Abstract
Recent advances in DNA sequencing technologies are revealing how human genetic variations associate with differential health risks, disease susceptibilities, and drug responses. Such information is now expected to help evaluate individual health risks, design personalized health plans and treat patients with precision. It is still challenging, however, to understand how such genetic variations cause the phenotypic alterations in pathobiologies and treatment response. Human induced pluripotent stem cell (iPSC) technologies are emerging as a promising strategy to fill the knowledge gaps between genetic association studies and underlying molecular mechanisms. Breakthroughs in genome editing technologies and continuous improvement in iPSC differentiation techniques are particularly making this research direction more realistic and practical. Pioneering studies have shown that iPSCs derived from a variety of monogenic diseases can faithfully recapitulate disease phenotypes in vitro when differentiated into disease-relevant cell types. It has been shown possible to partially recapitulate disease phenotypes, even with late onset and polygenic diseases. More recently, iPSCs have been shown to validate effects of disease and treatment-related single nucleotide polymorphisms identified through genome wide association analysis. In this review, we will discuss how iPSC research will further contribute to human health in the coming era of precision medicine.
Keywords: Genomics, Induced pluripotent stem cells, Experimental models
Recent Advances in Human Genome Research Leading to an Era of Precision Medicine
The first human reference genome was drafted in 2001 after an international collaborative effort between academic institutions, with the goal of characterizing genetic variations across the human genome [1, 2]. After completion of the human genome project, extended efforts to catalog these genetic variants have been made, the first of which was the international HapMap project, which aimed to build haplotype blocks of the most common genetic variations, namely, single nucleotide polymorphisms (SNPs) across human populations [3, 4]. The 1000 genomes project (http://www.1000genomes.org/) characterized common and rare genetic variations in 2,504 individuals from 26 different populations using next generation sequencing based methods and dense genotyping arrays [5]. With the influx of information and availability of genotyping platforms, it became possible to interrogate millions of SNPs simultaneously from hundreds to thousands of individuals through genome wide association analysis (GWAS). This approach has revolutionized the field of genetics, allowing for many genetic associations to be made through an agnostic, nonhypothesis driven approach. Since the first waves of GWAS publications in 2005, 23,058 SNP-trait associations have been published in the National Human Genome Research Institute - European (NHGRI–EBI) catalogue totaling 2,502 GWAS studies [6]
Pharmacogenomics is a field dedicated to identifying genetic determinants of drug response or adverse effects and is key to the concept of precision medicine, which entails utilizing genotype to guide selection of medication. Despite a fruitful era of GWAS findings in pharmacogenomics, many of these variants have not yet made it to clinical utilization. A major hurdle of pharmacogenomics implementation is the unknown underlying mechanistic link(s) between drug response phenotype and genotype. While some SNPs are located in biologically relevant genes for the phenotype being studied, the majority of variants lie in noncoding areas of the genome, where a direct connection to phenotype is unknown and a role in gene regulation is presumed. Deciphering the role of the associated genetic signals to reveal how these variants function at a molecular and cellular level is crucial for a clear understanding of the disease process and implementation of personalized medicine.
In 2015, the Obama administration announced the launch of a precision medicine initiative by the National Institutes of Health [7, 8] (https://www.nih.gov/precision-medicine-initiative-cohort-program) In this initiative, large scale cohort studies will be conducted to integrate individual lifestyle, environment, and genomic information, to build a comprehensive knowledge base which can predict individual disease risk and response to treatments. Genome sequencing and characterization of genetic variability were initial strides toward precision medicine goals of utilizing individual data to diagnose, treat, and predict response to medical treatments. With the advent of high throughput-next generation sequencing technologies and rapidly declining costs, it is feasible to perform whole genome, whole exome (protein coding), and transcriptome (RNA transcript) sequencing to probe the associations with phenotypic traits/disease conditions. Additionally, integrating multidimensionalomics data (e.g., genomics, transcriptomics, epigenomics, proteomics, and metabolomics) holds promise to elucidate biological interactions involved in complex diseases and to shed light on important genetic variants that may be missed with genetic approaches due to lack of strict statistical significance.
