Abstract
Epithelial-to-mesenchymal transition (EMT) is a process that plays essential roles in development and wound healing that is characterized by loss of homotypic adhesion and cell polarity and increased invasion and migration. At the molecular level, EMT is characterized by loss of E-cadherin and increased expression of several transcriptional repressors of E-cadherin expression (Zeb-1, Zeb-2, Twist, Snail, and Slug). Early work established that loss of E-cadherin and increased expression of MMP-9 was associated with a poor clinical outcome in patients with urothelial tumors, suggesting that EMT might also be associated with bladder cancer progression and metastasis. More recently, we have used global gene expression profiling to characterize the molecular heterogeneity in human urothelial cancer cell lines (n=20) and primary patient tumors, and unsupervised clustering analyses revealed that the cells naturally segregate into two discrete “epithelial” and “mes-enchymal” subsets, the latter consisting entirely of muscle-invasive tumors. Importantly, sensitivity to inhibitors of the epidermal growth factor receptor (EGFR) or type-3 fibroblast growth factor receptor (FGFR3) was confined to the “epithelial” subset, and sensitivity to EGFR inhibitors could be reestablished by micro-RNA-mediated molecular reversal of EMT. The results suggest that EMT coordinately regulates drug resistance and muscle invasion/metastasis in urothelial cancer and is a dominant feature of overall cancer biology.
Keywords: Epithelial-to-mesenchymal transition, Epidermal growth factor receptor, Type-3 fibroblast growth factor receptor, Micro-RNA-mediated molecular reversal, Urothelial tumor
1 Epithelial-to-mesenchymal transition-background and relevance to cancer progression
The term epithelial-to-mesenchymal transition (EMT) refers to the complex reprogramming of epithelial cells that occurs on occasion during embryonic development and tissue repair in the adult organism [1]. Epithelial cells mediate important barrier functions and form tight homotypic interactions that contribute to the formation of these permeability barriers. E-cadherin plays a central role through its interaction with β-catenin and the actin cytoskeleton [1]. Thus, the most familiar change that occurs during EMT is downregulation of surface E-cadherin expression, resulting in loss of homotypic adhesion [1]. Other common features of EMT include loss of basal-apical polarity genes and tight junction molecules and increased expression of vimentin, fibronectin, and the S100 proteins (Fig. 1) [1]. During wound healing, EMT facilitates repair by facilitating epithelial migration to the site of injury, and once repair is underway epithelial cells re-express E-cadherin and other epithelial markers via a process that is sometimes referred to as “mesenchymal-to-epithelial transition” (MET) [1]. There is evidence that cell division and invasion/migration are mutually exclusive processes [2, 3], so MET may be essential not only for reestablishing barrier function but also for enabling the cell proliferation that is required for complete wound closure.
Fig. 1.
Molecular markers of EMT. Epithelial markers are displayed in blue, mesenchymal in red. a Regulation of E-cadherin expression. Upstream signals, and in particular TGFβ, increase expression of transcriptional repressors of E-cadherin (Zeb-1, Zeb-2, Twist, Snail, and Slug) that function by binding to two so-called “E-box” elements that are located within the E-cadherin promoter. This results in recruitment of histone deacetylases and possibly histone and DNA methyltransferases to the E-cadherin promoter, resulting in transcriptional silencing. Members of the miR200 family of micro-RNAs directly antagonize this process by binding to multiple copies of specific “seed” sequences located within the mRNAs encoding Zeb-1 and Zeb-2, resulting in transcript degradation and inhibition of translation. b. Other representative markers of the “epithelial” and “mesenchymal” states. The lists provided are not comprehensive, and the potential mechanistic relationships between these markers and E-cadherin signaling has in most cases not been determined
EMT is controlled by a group of transcriptional repressors (Zeb-1, Zeb-2, Twist, Snail, and Slug) that recruit histone deacetylases to E-box elements that are located within the E-cadherin promoter and a variety of other genes [1]. Diverse upstream signals increase the expression of these E-cadherin repressors at the mRNA level. Of particular importance are the transforming growth factor-beta (TGFβ)/bone morphogenic protein (BMP) family of cytokines [1], although a variety of other developmental and inflammatory signals promote EMT as well (Fig. 1a). Upon binding their specific surface receptors, TGFβ and the BMPs promote the direct activation of a family of receptor-linked transcription factors (SMADs) [4], which translocate to the nucleus and regulate their target genes (including Snail) [5] in a phosphorylation-dependent manner. In addition, recent work demonstrated that SMADs 2 and 3 also exert effects on transcription and translation by regulating micro-RNA processing [6]. In normal epithelial cells, TGFβ simultaneously induces EMT by downregulating E-cadherin and upregulating Zeb-1, Zeb-2, and Snail and inhibits cell cycle progression by activating the p16/Rb checkpoint [1]. Increased TGFβ expression is a common feature of solid tumor progression. Although cancer progression often selects for the acquisition of defects on TGFβ signaling, in some cases, the mutations appear to selectively inactivate TGFβ's growth inhibitory effects without interfering with EMT signaling. An excellent example of this can be found in pancreatic cancer cells and other solid tumors that possess defects in SMAD4: recent work demonstrated that it is essential for TGFβ-induced growth inhibition but is dispensable for EMT [7].
Exciting recent work suggests that members of the miR200 family of micro-RNAs play central roles in mediating the effects of TGFβ and other EMT regulators in normal and malignant epithelial cells [8–11]. Members of the family bind directly to the mRNAs encoding Zeb-1 and Zeb-2, promoting their degradation and blocking translation. TGFβ downregulates miR200 family micro-RNAs via poorly characterized mechanism(s), leading to accumulation of Zeb-1 and Zeb-2, suppression of E-cadherin expression, and increased motility and invasiveness. Interestingly, Zeb-1 and Zeb-2 can also suppress miR200 family expression, indicating that feedback loops are in place to fine tune EMT-related signaling.
