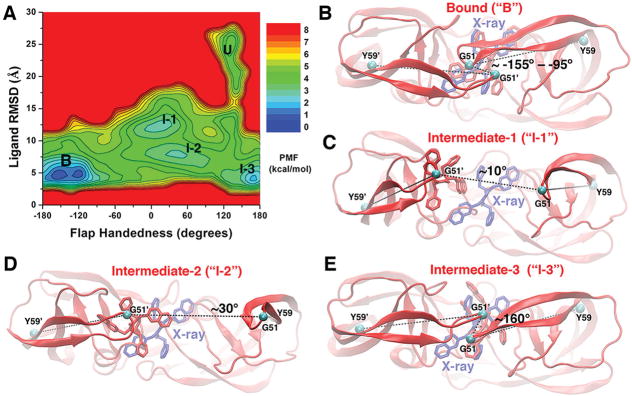
Figure 4.
(A) 2D PMF profile of the flap handedness and ligand molecule RMSD calculated by combining the “Sim1” and “Sim2” GaMD trajectories, during which the XK263 ligand molecule was observed to bind to the protein active site. Five low-energy conformational states are labeled, including the unbound (“U”), intermediate-1 (“I-1”), intermediate-2 (“I-2”), intermediate-3 (“I-3”), and bound (“B”) forms. (B–E) Structural conformations of the HIV protease in the bound (“B”), intermediate-1 (“I-1”), intermediate-2 (“I-2”), and intermediate-3 (“I-3”) states, respectively. The unbound (“U”) conformation is shown in Figure 1B. The evolving protein (ribbons) and ligand molecule (sticks) are colored red, and the X-ray conformation of the bound ligand molecule is colored blue. The dihedral angle of the Cα atoms (cyan spheres) of the Tyr59-Gly51-Gly51′-Tyr59′ motif is shown as in Figure 3.