Abstract
Catalytically activating mutations in Ptpn11, which encodes the protein tyrosine phosphatase SHP2, cause 50% of Noonan Syndrome (NS) cases, whereas inactivating mutations in Ptpn11 are responsible for nearly all cases of the similar, but distinct, developmental disorder Noonan Syndrome with Multiple Lentigines (NSML, formerly called LEOPARD Syndrome). Both types of disease mutations are gain-of-function mutations because they induce SHP2 to constitutively adopt an open conformation, yet they have opposing effects on SHP2 catalytic activity. Here, we report that the catalytic activity of SHP2 is required for the pathogenic effects of Ptpn11 gain-of-function disease-associated mutations on the development of hydrocephalus in the mouse. Targeted pan-neuronal knock-in of the Ptpn11 activating mutation E76K resulted in hydrocephalus due to aberrant development of ependymal cells and their cilia. These pathogenic effects of the E76K mutation were completely suppressed by the additional mutation C459S, which abolishes catalytic activity of SHP2. Moreover, ependymal cells in NSML mice bearing the Ptpn11 inactivating mutation Y279C were also unaffected. Mechanistically, the Ptpn11E76K mutation induced developmental defects in ependymal cells by enhancing dephosphorylation and inhibition of the transcriptional activator STAT3. Whereas STAT3 activity was reduced in Ptpn11E76K/+ cells, the activities of the kinases ERK and AKT were enhanced, and neural cell–specific Stat3 knockout mice also manifested developmental defects in ependymal cells and cilia. These genetic and biochemical data demonstrate a catalytic–dependent role of Ptpn11 gain-of-function disease mutations in the pathogenesis of hydrocephalus.
INTRODUCTION
Ptpn11 is ubiquitously expressed and encodes SH2 domain–containing phosphatase 2 (SHP2), a protein tyrosine phosphatase (PTP) implicated in multiple cell signaling processes, including the Rat Sarcoma (RAS)-Extracellular signal-Regulated Kinase (ERK), Janus Kinase (JAK)-Signal Transducer and Activator of Transcription (STAT), Phosphatidylinositol 3-Kinase (PI3K)-Protein Kinase B (PKB, also known as AKT), mouse Target Of Rapamycin (mTOR), and Nuclear Factor-κB (NF-κB) pathways (1, 2). Under baseline conditions, it is self-inhibited by hydrogen bonding of a loop on the back side of the N-terminal SH2 (N-SH2) domain with the deep pocket of the PTP domain (3, 4). SHP2 becomes activated upon binding to signaling partners that contain phosphorylated tyrosine residues, playing an overall positive role in transducing signals initiated from receptor and cytosolic kinases, particularly in the RAS-ERK pathway (1, 2). However, the signaling mechanisms of SHP2 are still not well understood; indeed, accumulating evidence suggests that SHP2 functions in cell signaling in both catalytic-dependent and -independent manners (5–8). Its roles that do not depend on catalytic activity are likely to involve SHP2 acting as a scaffolding protein.
The critical role for SHP2 in cell signaling is further underscored by the direct association of mutations in this phosphatase with human diseases. Catalytically activating heterozygous germline mutations in Ptpn11 are associated with 50% of cases of Noonan Syndrome (NS) (9), a developmental disorder characterized by congenital heart disease, dysmorphic facial and chest features, proportionate short stature, and intellectual disability (10). Furthermore, somatic mutations of Ptpn11 are found in various childhood leukemias (11–14). Ptpn11 mutations identified in NS and in leukemias typically result in amino acid changes in the region between the N-SH2 and PTP domains, which disrupt the autoinhibitory interaction and cause SHP2 to adopt an open conformation, leading to hyperactivation of SHP2 catalytic activity (9, 11, 15). Germline heterozygous mutations in Ptpn11 are also highly associated (>90%) with another rare developmental disorder called Noonan Syndrome with Multiple Lentigines (NSML, formerly known as LEOPARD Syndrome), which is characterized by multiple lentigines, cardiac abnormalities, facial dysmorphism, retardation of growth, and sensorineural deafness (16, 17). Intriguingly, in contrast to the NS mutations, Ptpn11 mutations identified in NSML eliminate the catalytic activity of SHP2 due to changes in the active site amino acid residues that are critical for hydrolyzing activity (18). However, both NS and NSML mutations cause SHP2 to adopt an open conformation and enhance SHP2 interactions with signaling partners despite having opposing effects on catalytic activity (9, 18–21), resulting in gain-of-function of SHP2. How allelic variants of Ptpn11 with opposing enzymatic activities lead to similar, yet distinct, syndromic disorders remains poorly understood (22). In the present study, we sought to determine the cellular and molecular mechanisms underlying the pathogenic effects of NS- and NSML-associated Ptpn11 mutations in brain development. Our results suggest that Ptpn11 disease mutations induce hydrocephalus in a manner that depends on the catalytic activity of SHP2.
RESULTS
Neural tissue–specific expression of Ptpn11E76K induces hydrocephalus and brain developmental defects
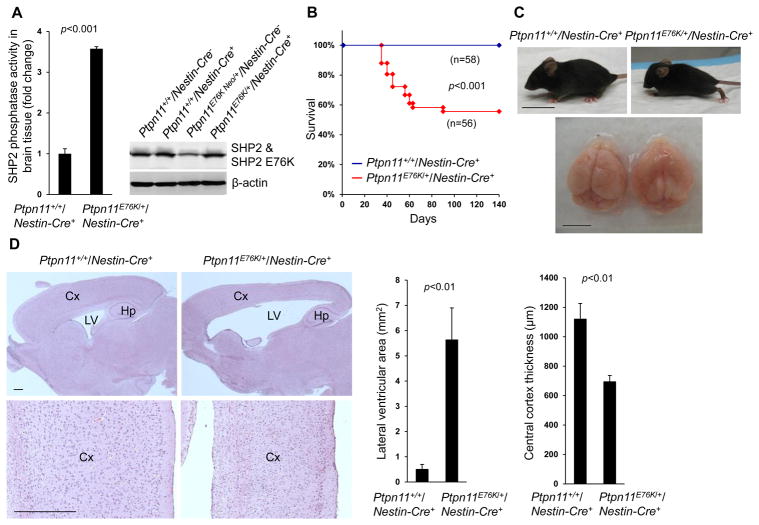
Germline and somatic activating mutations in Ptpn11 are associated with NS (9) and sporadic glioblastoma (23), respectively; one such mutation is E76K, in which substitution of Glu76 with Lys disrupts the intramolecular interaction that mediates SHP2 autoinhibition (11). To determine the potential impact of Ptpn11 activating mutations in neural cells, we generated neural tissue–specific Ptpn11E76K knock-in mice (Ptpn11E76K/+/Nestin-Cre+) by crossing activating mutation Ptpn11E76K conditional knock-in mice (Ptpn11E76K neo/+) (24, 25) with Nestin-Cre+ mice (26), which express Cre in neural precursor cells beginning at embryonic day 10.5 (E10.5). Following heterozygous induction of the SHP2 E76K mutant, SHP2 phosphatase activity in the brain increased by more than 3-fold compared to that in the brain of wild-type mice (Fig. 1A). No brain tumors developed in these animals during a 15 month follow-up, suggesting that Ptpn11 activating mutations found in human brain tumors are not driving mutations. Forty percent of Ptpn11E76K/+/Nestin-Cre+ mice died within two months of birth (Fig. 1B). They displayed obvious dome-shaped heads (Fig. 1C), and magnetic resonance imaging of live animals suggested that these mice had substantially enlarged cerebral ventricles (Fig. S1A). Autopsy revealed frank hydrocephalus of varying severity; specifically, the cerebral ventricles were filled with excess cerebrospinal fluid (Fig. 1C). Histopathological examination of brain tissues showed that the cerebral cortex was much thinner and lamination of the cortex was compromised, as compared to control tissues (Fig. 1D). The remaining 60% of Ptpn11E76K/+/Nestin-Cre+ mice survived, but still developed a mild form of hydrocephalus. In behavioral tests, these surviving Ptpn11E76K/+/Nestin-Cre+ mice exhibited reduced anxiety behavior as compared to Ptpn11+/+/Nestin-Cre+ control mice. They spent more time in the center and mid zone areas and less time in the outer zone in open field tests (Fig. S1B). Despite this apparent reduction in anxiety, they manifested hyperactivity, as indicated by an increase in the distance traveled (Fig. S1C), as compared to control mice. In addition, these mutant mice showed impaired motor function in grip strength tests (Fig. S1D) and reduced hanging times in wire hang tests (Fig. S1E). These aberrant behavior profiles verify developmental defects in the central nervous system in mice expressing SHP2E76K.