Although associations between human health and individual variabilities need to be validated in principle, it is still challenging to validate effects on actual biological processes. The use of induced pluripotent stem cells (iPSC) is an attractive system for modeling genetic variants to study molecular consequences in a relevant cell type. iPSC technology reprograms a fully mature somatic cell into a pluripotent stem cell that retains all the genetic characteristics of an individual patient. These iPSCs can then be differentiated into multiple different tissue types (for a growing list of validated tissue differentiation milestones, see Cell Stem Cell 18, March 2016) [9, 10]. Gene editing systems such as CRISPR-CAS9 or TALEN will expand studies that aim to unravel the mechanisms and functional consequences of genetic variations [11, 12]. This can be done through editing single nucleotides, introducing or reversing mutations in iPSCs and observing the phenotypic changes in terminally differentiated cells.
iPSCs to Find Cures of Monogenic Disorders
Diseases can have monogenic or polygenic etiologies. Monogenic diseases, caused by the inheritance of a single defective gene are considered rare because the prevalence of each disease is quite low, usually less than 1/10,000 at birth. The number of diseases with known causal genetic loci has doubled in the last 10 years as seen in Online Mendelian Inheritance in Man (OMIM) entry statistics. Improvement of genetic diagnostics and implementation of screening programs (e.g., newborn screening and high-risk screenings) make it possible to identify people with such rare disease. As a result, rare genetic diseases affect 350 million people worldwide and the global prevalence of all single gene diseases at birth is approximately 1/100. Since the establishment of human iPSCs in 2007 [13, 14], there has been an extraordinary expectation to utilize the cells to model human “diseases in a dish” [15, 16]. Pioneering studies have shown that iPSCs derived from a variety of monogenic disorders can faithfully recapitulate disease phenotypes in vitro when differentiated into disease-relevant cell types [17, 18]. Generating iPSC lines from patients with these monogenic diseases is a useful approach to establish an enduring in vitro human model and has been demonstrated in numerous published studies [19–22]. Collaborative efforts among research communities have yielded a variety of disease-specific iPSC lines readily available through iPSC banks [23] and researchers may be able to find stem cell lines of interest to conduct further mechanistic studies or directly apply the cells for drug screening.
Although many important discoveries have been made for monogenic diseases through iPSC research, one of the most exciting studies is a recent report on achondroplasia by Yamashita et al. [24]. Importantly, the authors carefully established a method to differentiate iPSCs into chondrocytes to form cartilaginous tissue. This was a critical step for Yamashita et al., because developing appropriate differentiation protocols for disease-relevant cell types can still be a limiting factor for iPSC research. They were able to successfully recapitulate abnormal cartilage formation during in vitro differentiation of iPSCs derived from patients with achondroplasia when compared to those from healthy controls. Furthermore, upon compound screening, they showed that statins, widely used lipid lowering medications, unexpectedly corrected the degraded cartilage in the iPSC model. This exemplary work clearly recapitulates disease processes in a dish and demonstrates the utility and promise of iPSC models to discover novel treatments for rare monogenic disorders.
In addition to two-dimensional monolayer differentiation or basic three-dimensional (3D) aggregate differentiation, several groups have developed sophisticated 3D differentiation protocols, often termed “organoid culture” due to their ability to form organized structures reminiscent of developing organs. Notably, for central nervous system organoid culture, Lancaster et al. demonstrated that iPSCs derived from a microcephalic patient indeed formed a smaller brain organoid than iPSCs from a healthy control [25]. Similarly, several organoid culture techniques for iPSCs have evolved to generate other tissue types and organs (optic cup, pituitary gland) [26, 27]. Undoubtedly, these breakthrough discoveries will provide necessary complexity to more accurately model disorders and allow for greater opportunity for preclinical testing of treatment options for human cells in vitro.
