Abstract
Alcoholism is a disorder categorized by significant impairment that is directly related to persistent and extreme use of alcohol. The effects of alcoholism on c-Myc protein expression in the brain have been scarcely studied. This is the first study to investigate the role of different characteristics of alcoholism in c-Myc protein levels in the brain. We analyzed c-Myc protein in the hypothalamus and amygdala from four different animal models of alcohol abuse. c-Myc protein was increased following alcohol exposure in acute, chronic and withdrawal models. We also observed increases in c-Myc protein exposure in animals that are genetically predisposed to alcohol and methamphetamine abuse. Lastly, c-Myc protein was increased in animals that were acutely exposed to methamphetamine when compared to control treated animals. These results suggest that in substance abuse c-Myc plays an important role in the brain’s response.
Keywords: c-Myc, alcoholism, withdrawal, preference, phosphorylation
1. Introduction
The present study focuses on the v-myc avian myelocytomatotsis viral oncogene homolog (c-myc) gene and its involvement in alcohol addiction and potentially other forms of substance abuse. Alcoholism combines the elements of both mental illness and physical disease (brain deterioration) and it is classified as a substance abuse disorder in the Diagnostic and Statistical Manual of Mental Disorders (DSM-V). Mental and emotional symptoms of alcoholism exist long before the physical complications of the disease appear. The mental symptoms consist of loss of control (taking in larger amounts of alcohol over a longer period of time than the person intends); continued drinking in spite of adverse social, occupational, or legal consequences; and frequent intoxication. These are behavioral or mental symptoms, which, unless treated, could lead to physical complications of alcoholism that include cancer (1-4), cardiovascular disease (5), anemia (6,7) and cirrhosis (8,9). A previous study has shown that c-Myc protein is increased in the prefrontal cortex postmortem tissues of chronic alcoholics (10). There are no further studies that examine what effects alcoholism plays on the expression of c-Myc protein in the brain. Therefore, it is important to further examine how c-Myc protein expression is regulated during alcohol abuse and how genetic predisposition to alcoholism might affect this.
Both acute and chronic alcohol consumption leads to costly healthcare treatments and preventable deaths. It is estimated that alcohol-related expenses cost federal, state, and local governments $223.5 billion. In spite of many efforts including therapies, detoxification and rehabilitation, alcoholism caused 88,000 deaths per year, approximately 2.5 million years of potential life lost (YPLL) each year between the years 2006-2010 in the United States (11). Twenty-five chronic diseases in the International Classification of Disease (ICD)-10 have been completely ascribed to alcohol. Alcohol use disorders play roles in certain cancers, psychiatric conditions, and numerous cardiovascular and digestive diseases. Furthermore, alcohol use disorders have damaging effects leading to diabetes, stroke and heart disease, depending on the overall volume of alcohol consumed (12-14).
The c-myc gene localizes to human chromosome 8q24 and contains three exons (15,16). Posttranslational modification of c-Myc protein has been studied at length (17,18). Phosphorylation of c-Myc protein at residues threonine 58 (T58) and serine 62 (S62) can regulate c-Myc protein stability (18). Phosphorylation of S62 transiently increases stability of c-Myc while phosphorylation of T58 triggers de-phosphorylation of S62 leading to ubiquitin-induced proteasomal degradation of the protein. As a transcription factor, c-Myc forms a heterodimer with Max protein, which is then translocated to the nucleus where it binds to target sites to activate genes (19-21). p21 (CIP1/WAF1) is a potent cyclin-dependent kinase inhibitor (CDKI) that binds to and inhibits the activity of cylcin dependent kinase (CDK) 4/6 complexes (22-24). It functions as a regulator of cell cycle progression at G1 and S phase. In CV1 cells, a direct interaction between c-Myc and p21 leads to inactivation of p21 and progression of the cell cycle. Also, p21 represses the transcriptional activity of c-Myc by inhibiting formation of the c-Myc/Max heterodimer (25).
c-myc is one of the most widely studied proto-oncogenes, and it is the best characterized member of the myc gene family. In general, c-myc expression is associated with cell proliferation and is down-regulated in quiescent and differentiated cells. c-Myc protein expression has been sparsely studied in brain in relation to alcohol abuse but has been studied in liver disease from alcoholism (26). Liver regeneration is important in the recovery from various forms of liver injury. The inhibition of liver regeneration from chronic ethanol exposure in humans may be an important factor in the pathogenesis of liver disease. Wen et al. found that hepatic osteopontin (OPN) protein levels were significantly elevated in wild-type (WT) mice after partial hepatectomy (PHx). Compared to WT mice, OPN knockout (KO) mice showed delayed liver regeneration after PHx and in these mice c-myc, was also suppressed post-PHx compared to WT mice (27). Another study done by Radaeva et al showed that microarray analysis of primary human hepatocytes show that IFN-alpha (possible hepatocellular carcinoma therapy) decreased c-myc gene expression by 50% (28). These studies suggest that liver damage induces c-Myc gene and protein expression in the liver and affects the ability of the liver to regenerate. Alcohol abuse also damages the liver which can lead to an increase in c-Myc expression.
Effects of alcohol exposure on c-Myc protein levels in the liver have been reported (26,27,29). In the context of alcohol exposure, c-Myc might also play a role in brain dysfunction. Postmortem tissues have shown c-Myc protein expression in the superior frontal cortex of chronic alcoholics is significantly higher compared to non- or social drinkers. Levels of β-Catenin, an upstream target of c-Myc, was also significantly higher in chronic alcoholics (10). Aside from this study there has been little research done to determine the effect that alcohol exposure or genetic predisposition to alcoholism has on c-Myc expression in the brain.
A genetic predisposition or genetic susceptibility is a genetic characteristic that influences the possible phenotypic development towards a particular disease. Research has shown that genes are responsible for about half of the risk for alcoholism. Multiple genes contribute to the risk for developing alcoholism. Mice genetically predisposed to a low ethanol withdrawal response are also often predisposed to high ethanol consumption and vice versa (30). These mice known as SOT and NOT were derived from C57BL/6J (B6) and DBA/2J (D2) inbred mouse strains, where consumption was measured using a standard two-bottle choice paradigm (10% ethanol vs water, continuous access) and withdrawal was measured using handling-induced convulsions (HICs) following injection or vapor inhalation (31). SOT mice were selected for high drinking and low withdrawal phenotype while the NOT mice were selected for a low drinking, high withdrawal phenotype. We hypothesized that mice with the genetic predisposition to higher alcohol consumption (SOT) will have higher levels of c-Myc protein in the amygdala and hypothalamus compared to the lower alcohol consumption mice (NOT).
Chronic drinking can have adverse effects on the nervous system causing dependence on alcohol. Withdrawal from alcohol can lead to alcohol withdrawal syndrome (AWS) (32-35). To mimic the effects of AWS in mice we obtained two mouse cell lines, withdrawal seizure prone (WSP) and the withdrawal seizure resistant (WSR), which were bi-directionally selectively bred, respectively, to have severe or mild ethanol withdrawal HICs after cessation of 3 days of ethanol vapor inhalation (36). Thus, correlated traits that are seen to differ between WSP vs WSR mice, particularly when differences are seen in both replicates in the same direction, are assumed to be influenced by genes underlying ethanol withdrawal severity. WSP mice from both replicates also display greater HIC responses after withdrawal from acute ethanol than their corresponding WSR lines, which provides strong evidence for a genetic correlation between withdrawal severity from acute and chronic ethanol exposure. We hypothesized that mice that suffer from less severe alcohol withdrawal symptoms express more c-Myc protein in the amygdala and hypothalamus when compared to those mice that have more severe withdrawal symptoms.