Fig. 1. Neural cell–specific expression of Ptpn11E76K induces hydrocephalus and brain developmental defects.
(A) Brain tissues dissected from one-month-old mice (n=3 mice per genotype) were lysed, and SHP2 catalytic activities in the lysates were assessed by the immunocomplex phosphatase assay. SHP2 abundance in the lysates was examined by immunoblotting. (B) Kaplan-Meier survival curves of mice of the indicated genotypes. (C) Representative images of one-month-old mice and their brains. (D) Sagittal brain sections were processed for H&E staining. Analyses in all panels were performed in 3 independent experiments, n=4 mice per genotype, and representative images are shown. Data are presented as mean±S.D. of biological replicates. Cx, cortex; Hp, hippocampus; LV, lateral ventricles. Scale bars, 2 cm (C, mice), 5 mm (C, brain), and 500 μm (D).
Ptpn11E76K reduces proliferation but enhances glial differentiation in neural stem and progenitor cells
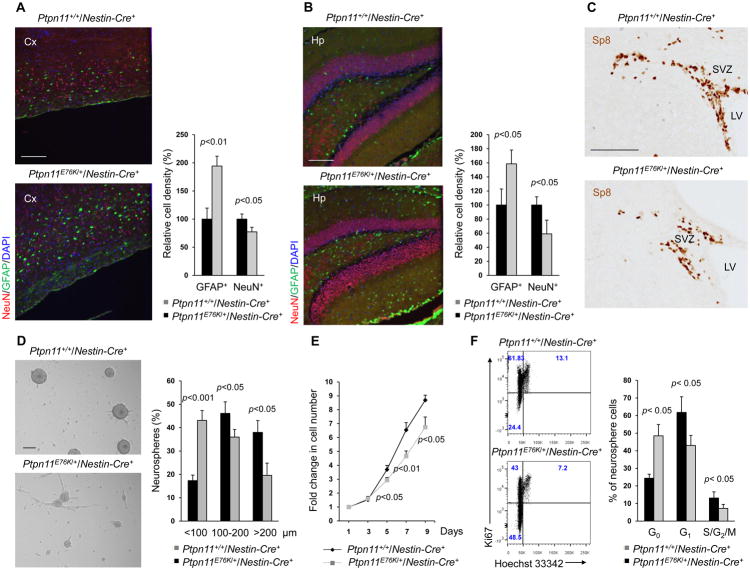
To further define the brain developmental defects in Ptpn11E76K/+/Nestin-Cre+ mice, we immunostained neurons and astrocytes in adult brain tissues and found that, relative to Ptpn11+/+/Nestin-Cre+ mice at the same developmental stage, the number of neurons decreased and the number of astrocytes markedly increased in both the cortex (Fig. 2A and Fig. S2A) and the hippocampus (Fig. 2B and Fig. S2B). We observed no obvious defects in the cerebellum (Fig. S2C). The cell composition and the structure of the mutant cortices were relatively normal at the neonatal stage (Fig. S2D), suggesting that the abnormal composition of neural cells in the adult mutant mice might be associated with indolent hydrocephalus, which was previously shown to cause enhanced gliogenesis and reduced neurogenesis (27). Specificity protein 8 (Sp8) is a member of the Sp1 family of zinc finger transcription factors that is enriched in migrating neuroblasts of the sub-ventricular zone (SVZ) (28), where neural stem and progenitor cells (NSPCs) and later-stage proliferative progenitors reside. Immunostaining for Sp8 illustrated that these precursor cells were decreased in the SVZ of Ptpn11E76K/+/Nestin-Cre+ mice relative to Ptpn11+/+/Nestin-Cre+ mice (Fig. 2C). This observation raised the possibility that NSPC activities might be altered by the Ptpn11E76K mutation. To test this possibility, we performed neurosphere assays for the cortices isolated from E14.5 embryos. Although cortices from Ptpn11E76K/+ mice and wild-type mice yielded similar total numbers of primary and secondary neurospheres (Fig. S3A), the actual size of mutant neurospheres was much smaller (Fig. 2D), and proliferation of mutant neurosphere cells was decreased, as compared to controls (Fig. 2E). The decrease in the size and growth of mutant neurospheres was not associated with reduced cell survival because we detected no significant changes in apoptosis (Fig. S3B). However, cell cycle analyses showed that the percentage of Ptpn11E76K/+ neurosphere cells in the G0 phase doubled, whereas the percentage of cells in the G1 and S, G2, or M phases decreased significantly (Fig. 2F). Furthermore, mutant neurosphere cells tended to spontaneously differentiate. Increased numbers of adherent glia-like cells developed in Ptpn11E76K/+ neurosphere cells during culture conditions that favored maintenance of NSPCs over differentiation (Fig. 2D).
Fig. 2. Ptpn11E76K decreases self-renewal in NSPCs.
Brain sections prepared from one-month-old mice (n=4 mice per genotype) were processed for immunofluorescence staining of NeuN (neurons) and GFAP (astrocytes) in the cortex (A) and hippocampus (B). (C) Brain sections (n=3 mice per genotype) were processed for immunohistochemistry staining for the migrating neuroblast marker Sp8. SVZ, sub-ventricular zone; LV, lateral ventricles. (D–F) Cerebral cortices dissected from E14.5 embryos (n=4 mice per genotype) were assessed by neurosphere assays. (D) Quantification of the size distribution of neurospheres formed by tissue from the indicated genotypes in culture. (E) Total cell numbers at the indicated time points following dissociation of neurospheres into single cells and culture in medium containing bFGF. (F) Cell cycle profiles of the dissociated neurosphere cells as determined by Ki67 and Hoechst 33342 staining followed by FACS analyses (F). Analyses in all panels were performed in 3–4 independent experiments. Data are presented as mean±S.D. of biological replicates. Representative images are shown. Scale bars, 200 μm (A–B upper, D), 100 μm (A–B lower, C).
