Abstract
Up to three gibberellin (GA) 20-oxidase genes have now been cloned from several species including Arabidopsis, bean (Phaseolus vulgaris), and potato (Solanum tuberosum). In each case the GA 20-oxidase genes exhibit different patterns of tissue expression. We have performed extensive northern analysis on one of the potato GA 20-oxidase genes (StGA20ox1), which is the only one that shows significant transcript levels in leaves. We show that levels of StGA20ox1 transcript are elevated in transgenic antisense plants that have reduced levels of phytochrome B (PHYB) compared with wild-type plants, implicating PHYB in the control of GA biosynthesis. We show that StGA20ox1 transcript levels vary in leaves of different age throughout the plant and cycle throughout the day, furthermore they are up-regulated by light and down-regulated in the dark. The degree of the response to the light-on signal is similar in potato plants deficient in phytochrome A or PHYB and wild-type plants. The induction of StGA20ox1 by blue light raises the possibility that a blue light receptor may be involved in the control of this gene by light.
Tuberization of certain lines of potato (Solanum tuberosum subsp. andigena) is a strict short-day (SD) photoperiodic response. Several lines of evidence implicate gibberellins (GAs) in the inhibition of the tuberization in this potato species in long days (LD); levels of GA-like activity decrease in leaves of potato upon transfer from LD to SD conditions (Railton and Wareing, 1973), treating plants with ancymidol, an inhibitor of GA biosynthesis, enables them to tuberize in LD (Jackson and Prat, 1996), and a dwarf mutant of potato that is partially blocked in the 13-hydroxylation of GA12-aldehyde to GA53 (Van den Berg et al., 1995b) is also able to tuberize in LD. The early 13-hydroxylation pathway has been shown to be the main pathway for GA biosynthesis in potato (Van den Berg et al., 1995a), thus the reduction in the levels of GAs subsequent to this step in the pathway must be the reason that the dwarf mutant is able to tuberize in LD. Phytochrome B (PHYB) deficient potato transgenic antisense plants are also able to tuberize in LD (Jackson et al., 1996), although these plants have elongated internodes and reduced chlorophyll levels, which is the converse phenotype to that of the dwarf mutant or of wild-type (WT) plants treated with GA inhibitors. PhyB mutants of sorghum and Brassica rapa are reported to have increased GA levels (Rood et al., 1990; Foster et al., 1994), whereas other studies of phyB mutants of pea, cucumber, and Arabidopsis suggest that GA sensitivity is affected (Weller et al., 1994; Lopez-Juez et al., 1995; Reed et al., 1996).
Genes for enzymes involved in several steps of the GA biosynthetic pathway have now been cloned from various different species and the expression of many of these genes is regulated by light (Hedden and Kamiya, 1997; Kamiya and Garcia-Martinez 1999). It has been shown that genes encoding 3β-hydroxylases from lettuce and Arabidopsis are under phytochrome control (Toyomasu et al., 1998; Yamaguchi et al., 1998) and in the case of GA4H from Arabidopsis it was shown to be under the control of PHYB. At least three GA 20-oxidase genes have been cloned from Arabidopsis, bean (Phaseolus vulgaris), and potato (Phillips et al., 1995; Garcia-Martinez et al., 1997; Carrera et al., 1999). In these species the individual GA 20-oxidase genes exhibit different levels and patterns of expression, indicating that they probably have separate roles to play in specific aspects of the growth and development of the plant such as stem elongation or fruit development. Red light did not induce the expression of either of two GA 20-oxidases from lettuce, and with one of the genes (Ls20ox2) it was found that red light reduced its expression suggesting that Pfr may inhibit expression of this GA 20-oxidase (Toyomasu et al., 1998). However, red light was found to induce the expression of a GA 20-oxidase in pea (Ait-Ali et al., 1999).
GA 20-oxidase is thought to be a key regulatory enzyme in the GA biosynthetic pathway, its expression is subject to feedback inhibition by GAs further down the pathway, suggesting that GA biosynthesis has an auto-regulatory component (Hedden and Croker, 1992; Phillips et al., 1995). It is regulated in the pericarp of peas by the presence of seeds or the shoot apex (Garcia-Martinez et al., 1997; Van Huizen et al., 1997), and transcript levels are reported to be higher in LD than SD in the LD plants spinach and Arabidopsis (Wu et al., 1996; Xu et al., 1997), although no evidence has yet been found for this in SD potato (Carrera et al., 1999). Evidence for photoperiodic regulation of the steps catalyzed by the GA 20-oxidase originally came from gas chromatography-mass spectrometry measurements of cell-free extracts and endogenous GA levels of spinach plants grown in either LD or SD conditions (Gilmour et al., 1986; Talon et al., 1991). These results showed that the enzyme activities catalyzing the conversion of GA53 to GA44, and GA19 to GA20, increased upon transfer from noninducing SD conditions to inducing LD conditions, whereas the activity of an enzyme catalyzing the intermediate conversion of GA44 to GA19 remains high in both photoperiods. These activities could be separated by HPLC (Gilmour et al., 1987), and thus there are at least two GA 20-oxidases in leaves of spinach plants, one whose activity is photoperiodically regulated and one with high constitutive activity.