Genetic factors may influence who is at risk for developing addiction to methamphetamine (MA) abuse. Several genetic variants in humans have been identified and associated with MA abuse and dependence (37). The heritability of MA drinking in mouse lines have been selectively bred for oral consumption of either high (MAHDR) or low (MALDR) amounts of MA (38,39). Selective breeding produced MAHDR lines that consume approximately 6 mg/kg of MA during an 18 hour MA access period, compared to 0.5 mg/kg MA intake in MALDR mice (38,39). These two lines were used to determine if c-Myc protein is modulated by alcohol exposure or can be regulated by other substances of abuse. We hypothesized that c-Myc protein expression in the amygdala and hypothalamus of MAHDR mice are higher than c-Myc protein levels in MALDR mice.
In addition to lines selected for alcohol traits, specific genes have been implicated in the acute effects of alcohol exposure. For example, the mGlu8 knockout mice have increased levels of anxiety (40,41). Also, the mGlu8 agonist (S)-3,4-dichlorophenylglycine ((S)-3,4-DCPG) suppressed alcohol self-administration as well as cue-induced reinstatement of alcohol-seeking behavior (42). Furthermore, the gene GRM8, a metabotropic glutamate receptor, is significantly associated with alcohol dependence in European Americans (43). In the brain, the amygdala might be particularly affected by acute alcohol exposure. When given placebos, people showed more activation to fearful than to neutral faces as measured by functional magnetic resonance imaging (fMRI) (44). When people were given alcohol, there was no significant difference in activation between fearful and neutral faces, indicating that alcohol can blunt emotional processing in the amygdala (45).
Non-human primates are genetically comparable to humans, sharing many anatomic, neurophysiological, and behavioral characteristics (46,47). One distinguishing feature that non-human primates and humans share is their brain structure and function, specifically expansion of the cerebral cortex, which enables complex cognitive processing and behavioral flexibility. Importantly, monkeys will voluntarily consume ethanol (EtOH) in levels and patterns of intake that closely parallel those observed in human alcoholics (48). We used non-human primates to study the effect of chronic alcoholism on c-Myc protein levels in the brain. We hypothesized that long-term alcohol exposure increases c-Myc protein levels in the amygdala and hypothalamus.
Chronic alcohol dependence is a complex condition that develops over many years and includes cycles of withdrawal, craving, and relapse. Chronic alcoholism is associated by structural brain damage that has been previously reported using neuropathological examinations, computed tomography (CT) and magnetic resonance imaging (MRI) (49-51). In addition, chronic alcohol consumption results in reduction of thalamic volume (50). The extended amygdala, particularly the bed nucleus of the stria terminalis have been shown to be affected by chronic alcohol exposure in nonhuman primates (52).
In the current study we used five animal models that are pertinent to drugs of abuse. Three animal models involved mice that were selectively bred for alcohol or methamphetamine intake and/or withdrawal symptoms. The fourth animal model involved acute alcohol exposure to wild-type mice and mice deficient in mGlu8. The fifth animal model involved a non-human primate model of chronic alcohol consumption. Protein levels of c-Myc were measured in pertinent brain regions and were increased in animal models that are genetically predisposed and in models that that have been treated with acute and chronic alcohol exposure.
2.Materials and Methods
2.1 Animals
Housing
Mice were maintained in a temperature-controlled room with 12:12 hour light:dark cycle (lights on at 0600) with free access to standard chow and water. Mice were group housed with up to 5 animals per cage. Animals were fed standard chow, except during breeding (when dietary fat was increased to 9%, Purina 5008, PMI Nutrition International, Brentwood, MO, USA). Water was available freely. All experiments were performed in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals and were approved by the OHSU and Portland VA Medical Center’s Institutional Animal Care and Use Committees.
SOT/NOT line (n=10 mice for each line)
Mice were sacrificed between the ages of 10 and 14 weeks of age. For the SOT line, in each generation, animals with the greatest drinking values and lowest withdrawal scores were used to select breeders to perpetuate the SOT line. Also, animals with the lowest drinking values and the greatest withdrawal severity scores were used to perpetuate the NOT line (30).
WSP/WSR line (n=8 mice for each line)
These selected lines and the MAHDR and MALDR lines described below were bred in the Portland VA Veterinary Medical Unit. All mice were between 7 and 14 weeks. Mice were maintained in standard plastic cages on Bed-o-cob bedding with stainless steel wire bar tops with a recess for chow. Rodent chow 5001 and tap water were available ad libitum and colonies and testing rooms were maintained on a 12 h:12 h light:dark schedule at a temperature of 21 ± 1 °C. For the WSP and WSR lines, mice presenting two independent selection replicates (WSP1, WSP2, WSR1, and WSR2) could be used as they are part of an ongoing breeding and selection effort in the laboratory of Dr Crabbe.
MAHDR/MALDR line (n=8 mice for each line)
All mice used in this study were aged 7 and 9 weeks. All behavioral testing occurred during the light phase between 0800 h and 1400 h. Mice had free access to water and standard rodent diet. Mice were weaned at age 20–22 days and thereafter group-housed with same-sex littermates in groups of two to five mice.
mGlu8 KO line and wild-type mice (n=10 mice for each line)
Mice were treated between the ages of 20 and 21 weeks of age. All tests were carried out between 0900 and 1000 h. Mice were treated with 2.5g/kg EtOH, 1mg/kg methamphetamine, or saline via intraperitoneal (IP) injection for one hour and killed by cervical dislocation and hypothalamic and amygdala tissues were dissected one hour after exposure.
Non-human primate tissues (n= 7 drinking animals, 3 non-drinking animals)
All non-human primates in this study were approximately 5 years old at the beginning of alcohol self-administration (53). Animals self-administered alcohol (n=7 animals) or an isocaloric sweetened solution (n=3 animals) under operant conditions previously described (54). All animals were healthy and treatment-naïve prior to entering the studies. No medications were administered to block or prevent reinstatement of ethanol. Only medications related to clinical care were administered. All procedures were approved by the Institutional Animal Care and Use Committees (IACUC) of Wake Forest School of Medicine (WFSM).
2.2 Materials
Primary antibodies, c-Myc (sc-764), p21 (sc-397) and GAPDH (sc-365062), were purchased from Santa Cruz (Dallas, TX). T58 p-c-Myc (ab28842) primary antibody was purchased from Abcam (Cambridge, MA). Both horseradish peroxidase-conjugated donkey anti-rabbit (sc-2004) and donkey anti-mouse antibodies (sc-2005) were from Santa Cruz. Ripa lysis buffer (R0278) was purchased from Sigma Aldrich (St. Louis, MO) and contained protease inhibitors (LC5925) purchased from Roche Diagnostics (Indianapolis, IN). Prestained protein ladder (NP0007) and 4× loading dye (LC5925) was purchased from Novex (Grand Island, NY). The MicroBCA protein assay kit was purchased from Thermo Scientific (Waltham, MA).
2.3 Methods
Western Blot Analysis
Amygdala and hypothalamus nonhuman primate and mouse tissues and hippocampal tissues of the MA drinking selected lines were homogenized in RIPA lysis buffer [0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40, 150 mM NaCl, 50 mM Tris pH 8.0] containing the phosphatase inhibitor sodium vanadate [NaV, 1mM]. Protein lysates were extracted by centrifugation and protein concentrations were calculated using the MicroBCA protein assay. Equal amounts of protein were separated on sodium dodecyl sulfate polyacrylamide (4-12% SDS-PAGE) gradient gels and transferred to a polyvinylidene difluoride (PVDF) membrane. Membrane blots were then blocked in tris-buffered saline and tween 20 (TBST) [1 X TBS, 0.1% Tween 20] containing 5% bovine serum albumin (BSA). The membranes were then incubated with primary antibody diluted (see table below for antibody dilutions) in TBST containing 1% bovine serum albumin (BSA) overnight at 4°C. Membranes were then washed in TBST [3×10min] before being incubated in secondary antibody [1:10000 dilution] for one hour. Membranes were incubated in enhanced chemiluminescence (ECL) reagent before being exposed to CL-XPosure Film to detect protein changes. We used both Actin and GAPDH antibodies as a loading control to ensure that potential changes in target proteins were not do to user error. Figure 1 shows a representative example of Western blot analysis with brain extracts of the MALDR and MAHDR animal models (Fig 1).