Hydrocephalus induced by Ptpn11E76K results from aberrant development of ependymal cells
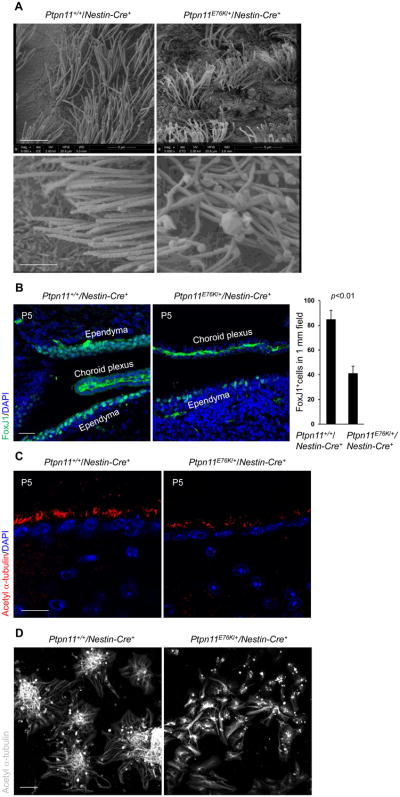
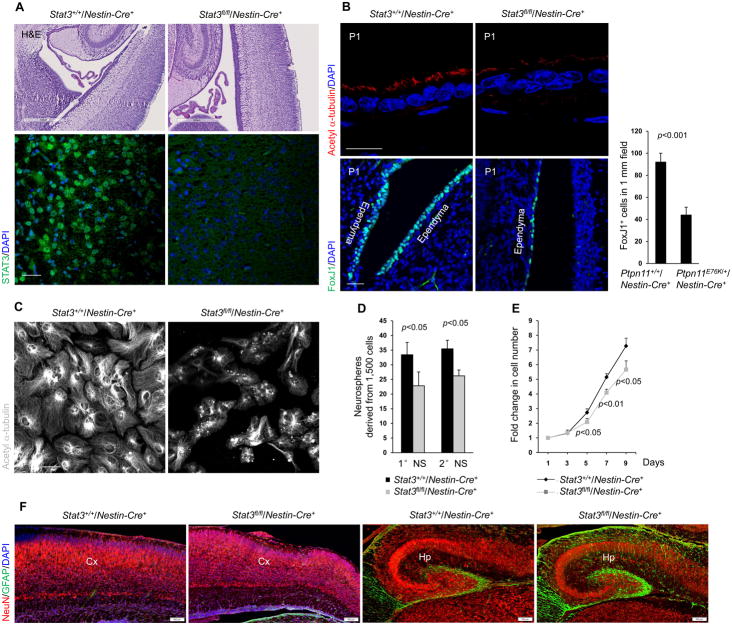
To identify the cellular mechanism underlying the hydrocephalus phenotype in Ptpn11E76K/+/Nestin-Cre+ mice, we examined the cerebrum, the cerebellar tonsils, and the cerebrospinal circulation system, but found no obstructions. We also carefully examined choroid plexuses, which are responsible for production of cerebrospinal fluid (29, 30), but no structural changes were observed there either. Scanning electron microscopy revealed that the cilia of ependymal cells, which play an essential role in the transport of cerebrospinal fluid (29, 30), developed aberrantly in Ptpn11E76K/+/Nestin-Cre+ mice; they were much shorter and apparently disorganized, as compared to those from control mice (Fig. 3A). Moreover, there was a reduction in the number of cells that stained positive for the ependymal cell marker Forkhead box J1 (FoxJ1) (Fig. 3B). The decrease in ependymal cells and the defects in ependymal cilia (marked by acetyl α-tubulin) (Fig. 3C) were readily detected in Ptpn11E76K/+/Nestin-Cre+ mice at the postnatal stage when no hydrocephalus had yet developed (ventricles were not enlarged), indicating that these defects in ependymal cells preceded the development of hydrocephalus. The aberrant ependymal cell development in Ptpn11E76K/+/Nestin-Cre+ mice appeared to be attributable to defects in Ptpn11E76K-expressing NSPCs. Indeed, the ability of Ptpn11E76K/+ NSPCs to differentiate into ependymal cells with cilia in vitro was substantially decreased (Fig. 3D). Quantitative reverse transcription PCR (qRT-PCR) analyses of cell lineage markers verified that ependymal cell and neuronal differentiation was decreased whereas astrocyte differentiation was increased in Ptpn11E76K/+/Nestin-Cre+ NSPCs (Fig. S4).
Fig. 3. Aberrant development of ependymal cells and cilia in Ptpn11E76K/+ mice.
(A) Brains dissected from one-month-old mice (n=3 mice per genotype) were processed for scanning electron microscopic analyses. Ependymal cilia on the lateral ventricular walls were examined. Scale bars, 5 μm (upper), 2 μm (lower). (B and C) Brain sections prepared from postnatal day 5 (P5) pups (n=3 mice per genotype) were processed for immunofluorescence staining for the ependymal cell marker FoxJ1 (B) and the cilium marker acetyl α-tubulin (C). The images show the ependymal cells (FoxJ1+, scale bar, 50 μm) and ependymal cilia (acetyl α-tubulin+, scale bar, 20 μm) on the walls of the lateral ventricles in brain sections. FoxJ1+ cells were quantified. Scale bars, 50 μm (B) and 20 μm (C). (D) Lateral ventricular walls dissected from newborn pups (n=3 mice per genotype) were processed for ependymal cell differentiation assays. Differentiated cells were immunostained for acetyl α-tubulin. Scale bar, 50 μm. Analyses in all panels were performed in 3 independent experiments. Data are presented as mean±S.D. of biological replicates. Representative images are shown.
Ptpn11E76K decreases STAT3 activity and enhances ERK and AKT signaling
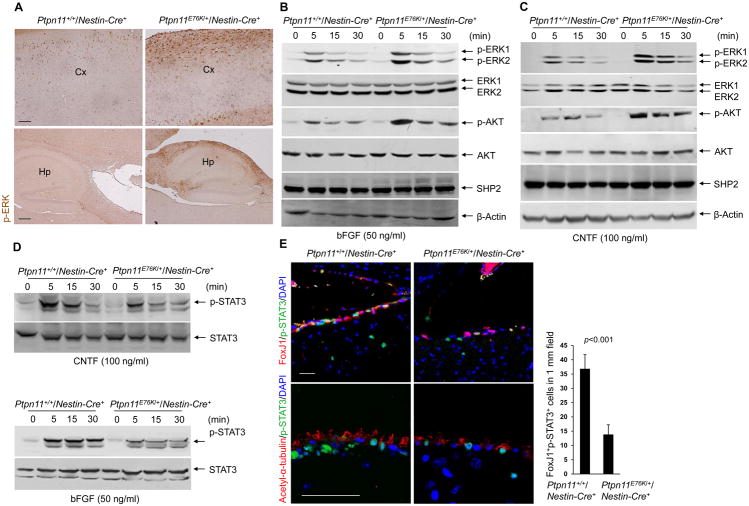
We next investigated the molecular mechanisms for the pathogenic effects of the Ptpn11E76K mutation. Consistent with the positive role that SHP2 plays in the RAS signaling pathway, ERK activity was increased in the cortex and hippocampus of Ptpn11E76K/+/Nestin-Cre+ mice, compared with that in control animals (Fig. 4A). ERK activity was also enhanced in forebrain progenitor cells of Ptpn11E76K/+/Nestin-Cre+ embryos, specifically in the lateral ganglionic eminence, which is the progenitor domain that gives rise to several cell types in the postnatal SVZ (31, 32) (Fig. S5). Unexpectedly, we observed no change in cell proliferation in this region, as measured by phosphorylated histone H3 staining (Fig. S5). Given that basic fibroblast growth factor (bFGF) plays an essential role in neural progenitor cell survival and proliferation, we examined bFGF signaling in neurosphere cells. Although the abundance of the bFGF receptor (bFGFR) was comparable in the mutant and control cells (Fig. S6A), bFGF-induced ERK activation was increased in the cells isolated from Ptpn11E76K/+ mice (Fig. 4B, S6B). In addition, AKT activation was enhanced in these mutant cells. Furthermore, we found that ciliary neurotrophic factor (CNTF), which promotes the survival and growth of neurons, induced similar increases in ERK and AKT activities in the mutant and control cells (Fig. 4C, S6C). In contrast to these effects on ERK and AKT, cells in neurospheres derived from Ptpn11E76K/+ mice showed a decrease in STAT3 activity in response to CNTF or bFGF activation (Fig. 4D, S6D). Indeed, tyrosine phosphorylation of STAT3 was markedly decreased in ependymal cells in Ptpn11E76K/+ mice (Fig. 4E). Because STAT3 is a substrate of SHP2 (33, 34), the diminished STAT3 activity was likely caused by enhanced dephosphorylation of STAT3 by hyperactive SHP2E76K.
Fig. 4. Ptpn11E76K decreases STAT3 activity and enhances ERK and AKT activity.
(A) Immunohistochemical staining for phosphorylated ERK (p-ERK) in the cortex (Cx) and hippocampus (Hp) of brain sections prepared from one-month-old Ptpn11+/+/Nestin-Cre+ and Ptpn11E76K/+/Nestin-Cre+ mice (n=3 mice per genotype). Scale bars, 100 μm (upper), 200 μm (lower). (B–D) Immunoblot showing p-ERK and p-AKT (B and C) or STAT3 phosphorylated at Tyr705 (p-STAT3, D) in whole cell lysates of neurospheres that were generated from Ptpn11+/+/Nestin-Cre+ and Ptpn11E76K/+/Nestin-Cre+ mice (n=3 mice per genotype), dissociated into single cells, and stimulated with bFGF or CNTF as indicated. Densitometric data of phosphoproteins (normalized to the total abundance of the respective proteins) are summarized in Fig. S6B–S6D. (E) Immunofluorescence staining for FoxJ1, acetyl α-tubulin, and phosphorylated STAT3 in brain sections prepared from one-month-old Ptpn11+/+/Nestin-Cre+ and Ptpn11E76K/+/Nestin-Cre+ mice (n=3 mice per genotype). Scale bar, 50 μm. Double-positive FoxJ1+p-STAT3+ cells were quantified. Analyses in all panels were performed in 3 independent experiments. Data are presented as mean±S.D. of biological replicates. Representative images are shown.