iPSCs to Define Further Phenotypic Variations in Monogenic Disorders
In human monogenic disorders, a single gene mutation is predominantly responsible for the phenotype of the disease. In many cases, we can predict how a specific mutation in a single gene affects protein function (e.g., residual enzyme activity), which correlates with severity and presentation of a disease. It is, however, still challenging to accurately predict clinical symptoms, severity and onset of the disease from the type of mutation. An example of this challenge is Gaucher disease (GD), an autosomal recessive disorder caused by mutations in GBA gene that encodes glucocerebrosidase (GCase) [28]. GCase is a lysosomal enzyme that catalyzes the hydrolysis of the glycolipid glucocerebroside to ceramide and glucose. Patients with GD show a broad spectrum of clinical symptoms including hepatosplenomegaly, bone deformity, hematological abnormality, and neurological symptoms. The N370S mutation in GBA is frequently found in type 1 GD, which presents with non-neuronal symptoms. On the other hand, the L444P mutation is frequently found in type 2 or 3 GD, which does present with neurological symptoms. Recombination events of the GBA locus with a neighboring pseudogene have also been linked to some unusual clinical presentations [29]. Phenotypic variabilities, however, have been observed among patients with identical GBA mutations, such as between affected sibling pairs, and even between identical twins. In a monozygotic twins case, one was affected with GD but the other had no clinical symptoms even with low GCase activity [30]. In GD, deficiency of GCase leads to accumulation of the intermediate metabolite glucosphingolipids glucosylceramide, which is further metabolized into sphingosine by an extra lysosomal GCase, GBA2. interestingly, deletion of GBA2 in a GD mouse model rescued visceral and bone symptoms, suggesting that GBA2 could potentially be targeted to ameliorate certain debilitating manifestations of GD [31].
In another study, Awad et al. uncovered involvement of lysosomal dysfunctions and an autophagy block during the neurodegenerative process of GD by using neuronal cells derived from the iPSCs of patients with type 2 GD (neuropathic form). Upon rapamycin treatment, neuronal death was preferentially induced in neurons from type 2 GD-iPSCs, but not type 1 GD-iPSCs. Although expression of the transcription factor EB (TFEB), the master regulator of lysosomal genes was downregulated, overexpression of TFEB only partially restored the neurodegenerative process in neurons from type 2 GD-iPSCs [32]. These findings represent a promising avenue to identify genetic and nongenetic (epigenetic and/or environmental) modulators that influence disease-causing mutations. Since iPSCs can be generated from individuals with various genetic backgrounds, and genomic loci can be targeted in iPSCs, disease-relevant cell types obtained from such iPSCs will be an indispensable tool to validate newly proposed disease mechanisms and to screen environmental factors/small compounds to modulate disease phenotypes.
iPSCs to Dissect the Roles of SNPs in Polygenic Disorders and Differential Drug Responses
Many common human diseases and traits are influenced by several genetic and environmental factors. Polygenic diseases result from the additive inheritance of multiple subtle polymorphisms, culminating in an affected phenotype. In 2016, nearly 5,000 disease phenotypes have been cataloged and linked with causal genetic loci in OMIM (http://omim.org/statistics/entry). GWAS have successfully identified hundreds of genetic variants associated with various conditions and have provided valuable insights into diagnostics, prognosis, and therapeutic optimization for complex human diseases [33]. An example of a common, complex and polyfactorial disease is hypertension (HTN). HTN is a major health burden in the U.S. that affects approximately 80 million people [34] and direct patient treatment totals nearly 40 billion dollars a year [35]. Additionally, HTN is increases the risk for advance cardiovascular diseases such as stroke and heart failure [34, 36] Numerous antihypertensive agents such as diuretics, ACE inhibitors, angiotensin receptor blockers, beta blockers and calcium channel inhibitors are currently available, but their effectiveness on blood pressure varies among individuals. GWAS and case studies for candidate genes have identified several genetic variants which may regulate blood pressure or contribute to a drug’s pharmacological pathway [37]. Animal models have been used intensively for studying systemic diseases like HTN, however they may not always be suitable for understanding the biological impact of human genetic variants. It is also difficult to obtain a large number of appropriate tissues of relevance for the phenotype of interest (e.g., vascular smooth muscle or endothelium) from a person with a specific genotype to test the biological or functional consequences of these genetic variations. To combat such challenges, Biel et al. constructed an iPSC repository from 17 HTN patients, whose genome-wide SNP variations as well as clinical responses to antihypertensive drugs were available [38]. The iPSCs were generated from a blood draw of peripheral blood mononuclear cells collected from participants of the Pharmacogenomic Evaluation of Antihypertensive Response (PEAR) study [39] (https://clinicaltrials.gov/NCT00246519). Biel et al. then differentiated these iPSCs into vascular smooth muscle cells and quantified their contraction in response to various physiological stimuli [38]. Furthermore, the study also demonstrated the ability of iPSCs to recapitulate a SNP-associated modification of PRKCA expression. The SNP rs16960228 has been well-documented in multiple GWAS cohorts to associate with a hypertensive drug response as well as differential expression levels of PRKCA. These data support the applicability and translational value of iPSCs in modeling GWAS findings.