Figure 1.
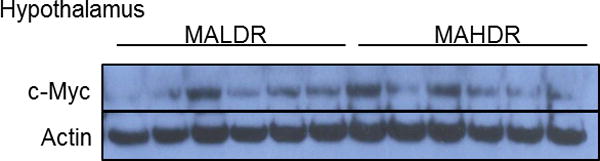
c-Myc protein in mice with genetic predisposition to methamphetamine abuse. Representative Western blot analysis of c-Myc protein in hypothalamic tissues of methamphetamine low drinking (MALDR) and methamphetamine high drinking (MAHDR) mice that were selectively bred and methamphetamine naïve.
| Antibody | Company | Source | Dilution |
|
| |||
| c-Myc | Santa Cruz | Rabbit | 1:5000 |
|
| |||
| phospho-c-Myc | Abcam | Rabbit | 1:1000 |
|
| |||
| p21 | Santa Cruz | Rabbit | 1:5000 |
|
| |||
| Actin | Santa Cruz | Mouse | 1:10000 |
|
| |||
| GAPDH | Santa Cruz | Mouse | 1:10000 |
Statistical Analyses
For quantitation of Western blot analysis, two-way analysis of variance was performed using the Graphpad Prism program (La Jolla, CA) to detect differences between treatment groups. A P value <0.05 was considered statistically significant. Each experiment was repeated at least three times. A representative blot is shown along with quantification.
3.Results
3.1 c-Myc protein levels in the amygdala increased in animals that have genetic predisposition to increased alcohol consumption
We first determined whether genetic predisposition affects c-Myc protein levels in the brain. Phosphorylated levels of c-Myc (T58) were significantly lower in the amygdala of SOT mice (Fig 2A). More c-Myc protein was observed in the amygdala of SOT mice when compared to NOT mice (Fig 2B). p21 protein was not changed in the amygdala or hypothalamus of SOT tissues when compared to NOT tissues indicating that c-Myc protein is increased but it doesn’t affect downstream targets (Fig 2C). (p < 0.05 vs SOT group)
Figure 2.
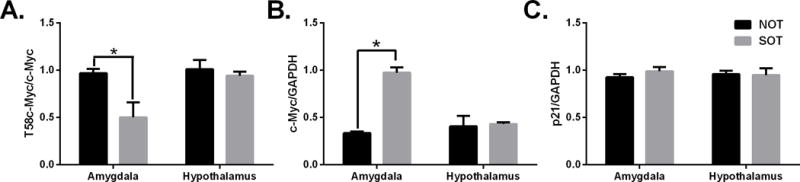
Protein levels in mice with genetic predisposition to increased or decreased alcohol consumption. Western blot quantification of SOT and NOT mice that were alcohol naïve. Panels show: (A) T58c-Myc, (B) total c-Myc, (C) p21 protein. *p<0.05 vs SOT group
3.2 c-Myc protein activation in the hypothalamus increased in animals that have predisposition to withdrawal symptoms
In the hypothalamus, we observed a significant decrease in T58 phosphorylated c-Myc protein in WSR mice when compared to WSP mice (Fig 3A). In contrast to the hypothalamus, there were no significant changes in T58 phosphorylated c-Myc in the amygdala between WSP and WSR mice. Unlike T58 phosphorylated c-Myc protein, there was no change in total c-Myc protein in either the hypothalamus or amygdala when we compared WSP mice to WSR mice (Fig 3B). However, we did observe significant decreases in p21 protein levels in the hypothalamus of WSR when compared to those in WSP mice (Fig 3C). There were no significant changes in p21 protein in the amygdala between the two lines. Although c-Myc protein is not changed between the two lines, these data together suggest that predisposition to severe alcohol withdrawal symptoms leads to increases in c-Myc protein activation (phosphorylation) as well as downstream targets. (p < 0.05 between lines.)
Figure 3.
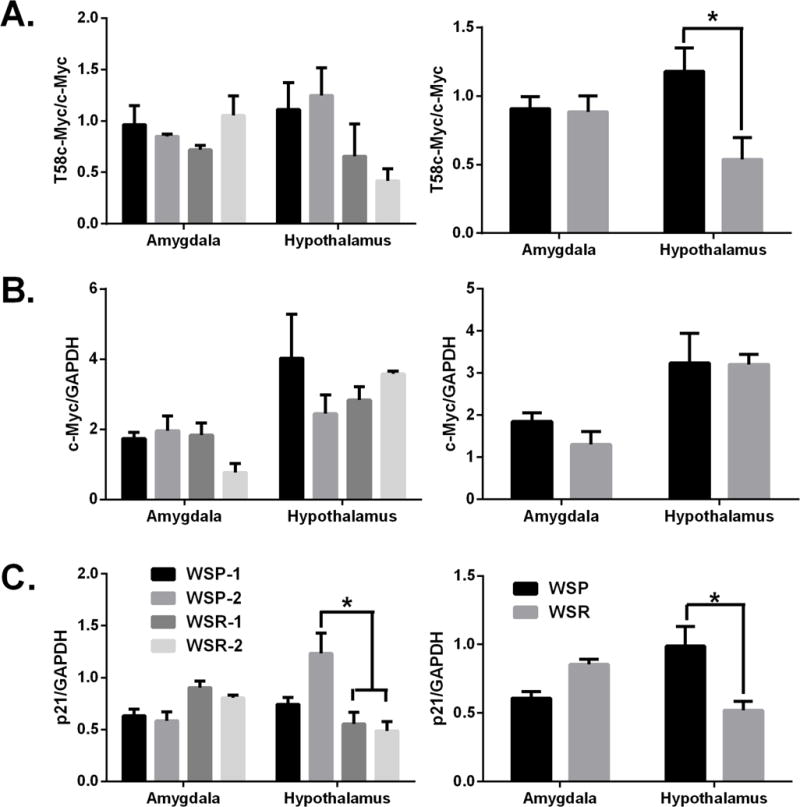
Effects of predisposition to withdrawal symptoms on c-Myc protein expression. Western blot quantification of WSP and WSR mice. Panels show: (A) T58c-Myc, (B) total c-Myc and (C) p21 protein. * p< 0.05 between lines.
3.3 c-Myc protein levels in the hypothalamus increased in mice that have genetic predisposition to methamphetamine abuse
To determine if the increase in c-Myc levels were specific for alcohol-related traits, c-Myc levels were analyzed in the hypothalamus and hippocampus of mice selectively bred for high or low methamphetamine intake in a two-bottle choice MA drinking. Mice that had a genetic predisposition to drink more methamphetamine (MAHDR) had higher levels of hypothalamic c-Myc than mice with a genetic predisposition to drink lower levels of methamphetamine (MALDR) (Fig 4). In contrast, there was no difference in c-Myc levels in hippocampus tissues in these lines (Fig 4). (p < 0.05 between strains)
Figure 4.