Depletion of STAT3 results in developmental defects in ependymal cilia
To test the possibility that diminished STAT3 activity might be responsible for the aberrant development of ependymal cells and hydrocephalus in Ptpn11E76K/+/Nestin-Cre+ mice, we generated neural cell–specific Stat3 knockout mice (Stat3fl/fl/Nestin-Cre+). These animals die shortly after birth due to defective heart development and respiratory failure (34). We were able, however, to examine the brains of Stat3fl/fl/Nestin-Cre+ neonates. These newborn pups did not exhibit obvious enlargement of the ventricular system, and the cerebrum appeared normal (Fig. 5A). However, immunostaining of brain sections revealed clear defects in ependymal cilia and a reduced number of ependymal cells in the mutant mice (Fig. 5B). Moreover, NSPCs from Stat3 knockout newborn pups failed to differentiate into ependymal cells in culture (Fig. 5C), similar to NSPCs from Ptpn11E76K/+ pups. The self-renewal capability of Stat3 knockout NSPCs was also reduced (Fig. 5D, 5E), implying that the decreased proliferation of Ptpn11E76K/+ NSPCs (Fig. 2D, 2E) is likely due to diminished STAT3 activity. In addition, although the number of neurons was not changed, the number of astrocytes was increased in the cerebral cortex and hippocampus of the Stat3 conditional knockout neonates (Fig. 5F).
Fig. 5. Depletion of STAT3 in neural cells results in developmental defects in ependymal cells and cilia.
Immunohistochemical and immunofluroescence staining showing (A) STAT3 and (B) acetyl α-tubulin and FoxJ1 in ependymal cells on the walls of the lateral ventricles in brain sections of newborn Stat3+/+/Nestin-Cre+ and Stat3fl/fl/Nestin-Cre+ pups (n=3 mice per genotype). FoxJ1+ cells were quantified (B). Sacle bars, 500 μm (A, upper), 50 μm (A, lower and B, lower), 20 μm (B, upper). (C) Acetyl α-tubulin in cells isolated from lateral ventricular walls dissected from newborn Stat3+/+/Nestin-Cre+ and Stat3fl/fl/Nestin-Cre+ pups (n=3 mice per genotype). Scale bar, 50 μm. (D) Quantification of the number of primary neurospheres (1 °NS) and secondary neurospheres (2° NS) derived from cerebral cortices dissected from E14.5 embryos (n=3 mice per genotype) of the indicated genotypes. (E) Quantification of the proliferation of cells in primary neurospheres derived from the indicated genotypes. (F) Immunofluorescence staining of brain sections from newborn Stat3+/+/Nestin-Cre+ and Stat3fl/fl/Nestin-Cre+ pups (n=3 mice per genotype) showing the neuron marker NeuN and the astrocyte marker GFAP. Scale bar, 100 μm. Analyses in all panels were performed in 3 independent experiments. Data are presented as mean±S.D. of biological replicates. Representative images are shown.
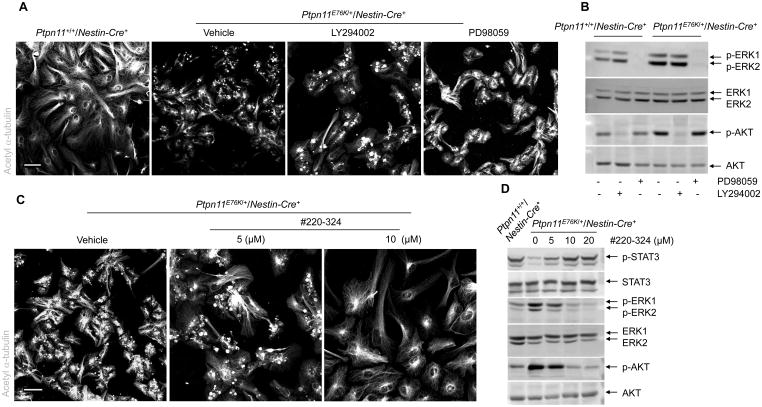
The Ptpn11E76K mutation impacted several signaling pathways. To further determine which altered pathway was responsible for the defective ependymal cell differentiation of Ptpn11E76K/+/Nestin-Cre+ NSPCs, we treated these cells with mitogen-activated protein kinase kinase 1 (MAPKK1, also known as MEK1) and PI3K inhibitors. Because MAPKK1 and P13K function upstream of ERK and AKT, respectively, inhibiting their activity should counteract the hyperactivation of ERK and AKT. Inhibiting neither MAPKK1 nor P13K activity with PD98059 or LY294002, respectively, rescued ependymal cell differentiation in explants from Ptpn11E76K/+/Nestin-Cre+ pups (Fig. 6A, 6B). In contrast, treatment with the SHP2 inhibitor #220-324 (35) largely rescued ependymal cell development, increased STAT3 activity, and decreased ERK and AKT activities (Fig. 6C, 6D) in explants from Ptpn11E76K/+ pups. These rescue data provide additional evidence that aberrant ependymal cell development and hydrocephalus manifested by Ptpn11E76K/+/Nestin-Cre+ mice were attributable to diminished STAT3 activity rather than increased ERK or AKT activity.
Fig. 6. Inhibition of SHP2, but not ERK or AKT, rescues ependymal cell differentiation of Ptpn11E76K/+ NSPCs.
(A–D) Lateral ventricular walls dissected from newborn pups (n=3 mice per genotype) were processed for ependymal cell differentiation assays in the presence of the MEK1 inhibitor PD98059, the PI3K inhibitor LY294002, the SHP2 inhibitor #220-324, or vehicle. Inhibitor-treated cells were immunostained for acetyl α-tubulin to mark cilia (A, C) and examined by immunoblotting for phosphorylated AKT (p-AKT), phosphorylated ERK (p-ERK), and phosphorylated STAT3 (p-STAT3) (B,D). Analyses in all panels were performed in 3 independent experiments. Representative images are shown. Scale bars, 50 μm.
The pathological effects of Ptpn11 disease mutations in hydrocephalus development depend on increased catalytic activity of SHP2
To verify that Ptpn11 mutations induce aberrant brain development through increased SHP2 catalytic activity, we examined a NS mouse model ubiquitously expressing the SHP2 activating mutation D61G (Ptpn11D61G/+) (36), which is less catalytically active than the E76K mutation (11, 15). None of the Ptpn11D61G/+ mice displayed overt hydrocephalus; however, ependymal cilia were found to be abnormal similarly to, but to a lesser degree than, those in Ptpn11E76K/+ mice. Ptpn11D61G/+ mice showed ependymal cilia defects only on the third ventricular walls (Table 1, Fig. S7). These observations suggest that the pathological effects of Ptpn11 activating mutations in the development of hydrocephalus correlate with the increased catalytic activity of mutant SHP2, although a potential systematic effect on the phenotypes cannot be completely excluded because Ptpn11E76K was expressed in neural tissues, whereas Ptpn11D61G was expressed in the whole body. Also, Ptpn11D61G/+ mice were the F4 generation from backcrosses with C57BL/6 mice, whereas Ptpn11E76K/+ mice were on pure C57BL/6 background, therefore potential effects of the genetic backgrounds on the phenotypes cannot be completely excluded. We next extended our studies to examine the effects of mutations that catalytically inactivate SHP2. To do this, we employed the NSML mouse model, in which the most common mutation found in NSML patients, Y279C, is ubiquitously expressed (Ptpn11Y279C/+) (20). This is an inactivating mutation because it affects a conserved residue important for PTP catalysis. In contrast to Ptpn11D61G/+ NS mice, none of the 17 Ptpn11Y279C/+ NSML mice (F4 generation from backcrosses with C57BL/6 mice) we examined developed hydrocephalus, nor did we observe defects in ependymal cilia (Table 1, Fig. S7).
Table 1. Ependymal cilia defects are proportionate to the catalytic activities of various mutant forms of SHP2.
Brain sections prepared from 12-month-old mice (n=3 mice per genotype) were immunofluorescently stained for acetyl α-tubulin to mark cilia, and the ependymal cilia on the walls of ventricles were examined.
| Mice | Catalytic activity of mutant SHP2 | Ependymal cilia |
|---|---|---|
| Ptpn11E76K/+/Nestin-Cre+ | Substantially enhanced | Severely abnormal |
| Ptpn11D61G/+ (NS mice) | Enhanced | Abnormal in some areas * |
| Ptpn11Y279C/+ (NSML mice) | No catalytic activity | Normal |
Ependymal cilia of the third ventricle, but not the lateral ventricles, were abnormal.