Another example of cardiovascular disease modeling using iPSCs is presented by Ebert et al. [40]. Ebert et al. studied a SNP in the gene coding for aldehydronease 2 enzyme, which confers a loss of cardioprotective effects and increases risk for coronary artery and ischemic heart disease. Cardiomyocytes (CM) differentiated from iPSCs derived from an east Asian population genotyped for a common ALDH2* SNP (MAF = 0.08), demonstrated that CMs carrying the ALDH2* genotype had increased levels of oxidative stress and aldehyde byproduct 4HNE buildup. Accumulation of these two byproducts resulted in dysregulated cell cycle and apoptosis signaling, which exacerbated damage and reduced cellular recovery to ischemic challenge in the CMs of ALDH2* carriers, thereby establishing the cellular mechanisms for increased disease susceptibility for a single SNP.
Finally, it is well established that differences in susceptibility and drug response to HTN and multiple polygenic diseases varies by ethnic group (i.e., African American vs. Western European American), it will be important to understand the utility of such iPSCs libraries based on ethnic background. To address the challenge of diversity in disease genetics using iPSCs, Chang et al. reported the construction of an iPSC bank from ethnically diverse populations [41]. Taken together, these studies demonstrate that an iPSC library with defined SNPs and phenotypic data will be a useful resource to validate the effects of GWAS-identified SNPs and to facilitate mechanistic understanding of human physiological and pathological conditions.
It is increasingly important to understand how specific risk variants functionally contribute to underlying pathogenesis. Compared with single gene mutation found in monogenic diseases, the effects of SNP variants can often be minor or subtle. It is important to utilize isogenic cells to decode the significance of such gene variants. Recent advances in genome-editing technology (e.g., CRISPR/Cas9 systems) have simplified the ability to target specific genetic loci for functional studies. Gene-editing methods in iPSC’s has been reviewed in detail elsewhere [42, 43]. Soldner et al. demonstrated functional connect of GWAS-identified risk variants of Parkinson’s disease in neurons derived from human iPSCs [44]. They focused on Parkinson’s disease associated risk SNPs, which were located in an α-synuclein (SNCA) regulatory region based on genome-wide epigenetic information. By establishing TaqMan SNP genotyping assays for quantitative reverse transcription polymerase chain reaction, they were able to monitor subtle changes in allele-specific transcription of SNCA between two SNPs located in the SCNA enhancer region. As a follow up approach, they knocked-out the single allele of the SNPs using the CRISPR/Cas9 system to see how the SNPs affect SNCA expression. They found that allele-specific expression roughly translated to an increase of total SNCA expression of 1.06 times in neurons and 1.18 times in neural precursors. Furthermore, sequence-dependent binding of the brain-specific transcription factors EMX2 and NKX6-1 on this locus was revealed.
As part of the Next Generation Genetic Association Studies (Next Gen) Program, various fields of researchers are now depositing iPSC resources, generated from individuals representing various conditions as well as healthy controls, with the goal of following up findings from functional genomics with mechanistic investigations. The program is aimed at generating iPSC lines from more than 1,500 individuals some of which are available through a public iPSC bank (http://www.wicell.org/home/stem-cell-lines/collections/collections.cmsx). Each iPSC line is linked with clinical data (e.g., lipid condition, QT interval and ECG cardiac trait, pulmonary HTN) as well as age, gender and ethnic background. SNP genotyping, gene expression, and -omics analysis data will be available for these lines in the future.