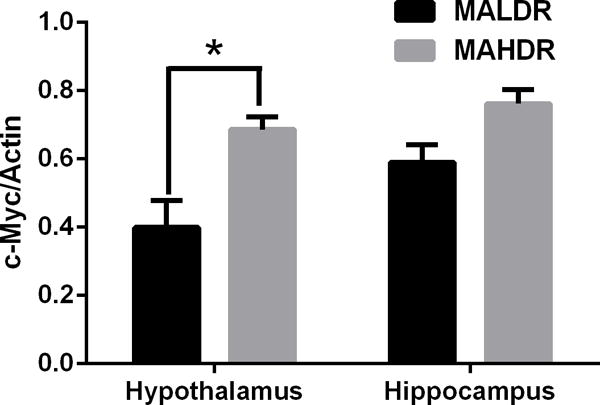
c-Myc protein in mice with genetic predisposition to methamphetamine abuse. Western blot quantification of methamphetamine high drinking (MAHDR) and methamphetamine low drinking (MALDR) mice that were selectively bred and methamphetamine naïve. Upper panel: hypothalamic tissues, lower panel: hippocampus tissues. *p< 0.05 between strains
3.4 c-Myc protein in the amygdala and the hypothalamus increased following acute alcohol exposure in mGlu8 KO and wild-type mice
We first determined the effects of acute alcohol intraperitoneal injection (IP) (2.5g/kg) on c-Myc protein levels in wildtype mice. Phosphorylated (T58) c-Myc protein expression was decreased in amygdala tissues following alcohol treatment (Fig 5A). There were no significant changes seen in the hypothalamus. We were also only able to observe increased levels of total c-Myc levels in the amygdala in wild-type mice. In contrast to wild-type mice, phosphorylated levels of c-Myc protein were decreased in the hypothalamus and total c-Myc levels were increased in the amygdala and hypothalamus of mGlu8 KO mice treated with ethanol when compared to genotype-matched mice treated with saline (Fig 5B). The p21 protein levels were significantly decreased in mGlu8KO mice in both the amygdala and hypothalamus following IP ethanol injection (Fig 5C). These data support an important role for mGlu8 in the hypothalamus in c-Myc expression following alcohol exposure. In order to determine if c-Myc protein is uniquely regulated by alcohol we treated mGlu8KO mice by IP injection with methamphetamine (1mg/kg). c-Myc protein expression in the amygdala of mGlu8 KO mice were also increased following methamphetamine exposure (Fig 5D). We did not observe any significant changes in c-Myc protein in the hypothalamus of mGlu8KO mice or in the amygdala of wildtype mice as observed in mice injected with alcohol. Thus, the increase in c-Myc protein levels is not specific for acute alcohol injections in the amygdala of mGlu8KO mice as it is seen following acute methamphetamine injections as well. (p < 0.05 versus treated animals)
Figure 5.
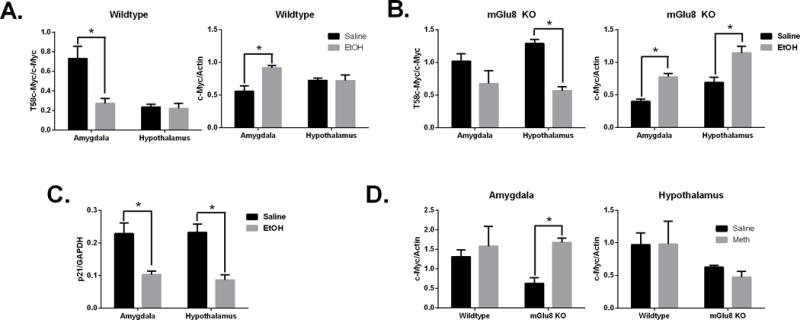
Acute alcohol or methamphetamine injection in mGlu8 KO and WT mice increases c-Myc protein expression. Western quantification of mGlu8 or wildtype mice following ethanol or saline treatment (Panels A-C). Methamphetamine or saline treatment was also examined (Panel D). Panels show: (A-B) T58c-Myc and c-Myc protein, (C) p21 protein, (D) c-Myc protein. *p <0.05 versus treated animals.
3.5 Chronic alcohol drinking increases c-Myc protein in the hypothalamic tissues of non-human primates
To measure the effect of chronic drinking on c-Myc protein expression in the brain we studied male vervet monkeys that were given free access to an isocaloric maltose dextrin solution (control) and water or 4% ethanol and water for 22hr/day for 442 days. Phosphorylated (T58) c-Myc levels were not changed in monkeys that were exposed to ethanol (Fig 6A). Though there were no changes in phosphorylated c-Myc protein we did detect an increase in total c-Myc protein in the hypothalamus of ethanol-exposed primates (Fig 6B). The levels of p21 protein were also lower in hypothalamic tissues of animals that were given free access to alcohol than controls (Fig 6C) (p < 0.05 versus vehicle-treated animals). Of the seven alcohol-drinking monkeys, five were classified as low drinking, one was categorized as high drinking and one was determined to be a binge drinker. The two monkeys that were categorized as high and binge drinkers also had the highest c-Myc protein expression and are indicated as open circles (Fig 7A). When c-Myc protein expression was analyzed in relation to the interval of time between last drink of ethanol and tissue collection, there was an increase in c-Myc with a longer interval until approximately 127 minutes post drinking. Following intervals longer than 127 minutes, c-Myc protein levels fell with the exception of the monkey that was classified as a binge drinker who showed higher c-Myc levels and is also indicated as an open circle (Fig 7B).
Figure 6.

Chronic alcohol drinking increases c-Myc protein activity. Western blot quantification of protein levels in vervet monkeys that drank alcohol or maltose-dextrin solution (control) for one year. Panels show: (A) T58c-Myc, (B) total c-Myc and (C) p21 protein. *p< 0.05 versus control treated animals.
Figure 7.
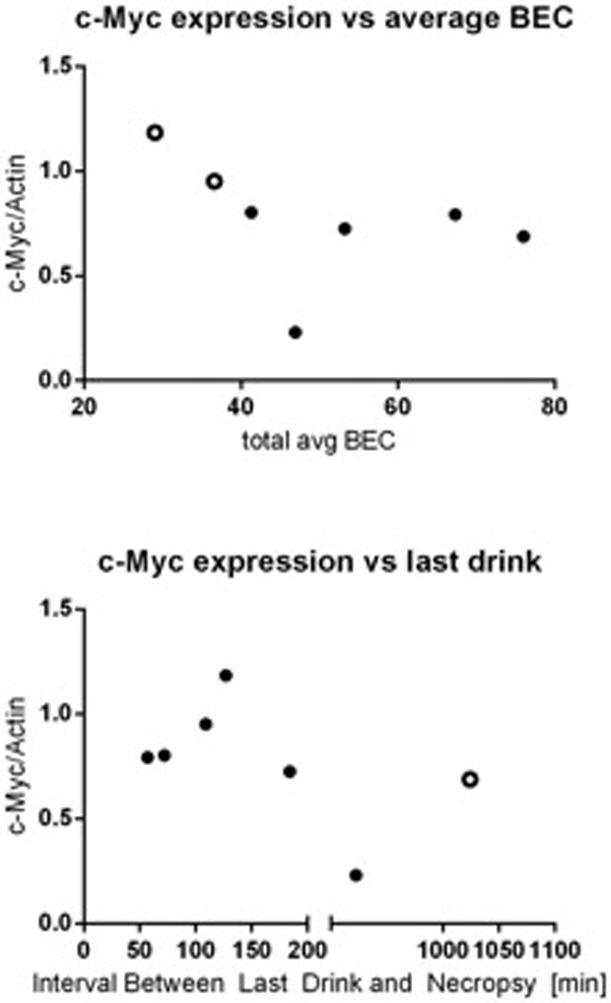
Chronic alcohol drinking data in vervet monkeys. (A) c-Myc protein expression was compared to average blood ethanol concentration and (B) c-Myc protein expression was also compared to the time between last drink and necropsy
4.Discussion
To our knowledge, this is the first study that defines the effects of alcohol exposure and predisposition to alcoholism on c-Myc protein levels in the brain. The effects of alcohol on c-Myc and p21 protein levels in the amygdala and hypothalamus are summarized in Table 1. The causes that lead to predisposition to alcohol dependence remain largely unknown. One major risk factor for developing alcohol abuse disorder is having a family history of alcoholism (55). There has been research done to identify genes and biomarkers involved in alcoholism neuropathology. These markers reflect complex overlapping and competing effects of possibly hundreds of genes that impact brain structure, function, biochemical alcohol processing, sensitivity and risk for dependence. In this study, we show that naïve mice with a genetic predisposition towards high dinking and low withdrawal from alcohol (SOT) exhibit higher expression levels of c-Myc protein in the amygdala when compared to the mice (NOT) that have been genetically bred to have lower drinking and higher withdrawal symptoms from alcohol. SOT mice have also been selectively bred to exhibit lower withdrawal symptoms. It is possible that similar mechanisms that cause an increase in c-Myc protein in mice that have a genetic predisposition to alcohol may also play a role in withdrawal from alcohol. Based on our findings elevated c-Myc protein in the amygdala may lead to increased risk of dependence of alcohol.