In addition to the existence of multiple GA 20-oxidases, which have different activities and/or are subject to different tissue and light regulation, several time course studies show that GA levels fluctuate throughout the day (Talon et al., 1991; Foster and Morgan, 1995; Lee et al., 1998), indicating that the activity or expression of genes involved in the biosynthetic pathway might also fluctuate throughout the day. This has been observed to some extent for a GA 20-oxidase and a 3β-hydroxylase from pea (Ait-Ali et al., 1999). It is thus difficult to draw general conclusions from measurements of GA levels or GA 20-oxidase expression levels from samples harvested at just a single time point.
In an attempt to understand some of the factors affecting GA biosynthesis and the photoperiodic control of tuber induction in potato, we have performed some detailed northern analysis on one of the three GA 20-oxidases (StGA20ox1) that were recently cloned from potato (Carrera et al., 1999). As the principle site of photoperiodic perception is young mature leaves, rather than the apex or other parts of the plant, we chose to analyze StGA20ox1 because it is the only one of the three clones that shows significant expression in the leaves. Through comparisons between WT plants and transgenic plants antisensed for the potato PHYB1 gene (that have reduced levels of PHYB1 and possibly also PHYB2), we show that PHYB and possibly a blue-light photoreceptor are involved in regulating the transcript levels of this gene.
RESULTS
StGA20ox1 Transcript Levels Are Higher in PHYB Antisense Plants Than WT
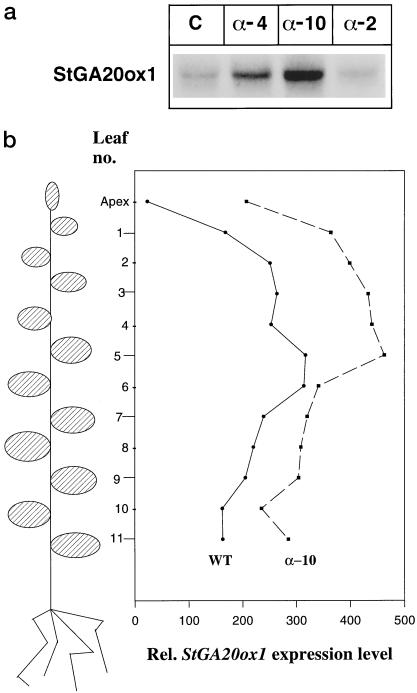
In a preliminary experiment the levels of StGA20ox1 transcript were examined on a northern blot of leaf tissue harvested around mid-day from WT control, antisense PHYB 4 (α-4), antisense PHYB 10 (α-10), and antisense PHYB 2 (α-2) plants. α-4 and α-10 are transgenic plants that have greatly reduced levels of PHYB and which are able to tuberize in LD (Jackson et al., 1996). α-2 is a transgenic plant that has a slight reduction in PHYB levels yet is unable to tuberize in LD and thus behaves as WT control plants. Figure 1a shows the levels of StGA20ox1 transcript in those plants at that particular time point, the levels in α-4 and α-10 being much higher than in WT and α-2 plants.
Figure 1.
a, Expression of StGA20ox1 in WT control (C), antisense PHYB 4 (α-4), antisense PHYB 10 (α-10), and antisense PHYB 2 (α-2) plants. b, Expression levels of StGA20ox1 relative to the S4 ribosomal protein gene in the apex and in leaves of increasing age down the plant.
Looking in more detail at transcript levels throughout the plant we harvested samples from the apex and then from individual leaves all the way down the plant from WT and α-10 plants. The relative levels of StGA20ox1 transcript compared with a constitutively expressed ribosomal protein gene (S4) were calculated. As shown in Figure 1b, higher StGA20ox1 transcript levels were observed in the antisense PHYB plants compared with WT plants all the way down the plant from the apex to leaves that had just started to senesce. Furthermore, transcript levels were found to vary with the age of the leaf in both types of plant. These results demonstrate that the variation in transcript levels in leaves of different ages is an important factor to be considered in the analysis of StGA20ox1 expression, therefore in all subsequent experiments we only harvested the first three fully mature leaves (usually leaf nos. 3, 4, and 5 counting down from the apex).
The Levels of StGA20ox1 Transcript Fluctuate throughout the Day
As previous time course analysis of the StGA20ox1 gene (Carrera et al., 1999), and also of a GA 20-oxidase from pea (Ait-Ali et al., 1999), had found differences in transcript levels over a 24-h time period we decided to look at fluctuations in StGA20ox1 transcript levels over the course of a day.