It is critically important that high quality iPSC lines are also paired with high quality genetic and clinical data. This can be facilitated through large collaborations that generate harmonized phenotypes through established criteria for diagnosis and accurate phenotype definition, with an ultimate goal of reducing phenotype variability. The more accurately a phenotype is defined, the higher the likelihood of identifying the culprit gene and genetic variants [45]. With such standardized phenotypes, advancement of genetic discoveries and their replication can be made, which can be carried forward to iPSC studies using the tissues of relevance. A study by Akawi et al. shows that the value of deep sequencing information is decreased if it is not coupled with high quality phenotype data from patients [46]. An analogy can be made here as we think of the diminished value of iPSC if we do not have an accurately defined clinical phenotype that will be ultimately translated into a cellular phenotype in a dish. Therefore, it is increasingly important for the collaborative genetic consortia to establish procedures for phenotype ascertainment to reap the maximum benefit of iPSC modeling.
iPSCs to Understand Genetic and Phenotypic Variations Beyond GWAS
It has been recently shown that especially rare genetic variants, such as homozygous variant defects resulting in rare pathologies, can associate for increased risk of more common maladies as well. For example, having a pathogenic GBA mutation for Gaucher’s Disease (GD) [28] (e.g., N370S, L444P) in one allele (carrier) will not usually manifest the full symptoms of GD, but does increase risk for Parkinson’s disease [47, 48]. The odds ratio for the GBA mutation in PD was greater than 5, which is unusually high compared to risk loci found from GWAS [49]. On the other hand, there is an example where a rare genetic variant has a protective effect on a complex disease. SLC30A8 encodes an islet zinc transporter (ZnT8) and ZnT8 has been known as a key regulator of insulin secretion in pancreatic beta cells. [50]. Furthermore, large scale GWAS identified a common variant (p.Trp325Arg) on SLC30A8 that results in an increased risk for type 2 diabetes (T2D) [51–53]. Animal studies with this variant, however, showed conflicting results for pathogenesis of T2D. Breakthrough have been made through international collaborative studies, which aimed to find loss-of-function variants protective against T2D. Sequencing data from more than 150,000 people identified heterozygous individuals for a nonsense variant (p. Arg 138*) in a Finnish cohort exhibited a 60% reduced risk of type 2 diabetes [54]. Recently, Chen et al. proposed the reverse approach to find healthy individuals resilient to highly penetrant forms of genetic childhood disorders. They sequenced 874 genes in 589,306 genomes and found 13 adults carried mutations for 8 severe Mendelian conditions with no reported clinical manifestation of the indicated disease [55]. This could be a first step toward uncovering protective genetic variants, and further mechanistic studies are anticipated. As discussed above, iPSCs will serve as a powerful tool here as well to dissect molecular mechanisms of the genetic associations, hopefully leading to novel therapeutic discoveries.
Conclusion
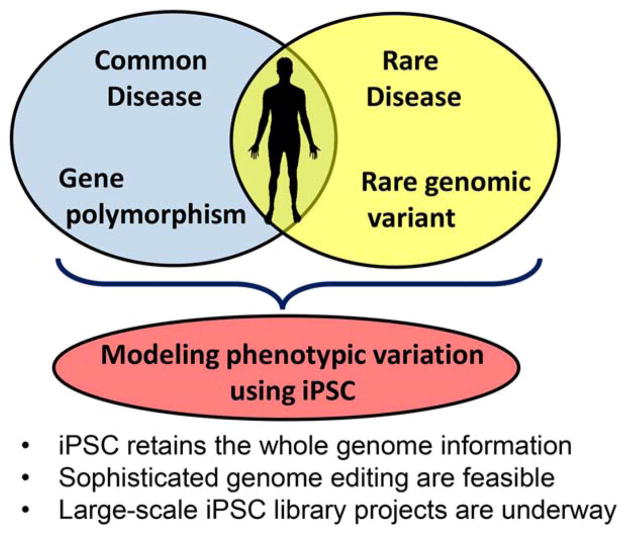
Gathering our knowledge of human disease genetics, we start to realize that each person has a unique set of variants that contribute to susceptibility and protection for a variety to disorders. Phenotypes vary even within rare monogenic diseases based on their mutation types, genetic background and environmental factors. To further advance precision medicine, it will become increasingly important to dissect molecular mechanisms underlying these genotype-phenotype associations. Human iPSCs provide a unique opportunity to fill these knowledge gaps, and their anticipated increase in utilization by researchers via cellular repositories position them as a crucial reagent for the next generation of disease genomics studies (Fig. 1).