The parallels between EMT and the processes that underlie various aspects of cancer progression and metastasis have not been lost on cancer researchers [1]. It has long been recognized that E-cadherin expression is decreased in metastatic cancers [12]. Fidler's group demonstrated that loss of E-cadherin accompanied by increased expression of matrix metalloproteinases (which mediate invasion and are markers of EMT) (Fig. 1b) was a better predictor of poor prognosis in several models as compared to downregulation of E-cadherin or upregulation of MMPs alone [13–22] (based on this work, Slaton et al. demonstrated that a low E-cadherin-to-MMP9 ratio predicted poor outcome in patients with bladder cancer [23]). Studies in preclinical models demonstrated that loss of E-cadherin directly promoted an EMT phenotype and reprogrammed global gene expression [24]. Aside from promoting adhesion-related signaling, E-cadherin re-expression also promotes relocalization of β-catenin, which can function as a transcriptional co-activator, from the nucleus to the plasma membrane [25]. Finally, recent analyses of the changes in micro-RNA expression in a transgenic mouse model of lung cancer demonstrated that downregulation of miR200 family expression plays a major role in disease progression [26]. These observations have stimulated strong interest in defining the molecular mechanisms underlying this epithelial-to-mesenchymal “switch” during tumor progression. Although autocrine or paracrine cytokine (especially TGFβ family) signaling seems an obvious place to start, the E-cadherin promoter is commonly silenced by chromatin methylation in solid tumor cells [27], and whether this is facilitated by the same transcriptional repressors that down-regulate E-cadherin is not yet clear. In addition, E-cadherin is infrequently inactivated by mutation [27], and under these circumstances, there may or may not be added selective pressure on the miR200 family or the EMT-mediating E-cadherin repressors.
2 EMT and the two tracks of bladder cancer progression
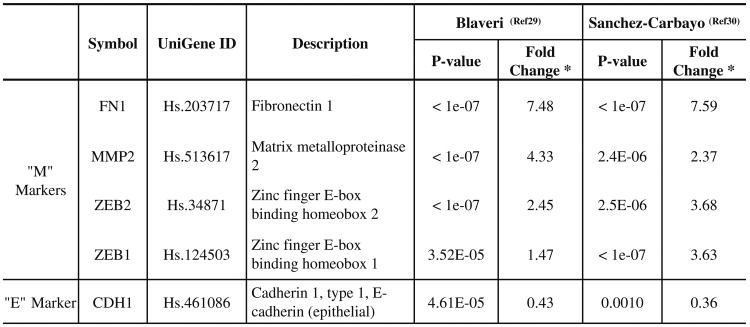
As discussed throughout this themed issue, one of the distinguishing features of urothelial cancer is that it progresses along two pathways that appear to be distinct at the phenotypic and molecular levels [28]. The fact that they are distinguished by their relative propensities to become muscle-invasive strongly suggests that the EMT-related programming of the two types of cancer is vastly different. Indeed, independent global gene expression profiling studies by Waldman's [29] and Cordon-Cardo's [30] groups demonstrated that superficial and muscle-invasive tumors fall into two distinct subgroups when analyzed by unsupervised clustering. We downloaded their data and performed an in silico analysis of the relationship between EMT marker expression (E-cadherin, Zeb-1, and Zeb-2) and disease subtype. The results confirmed that there was a highly significant enrichment for Zeb-1 and Zeb-2 expression in the muscle-invasive tumors (Fig. 2) (W. Choi et al., manuscript submitted), supporting the concept that EMT might underlie urothelial cancer invasion and metastasis.
Fig. 2.
Relationship of EMT to the two tracks of bladder cancer progression. We compared the expression of four “mesenchymal” markers and E-cadherin in gene expression profiles obtained from superficial and muscle-invasive urothelial cancers using data from two publically available datasets. Fold changes and statistical signficance are indicated. Note that upregulation of the mesenchymal markers and downregulation of E-cadherin are significantly associated with muscle-invasive disease
*Fold change: Invasive tumors/Superficial tumors.
Two other recent studies provide further support for this concept. In the first, Baumgart and coworkers characterized the expression of E-cadherin, β-catenin, plakoglobin, and vimentin in a series of 825 primary tumors displayed on 10 tissue microarrays. Downregulation of epithelial markers was associated with disease progression in terms of both grade and stage (i.e., superficial versus muscle-invasive cancer), and in univariate analyses, downregulation of either β-catenin or plakoglobin was associated with shorter disease-specific survival [31]. In the second study, Sayan and coworkers assessed expression of the EMT regulators Zeb-1 and Zeb-2 in a panel of human urothelial cancer cell lines and primary human tumors. They confirmed that Zeb-1 protein expression was associated with increased invasion/ migration in vitro and Zeb-2 was enriched in muscle-invasive cancers [32]. Importantly, overexpression of Zeb-2 in “epithelial” cell lines rendered them resistant to radiation-induced apoptosis, and overexpression of Zeb-2 in radiation-exposed primary tumors was also associated with poor clinical outcome [32]. Radiation resistance was linked to increased DNA repair capacity as measured by clearance of radiation-induced H2AX foci in cell lines in vitro [32].
Prompted by these observations, we recently measured EMT marker expression in a panel of 20 human urothelial cell lines and a set of 114 primary urothelial tumors for which outstanding clinical outcome information was available (W. Choi et al., manuscript submitted). We observed a strong inverse correlation between the expression of E-cadherin and Zeb-1, Zeb-2 and vimentin in the cell lines and the tumors, and expression of the mesenchymal markers was confined to muscle-invasive disease. Included in our panel of markers was p63 (Fig. 1b), a member of the p53 family that Cordon-Cardo's group [33, 34] and others [35–41] showed was downregulated as a function of progression, particularly in muscle-invasive tumors. Expression of p63 and E-cadherin were closely correlated in the cell lines and the primary tumors, strongly suggesting that it is a marker of the “epithelial” phenotype (W Choi et al., manuscript submitted). Strikingly, however, high-level expression of p63 in muscle-invasive (T2–T4) tumors was significantly associated with poor clinical outcome. This relationship was even apparent in the T1 tumors, a transitional subset in which clinical outcome is particularly difficult to predict. Our data create an important paradox and a conceptual inconsistency with the recent published literature [31, 32]. On the one hand, our data strongly support the previous implication of EMT in muscle-invasive disease, the “track” that is responsible for essentially all bladder cancer mortality. However, they also suggest that within this subset, retention, or reversion to a more “epithelial” phenotype is associated with even worse outcome. Whether this represents a “MET” that selects for increased proliferation once cancer cells have metastasized is unclear. In ongoing studies, we are aggressively pursuing the mechanistic basis for the link between p63 expression and the “bad” biology observed within the muscle-invasive subset. One possibility is that it is somehow linked to urothelial cancer “stemness,” since p63 expression tracks closely with markers such as cytokeratins 5 and 17 that identify the “basal” cells within the bladder (M. Tran, unpublished observations; see chapter by Brandt et al. in this volume), and p63 clearly plays an essential role in maintaining “stemness” in the epithelial (basal) compartments of the prostate [42–44] and skin [45], and it also localizes to the basal layer of the normal urothelium [46] (M. Tran, unpublished observations).