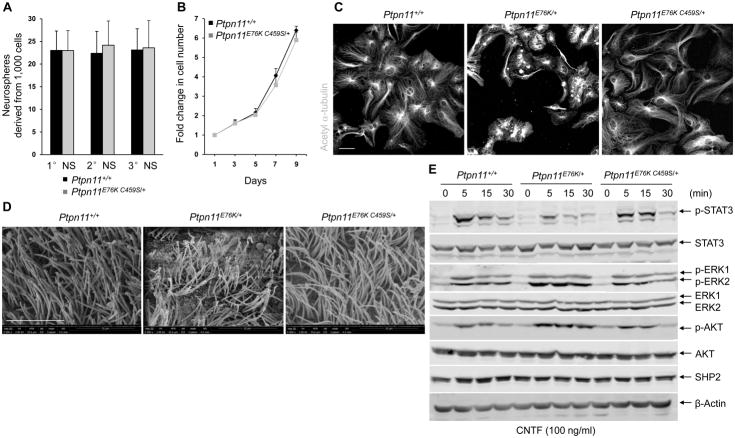
Inspired by the above observations and also to further confirm that Ptpn11 activating mutations induce hydrocephalus through enhanced catalytic activity of SHP2, we generated Ptpn11E76K,C459S double mutant knock-in mice by gene targeting (Fig. S8A–S8C). The Cys residue at amino acid position 459 in the catalytic core is critical for the enzymatic activity of SHP2 (37, 38). Replacement of Cys with Ser at this site completely abolishes SHP2 enzymatic activity (37, 38). Therefore, the E76K C459S double mutant form of SHP2 does not have catalytic activity but retains the effect of the E76K mutation in the N-SH2 domain, which confers an open conformation that enhances the ability of SHP2 to participate in protein-protein interactions, although a potential substrate-trapping activity of the C459S mutation might also affect protein-protein interactions. Similar to NSML mice (Ptpn11Y279C/+), which harbor open but catalytically-inactive mutant SHP2, none of 56 double mutation knock-in mice (Ptpn11E76K, C459S/+) developed hydrocephalus during a 15 month follow-up (Table 2). We also assessed Ptpn11E76K,C459S/+ NSPCs by the neurosphere assay; neither the number nor the size of the primary, secondary, and tertiary neurospheres derived from Ptpn11E76K,C459S/+ mice showed significant differences from wild-type controls (Fig. 7A). The growth of these double mutant neurosphere cells was essentially normal (Fig. 7B). Moreover, the ability of Ptpn11E76K,C459S/+ NSPCs to differentiate into ependymal cells in vitro was restored (Fig. 7C), and the ependymal cilia in Ptpn11E76K,C459S/+ mice developed without noticeable defects (Fig. 7D). Cell signaling, especially STAT3 activity, in Ptpn11E76K,C459S/+ double mutant neurosphere cells was restored to that of controls (Fig. 7E, S8D, S8E, and S8F). The ability of the inactivating C459S mutation to rescue ependymal cell development in double mutants reaffirmed that Ptpn11 activating mutations in NS induced hydrocephalus in a catalytically-dependent manner.
Table 2. The increased catalytic activity of SHP2E76K is required for its detrimental effect on ependymal cell development.
Ptpn11E76K,C459S/+ double mutation knock-in and Ptpn11E76K/+ single mutation knock-in mice were monitored for 15 months. The incidences of frank hydrocephalus in the euthanized animals were documented.
| Mice * | Incidence of frank hydrocephalus |
|---|---|
| Ptpn11E76K/+/Nestin-Cre+ | 26/56 |
| Ptpn11+/+/Nestin-Cre+ | 0/58 |
| Ptpn11E76K C459S/+ | 0/56 |
| Ptpn11+/+ | 0/44 |
Mice were monitored for 15 months.
Fig. 7. Pathological effects of Ptpn11E76K/+ on NSPCs and brain development depend on SHP2 catalytic activity.
(A and B) Cerebral cortices dissected from E14.5 embryos (n=4 mice per genotype) were assessed by neurosphere assays. Quantification of total numbers of primary (1° NS), secondary (2° NS), and tertiary (3° NS) neurospheres (A) and proliferation of cells in primary neurospheres (B). (C) Lateral ventricular walls dissected from newborn pups (n=4 mice per genotype) were processed for ependymal cell differentiation assays and stained with acetyl α-tubulin to mark cilia. Scale bar, 50 μm. (D) Scanning electron microscopy showing ependymal cells in brains dissected from 1–2 month-old mice (n=3 mice per genotype) of the indicated genotypes. Scale bar, 10 μm. (E) Immunoblot showing phosphorylated AKT (p-AKT), phosphorylated ERK (p-ERK), and phosphorylated STAT3 (p-STAT3) in neurosphere cells generated from mice of the indicated genotypes (n=3 mice per genotype) and treated with CNTF. Densitometric data of phosphoproteins (normalized to the respective total proteins) are summarized in Fig. S8D. Assays in all panels were performed in 3 independent experiments. Data are presented as mean±S.D. of biological replicates. Representative images are shown.
DISCUSSION
Germline activating and inactivating mutations of Ptpn11 (SHP2) are associated with two highly related developmental disorders, NS and NSML, respectively. In this report, we demonstrate that Ptpn11 mutations impact brain development in a manner that depends on the catalytic activity of SHP2. Neural expression of the activating mutation E76K in mice (Ptpn11E76K/+) caused hydrocephalus due to aberrant development of ependymal cells and their cilia, whereas ubiquitous expression of the inactivating mutation Y279C in mice (Ptpn11Y279C/+) did not induce hydrocephalus and had no effect on the ependymal cells. In addition, the severity of hydrocephalus caused by Ptpn11 activating mutations correlated with the potency of the enhanced SHP2 catalytic activity. More importantly, the pathogenic effect of the activating mutation E76K in Ptpn11 was completely abolished by the addition of the catalytically inactivating mutation C459S. None of the Ptpn11E76K,C459S/+ double mutation knock-in mice developed hydrocephalus. Given that Ptpn11 activating mutations induced hydrocephalus but inactivating mutations did not, this phenotype may be used for differential diagnosis or prognosis, or both, in NS and NSML patients. Indeed, hydrocephalus has been observed in NS patients (39–41). Because hydrocephalus development depends on increased catalytic activity of mutant SHP2, our mouse genetics data additionally imply that increased catalytic activitiy of mutant SHP2 may be a useful therapeutic target for controlling the progression of hydrocephalus in NS patients.
Ptpn11 disease mutations appear to perturb neural cell function and brain development through various signaling pathways. SHP2 is involved in several cell signaling processes, and it can also function at multiple steps in a single pathway (1, 2). This phosphatase has functions that depend on its catalytic activity and other functions that depend only on its ability to interact with other proteins and act as a scaffolding protein. SHP2 has been shown to play multiple roles in NSPCs, corticogenesis, astroglia cell fate decisions, and oligodendrocyte development in the telencephalon (42–44). NS mutations in SHP2 cause aberrant lineage specification in cultured NSPCs and in neonatal mice (45). Our data in this report provide insights into how Ptpn11 activating mutations can cause hydrocephalus, specifically by affecting the development of ependymal cells from NSPCs. The potency of the mutations in activating SHP2 catalytic activity correlates with the developmental defects in ependymal cells and the hydrocephalus phenotype. Therefore, “weaker” SHP2 activating mutations might not have the necessary threshold of SHP2 catalytic activity to induce hydrocephalus but may still have an impact in other central nervous system cell lineages.