Table 1.
Summary of the effect of ethanol on c-Myc and p21 protein in the brain.
| Model | Rx | c-Myc Amygdala | c-Myc Hypothalamus | Model | Rx | p21 Amygdala |
p21 Hypothalamus |
|---|---|---|---|---|---|---|---|
| SOT > NOT | None |

|
– | SOT > NOT | None |

|
– |
| WSP> WSR | None | – |

|
WSP> WSR | None | – |

|
| Vervet Monkey | Free Access | – |

|
Vervet Monkey | Free Access | – | – |
| Wild-type | 2.5g/kg |

|
— | Wild-type | 2.5g/kg |

|
– |
| mGlu8 KO | 2.5g/kg |

|

|
mGlu8 KO | 2.5g/kg |

|

|
Since c-Myc protein expression was higher in SOT mice which were bred to have lower withdrawal symptoms, we further examined the role c-Myc protein plays in withdrawal from alcohol abuse. Alcohol withdrawal syndrome (AWS) is the name for the symptoms that occur when a heavy drinker stops or reduces their alcohol intake. With AWS, a person may experience a combination of physical and emotional symptoms, from mild anxiety and fatigue to nausea and seizures (56). To mimic AWS in humans we used two different mouse models, WSP mice that are selectively bred to have severe ethanol withdrawal HICs after cessation of 3 days of ethanol vapor inhalation and WSR mice that have mild HICs after high ethanol inhalation (57). We did not detect changes in total c-Myc protein when we compared the WSP and WSR mice but we did observe higher T58 phosphorylated c-Myc protein expression in the hypothalamus of WSP mice when compared to WSR mice. We also observed a significant increase in the hypothalamus of the downstream target p21 when we compared WSR mice to WSP mice. Although there was no significant change in c-Myc protein between the two lines, the significant increase in the T58 phosphorylated c-Myc suggests that the protein is being activated. No changes in total c-Myc protein while observing significant changes in p21 protein lends to the premise that the changes in p21 protein may be occurring through a different regulator. Anxiety is one of the early signs of withdrawal. Among the various limbic structures involved in emotional processing, the hypothalamus is thought to play a crucial role in modulating anxiety-related behaviors (58). Alcohol exposure results in various neurochemical adaptations in the brain (59) and as a result of these adaptations, manifestation of withdrawal symptoms during abstinence (60) from alcohol occurs. One of these neuroadaptations that may occur during withdrawal symptoms is changes in c-Myc protein expression within the hypothalamus.
The effects of chronic alcohol consumption include malnutrition, chronic pancreatitis, alcoholic liver disease and cancer. In addition, damage to the central nervous system and peripheral nervous system can occur from chronic alcohol abuse. Previous research has shown that chronic alcohol abuse elevates c-Myc protein in the superior frontal cortex (10). Alcohol can act as a stressor, activating the hypothalamic/pituitary/adrenocortical (HPA) axis, which combines a major component of the hormonal stress response (61). Furthermore, chronic alcohol exposure and withdrawal not only produce agitations in the HPA axis but also causes neuroendocrine-independent (example: hypothalamic) brain stress systems that influence drinking behavior in a complex manner (62). In this study, we showed that chronic daily drinking over the course of a year increases c-Myc protein in the hypothalamus of non-human primates. These non-human primates were given free access to alcohol and water and the primates that were classified as high and binge drinkers also had the highest average blood ethanol concentration. We also observed an increase in c-Myc protein in these primates when compared to the length of time before necropsy. It is possible that this increase is correlated to withdrawal symptoms as seen in our withdrawal model [WSP/WSR mice] (Fig 3B). This would explain why T58 phosphorylated c-Myc protein in both of these animal models was not changed following alcohol exposure. The binge drinking non-human primate did show higher total c-Myc protein levels past the point of withdrawal. The post-withdrawal decline in c-Myc protein may be ablated by binge drinking but not by high or low level drinking. During chronic drinking, c-Myc protein levels are elevated immediately following drinking and continue to remain high until withdrawal symptoms decline. c-Myc protein regulates cell proliferation as well as apoptosis which may impact neurogenesis. Adult neurogenesis is affected in many neurological and neurodegenerative diseases and alcohol abuse can result in a neurodegenerative condition called alcohol-related brain damage. The exact mechanism linking chronic alcohol intoxication with alcohol-related brain damage remains largely unknown. Further studies need to be done to determine if it is possible that c-Myc mediates the effect that chronic alcohol abuse has on neurodegenerative diseases.
Acute alcohol intoxication is the result of a single event of excessive drinking. Ethanol accumulates in the body and the brain, causing central nervous system suppression after initial excitement. Clinical manifestations of acute alcohol intoxication are varied and involve different organs, with behavioral, cardiac, gastrointestinal, neurological, metabolic, and especially hepatic effects (63-67). The brain is one of the major target organs of ethanol actions. To measure the effects of acute alcohol exposure on c-Myc protein expression we used mGlu8 knockout mice compared to wildtype mice. mGlu8 knockout mice were used as a model because the GRM8 receptor significantly is associated with alcohol dependence in European Americans (43). We observed a significant increase in c-Myc protein in the amygdala and hypothalamus of mGlu8 knockout mice following one-hour acute alcohol exposure. This significant increase was observed in the amygdala of wild-type mice but there was no change in c-Myc protein expression in the hypothalamus following acute alcohol exposure. This alcohol-induced increase in c-Myc protein in the hypothalamus of mGlu8 knockout mice but not in wild-type mice leads us to believe that the hypothalamus may be a brain area of interest in regard to acute alcohol exposure. We also observed increases to c-Myc protein expression in the hypothalamus of non-human primates that were chronically exposed to alcohol and in mice that had less pronounced withdrawal symptoms (WSR). It is possible that acute, chronic and withdrawal from alcohol exposure can activate the hypothalamic–pituitary–adrenocortical (HPA) axis, which creates a major component of the hormonal stress response (61). Further studies need to be done to establish if this occurs within our animal models.
Drug abuse is a biological, pathological process that alters the way in which the pleasure center, as well as other parts of the brain, functions. To understand this process, it is necessary to examine the effects of substance abuse on different pathways within the brain. We not only observed an increase in c-Myc protein following alcohol exposure or genetic predisposition but we also detected similar increases in c-Myc protein expression following methamphetamine exposure and genetic predisposition to methamphetamine. Based on these results, the brain’s response following exposure to substances of abuse might be to increase cell proliferation and neurogenesis and this response may not be limited to alcohol. Further studies are needed to determine the exact pathway in which substances of abuse increases c-Myc protein and the role that this protein may have on addictive behaviors.
Constitutively expressed c-Myc leads to the unregulated expression of many genes, some of which are involved in cell proliferation, and mutations in c-Myc is associated with cancer (68-70). Several mechanisms activate the c-myc gene in human cancers. The c-myc gene is amplified in various human cancers, including lung carcinoma, breast carcinoma, cervical carcinoma, ovarian carcinoma, and prostate carcinoma (71-79).
Highlights.
Genetic predisposition to alcohol-related features leads to c-Myc protein increase in the amygdala.
Chronic alcoholism increases c-Myc protein in the hypothalamus.
Methamphetamine abuse increases c-Myc protein in the amygdala.
p21 protein is decreased in withdrawal and chronic drinking.
Acknowledgments
Thanks are extended to Tamara Phillips from Oregon Health and Science University for providing the MAHDR/MALDR mice and to Robert Duvoisin for providing the mGlu8 knockout and wild-type littermates. The authors gratefully acknowledge the support of Drs. John Crabbe and Pamela Metten for providing the WSR and WSP mice and for assistance with chronic alcohol exposure, and Melissa Andrew for assistance with the chronic exposure studies. WSR and WSP treatments were performed in facilities provided by the Department of Veteran Affairs.