3 Role of EMT in EGFR dependency
Inhibitors of the epidermal growth factor receptor (EGFR) were among the first targeted agents to be developed for cancer therapy [47]. The rationale underlying this effort came from a large number of studies that documented that the EGFR tends to be overexpressed as a function of progression in solid tumors, including urothelial cancer [48, 49]. Preclinical studies provided further support for the idea that EGFR inhibitors might exert unique potency in bladder cancer. For example, work from our group demonstrated that EGFR blockade inhibited the growth and metastasis of orthotopic human 253J B-V bladder xenografts, effects that were linked to inhibition of tumor production of angiogenic factors (VEGF, bFGF, and IL-8) [50], and overexpression of the EGFR promotes tumorigenesis in transgenic mice [51]. These studies and others prompted the clinical evaluation of small molecule and antibody-based inhibitors of the EGFR in patients with muscle-invasive urothelial cancers. Although it is still too early to evaluate whether or not the approach has been successful, the preliminary results suggest that benefit will be limited to a relatively small subset of patients, if indeed the approach proves to add benefit at all (discussed in a subsequent article by Dovedi and Davies). One of us (A. Siefker-Radtke) is currently leading three such studies. The first is an MD Anderson-based, SPORE-supported investigator-initiated trial in which the small molecule EGFR tyrosine kinase inhibitor gefitinib is given only after conventional combination chemotherapy has been administered to maximum benefit (i.e., in the setting of minimal residual disease), and the second is a Southwest Oncology Group trial in which the blocking anti-EGFR antibody cetuximab is administered in combination with gemcitabine and cisplatin, where we will be measuring EMT markers as a component of the correlative studies attached to the trial. Finally, in another SPORE trial we are performing a neoadjuvant study with erlotinib, where the main objective is to determine whether EMT markers correlate with biological response to the drug (measured by Ki-67 and p27 immunohistochemistry).
However, our expectations for the potential efficacy of EGFR-directed therapy in muscle-invasive disease have changed markedly since we conducted some of the first xenograft studies with this class of drugs almost a decade ago. This change has been driven largely by the results of clinical trials with EGFR inhibitors in other solid malignancies [52–54]. The EGFR is also overexpressed in advanced lung, colon, pancreas, and head and neck cancers, and small molecules (gefitinib, erlotinib) and blocking antibodies have been evaluated extensively in patients with lung and colon cancer. Originally, patient enrollment was guided by immunohistochemical detection of high level EGFR expression in pretreatment tumor tissues, but it has become clear that EGFR expression does not correlate clearly with response [53, 54]. Rather, in lung cancer, the presence of activating kinase domain mutations, which are found in approximately 15% of patients in the USA (closer to 25% in Japan), are clearly associated with response to the small molecule inhibitors [53, 55], and there is also evidence that lung and colon cancers that display EGFR gene amplification are also sensitive [56–59]. Conversely, the presence of activating Kras mutations renders lung and colon cancers resistant to EGFR-directed therapy [53, 60, 61]. Overall, what is becoming increasingly apparent is that sensitivity or resistance to targeted therapy is complex and not tightly linked to overexpression of the target.
We, therefore, designed experiments to define the potential heterogeneity in EGFR inhibitor responsiveness more comprehensively. We assembled a relatively large set of unique human urothelial cancer lines (n=20) and screened them for sensitivity to gefitinib-induced growth arrest (as measured by 3H-thymidine incorporation) [62, 63]. In parallel, we characterized their baseline gene expression profiles using the Illumina platform. We found that approximately 30% of the lines displayed at least 50% growth arrest at clinically relevant concentrations of the drug (<1 μM) [64]. Sensitivity was only loosely associated with surface EGFR levels but was clearly linked to autocrine signaling, as measured by the coupling of EGFR tyrosine kinase activity to downstream ERK and AKT pathway activation [62, 64]. Strikingly, all of the drug-sensitive cells expressed E-cadherin [64] and the panel displayed indistinguishable patterns of sensitivity and resistance to cetuximab [65], demonstrating that the effects of gefitinib were related to intrinsic EGFR dependency and not to other effects of the drug. Importantly, RNAi-mediated knockdown of E-cadherin rendered cells resistant to cetuximab [65], demonstrating that E-cadherin plays a causal role in maintaining EGFR inhibitor sensitivity.
Not all of the “epithelial” bladder cancer cell lines in our panel responded to EGFR tyrosine kinase inhibitors or cetuximab. Reanalysis of our gene expression profiling data revealed that the levels of FGFR3 were higher in the “epithelial” as compared to the “mesenchymal” cells [66] (W. Choi, unpublished observation), and direct sequencing of FGFR3 demonstrated that four of our cell lines possessed activating FGFR3 mutations (P. Black, manuscript in preparation). As discussed in previous chapters of this volume, activating FGFR3 mutations are very common in superficial tumors [67], and more recent work has demonstrated that RNAi-mediated knockdown of FGFR3 [68] or exposure to a clinical FGFR3 inhibitor [69] blocked proliferation in these cells. We have confirmed that several of the “epithelial” lines that do not respond to EGFR antagonists are strongly inhibited by a selective small molecule inhibitor of FGFR3 (A. Kwan, I. Lee, unpublished observations). Interestingly, some of the inhibitor-sensitive cell lines (i.e., RT4, UM-UC1, and RT112) express only wild-type FGFR3 [69], indicating that FGFR3 inhibitor sensitivity is not limited to the mutant tumors. However, as is true for EGFR antagonists, all of the cells that are sensitive to FGFR3 inhibitors are confined to the “epithelial” subset of cell lines. We are currently in the process of determining whether a molecular signature for FGFR3 dependency can be identified that can distinguish FGFR3-dependent from EGFR-dependent cells.