Activating mutations in Ptpn11 enhance growth factor-induced ERK and AKT activities in neurosphere cells, which may account for their accelerated or spontaneous differentiation toward the glial lineage. This perturbation in neurogenesis and gliogenesis is likely responsible for the aberrant behavior of Ptpn11E76K knock-in mice. However, the the increased ERK and AKT activities do not explain the reduced growth rate of Ptpn11E76K/+ neurosphere cells. Rather, suppression of STAT3 activity in response to catalytically hyperactive SHP2 may account for this pathological effect. The inhibition of STAT3 possibly also contributes to the defects in ependymal cells and cilia in these mutant mice. This notion is supported by the fact that Stat3 knockouts (Stat3fl/fl/Nestin-Cre+) displayed similar defects in ependymal cells and cilia as did Ptpn11E76K/+/Nestin-Cre+ mice, and that STAT3 deletion and the Ptpn11E76K mutation similarly resulted in a decrease in the differentiation of NSPCs toward ependymal cells. In addition, the correlation of rescued ependymal cell development with restored STAT3 activity in SHP2 inhibitor–treated Ptpn11E76K/+ cells and in Ptpn11E76K,C459S/+ double mutant mice also supports that diminished STAT3 activity is likely responsible for the developmental defects in ependymal cells and hydrocephalus associated with Ptpn11 (SHP2) activating mutations. Nevertheless, it remains to be determined how diminished STAT3 activity caused a decrease in the ependymal cell differentiation from NSPCs and the malformation of ependymal cilia. A previous study showed that STAT3 also plays a critical role in the generation of airway ciliated cells from basal stem cells although the mechanism is unclear (46). Given that STAT3 is a transcription factor critical for the expression of many downstream genes, it is likely that one or more of these targets mediate the essential role of STAT3 in promoting the generation of ependymal cells and the maturation of post-mitotic ependymal cells with motile cilia. Future studies are required to identify these downstream mediators.
Finally, it should be noted that SHP2 plays complicated roles in cell signaling; the cellular and molecular mechanisms responsible for Ptpn11 mutation-associated diseases and symptoms are likely to be complex. Despite having opposing effects on catalytic function, both NS and NSML-associated Ptpn11 mutations impact the protein-protein interactions of SHP2. Similar to NS Ptpn11 mutations (9, 11, 15, 18), NSML-associated Ptpn11 mutations also disrupt the intramolecular N-SH2-PTP interaction and enhance the binding between SHP2 and tyrosine phosphorylated signaling partners (9, 18–20), due to similar changes in SHP2 protein conformation. NSML mutants function as dominant negative molecules that interfere with wild-type SHP2-mediated signaling. NSML-associated Ptpn11 inactivating mutations greatly decrease growth factor-induced activation of the RAS-ERK pathway (18), but increase AKT-mTOR signaling (19, 20), due to enhanced binding of SHP2 to insulin receptor substrate 1 (IRS-1) and decreased ability to dephosphorylate and inactivate the downstream pathway. Conceivably, both of the deregulated catalytic activities of SHP2 mutants, combined with their enhanced protein-protein interaction capabilities, determine the differing pathological effects that distinguish NS from NSML.
MATERIALS AND METHODS
Mice
Ptpn11E76K,C459S/+ mice were generated by gene targeting. In brief, a full length SHP2 E76K C459S cDNA was used to replace start codon ATG-containing the first exon and part of the first intron of the Ptpn11 locus in the same reading frame through homologous recombination. Ptpn11E76K neo/+ conditional knock-in mice were generated in our previous study (24). A loxP flanked neo cassette with a stop codon prevents the expression of the targeted allele (Ptpn11E76K neo). Upon deletion of neo by Cre DNA recombinase, the mutant allele (Ptpn11E76K) is reactivated, producing SHP2 E76K at physiological levels (24). Ptpn11E76K,C459S/+ and Ptpn11E76K neo/+ mice were backcrossed with C57BL6 mice for more than 10 generations. Ptpn11D61G/+ (36) and Ptpn11Y279C/+ (20) mice originally obtained from Beth Israel Deaconess Medical Center were backcrossed with C57BL/6 mice for 4 generations. Stat3fl/+ mice and Nestin-Cre+ mice were purchased from the Jackson Laboratory. Mice of the same age, sex, and genotype were mixed and then randomly grouped for subsequent analyses (investigators were not blinded). All mice were kept under specific pathogen-free conditions at Emory University Division of Animal Resources. All animal procedures complied with the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee.
Immunohistochemistry and immunofluorescence staining
Paraffin sections were prepared at 5 μm thickness. Before incubation in 3% H2O2 in methanol for 10 min abolishing endogenous peroxidases, sections were dewaxed, rehydrated, and heated in antigen unmasking buffer (10 mM Citric acid, 0.05% Tween 20, pH 6.0). After blocking sections with 10% normal goat serum in Tris-buffered saline (TBS) for 30 min, primary antibodies were applied for 16 hours at 4 °C. Sections were then rinsed in 1% NGS in TBS, followed by 10 min in 10% NGS, and a species-specific secondary antibody made in goat was applied for 30 min. After rinsing again as above, the species-specific peroxidase-anti-peroxidase complex was applied for 1 hour at room temperature. Sections were rinsed twice in Tris buffer (pH 7.6) and developed with 3′-3′-diaminobenzidine following the manufacturer’s protocol. For fluorescent signals of immunoreactivity, FITC-or PE-conjugated secondary antibodies were used and counterstained with 4′6′-diamidino-2-phenylindole (DAPI). Sections were mounted with Fluoromount-G. To quantify ependymal cells, FoxJ1+ cells or FoxJ1+STAT3+ cells in 1 mm ependymal field were counted. Three random different fields (technical replicates) were quantified and averaged for each mouse sample. The averaged numbers from all mice (biological replicates) were then subject to statistical analysis.
Immunocomplex phosphatase assay
Whole cell lysates (500 μg) were immunoprecipitated with 2 μg of an antibody recognizing SHP2. Immune complexes were washed twice with washing buffer (20 mM HEPES pH 7.4, 50 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA, and 0.05% Triton X-100), once with phosphatase assay buffer containing 20 mM HEPES pH 7.4, 50 mM NaCl, 2.5 mM EDTA, 5 mM DTT, and 100 μg/ml BSA. p-nitrophenyl phosphate (pNPP) was then added to each sample (3.4 mM) and incubated at 30 °C for 1 hour. Supernatants were transferred to 96-well plates and absorbance at 405 nm was measured.
Neurosphere assay
The neurosphere assay was performed to determine self-renewal capacity of NSPCs, as previously described (42, 47). Cerebral cortices dissected from E14.5 embryos were dissociated into single cells by trypsinization and mechanical dissociation. These cells were suspended and maintained in B27/neurobasal medium supplemented with bFGF (20 ng/ml), EGF (20 ng/ml), L-glutamine (1mM), and Penicillin/Streptomycin in a 96-well plate at the density of 5,000 cells/ml (200 μl per well). After 7 days of incubation, neurospheres formed in the culture were counted under an optical microscope, and a minimum of 10 wells were counted. For the secondary neurosphere assay, primary neurospheres were collected and dissociated into a single-cell suspension and reseeded in a 96-well plate. Secondary spheres derived were enumerated 7 days later as above.
Western blotting
Neurospheres were generated, dissociated into single cells, and cultured in the presence of bFGF (20 ng/ml) for 5 days. Cells were harvested and subjected to real-time quantitative PCR analyses for mRNA abundance of bFGFR and CNTFR. The PCR primers used were: 5′-GCAGAGCATCAACTGGCTG-3′ and 5′-GGTCACGCAAGCGTAGAGG-3′for FGFR1; 5′-TGTCTACACGCAGAAACACAG-3′ and 5′-CCCAGACGCTCATACTGCAC-3′for CNTFR. To examine bFGF and CNTF signaling, cultured neurosphere cells were starved in neural basal medium without B27 supplement and growth factors for 4 hours. Cells were then stimulated with CNTF (100 ng/ml) or bFGF (50 ng/ml) for the indicated periods of time. Cells were washed and lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 2 mM Na3VO4, 10 μg/mL leupeptin, 10 μg/mL aprotin, and 1 mM PMSF). Whole cell lysates (50 μg) were resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies.
Neurosphere cell cycle and apoptosis analyses
Neurospheres were harvested and dissociated into single cells. These cells were fixed at 4 °C overnight, permeabilized, and then stained with an FITC-labeled antibody recognizing Ki67 followed by Hoechst 33342 (20 μg/ml) staining. Percentages of the cells in G0, G1, and S/G2/M phases were quantified by FACS. For apoptosis analysis, neurosphere cells were stained with Annexin V-FITC and 7-amino-actinomycin D (7-AAD) using an Annexin V-FITC apoptosis Detection Kit I (BD Biosciences). Percentages of the Annexin V positive (early apoptotic cells) and Annexin V/7-ADD double positive cells (late apoptotic cells) were quantified by FACS.