Formatting of funding sources
This work was supported by the National Institutes of Health [NIAAA T32AA007290-33, AA014106/AA/NIAAA, R24 AA019431/AA/NIAAA, R01 AA021468 and NIAAA R24AA020245].
Footnotes
Conflicts of Interest
The authors have no conflict of interest to report.
Author Contributions
TA: performance of the majority of the experiments and writing the first draft of the document.
TA and JR: conception and design of the study, analysis, and interpretation of data, revising, and final approval of the manuscript.
SJW: assistance with experimental procedures and manuscript revision.
MG, DF, AD, EB and JD: revision of the manuscript.
AD, JD, EB: conducted ethanol self-administration study in vervet monkeys at Wake Forest School of Medicine and provided brain tissues.
References
- 1.Bagnardi V, Blangiardo M, La Vecchia C, Corrao G. A meta-analysis of alcohol drinking and cancer risk. British journal of cancer. 2001;85:1700–1705. doi: 10.1054/bjoc.2001.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donato F, Tagger A, Gelatti U, Parrinello G, Boffetta P, Albertini A, Decarli A, Trevisi P, Ribero ML, Martelli C, Porru S, Nardi G. Alcohol and hepatocellular carcinoma: the effect of lifetime intake and hepatitis virus infections in men and women. American journal of epidemiology. 2002;155:323–331. doi: 10.1093/aje/155.4.323. [DOI] [PubMed] [Google Scholar]
- 3.Dong C, Yoon YH, Chen CM, Yi HY. Heavy alcohol use and premature death from hepatocellular carcinoma in the United States, 1999-2006. Journal of studies on alcohol and drugs. 2011;72:892–902. doi: 10.15288/jsad.2011.72.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marrero JA, Fontana RJ, Fu S, Conjeevaram HS, Su GL, Lok AS. Alcohol, tobacco and obesity are synergistic risk factors for hepatocellular carcinoma. Journal of hepatology. 2005;42:218–224. doi: 10.1016/j.jhep.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Roerecke M, Rehm J. Alcohol consumption, drinking patterns, and ischemic heart disease: a narrative review of meta-analyses and a systematic review and meta-analysis of the impact of heavy drinking occasions on risk for moderate drinkers. BMC medicine. 2014;12:182. doi: 10.1186/s12916-014-0182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashmi S, Allison MG, McCurdy MT, Reed RM. Hyperbilirubinaemia and haemolytic anaemia in acute alcoholic hepatitis: there’s oil in them thar veins. BMJ case reports. 2014;2014 doi: 10.1136/bcr-2014-203804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koike H, Hama T, Kawagashira Y, Hashimoto R, Tomita M, Iijima M, Sobue G. The significance of folate deficiency in alcoholic and nutritional neuropathies: analysis of a case. Nutrition (Burbank, Los Angeles County, Calif) 2012;28:821–824. doi: 10.1016/j.nut.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 8.Bataller R, Gao B. Liver fibrosis in alcoholic liver disease. Seminars in liver disease. 2015;35:146–156. doi: 10.1055/s-0035-1550054. [DOI] [PubMed] [Google Scholar]
- 9.Guirguis J, Chhatwal J, Dasarathy J, Rivas J, McMichael D, Nagy LE, McCullough AJ, Dasarathy S. Clinical impact of alcohol-related cirrhosis in the next decade: estimates based on current epidemiological trends in the United States. Alcoholism, clinical and experimental research. 2015;39:2085–2094. doi: 10.1111/acer.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Housseini AM, Sivanandam TM, Bradbury EL, Tannenberg RK, Dodd PR, Gu Q. Upregulation of beta-catenin levels in superior frontal cortex of chronic alcoholics. Alcoholism, clinical and experimental research. 2008;32:1080–1090. doi: 10.1111/j.1530-0277.2008.00670.x. [DOI] [PubMed] [Google Scholar]
- 11.Prevention C. f. D. C. a. Alcohol and Public Health: Alcohol-Related Disease Impact (ARDI). Center for Disease Control and Prevention.
- 12.Hong SW, Linton JA, Shim JY, Kang HT. High-risk drinking is associated with a higher risk of diabetes mellitus in Korean men, based on the 2010-2012 KNHANES. Alcohol (Fayetteville, NY) 2015;49:275–281. doi: 10.1016/j.alcohol.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Ikehara S, Iso H, Yamagishi K, Yamamoto S, Inoue M, Tsugane S. Alcohol consumption, social support, and risk of stroke and coronary heart disease among Japanese men: the JPHC Study. Alcoholism, clinical and experimental research. 2009;33:1025–1032. doi: 10.1111/j.1530-0277.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- 14.Miyake T, Kumagi T, Hirooka M, Furukawa S, Yoshida O, Koizumi M, Yamamoto S, Watanabe T, Yamamoto Y, Tokumoto Y, Takeshita E, Abe M, Kitai K, Matsuura B, Hiasa Y. Low alcohol consumption increases the risk of impaired glucose tolerance in patients with non-alcoholic fatty liver disease. Journal of gastroenterology. 2016 doi: 10.1007/s00535-016-1194-0. [DOI] [PubMed] [Google Scholar]
- 15.Neel BG, Jhanwar SC, Chaganti RS, Hayward WS. Two human c-onc genes are located on the long arm of chromosome 8. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:7842–7846. doi: 10.1073/pnas.79.24.7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rappold GA, Hameister H, Cremer T, Adolph S, Henglein B, Freese UK, Lenoire GM, Bornkamm GW. c-myc and immunoglobulin kappa light chain constant genes are on the 8q+ chromosome of three Burkitt lymphoma lines with t(2;8) translocations. The EMBO journal. 1984;3:2951–2955. doi: 10.1002/j.1460-2075.1984.tb02239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escamilla-Powers JR, Sears RC. A conserved pathway that controls c-Myc protein stability through opposing phosphorylation events occurs in yeast. The Journal of biological chemistry. 2007;282:5432–5442. doi: 10.1074/jbc.M611437200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Seminars in cancer biology. 2006;16:288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Muhle-Goll C, Nilges M, Pastore A. The leucine zippers of the HLH-LZ proteins Max and c-Myc preferentially form heterodimers. Biochemistry. 1995;34:13554–13564. doi: 10.1021/bi00041a035. [DOI] [PubMed] [Google Scholar]
- 20.Reddy CD, Dasgupta P, Saikumar P, Dudek H, Rauscher FJ, 3rd, Reddy EP. Mutational analysis of Max: role of basic, helix-loop-helix/leucine zipper domains in DNA binding, dimerization and regulation of Myc-mediated transcriptional activation. Oncogene. 1992;7:2085–2092. [PubMed] [Google Scholar]
- 21.Walhout AJ, Gubbels JM, Bernards R, van der Vliet PC, Timmers HT. c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic acids research. 1997;25:1493–1501. doi: 10.1093/nar/25.8.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper JW, Elledge SJ, Keyomarsi K, Dynlacht B, Tsai LH, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, et al. Inhibition of cyclin-dependent kinases by p21. Molecular biology of the cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes & development. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 24.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 25.Kitaura H, Shinshi M, Uchikoshi Y, Ono T, Iguchi-Ariga SM, Ariga H. Reciprocal regulation via protein-protein interaction between c-Myc and p21(cip1/waf1/sdi1) in DNA replication and transcription. The Journal of biological chemistry. 2000;275:10477–10483. doi: 10.1074/jbc.275.14.10477. [DOI] [PubMed] [Google Scholar]