We also screened our cell lines and a relatively large set of primary human tumors for the presence of EGFR gene amplification, the presence of activating kinase domain mutations (akin to those described in lung cancer), and expression of a variant form of the EGFR (type III) that has been implicated in EGFR dependency in gliomas [70, 71]. We found only one cell line (UMUC5) that possessed high level amplification of the EGFR, but otherwise, none of the other specimens possessed these features of EGFR dependency [72]. We also found that two cell lines that possess activating Ras mutations (T24-H-Ras, UMUC3-K-Ras) are completely resistant to EGFR or FGFR3 inhibitors and cluster with our “mesenchymal” cell lines. Therefore, it is not clear to us that activating Ras mutations and activation of the EGFR or FGFR3 play redundant roles in driving the growth of the “epithelial” subset of urothelial cancer cells.
4 Relationship between EMT and TRAIL sensitivity
As discussed by Rosevear and colleagues in a subsequent article, Bacillus Calmette-Guerin (BCG) is the current frontline therapy for high-risk superficial bladder cancer and carcinoma in situ, but many patients ultimately fail BCG therapy and no molecular markers are available that can distinguish BCG-responsive from BCG-refractory disease. BCG appears to act (at least in part) by triggering neutrophils to secrete TNF-related apoptosis-inducing ligand (TRAIL) [73–77]. Another immunomodulator that has promising anti-tumor activity in bladder cancer is interferon-α (IFNα) [78, 79], which blocks tumor growth in preclinical models via direct induction of tumor cell death and inhibition of angiogenesis [80–86]. We, therefore, performed experiments to identify the molecular mechanisms that underlie IFNα's anti-tumoral effects. Our results demonstrate that like BCG, the direct tumoricidal effects of IFNα are dependent on autocrine TRAIL production [80–82], whereas the inhibition of angiogenesis involves parallel downregulation of bFGF and IL-8 [85, 86].
With these observations in mind, we have also characterized the heterogeneity in TRAIL responsiveness across our panel of human urothelial cancer cell lines. The results demonstrate again that sensitivity to TRAIL tracks closely with an “epithelial” phenotype (T24, which is TRAIL-sensitive but mesenchymal, is the “outlier”) (F. Martin, manuscript in preparation). We have not yet identified a molecular explanation for the link between EMT and TRAIL resistance, but the “mesenchymal” lines express higher levels of the apoptosis inhibitors XIAP and BCL-2 and compounds that target these factors increase TRAIL sensitivity in the resistant cells. Notably, a recent screen by a group from Genentech demonstrated that expression of Golgi enzymes involved in protein glycosylation and fucosylation correlated with TRAIL sensitivity in a much larger set of human solid tumor cell lines [87]. Strikingly, the expression of two of these enzymes (FUT1 and GALNT14) correlates closely with E-cadherin and inversely with Zeb-1 in our cell lines and the public gene expression profiling datasets (W. Choi, unpublished observations), suggesting that in addition to their potential direct roles in regulating TRAIL sensitivity, the fucosylation and glycosylation enzymes identified by the Genentech group are also excellent surrogate markers of an “epithelial” phenotype and that this programming probably also plays a central role in maintaining TRAIL sensitivity.
5 Molecular control of EMT in urothelial cancer cells
As discussed above, EMT is controlled by antagonistic interactions between members of the miR200 family of micro-RNAs and transcriptional repressors of E-cadherin expression including the direct miR200 targets Zeb-1 and Zeb-2 [9, 10]. We are, therefore, exploring how these proteins regulate invasion/migration and EGFR dependency in urothelial cancer cells. As discussed earlier, overexpression of Zeb-2 in “epithelial” urothelial cells renders them more invasive and resistant to radiation-induced apoptosis, effects that are associated with enhanced DNA repair [32]. Conversely, we have found that knockdown of Zeb-1 in “mesenchymal” cell lines inhibits tumor cell invasion and migration, although the effects are highly cell type-dependent. For example, in UMUC3 cells, Zeb-1 knockdown results in strong upregulation of E-cadherin mRNA and protein levels and parallel strong inhibition of invasion (A. Das et al, manuscript in preparation). However, in KU7 or T24 cells, Zeb-1 knockdown weakly upregulates E-cadherin mRNA expression and does not appreciably affect protein expression, and as a result, invasion is not attenuated. Importantly, enforced overexpression of E-cadherin does inhibit invasion in the KU7 cells, indicating that the block in E-cadherin expression is probably responsible for the inability of Zeb-1 knockdown to decrease invasion and migration. In ongoing studies, we are investigating whether other known EMT-related repressors (Zeb-2, Snail) are bound to the E-cadherin promoter in the KU7 and T24 cells. Intriguingly, our preliminary studies suggest that additional chromatin-modifying enzymes may also be involved. By analyzing E-cadherin promoter histone methylation by chromatin immunoprecipitation, we have found that a repressive histone methylation mark (H3K27me3) is present at much higher levels in the KU7 and T24 cells as compared to UMUC3 (A. Das, manuscript in preparation). Therefore, it seems likely that enzymes that regulate histone methylation reinforce the EMT phenotype in the KU7 and T24 cells.
In a parallel study, we compared the global micro-RNA expression profiles in representative “epithelial,” EGFR inhibitor-sensitive (UMUC5) and “mesenchymal,” EGFR inhibitor-resistant (KU7) cell lines to define the potential roles of differential miRNA expression in the regulation of EGFR inhibitor sensitivity [11]. The results demonstrated that members of the miR200 family were among the most differentially expressed micro-RNAs [11], present at very high levels in UMUC5 and almost undetectable levels in KU7. We have since measured all five members of the miR200 family by quantitative real-time PCR in our whole panel of cell lines and have confirmed that there is a strong inverse association between miR200 expression and the “mesenchymal” phenotype (M. Williams, manuscript in preparation). Overexpression of miR200c in the UMUC3 cells suppressed expression of Zeb-1 and Zeb-2, restored E-cadherin expression, and inhibited invasion and migration, effects that were similar to those observed with Zeb-1 knockdown [11]. Strikingly, however, overexpression of miR200c in UMUC3 or miR200b in T24 also increased cell sensitivity to EGFR inhibitors [11], effects that we have not been able to reproduce in the Zeb-1-silenced cells. Because E-cadherin restoration was similar in the miR200c and Zeb-1 knockdown cells, the most attractive explanation for the discrepancy in phenotypes is that there are additional miR200 targets (aside from Zeb-1 and Zeb-2) that play important role(s) in regulating EGFR-mediated signal transduction. Consistent with this idea, we have shown that miR200c directly targets the mRNA encoding ERRFI-1, an inhibitor of EGFR signaling, and that knockdown of ERRFI-1 in UMUC3 also increases cellular sensitivity to EGFR inhibitors [11].