Ependymal cell differentiation
Lateral ventricular walls were dissected from newborn pups and dissociated into single cells by trypsinization and mechanical dissociation. Cells were cultured in DMEM/GluMAX supplemented with 10% fatal bovine serum (FBS) and penicillin-streptomycin in poly-L-lysine (50 μg/ml) coated flasks for 4 days. When the cells reached confluence, the flasks were closed tightly and shaken at 250 rpm for 30 min to remove weakly attached cells (mainly differentiated oligodendrocytes and neurons). The remaining cells were collected by trypsinization and resuspended in the same medium. Aliquots of the cell suspension (2×105 cells/50 μl) were dropped onto poly-L-lysine coated, dried coverslips. The cells were incubated for 1 hour to allow the cells to adhere at high density. Cells on the coverslips were then cultured in tissue culture plates with the same medium for 24 hours. The medium was changed to FBS-free DMEM/GluMAX and the cells were continuously cultured for 7 days to allow ependymal progenitors to progressively differentiate into ependymal cells. Coverslips were washed twice with phosphate buffered saline, fixed in 4% paraformaldehyde and processed for immunostaining with acetyl α-tubulin.
The differentiated cells were also harvested and subject to quantitative reverse transcription PCR analyses for mRNA abundance of cell lineage markers. The PCR primers used were: 5′-GCCAGCCTCAGAACAAACAG-3′and 5′-AAGGTCTTGGGAGGGAAGAAC-3′for Mtap2; 5′-GGCAAATGTTCGGGCAATTCG-3′ and 5′-TCAATTTTCCGTCCCTCTACGAT-3′for Rbfox3; 5′-TAGACCCCAGCGGCAACTAT-3′ and 5′-GTTCCAGGTTCCAAGTCCACC-3′ for Tubb3; 5′-CCCTGGCTCGTGTGGATTT-3′ and 5′-GACCGATACCACTCCTCTGTC-3′ for Gfap; 5′-TGGTTGCCCTCATTGATGTCT-3′ and 5′-CCCATCCCCATCTTCGTCC-3′ for S100b; 5′-GGACACAATGCACATTCAAGAAC-3′ and 5′-CCGAACCACAGACTTACAGTTT-3′ for Fabp7; 5′-CCTTGTGGTTCTTACGTTTGTTG-3′ and 5′-CGTTGACGACATTCTCAAGCTG-3′ for Prom1; 5′-GTTGCACCGTTTCCCGGTAA-3′ and 5′-CCCCTCTGGTGGTAGCGTTA-3′ for Cd24a. To determine the rescue effects of inhibition of ERK, AKT, and SHP2 on the ependymal differentiation of Ptpn11E76K/+ NSPCs, the MAPKK1 inhibitor PD98059 (25 μM), the PI3K inhibitor LY294002 (10 μM), the SHP2 inhibitor #220-324 (5 and 10 μM), or vehicle was included in the ependymal cell differentiation assay.
Electron microscopy
Brains were fixed with 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer (PB; pH 7.4) for 1 hour, and then the ventral region of the brain, the septum, and the hippocampus were removed and fixed overnight at 4°C in the same solution. Samples were washed with 0.1 M PB 3 times on ice at 5-min intervals and postfixed with 1% osmium tetroxide in 0.1 M PB for 2 hours. The samples were then washed with distilled water 5 times on ice at 5-min intervals, and dehydrated 3 times in an ethanol series. The ethanol was cleared with tert-butyl alcohol, and the samples were freeze-dried (ES-2030 Freeze Dryer; Hitachi), vapor-deposited with an HPC-1S osmium coater (Vacuum Devices) and examined with a field-emission scanning electron microscope (Model S4500; Hitachi).
Magnetic resonance imaging (MRI)
The mouse MR images were acquired using a high-field (9.4-T) small-animal MR scanner (Bruker BioSpin GmbH, Rheinstetten, Germany). T2-weighted MRI scans using a multi-slice multi-echo (MSME) sequence were acquired, producing a set of sagittal and axial images [repetition time (TR) = 1250 ms, echo time (TE) =15 ms, flip angle = 90u, voxel size = 0.031260.031260.05 cm] while each mouse was under isoflurane-induced anesthesia at the University Hospital, Case Western Reserve University Medical Center. The animals were allowed to recover from anesthesia in their cages for at least 1 hour before imaging.
Animal behavior test
Animal behavior tests were performed at the rodent behavior core facility of Case Western Reserve University following standard protocols. The open field test was used to assess anxiety of animals (48). The muscle strengths of forelimbs and hindlimbs of mice were measured using a commercially available grip strength meter apparatus that measures the gripping strength of mice (49). The global neuromuscular strength and motor function were also determined by wire hang tests. In addition, the rotarod test was used to determine motor coordination (50).
Statistical analysis
Experiments were repeated three to four times with the indicated numbers of mice (biological replicates). Data are presented as mean±S.D. Statistical significance was determined using unpaired two-tailed Student’s t test. p<0.05 is considered statistically significant.
Supplementary Material
Fig. S1. The Ptpn11E76K mutation induces brain developmental defects with aberrant behaviors.
Fig. S2. Neurons are reduced and astrocytes are increased in the cerebral cortex and hippocampus of adult Ptpn11E76K/+/Nestin-Cre+ mice.
Fig. S3. Total number and survival of NSPCs in the developing brain of Ptpn11E76K/+/Nestin-Cre+ mice are not significantly changed.
Fig. S4. Neuron and ependymal cell differentiation is decreased whereas astrocyte differentiation is increased in Ptpn11E76K/+/Nestin-Cre+ NSPCs.
Fig. S5. ERK activity in the developing brain of Ptpn11E76K/+/Nestin-Cre+ mice is enhanced, but cell proliferation does not change.
Fig. S6. Similar abundance of bFGF and CNTF receptors in Ptpn11E76K/+/Nestin-Cre+ and Ptpn11+/+/Nestin-Cre+ NSPCs.
Fig. S7. Defects of ependymal cilia are proportionate to the catalytic activity of various mutant forms of SHP2.
Fig. S8. Generation and characterization of Ptpn11E76K,C459S/+ mice.
Acknowledgments
Funding: This work was supported by National Institutes of Health grants HL130995, DK092722 and HD087760 (to C.K.Q.), NS088529 (to R.R.W.), and HL114775 (to M.I.K.).
Footnotes
Author contributions: H.Z., W.M.Y., and R.R.W. conducted the research and summarized the data. M.I.K. and B.G.N. provided critical reagents and experimental tools, discussed the work, and edited the manuscript. C.K.Q. designed the experiments and provided technical training to the first two authors. H.Z. and C.K.Q. wrote the manuscript with input from all authors.
Competing interests: The authors declare no competing financial interests.
Data and materials availability: Requests for the plasmids and genetically modified mice require a material transfer agreement from Emory University, U.S.A.