Figure 1.
Stem cell tactic to advance precision medicine. Each person has a unique set of gene variations that affect susceptibility to and protection from both common & rare disorders. Human iPSCs provide a unique opportunity to dissect the roles of genetic variants for pathogenesis. Abbreviation: iPSCs, induced pluripotent stem cells.
Significance Statement.
Each person has a unique set of gene variations that affect susceptibility to and protection from both common and rare disorders. Although associations between human health and individual variabilities need to be validated in principle, it is still challenging to validate effects on actual biological processes. Human induced pluripotent stem cells provide a unique opportunity to dissect the roles of genetic variants for pathogenesis. This review overviews recent developments how induced pluripotent stem cell research will further contribute to human health in the coming era of precision medicine.
Acknowledgments
This work was supported in part by Japan Agency for Medical Research and Development, AMED, Practical Research Project for Rare/Intractable Diseases, National Institutes of Health (GM119977 and DK104194), American Heart Association (16GRNT30980002), and the University of Florida Clinical and Translational Science Institute (UL1TR001427). NCF is a recipient of postdoctoral fellowship T32 DK074367.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Author Contributions
N.E., N.F., and K.S.: Conception and design and manuscript writing; T.H. and N.T: Conception and design, manuscript writing, financial support, and administrative support.
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.International HapMap Consortium. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 4.International HapMap Consortium. Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3. 1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.1000 Genomes Project Consortium. Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarthy M. Obama seeks $213m to fund “precision medicine? BMJ. 2015;350:h587. doi: 10.1136/bmj.h587. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka S. Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Wilson KD, Wu JC. Induced pluripotent stem cells. JAMA. 2015;313:1613–1614. doi: 10.1001/jama.2015.1846. [DOI] [PubMed] [Google Scholar]
- 11.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 15.Hankowski KE, Hamazaki T, Umezawa A, et al. Induced pluripotent stem cells as a next-generation biomedical interface. Lab Invest. 2011;91:972–977. doi: 10.1038/labinvest.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebert AD, Yu J, Rose FF, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itzhaki I, Maizels L, Huber I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 19.Juopperi TA, Kim WR, Chiang C-H, et al. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol Brain. 2012;5:17. doi: 10.1186/1756-6606-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du J, Campau E, Soragni E, et al. Length-dependent CTG·CAG triplet-repeat expansion in myotonic dystrophy patient-derived induced pluripotent stem cells. Hum Mol Genet. 2013;22:5276–5287. doi: 10.1093/hmg/ddt386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cayo MA, Cai J, DeLaForest A, et al. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 2012;56:2163–2171. doi: 10.1002/hep.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Y, Luo Y, Chen X, et al. Generation of human β-thalassemia induced pluripotent stem cells from amniotic fluid cells using a single excisable lentiviral stem cell cassette. J Reprod Dev. 2012;58:404–409. doi: 10.1262/jrd.2011-046. [DOI] [PubMed] [Google Scholar]
- 23.Santostefano KE, Hamazaki T, Biel NM, et al. A practical guide to induced pluripotent stem cell research using patient samples. Lab Invest. 2015;95:4–13. doi: 10.1038/labinvest.2014.104. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita A, Morioka M, Kishi H, et al. Statin treatment rescues FGFR3 skeletal dysplasia phenotypes. Nature. 2014;513:507–511. doi: 10.1038/nature13775. [DOI] [PubMed] [Google Scholar]
- 25.Lancaster M, Renner AM, Martin C-A, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakano T, Ando S, Takata N, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Ozone C, Suga H, Eiraku M, et al. Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat Commun. 2016;7:10351. doi: 10.1038/ncomms10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pastores GM, Hughes DA. [Accessed August 18, 2016];Gaucher disease. 1993 Available at http://www.ncbi.nlm.nih.gov/pubmed/20301446.