6 Conclusions and future directions
Our experience with urothelial cancer cell lines indicates that “epithelial” and “mesenchymal” gene expression signatures exert extremely strong, global effects on cell biology. We have made progress in defining the molecular mechanisms underlying the stable signatures but much more work needs to be done. We do not yet know if TGFβ family cytokines or other autocrine mechanisms maintain EMT-related gene expression in the “mesenchymal” urothelial cancer cell lines. This seems to be a likely possibility given the central roles these cytokines play in normal tissue biology and cancer metastasis in other models, but it is also possible that the genetic alterations that underlie the two tracks of bladder cancer are involved as well. Related to this question, it appears clear that members of the miR200 family of micro-RNAs control the EMT phenotype in bladder cancer cells, but what controls expression of the miR200 family itself needs to be defined. Again, the TGFβ family of cytokines is an attractive place to start.
There is also a need to place EMT within the context of other processes that are known to regulate urothelial cancer progression and metastasis. The earlier studies demonstated that VEGF levels and the ratio of E-cadherin to MMP9 expression predicted poor outcome [23], and our real-time quantitative RT-PCR assessment of MMP2 and MMP9 levels in our cohort of patient tumors confirms that both are elevated in muscle-invasive disease. However, what is not clear is whether the global regulators of EMT also control expression of VEGF, the MMPs, and possibly other factors relevant to the biology of this disease subset. With respect to EGFR signaling, our preliminary assessments suggest that ligand (TGFα) expression correlates with an “epithelial” phenotype (Fig. 1b) but whether this is restricted to EGFR-dependent (as opposed to FGFR3-dependent) “epithelial” lines requires additional investigation.
Another important objective for future study is to determine how having tumor EMT information can be used to improve clinical outcome. There is arguably limited value in being able to distinguish superficial from muscle-invasive disease using primary tumor tissue. However, if confirmed in patients, the observation that EGFR inhibitors selectively target “epithelial” urothelial cells could be used to identify the subset of muscle-invasive tumors that will respond better to EGFR-directed therapy. The information could also prove valuable within the setting of high-risk superficial disease and CIS, where an EMT signature would also predict de novo resistance to BCG and other TRAIL-based therapies. Here these agents should probably be combined either with another modality that inactivates the specific resistance mechanism(s) that are in place (i.e., combination therapy with BCG and an XIAP or BCL-2 inhibitor) or an agent that selectively kills the “mesenchymal” cells, or some attempt could be made to reverse the EMT signature altogether. Along these lines, recent work has demonstrated that clinical histone deacetylase (HDAC) inhibitors are capable of restoring E-cadherin expression and EGFR inhibitor sensitivity in lung cancer cells [88], prompting interest in performing clinical trials with EGFR inhibitor/HDAC inhibitor combinations in patients.
Finally, it will be important to determine how EMT affects tumor responses to other conventional and investigational therapies. The recently published work demonstrating that Zeb-2 expression is associated with poor outcome in patients receiving radiation therapy [32] suggests that EMT might contribute to a more global therapeutic resistance than has been described here, or it may at least suppress DNA damage-induced cell death. The observation that Zeb-2 promotes DNA repair and suppresses radiation-induced apoptosis in urothelial cancer cells in vitro provides further support for this concept [32], as does recent work in other models directly linking EMT to cancer “stemness” [89] and resistance to conventional chemotherapy [90, 91]. It should be possible to identify agents that selectively target “mesenchymal” cancer cells, as was done recently by another group [91]. Overall, understanding how EMT and other developmental pathways control cancer cell biology should greatly facilitate the objective of designing more personalized cancer therapy.
Contributor Information
David J. McConkey, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA; Department of Cancer Biology, U.T. M.D. Anderson Cancer Center, Houston, TX 77030, USA
Woonyoung Choi, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA; Department of Cancer Biology, U.T. M.D. Anderson Cancer Center, Houston, TX 77030, USA.
Lauren Marquis, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA; Department of Cancer Biology, U.T. M.D. Anderson Cancer Center, Houston, TX 77030, USA.
Frances Martin, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
Michael B. Williams, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA
Jay Shah, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
Robert Svatek, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
Aditi Das, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA; Department of Cancer Biology, U.T. M.D. Anderson Cancer Center, Houston, TX 77030, USA.
Liana Adam, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA; Department of Cancer Biology, U.T. M.D. Anderson Cancer Center, Houston, TX 77030, USA.
Ashish Kamat, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
Arlene Siefker-Radtke, Department of Genitourinary Medical Oncology, U.T. M.D. Anderson Cancer Center, Houston, TX 77030, USA.