- 26.Nevzorova YA, Cubero FJ, Hu W, Hao F, Haas U, Ramadori P, Gassler N, Hoss M, Strnad P, Zimmermann HW, Tacke F, Trautwein C, Liedtke C. Enhanced expression of c-myc in hepatocytes promotes initiation and progression of alcoholic liver disease. Journal of hepatology. 2016;64:628–640. doi: 10.1016/j.jhep.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Wen Y, Feng D, Wu H, Liu W, Li H, Wang F, Xia Q, Gao WQ, Kong X. Defective Initiation of Liver Regeneration in Osteopontin-Deficient Mice after Partial Hepatectomy due to Insufficient Activation of IL-6/Stat3 Pathway. International journal of biological sciences. 2015;11:1236–1247. doi: 10.7150/ijbs.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radaeva S, Jaruga B, Hong F, Kim WH, Fan S, Cai H, Strom S, Liu Y, El-Assal O, Gao B. Interferon-alpha activates multiple STAT signals and down-regulates c-Met in primary human hepatocytes. Gastroenterology. 2002;122:1020–1034. doi: 10.1053/gast.2002.32388. [DOI] [PubMed] [Google Scholar]
- 29.Tsuchishima M, George J, Shiroeda H, Arisawa T, Takegami T, Tsutsumi M. Chronic ingestion of ethanol induces hepatocellular carcinoma in mice without additional hepatic insult. Digestive diseases and sciences. 2013;58:1923–1933. doi: 10.1007/s10620-013-2574-4. [DOI] [PubMed] [Google Scholar]
- 30.Metten P, Iancu OD, Spence SE, Walter NA, Oberbeck D, Harrington CA, Colville A, McWeeney S, Phillips TJ, Buck KJ, Crabbe JC, Belknap JK, Hitzemann RJ. Dual-trait selection for ethanol consumption and withdrawal: genetic and transcriptional network effects. Alcoholism, clinical and experimental research. 2014;38:2915–2924. doi: 10.1111/acer.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, Erwin VG, Belknap JK. High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mammalian genome : official journal of the International Mammalian Genome Society. 1998;9:983–990. doi: 10.1007/s003359900911. [DOI] [PubMed] [Google Scholar]
- 32.Aarabi P, Norouzi N, Dear T, Carver S, Bromberg S, Gray S, Kahan M, Borgundvaag B. A quantitative evaluation of alcohol withdrawal tremors. Conference proceedings : … Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society. Annual Conference. 2015;2015:6215–6218. doi: 10.1109/EMBC.2015.7319812. [DOI] [PubMed] [Google Scholar]
- 33.Das SC, Althobaiti YS, Alshehri FS, Sari Y. Binge ethanol withdrawal: Effects on post-withdrawal ethanol intake, glutamate-glutamine cycle and monoamine tissue content in P rat model. Behavioural brain research. 2016;303:120–125. doi: 10.1016/j.bbr.2016.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gortney JS, Raub JN, Patel P, Kokoska L, Hannawa M, Argyris A. Alcohol withdrawal syndrome in medical patients. Cleveland Clinic journal of medicine. 2016;83:67–79. doi: 10.3949/ccjm.83a.14061. [DOI] [PubMed] [Google Scholar]
- 35.Kim DW, Kim HK, Bae EK, Park SH, Kim KK. Clinical predictors for delirium tremens in patients with alcohol withdrawal seizures. The American journal of emergency medicine. 2015;33:701–704. doi: 10.1016/j.ajem.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 36.Metten P, Crabbe JC. Alcohol withdrawal severity in inbred mouse (Mus musculus) strains. Behavioral neuroscience. 2005;119:911–925. doi: 10.1037/0735-7044.119.4.911. [DOI] [PubMed] [Google Scholar]
- 37.Bousman CA, Glatt SJ, Everall IP, Tsuang MT. Genetic association studies of methamphetamine use disorders: A systematic review and synthesis. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2009;150b:1025–1049. doi: 10.1002/ajmg.b.30936. [DOI] [PubMed] [Google Scholar]
- 38.Shabani S, McKinnon CS, Reed C, Cunningham CL, Phillips TJ. Sensitivity to rewarding or aversive effects of methamphetamine determines methamphetamine intake. Genes, brain, and behavior. 2011;10:625–636. doi: 10.1111/j.1601-183X.2011.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler JM, Reed C, Burkhart-Kasch S, Li N, Cunningham CL, Janowsky A, Franken FH, Wiren KM, Hashimoto JG, Scibelli AC, Phillips TJ. Genetically correlated effects of selective breeding for high and low methamphetamine consumption. Genes, brain, and behavior. 2009;8:758–771. doi: 10.1111/j.1601-183X.2009.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linden AM, Bergeron M, Baez M, Schoepp DD. Systemic administration of the potent mGlu8 receptor agonist (S)-3,4-DCPG induces c-Fos in stress-related brain regions in wild-type, but not mGlu8 receptor knockout mice. Neuropharmacology. 2003;45:473–483. doi: 10.1016/s0028-3908(03)00200-4. [DOI] [PubMed] [Google Scholar]
- 41.Linden AM, Johnson BG, Peters SC, Shannon HE, Tian M, Wang Y, Yu JL, Koster A, Baez M, Schoepp DD. Increased anxiety-related behavior in mice deficient for metabotropic glutamate 8 (mGlu8) receptor. Neuropharmacology. 2002;43:251–259. doi: 10.1016/s0028-3908(02)00079-5. [DOI] [PubMed] [Google Scholar]
- 42.Backstrom P, Hyytia P. Suppression of alcohol self-administration and cue-induced reinstatement of alcohol seeking by the mGlu2/3 receptor agonist LY379268 and the mGlu8 receptor agonist (S)-3,4-DCPG. European journal of pharmacology. 2005;528:110–118. doi: 10.1016/j.ejphar.2005.10.051. [DOI] [PubMed] [Google Scholar]
- 43.Long EC, Aliev F, Wang JC, Edenberg HJ, Nurnberger J, Jr, Hesselbrock V, Porjesz B, Dick DM. Further Analyses of Genetic Association Between GRM8 and Alcohol Dependence Symptoms Among Young Adults. Journal of studies on alcohol and drugs. 2015;76:414–418. doi: 10.15288/jsad.2015.76.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilman JM, Hommer DW. Modulation of brain response to emotional images by alcohol cues in alcohol-dependent patients. Addiction biology. 2008;13:423–434. doi: 10.1111/j.1369-1600.2008.00111.x. [DOI] [PubMed] [Google Scholar]
- 45.Bjork JM, Gilman JM. The effects of acute alcohol administration on the human brain: insights from neuroimaging. Neuropharmacology. 2014;84:101–110. doi: 10.1016/j.neuropharm.2013.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 47.Hurle B, Swanson W, Green ED. Comparative sequence analyses reveal rapid and divergent evolutionary changes of the WFDC locus in the primate lineage. Genome research. 2007;17:276–286. doi: 10.1101/gr.6004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuzon Carlson VC, Seabold GK, Helms CM, Garg N, Odagiri M, Rau AR, Daunais J, Alvarez VA, Lovinger DM, Grant KA. Synaptic and morphological neuroadaptations in the putamen associated with long-term, relapsing alcohol drinking in primates. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2011;36:2513–2528. doi: 10.1038/npp.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilman S, Adams K, Koeppe RA, Berent S, Kluin KJ, Modell JG, Kroll P, Brunberg JA. Cerebellar and frontal hypometabolism in alcoholic cerebellar degeneration studied with positron emission tomography. Annals of neurology. 1990;28:775–785. doi: 10.1002/ana.410280608. [DOI] [PubMed] [Google Scholar]
- 50.Pitel AL, Chetelat G, Le Berre AP, Desgranges B, Eustache F, Beaunieux H. Macrostructural abnormalities in Korsakoff syndrome compared with uncomplicated alcoholism. Neurology. 2012;78:1330–1333. doi: 10.1212/WNL.0b013e318251834e. [DOI] [PubMed] [Google Scholar]