- 29.Tayebi N, Stubblefield BK, Park JK, et al. Reciprocal and nonreciprocal recombination at the glucocerebrosidase gene region: Implications for complexity in Gaucher disease. Am J Hum Genet. 2003;72:519–534. doi: 10.1086/367850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lachmann RH, Grant IR, Halsall D, et al. Twin pairs showing discordance of phenotype in adult Gaucher’s disease. QJM. 2004;97:199–204. doi: 10.1093/qjmed/hch036. [DOI] [PubMed] [Google Scholar]
- 31.Mistry PK, Liu J, Sun L, et al. Glucocerebrosidase 2 gene deletion rescues type 1 Gaucher disease. Proc Natl Acad Sci USA. 2014;111:4934–4939. doi: 10.1073/pnas.1400768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Awad O, Sarkar C, Panicker LM, et al. Altered TFEB-mediated lysosomal biogenesis in Gaucher disease iPSC-derived neuronal cells. Hum Mol Genet. 2015;24:5775–5788. doi: 10.1093/hmg/ddv297. [DOI] [PubMed] [Google Scholar]
- 33.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mozaffarian D, Benjamin EJ, Go AS, et al. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. 2015;131:e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 35.Moran AE, Odden MC, Thanataveerat A, et al. Cost-effectiveness of hypertension therapy according to 2014 guidelines. N Engl J Med. 2015;372:447–455. doi: 10.1056/NEJMsa1406751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rapsomaniki E, Timmis A, George J, et al. Blood pressure and incidence of twelve cardiovascular diseases: Lifetime risks, healthy life-years lost, and age-specific associations in 1·25 million people. Lancet. 2014;383:1899–1911. doi: 10.1016/S0140-6736(14)60685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mellen PB, Herrington DM. Pharmacogenomics of blood pressure response to antihypertensive treatment. J Hypertens. 2005;23:1311–1325. doi: 10.1097/01.hjh.0000173510.52987.68. [DOI] [PubMed] [Google Scholar]
- 38.Biel NM, Santostefano KE, DiVita BB, et al. Vascular smooth muscle cells from hypertensive patient-derived induced pluripotent stem cells to advance hypertension pharmacogenomics. Stem Cells Transl Med. 2015;4:1380–1390. doi: 10.5966/sctm.2015-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JA, Boerwinkle E, Zineh I, et al. Pharmacogenomics of antihypertensive drugs: Rationale and design of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study. Am Heart J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebert AD, Kodo K, Liang P, et al. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci Transl Med. 2014;6:255ra130. doi: 10.1126/scitranslmed.3009027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang EA, Tomov ML, Suhr ST, et al. Derivation of ethnically diverse human induced pluripotent stem cell lines. Sci Rep. 2015;5:15234. doi: 10.1038/srep15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18:573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deleidi M, Yu C. Genome editing in pluripotent stem cells: Research and therapeutic applications. Biochem Biophys Res Commun. 2016;473:665–674. doi: 10.1016/j.bbrc.2016.02.113. [DOI] [PubMed] [Google Scholar]
- 44.Soldner F, Stelzer Y, Shivalila CS, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vissers LELM, Veltman JA. Standardized phenotyping enhances Mendelian disease gene identification. Nat Genet. 2015;47:1222–1224. doi: 10.1038/ng.3425. [DOI] [PubMed] [Google Scholar]
- 46.Akawi N, McRae J, Ansari M, et al. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat Genet. 2015;47:1363–1369. doi: 10.1038/ng.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gatto EM, Etcheverry JL, Sanguinetti A, et al. Prodromal clinical markers of Parkinson disease in Gaucher disease individuals. Eur Neurol. 2016;76:19–21. doi: 10.1159/000447510. [DOI] [PubMed] [Google Scholar]
- 48.Ran C, Brodin L, Forsgren L, et al. Strong association between glucocerebrosidase mutations and Parkinson’s disease in Sweden. Neurobiol Aging. 2016;45:212e5–212.e11. doi: 10.1016/j.neurobiolaging.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sidransky E, Nalls MA, Aasly JO, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chimienti F, Devergnas S, Pattou F, et al. In vivo expression and functional characterization of the zinc transporter ZnT8 in glucose-induced insulin secretion. J Cell Sci. 2006;119:4199–4206. doi: 10.1242/jcs.03164. [DOI] [PubMed] [Google Scholar]
- 51.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strawbridge RJ, Dupuis J, Prokopenko I, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flannick J, Thorleifsson G, Beer NL, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen R, Shi L, Hakenberg J, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]