Colin Dinney, Department of Urology, U.T. M.D. Anderson Cancer Center, P.O. Box 1373, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
References
- 1.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? ature Reviews Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 2.De Donatis A, Comito G, Buricchi F, et al. Proliferation versus migration in platelet-derived growth factor signaling: the key role of endocytosis. Journal of Biological Chemistry. 2008;283:19948–19956. doi: 10.1074/jbc.M709428200. [DOI] [PubMed] [Google Scholar]
- 3.Giese A, Loo MA, Tran N, Haskett D, Coons SW, Berens ME. Dichotomy of astrocytoma migration and proliferation. International Journal of Cancer. 1996;67:275–282. doi: 10.1002/(SICI)1097-0215(19960717)67:2<275::AID-IJC20>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 4.Engel ME, Datta PK, Moses HL. Signal transduction by transforming growth factor-beta: a cooperative paradigm with extensive negative regulation. Journal of Cellular Biochemistry Supplement. 1998;30–31:111–122. [PubMed] [Google Scholar]
- 5.Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, Saitoh M. Role of Ras signaling in the induction of snail by transforming growth factor-beta. Journal of Biological Chemistry. 2009;284:245–253. doi: 10.1074/jbc.M804777200. [DOI] [PubMed] [Google Scholar]
- 6.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor beta (TGF-{beta}) target genes and distinguishes TGF-{beta}-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Molecular and Cellular Biology. 2005;25:8108–8125. doi: 10.1128/MCB.25.18.8108-8125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Research. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 9.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 10.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes and Development. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adam L, Zhong M, Choi W, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clinical Cancer Research. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honn KV, Tang DG. Adhesion molecules and tumor cell interaction with endothelium and subendothelial matrix. Cancer and Metastasis Reviews. 1992;11:353–375. doi: 10.1007/BF01307187. [DOI] [PubMed] [Google Scholar]
- 13.Herrera CA, Xu L, Bucana CD, et al. Expression of metastasis-related genes in human epithelial ovarian tumors. International Journal of Oncology. 2002;20:5–13. [PubMed] [Google Scholar]
- 14.Kim SJ, Uehara H, Karashima T, McCarty M, Shih N, Fidler IJ. Expression of interleukin-8 correlates with angiogenesis, tumorigenicity, and metastasis of human prostate cancer cells implanted orthotopically in nude mice. Neoplasia. 2001;3:33–42. doi: 10.1038/sj.neo.7900124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slaton JW, Inoue K, Perrotte P, et al. Expression levels of genes that regulate metastasis and angiogenesis correlate with advanced pathological stage of renal cell carcinoma. American Journal of Pathology. 2001;158:735–743. doi: 10.1016/S0002-9440(10)64016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuniyasu H, Troncoso P, Johnston D, et al. Relative expression of type IV collagenase, E-cadherin, and vascular endothelial growth factor/vascular permeability factor in prostatectomy specimens distinguishes organ-confined from pathologically advanced prostate cancers. Clinical Cancer Research. 2000;6:2295–2308. [PubMed] [Google Scholar]
- 17.Herbst RS, Yano S, Kuniyasu H, et al. Differential expression of E-cadherin and type IV collagenase genes predicts outcome in patients with stage I non-small cell lung carcinoma. Clinical Cancer Research. 2000;6:790–797. [PubMed] [Google Scholar]
- 18.Kuniyasu H, Ellis LM, Evans DB, et al. Relative expression of E-cadherin and type IV collagenase genes predicts disease outcome in patients with resectable pancreatic carcinoma. Clinical Cancer Research. 1999;5:25–33. [PubMed] [Google Scholar]
- 19.Anzai H, Kitadai Y, Bucana CD, Sanchez R, Omoto R, Fidler IJ. Expression of metastasis-related genes in surgical specimens of human gastric cancer can predict disease recurrence. European Journal of Cancer. 1998;34:558–565. doi: 10.1016/s0959-8049(97)10075-2. [DOI] [PubMed] [Google Scholar]
- 20.Greene GF, Kitadai Y, Pettaway CA, von Eschenbach AC, Bucana CD, Fidler IJ. Correlation of metastasis-related gene expression with metastatic potential in human prostate carcinoma cells implanted in nude mice using an in situ messenger RNA hybridization technique. American Journal of Pathology. 1997;150:1571–1582. [PMC free article] [PubMed] [Google Scholar]
- 21.Kitadai Y, Ellis LM, Tucker SL, et al. Multi-parametric in situ mRNA hybridization analysis to predict disease recurrence in patients with colon carcinoma. American Journal of Pathology. 1996;149:1541–1551. [PMC free article] [PubMed] [Google Scholar]
- 22.Kitadai Y, Ellis LM, Takahashi Y, et al. Multi-parametric in situ messenger RNA hybridization analysis to detect metastasis-related genes in surgical specimens of human colon carcinomas. Clinical Cancer Research. 1995;1:1095–1102. [PubMed] [Google Scholar]
- 23.Slaton JW, Millikan R, Inoue K, et al. Correlation of metastasis related gene expression and relapse-free survival in patients with locally advanced bladder cancer treated with cystectomy and chemotherapy. Journal of Urology. 2004;171:570–574. doi: 10.1097/01.ju.0000108845.91485.20. [DOI] [PubMed] [Google Scholar]
- 24.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Research. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 25.Hajra KM, Fearon ER. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer. 2002;34:255–268. doi: 10.1002/gcc.10083. [DOI] [PubMed] [Google Scholar]
- 26.Gibbons DL, Lin W, Creighton CJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes and Development. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strathdee G. Epigenetic versus genetic alterations in the inactivation of E-cadherin. Seminars in Cancer Biology. 2002;12:373–379. doi: 10.1016/s1044-579x(02)00057-3. [DOI] [PubMed] [Google Scholar]
- 28.Dinney CP, McConkey DJ, Millikan RE, et al. Focus on bladder cancer. Cancer Cell. 2004;6:111–116. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Blaveri E, Simko JP, Korkola JE, et al. Bladder cancer outcome and subtype classification by gene expression. Clinical Cancer Research. 2005;11:4044–4055. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonu-cleotide microarrays. Journal of Clinical Oncology. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 31.Baumgart E, Cohen MS, Silva Neto B, et al. Identification and prognostic significance of an epithelial-mesenchymal transition expression profile in human bladder tumors. Clinical Cancer Research. 2007;13:1685–1694. doi: 10.1158/1078-0432.CCR-06-2330. [DOI] [PubMed] [Google Scholar]