- 51.Sullivan EV, Harding AJ, Pentney R, Dlugos C, Martin PR, Parks MH, Desmond JE, Chen SH, Pryor MR, De Rosa E, Pfefferbaum A. Disruption of frontocerebellar circuitry and function in alcoholism. Alcoholism, clinical and experimental research. 2003;27:301–309. doi: 10.1097/01.ALC.0000052584.05305.98. [DOI] [PubMed] [Google Scholar]
- 52.Pleil KE, Lowery-Gionta EG, Crowley NA, Li C, Marcinkiewcz CA, Rose JH, McCall NM, Maldonado-Devincci AM, Morrow AL, Jones SR, Kash TL. Effects of chronic ethanol exposure on neuronal function in the prefrontal cortex and extended amygdala. Neuropharmacology. 2015;99:735–749. doi: 10.1016/j.neuropharm.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daunais JB, Davenport AT, Helms CM, Gonzales SW, Hemby SE, Friedman DP, Farro JP, Baker EJ, Grant KA. Monkey alcohol tissue research resource: banking tissues for alcohol research. Alcoholism, clinical and experimental research. 2014;38:1973–1981. doi: 10.1111/acer.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grant KA, Leng X, Green HL, Szeliga KT, Rogers LS, Gonzales SW. Drinking typography established by scheduled induction predicts chronic heavy drinking in a monkey model of ethanol self-administration. Alcoholism, clinical and experimental research. 2008;32:1824–1838. doi: 10.1111/j.1530-0277.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lieb R, Merikangas KR, Hofler M, Pfister H, Isensee B, Wittchen HU. Parental alcohol use disorders and alcohol use and disorders in offspring: a community study. Psychological medicine. 2002;32:63–78. doi: 10.1017/s0033291701004883. [DOI] [PubMed] [Google Scholar]
- 56.Feeney C, Alter HJ, Jacobsen E, Rehrer M, Shao S, Subramanian I, Clements RC. A Simplified Protocol for the Treatment of Alcohol Withdrawal. Journal of addiction medicine. 2015;9:485–490. doi: 10.1097/ADM.0000000000000167. [DOI] [PubMed] [Google Scholar]
- 57.Crabbe JC, Kosobud A, Young ER. Genetic selection for ethanol withdrawal severity: differences in replicate mouse lines. Life sciences. 1983;33:955–962. doi: 10.1016/0024-3205(83)90751-8. [DOI] [PubMed] [Google Scholar]
- 58.Anglada-Figueroa D, Quirk GJ. Lesions of the basal amygdala block expression of conditioned fear but not extinction. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:9680–9685. doi: 10.1523/JNEUROSCI.2600-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Becker HC, Mulholland PJ. Neurochemical mechanisms of alcohol withdrawal. Handbook of clinical neurology. 2014;125:133–156. doi: 10.1016/B978-0-444-62619-6.00009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koob GF, Le Moal M. Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2001;24:97–129. doi: 10.1016/S0893-133X(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 61.Smith SM, Vale WW. The role of the hypothalamic-pituitary-adrenal axis in neuroendocrine responses to stress. Dialogues in clinical neuroscience. 2006;8:383–395. doi: 10.31887/DCNS.2006.8.4/ssmith. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. The American journal of psychiatry. 2007;164:1149–1159. doi: 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonitenko EY, Grebenyuk AN, Basharin VA, Ivanov MB, Makarova NV. Evaluation of the neurological status in experimental acute alcohol intoxication. Bulletin of experimental biology and medicine. 2010;149:324–327. doi: 10.1007/s10517-010-0938-8. [DOI] [PubMed] [Google Scholar]
- 64.Darke S, Duflou J, Torok M, Prolov T. Toxicology, circumstances and pathology of deaths from acute alcohol toxicity. Journal of forensic and legal medicine. 2013;20:1122–1125. doi: 10.1016/j.jflm.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 65.Karkhanis AN, Locke JL, McCool BA, Weiner JL, Jones SR. Social isolation rearing increases nucleus accumbens dopamine and norepinephrine responses to acute ethanol in adulthood. Alcoholism, clinical and experimental research. 2014;38:2770–2779. doi: 10.1111/acer.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karkkainen JM, Miilunpohja S, Rantanen T, Koskela JM, Jyrkka J, Hartikainen J, Paajanen H. Alcohol Abuse Increases Rebleeding Risk and Mortality in Patients with Non-variceal Upper Gastrointestinal Bleeding. Digestive diseases and sciences. 2015;60:3707–3715. doi: 10.1007/s10620-015-3806-6. [DOI] [PubMed] [Google Scholar]
- 67.Mehta AJ. Alcoholism and critical illness: A review. World journal of critical care medicine. 2016;5:27–35. doi: 10.5492/wjccm.v5.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niu Z, Liu H, Zhou M, Wang H, Liu Y, Li X, Xiong W, Ma J, Li X, Li G. Knockdown of c-Myc inhibits cell proliferation by negatively regulating the Cdk/Rb/E2F pathway in nasopharyngeal carcinoma cells. Acta biochimica et biophysica Sinica. 2015;47:183–191. doi: 10.1093/abbs/gmu129. [DOI] [PubMed] [Google Scholar]
- 69.Wong PP, Miranda F, Chan KV, Berlato C, Hurst HC, Scibetta AG. Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21(cip) (CDKN1A) Molecular and cellular biology. 2012;32:1633–1644. doi: 10.1128/MCB.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang D, Qi J, Liu R, Dai B, Ma W, Zhan Y, Zhang Y. c-Myc plays a key role in TADs-induced apoptosis and cell cycle arrest in human hepatocellular carcinoma cells. American journal of cancer research. 2015;5:1076–1088. [PMC free article] [PubMed] [Google Scholar]
- 71.Covington M, Sikora K, Turner MJ, White JO, Moore P, Soutter WP. C-myc expression in cervical cancer. Lancet (London, England) 1987;1:1260–1261. doi: 10.1016/s0140-6736(87)92706-1. [DOI] [PubMed] [Google Scholar]
- 72.Fleming WH, Hamel A, MacDonald R, Ramsey E, Pettigrew NM, Johnston B, Dodd JG, Matusik RJ. Expression of the c-myc protooncogene in human prostatic carcinoma and benign prostatic hyperplasia. Cancer research. 1986;46:1535–1538. [PubMed] [Google Scholar]
- 73.Fox SB, Persad RA, Royds J, Kore RN, Silcocks PB, Collins CC. p53 and c-myc expression in stage A1 prostatic adenocarcinoma: useful prognostic determinants? The Journal of urology. 1993;150:490–494. doi: 10.1016/s0022-5347(17)35533-7. [DOI] [PubMed] [Google Scholar]
- 74.He M, Fu Y, Yan Y, Xiao Q, Wu H, Yao W, Zhao H, Zhao L, Jiang Q, Yu Z, Jin F, Mi X, Wang E, Cui Z, Fu L, Chen J, Wei M. The Hedgehog signalling pathway mediates drug response of MCF-7 mammosphere cells in breast cancer patients. Clinical science (London, England : 1979) 2015;129:809–822. doi: 10.1042/CS20140592. [DOI] [PubMed] [Google Scholar]
- 75.Kacinski BM, Carter D, Kohorn EI, Mittal K, Bloodgood RS, Donahue J, Kramer CA, Fischer D, Edwards R, Chambers SK, et al. Oncogene expression in vivo by ovarian adenocarcinomas and mixed-mullerian tumors. The Yale journal of biology and medicine. 1989;62:379–392. [PMC free article] [PubMed] [Google Scholar]
- 76.Kozbor D, Croce CM. Amplification of the c-myc oncogene in one of five human breast carcinoma cell lines. Cancer research. 1984;44:438–441. [PubMed] [Google Scholar]
- 77.Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 78.Riou G, Barrois M, Le MG, George M, Le Doussal V, Haie C. C-myc proto-oncogene expression and prognosis in early carcinoma of the uterine cervix. Lancet (London, England) 1987;1:761–763. doi: 10.1016/s0140-6736(87)92795-4. [DOI] [PubMed] [Google Scholar]
- 79.Yasue H, Takeda A, Ishibashi M. Amplification of the c-myc gene and the elevation of its transcripts in human ovarian tumor lines. Cell structure and function. 1987;12:121–125. doi: 10.1247/csf.12.121. [DOI] [PubMed] [Google Scholar]