REFERENCES AND NOTES
- 1.Neel BG, Gu H, Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 2.Xu D, Qu CK. Protein tyrosine phosphatases in the JAK/STAT pathway. Front Biosci. 2008;13:4925–4932. doi: 10.2741/3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 4.Barford D, Neel BG. Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure. 1998;6:249–254. doi: 10.1016/s0969-2126(98)00027-6. [DOI] [PubMed] [Google Scholar]
- 5.Bennett AM, Tang TL, Sugimoto S, Walsh CT, Neel BG. Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc Natl Acad Sci U S A. 1994;91:7335–7339. doi: 10.1073/pnas.91.15.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W, et al. A new function for a phosphotyrosine phosphatase: linking GRB2-Sos to a receptor tyrosine kinase. Mol Cell Biol. 1994;14:509–517. doi: 10.1128/mcb.14.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart RA, et al. Phosphatase-dependent and -independent functions of Shp2 in neural crest cells underlie LEOPARD syndrome pathogenesis. Dev Cell. 2010;18:750–762. doi: 10.1016/j.devcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu WM, Hawley TS, Hawley RG, Qu CK. Catalytic-dependent and -independent roles of SHP-2 tyrosine phosphatase in interleukin-3 signaling. Oncogene. 2003;22:5995–6004. doi: 10.1038/sj.onc.1206846. [DOI] [PubMed] [Google Scholar]
- 9.Tartaglia M, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 10.Roberts AE, Allanson JE, Tartaglia M, Gelb BD. Noonan syndrome. Lancet. 2013;381:333–342. doi: 10.1016/S0140-6736(12)61023-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tartaglia M, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 12.Loh ML, et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood. 2004;103:2325–2331. doi: 10.1182/blood-2003-09-3287. [DOI] [PubMed] [Google Scholar]
- 13.Tartaglia M, et al. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 2004;104:307–313. doi: 10.1182/blood-2003-11-3876. [DOI] [PubMed] [Google Scholar]
- 14.Loh ML, et al. PTPN11 mutations in pediatric patients with acute myeloid leukemia: results from the Children’s Cancer Group. Leukemia. 2004;18:1831–1834. doi: 10.1038/sj.leu.2403492. [DOI] [PubMed] [Google Scholar]
- 15.Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J Biol Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- 16.Legius E, et al. PTPN11 mutations in LEOPARD syndrome. J Med Genet. 2002;39:571–574. doi: 10.1136/jmg.39.8.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Digilio MC, et al. Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am J Hum Genet. 2002;71:389–394. doi: 10.1086/341528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 19.Edouard T, et al. Functional effects of PTPN11 (SHP2) mutations causing LEOPARD syndrome on epidermal growth factor-induced phosphoinositide 3-kinase/AKT/glycogen synthase kinase 3beta signaling. Mol Cell Biol. 2010;30:2498–2507. doi: 10.1128/MCB.00646-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marin TM, et al. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest. 2011;121:1026–1043. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu WM, Daino H, Chen J, Bunting KD, Qu CK. Effects of a Leukemia-associated Gain-of-Function Mutation of SHP-2 Phosphatase on Interleukin-3 Signaling. J Biol Chem. 2006;281:5426–5434. doi: 10.1074/jbc.M507622200. [DOI] [PubMed] [Google Scholar]
- 22.Lauriol J, Kontaridis MI. PTPN11-associated mutations in the heart: has LEOPARD changed Its RASpots? Trends Cardiovasc Med. 2011;21:97–104. doi: 10.1016/j.tcm.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.N. Cancer Genome Atlas Research. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu D, et al. Non-lineage/stage-restricted effects of a gain-of-function mutation in tyrosine phosphatase Ptpn11 (Shp2) on malignant transformation of hematopoietic cells. J Exp Med. 2011;208:1977–1988. doi: 10.1084/jem.20110450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong L, et al. Leukaemogenic effects of Ptpn11 activating mutations in the stem cell microenvironment. Nature. 2016;539:304–308. doi: 10.1038/nature20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 27.Qin S, Liu M, Niu W, Zhang CL. Dysregulation of Kruppel-like factor 4 during brain development leads to hydrocephalus in mice. Proc Natl Acad Sci U S A. 2011;108:21117–21121. doi: 10.1073/pnas.1112351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waclaw RR, et al. The zinc finger transcription factor Sp8 regulates the generation and diversity of olfactory bulb interneurons. Neuron. 2006;49:503–516. doi: 10.1016/j.neuron.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 29.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 30.Kahle KT, Kulkarni AV, Limbrick DD, Jr, Warf BC. Hydrocephalus in children. Lancet. 2016;387:788–799. doi: 10.1016/S0140-6736(15)60694-8. [DOI] [PubMed] [Google Scholar]
- 31.Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young KM, Fogarty M, Kessaris N, Richardson WD. Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J Neurosci. 2007;27:8286–8296. doi: 10.1523/JNEUROSCI.0476-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bard-Chapeau EA, et al. Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell. 2011;19:629–639. doi: 10.1016/j.ccr.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, et al. Negative regulation of Stat3 by activating PTPN11 mutants contributes to the pathogenesis of Noonan syndrome and juvenile myelomonocytic leukemia. J Biol Chem. 2009;284:22353–22363. doi: 10.1074/jbc.M109.020495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu B, et al. Targeting protein tyrosine phosphatase SHP2 for the treatment of PTPN11-associated malignancies. Mol Cancer Ther. 2013;12:1738–1748. doi: 10.1158/1535-7163.MCT-13-0049-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Araki T, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 37.Bennett AM, Hausdorff SF, O’Reilly AM, Freeman RM, Neel BG. Multiple requirements for SHPTP2 in epidermal growth factor-mediated cell cycle progression. Mol Cell Biol. 1996;16:1189–1202. doi: 10.1128/mcb.16.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamauchi K, Milarski KL, Saltiel AR, Pessin JE. Protein-tyrosine-phosphatase SHPTP2 is a required positive effector for insulin downstream signaling. Proc Natl Acad Sci U S A. 1995;92:664–668. doi: 10.1073/pnas.92.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henn W, et al. Progressive hydrocephalus in two members of a family with autosomal dominant Noonan phenotype. Clin Dysmorphol. 1997;6:153–156. [PubMed] [Google Scholar]
- 40.Heye N, Dunne JW. Noonan’s syndrome with hydrocephalus, hindbrain herniation, and upper cervical intracord cyst. J Neurol Neurosurg Psychiatry. 1995;59:338–339. doi: 10.1136/jnnp.59.3.338-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fryns JP. Progressive hydrocephalus in Noonan syndrome. Clin Dysmorphol. 1997;6:379. doi: 10.1097/00019605-199710000-00014. [DOI] [PubMed] [Google Scholar]
- 42.Ke Y, et al. Deletion of Shp2 in the brain leads to defective proliferation and differentiation in neural stem cells and early postnatal lethality. Mol Cell Biol. 2007;27:6706–6717. doi: 10.1128/MCB.01225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li K, Leung AW, Guo Q, Yang W, Li JY. Shp2-dependent ERK signaling is essential for induction of Bergmann glia and foliation of the cerebellum. J Neurosci. 2014;34:922–931. doi: 10.1523/JNEUROSCI.3476-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehrman LA, et al. The protein tyrosine phosphatase Shp2 is required for the generation of oligodendrocyte progenitor cells and myelination in the mouse telencephalon. J Neurosci. 2014;34:3767–3778. doi: 10.1523/JNEUROSCI.3515-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gauthier AS, et al. Control of CNS cell-fate decisions by SHP-2 and its dysregulation in Noonan syndrome. Neuron. 2007;54:245–262. doi: 10.1016/j.neuron.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tadokoro T, et al. IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc Natl Acad Sci U S A. 2014;111:E3641–3649. doi: 10.1073/pnas.1409781111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ethell IM, Yamaguchi Y. Cell surface heparan sulfate proteoglycan syndecan-2 induces the maturation of dendritic spines in rat hippocampal neurons. J Cell Biol. 1999;144:575–586. doi: 10.1083/jcb.144.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deacon RM. Housing, husbandry and handling of rodents for behavioral experiments. Nat Protoc. 2006;1:936–946. doi: 10.1038/nprot.2006.120. [DOI] [PubMed] [Google Scholar]
- 49.Tucci V, et al. Reaching and grasping phenotypes in the mouse (Mus musculus): a characterization of inbred strains and mutant lines. Neuroscience. 2007;147:573–582. doi: 10.1016/j.neuroscience.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 50.Carter RJ, et al. Characterization of progressive motor deficits in mice transgenic for the human Huntington’s disease mutation. J Neurosci. 1999;19:3248–3257. doi: 10.1523/JNEUROSCI.19-08-03248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The Ptpn11E76K mutation induces brain developmental defects with aberrant behaviors.
Fig. S2. Neurons are reduced and astrocytes are increased in the cerebral cortex and hippocampus of adult Ptpn11E76K/+/Nestin-Cre+ mice.
Fig. S3. Total number and survival of NSPCs in the developing brain of Ptpn11E76K/+/Nestin-Cre+ mice are not significantly changed.
Fig. S4. Neuron and ependymal cell differentiation is decreased whereas astrocyte differentiation is increased in Ptpn11E76K/+/Nestin-Cre+ NSPCs.
Fig. S5. ERK activity in the developing brain of Ptpn11E76K/+/Nestin-Cre+ mice is enhanced, but cell proliferation does not change.
Fig. S6. Similar abundance of bFGF and CNTF receptors in Ptpn11E76K/+/Nestin-Cre+ and Ptpn11+/+/Nestin-Cre+ NSPCs.
Fig. S7. Defects of ependymal cilia are proportionate to the catalytic activity of various mutant forms of SHP2.
Fig. S8. Generation and characterization of Ptpn11E76K,C459S/+ mice.