- 32.Sayan AE, Griffiths TR, Pal R, et al. SIP1 protein protects cells from DNA damage-induced apoptosis and has independent prognostic value in bladder cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14884–14889. doi: 10.1073/pnas.0902042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urist MJ, Di Como CJ, Lu ML, et al. Loss of p63 expression is associated with tumor progression in bladder cancer. American Journal of Pathology. 2002;161:1199–1206. doi: 10.1016/S0002-9440(10)64396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Como CJ, Urist MJ, Babayan I, et al. p63 expression profiles in human normal and tumor tissues. Clinical Cancer Research. 2002;8:494–501. [PubMed] [Google Scholar]
- 35.Comperat E, Camparo P, Haus R, et al. Immunohis-tochemical expression of p63, p53 and MIB-1 in urinary bladder carcinoma. A tissue microarray study of 158 cases. Virchows Archiv. 2006;448:319–324. doi: 10.1007/s00428-005-0092-2. [DOI] [PubMed] [Google Scholar]
- 36.Koga F, Kawakami S, Fujii Y, et al. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clinical Cancer Research. 2003;9:5501–5507. [PubMed] [Google Scholar]
- 37.Moll UM. The Role of p63 and p73 in tumor formation and progression: coming of age toward clinical usefulness. Commentary re: F. Koga et al. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clinical Cancer Research. 2003;9:5501–5507. [PubMed] [Google Scholar]
- 38.Puig P, et al. p73 Expression in human normal and tumor tissues: loss of p73alpha expression is associated with tumor progression in bladder Cancer. Clinical Cancer Research. 2003;9:5642–5651. Clin Cancer Res 2003; 9: 5437–5441. [PubMed] [Google Scholar]
- 39.Reis-Filho JS, Simpson PT, Martins A, Preto A, Gartner F, Schmitt FC. Distribution of p63, cytokeratins 5/6 and cytokeratin 14 in 51 normal and 400 neoplastic human tissue samples using TARP-4 multi-tumor tissue microarray. Virchows Archiv. 2003;443:122–132. doi: 10.1007/s00428-003-0859-2. [DOI] [PubMed] [Google Scholar]
- 40.Koga F, Kawakami S, Kumagai J, et al. Impaired Delta Np63 expression associates with reduced beta-catenin and aggressive phenotypes of urothelial neoplasms. British Journal of Cancer. 2003;88:740–747. doi: 10.1038/sj.bjc.6600764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park BJ, Lee SJ, Kim JI, et al. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Research. 2000;60:3370–3374. [PubMed] [Google Scholar]
- 42.Signoretti S, Loda M. Defining cell lineages in the prostate epithelium. Cell Cycle. 2006;5:138–141. doi: 10.4161/cc.5.2.2340. [DOI] [PubMed] [Google Scholar]
- 43.Signoretti S, Pires MM, Lindauer M, et al. p63 regulates commitment to the prostate cell lineage. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11355–11360. doi: 10.1073/pnas.0500165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Signoretti S, Waltregny D, Dilks J, et al. p63 is a prostate basal cell marker and is required for prostate development. American Journal of Pathology. 2000;157:1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanpain C, Fuchs E. p63: revving up epithelial stem-cell potential. Nature Cell Biology. 2007;9:731–733. doi: 10.1038/ncb0707-731. [DOI] [PubMed] [Google Scholar]
- 46.Kurzrock EA, Lieu DK, Degraffenried LA, Chan CW, Isseroff RR. Label-retaining cells of the bladder: candidate urothelial stem cells. American Journal of Physiology Renal Physiology. 2008;294:F1415–F1421. doi: 10.1152/ajprenal.00533.2007. [DOI] [PubMed] [Google Scholar]
- 47.Mendelsohn J, Baselga J. Epidermal growth factor receptor targeting in cancer. Seminars in Oncology. 2006;33:369–385. doi: 10.1053/j.seminoncol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Lipponen P, Eskelinen M. Expression of epidermal growth factor receptor in bladder cancer as related to established prognostic factors, oncoprotein (c-erbB-2, p53) expression and long-term prognosis. British Journal of Cancer. 1994;69:1120–1125. doi: 10.1038/bjc.1994.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Izawa JI, Slaton JW, Kedar D, et al. Differential expression of progression-related genes in the evolution of superficial to invasive transitional cell carcinoma of the bladder. Oncology Reports. 2001;8:9–15. doi: 10.3892/or.8.1.9. [DOI] [PubMed] [Google Scholar]
- 50.Perrotte P, Matsumoto T, Inoue K, et al. Anti-epidermal growth factor receptor antibody C225 inhibits angiogenesis in human transitional cell carcinoma growing orthotopically in nude mice. Clinical Cancer Research. 1999;5:257–265. [PubMed] [Google Scholar]
- 51.Cheng J, Huang H, Zhang ZT, et al. Overexpression of epidermal growth factor receptor in urothelium elicits urothelial hyperplasia and promotes bladder tumor growth. Cancer Research. 2002;62:4157–4163. [PubMed] [Google Scholar]
- 52.Janne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. Journal of Clinical Oncology. 2005;23:3227–3234. doi: 10.1200/JCO.2005.09.985. [DOI] [PubMed] [Google Scholar]
- 53.Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. Journal of Clinical Oncology. 2008;26:983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 54.Heymach JV, Nilsson M, Blumenschein G, Papadimitrakopoulou V, Herbst R. Epidermal growth factor receptor inhibitors in development for the treatment of non-small cell lung cancer. Clinical Cancer Research. 2006;12:4441s–4445s. doi: 10.1158/1078-0432.CCR-06-0286. [DOI] [PubMed] [Google Scholar]
- 55.Lara-Guerra H, Waddell TK, Salvarrey MA, et al. Phase II study of preoperative gefitinib in clinical stage i non-small-cell lung cancer. Journal of Clinical Oncology. 2009 doi: 10.1200/JCO.2009.22.3370. in press. [DOI] [PubMed] [Google Scholar]
- 56.Hirsch FR, Scagliotti GV, Langer CJ, Varella-Garcia M, Franklin WA. Epidermal growth factor family of receptors in preneoplasia and lung cancer: perspectives for targeted therapies. Lung Cancer. 2003;41(Suppl 1):S29–S42. doi: 10.1016/s0169-5002(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 57.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. Journal of Clinical Oncology. 2009;27(35):5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 58.Zhu CQ, da Cunha Santos G, Ding K, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. Journal of Clinical Oncology. 2008;26:4268–4275. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 59.Miller VA, Riely GJ, Zakowski MF, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. Journal of Clinical Oncology. 2008;26:1472–1478. doi: 10.1200/JCO.2007.13.0062. [DOI] [PubMed] [Google Scholar]
- 60.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. Journal of Clinical Oncology. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 61.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. Journal of Clinical Oncology. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 62.Kassouf W, Dinney CP, Brown G, et al. Uncoupling between epidermal growth factor receptor and downstream signals defines resistance to the antiproliferative effect of Gefitinib in bladder cancer cells. Cancer Research. 2005;65:10524–10535. doi: 10.1158/0008-5472.CAN-05-1536. [DOI] [PubMed] [Google Scholar]
- 63.Shrader M, Pino MS, Lashinger L, et al. Gefitinib reverses TRAIL resistance in human bladder cancer cell lines via inhibition of AKT-mediated X-linked inhibitor of apoptosis protein expression. Cancer Research. 2007;67:1430–1435. doi: 10.1158/0008-5472.CAN-06-1224. [DOI] [PubMed] [Google Scholar]
- 64.Shrader M, Pino MS, Brown G, et al. Molecular correlates of gefitinib responsiveness in human bladder cancer cells. Molecular Cancer Therapeutics. 2007;6:277–285. doi: 10.1158/1535-7163.MCT-06-0513. [DOI] [PubMed] [Google Scholar]
- 65.Black PC, Brown GA, Inamoto T, et al. Sensitivity to epidermal growth factor receptor inhibitor requires E-cadherin expression in urothelial carcinoma cells. Clinical Cancer Research. 2008;14:1478–1486. doi: 10.1158/1078-0432.CCR-07-1593. [DOI] [PubMed] [Google Scholar]
- 66.Tomlinson DC, Baldo O, Harnden P, Knowles MA. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. Journal of Pathology. 2007;213:91–98. doi: 10.1002/path.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sibley K, Stern P, Knowles MA. Frequency of fibroblast growth factor receptor 3 mutations in sporadic tumours. Oncogene. 2001;20:4416–4418. doi: 10.1038/sj.onc.1204543. [DOI] [PubMed] [Google Scholar]
- 68.Tomlinson DC, Hurst CD, Knowles MA. Knockdown by shRNA identifies S249C mutant FGFR3 as a potential therapeutic target in bladder cancer. Oncogene. 2007;26:5889–5899. doi: 10.1038/sj.onc.1210399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qing J, Du X, Chen Y, et al. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. Journal of Clinical Investigation. 2009;119:1216–1229. doi: 10.1172/JCI38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang PH, Mukasa A, Bonavia R, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. New England Journal of Medicine. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 72.Blehm KN, Spiess PE, Bondaruk JE, et al. Mutations within the kinase domain and truncations of the epidermal growth factor receptor are rare events in bladder cancer: implications for therapy. Clinical Cancer Research. 2006;12:4671–4677. doi: 10.1158/1078-0432.CCR-06-0407. [DOI] [PubMed] [Google Scholar]
- 73.Simons MP, O'Donnell MA, Griffith TS. Role of neutrophils in BCG immunotherapy for bladder cancer. Urologic Oncology. 2008;26:341–345. doi: 10.1016/j.urolonc.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simons MP, Nauseef WM, Griffith TS. Neutrophils and TRAIL: insights into BCG immunotherapy for bladder cancer. Immunologic Research. 2007;39:79–93. doi: 10.1007/s12026-007-0084-1. [DOI] [PubMed] [Google Scholar]
- 75.Simons MP, Moore JM, Kemp TJ, Griffith TS. Identification of the mycobacterial subcomponents involved in the release of tumor necrosis factor-related apoptosis-inducing ligand from human neutrophils. Infection and Immunity. 2007;75:1265–1271. doi: 10.1128/IAI.00938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kemp TJ, Ludwig AT, Earel JK, et al. Neutrophil stimulation with Mycobacterium bovis bacillus Calmette-Guerin (BCG) results in the release of functional soluble TRAIL/Apo-2L. Blood. 2005;106:3474–3482. doi: 10.1182/blood-2005-03-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ludwig AT, Moore JM, Luo Y, et al. Tumor necrosis factor-related apoptosis-inducing ligand: a novel mechanism for Bacillus Calmette-Guerin-induced antitumor activity. Cancer Research. 2004;64:3386–3390. doi: 10.1158/0008-5472.CAN-04-0374. [DOI] [PubMed] [Google Scholar]
- 78.Logothetis CJ, Hossan E, Recondo G, et al. 5-Fluorouracil and interferon-alpha in chemotherapy refractory bladder carcinoma: an effective regimen. Anticancer Research. 1994;14:1265–1269. [PubMed] [Google Scholar]
- 79.Logothetis CJ, Hossan E, Sella A, Dexeus FH, Amato RJ. Fluorouracil and recombinant human interferon alfa-2a in the treatment of metastatic chemotherapy-refractory urothelial tumors. Journal of the National Cancer Institute. 1991;83:285–288. doi: 10.1093/jnci/83.4.285. [DOI] [PubMed] [Google Scholar]
- 80.Papageorgiou A, Dinney CP, McConkey DJ. Interferon-alpha induces TRAIL expression and cell death via an IRF-1-dependent mechanism in human bladder cancer cells. Cancer Biology and Therapy. 2007;6:872–879. doi: 10.4161/cbt.6.6.4088. [DOI] [PubMed] [Google Scholar]
- 81.Papageorgiou A, Kamat A, Benedict WF, Dinney C, McConkey DJ. Combination therapy with IFN-alpha plus bortezomib induces apoptosis and inhibits angiogenesis in human bladder cancer cells. Molecular Cancer Therapeutics. 2006;5:3032–3041. doi: 10.1158/1535-7163.MCT-05-0474. [DOI] [PubMed] [Google Scholar]
- 82.Papageorgiou A, Lashinger L, Millikan R, et al. Role of tumor necrosis factor-related apoptosis-inducing ligand in interferon-induced apoptosis in human bladder cancer cells. Cancer Research. 2004;64:8973–8979. doi: 10.1158/0008-5472.CAN-04-1909. [DOI] [PubMed] [Google Scholar]
- 83.Izawa JI, Sweeney P, Perrotte P, et al. Inhibition of tumorigenicity and metastasis of human bladder cancer growing in athymic mice by interferon-beta gene therapy results partially from various antiangiogenic effects including endothelial cell apoptosis. Clinical Cancer Research. 2002;8:1258–1270. [PubMed] [Google Scholar]
- 84.Slaton JW, Karashima T, Perrotte P, et al. Treatment with low-dose interferon-alpha restores the balance between matrix metalloproteinase-9 and E-cadherin expression in human transitional cell carcinoma of the bladder. Clinical Cancer Research. 2001;7:2840–2853. [PubMed] [Google Scholar]
- 85.Slaton JW, Perrotte P, Inoue K, Dinney CP, Fidler IJ. Interferon-alpha-mediated down-regulation of angiogenesis-related genes and therapy of bladder cancer are dependent on optimization of biological dose and schedule. Clinical Cancer Research. 1999;5:2726–2734. [PubMed] [Google Scholar]
- 86.Dinney CP, Bielenberg DR, Perrotte P, et al. Inhibition of basic fibroblast growth factor expression, angiogenesis, and growth of human bladder carcinoma in mice by systemic interferon-alpha administration. Cancer Research. 1998;58:808–814. [PubMed] [Google Scholar]
- 87.Wagner KW, Punnoose EA, Januario T, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nature Medicine. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- 88.Witta SE, Gemmill RM, Hirsch FR, et al. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Research. 2006;66:944–950. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 89.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arumugam T, Ramachandran V, Fournier KF, et al. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Research. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]