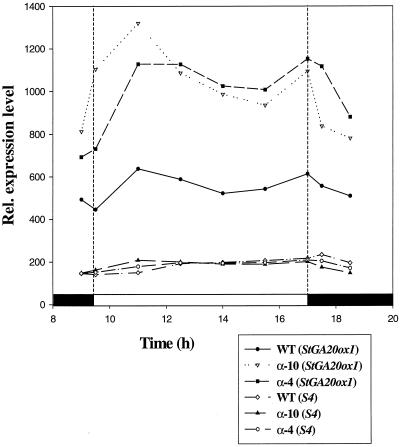
In the first experiment we looked at StGA20ox1 transcript levels in WT and antisense PHYB plants (both α-4 and α-10) over a short (8 h) photoperiod. Harvesting every 1.5 h, we obtained the time course shown in Figure 2 starting before lights on and continuing through into the following dark period. Samples for time points during the dark period were harvested using a dim green safe light (for durations of less than 5 min), which does not elicit phytochrome responses. Consistent with our previous observations the transcript levels of StGA20ox1 were much higher in the antisense PHYB plants compared with WT plants throughout the time-course experiment. Transcript levels showed a strong induction by the light-on signal, and this occurred in the antisense PHYB plants as well as the WT plants suggesting that PHYB may not be necessary for this induction to occur. After this initial induction the transcript levels fall and then start to rise again until lights off when they fall again. This cycling in the levels of StGA20ox1 transcript and the fairly rapid down-regulation in the dark implies that the transcript is turned over rapidly and is amenable to fine control.
Figure 2.
Time course of the relative expression level of StGA20ox1 in WT, α-4, and α-10 plants over a short (8-h) photoperiod. The white bar denotes the light period. Also shown is the variation in S4 transcript levels in these tissues in the different plants over the time course.
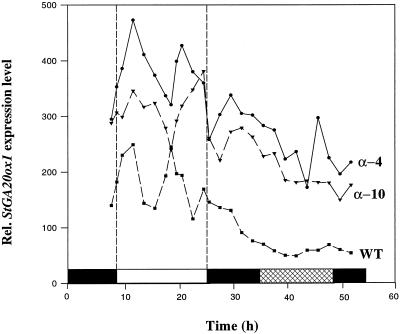
The cycling of StGA20ox1 transcript levels that we observed in the SD time course is even more apparent in the second experiment where the time course consisted of a LD followed by continuous darkness. Samples from WT, α-4, and α-10 plants were harvested as before, and the StGA20ox1 transcript levels relative to the S4 gene were calculated and are shown in Figure 3. The levels in α-4 and α-10 are very similar, and consistent with previous observations, the transcript levels of StGA20ox1 are much higher in the antisense PHYB plants compared with WT. Again we observe the induction by the light-on signal in both types of plant and also the down-regulation in the dark. During the extra 8 h of light in this LD time course, compared with the previous SD time course, we observe a second peak in levels and possibly the start of a third peak in WT plants. Being able to distinguish two complete peaks in the longer time course enables a second difference between WT and antisense PHYB plants to be seen. The period of the cycles of StGA20ox1 transcript levels appears to be longer in the antisense PHYB plants, the second peak in levels in WT plants occurring at the same time as the first trough in the antisense PHYB plants. To verify this, however, a more sensitive assay would probably be needed such as promoter-reporter gene analysis.
Figure 3.
Time course of the relative expression level of StGA20ox1 (in WT, α-4, and α-10 plants) over a long (16-h) photoperiod followed by continuous darkness. The white bar denotes the light period, the hatched bar represents the next subjective day during the subsequent dark period.
What Is the Cause of the Cycling of StGA20ox1 Transcript Levels?
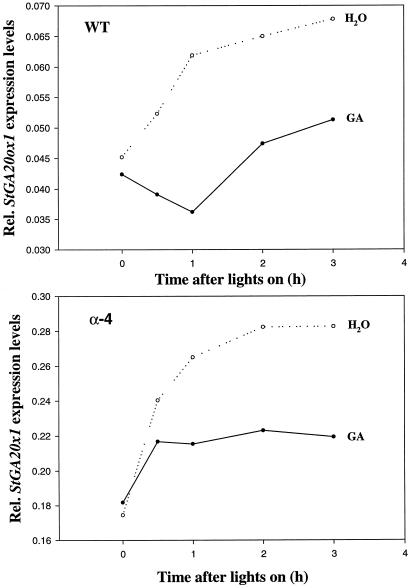
The levels of StGA20ox1 transcript are down-regulated in the dark although some cycling does appear to persist (Fig. 3), especially in the antisense PHYB plants. As StGA20ox1 transcript levels cycle in both antisense PHYB and WT plants, the cause of the rhythm is present in both types of plant even though the period of the rhythm appears to be different. The cycling of StGA20ox1 transcript levels may be explained by the feedback inhibition of GA 20-oxidases by GA1. The fact that cycling is observed in the antisense PHYB plants as well as WT plants implies that if the cycling is due to negative feedback by GA1 then this feedback control is still present in the antisense PHYB plants. This was shown in an experiment where WT and α-4 transgenic plants were sprayed to run off with 50 μm gibberellic acid (GA3), or water, on d 1 and then again 30 mins before the start of the light period on the following day. The levels of StGA20ox1 transcript were monitored after spraying on d 2 just before lights on and for the first 3 h of the light period. The reduced levels of StGA20ox1 transcript in WT and α-4 plants sprayed with GA3 is shown in Figure 4, confirming that the negative feedback mechanism is still present in the PHYB-deficient plants.
Figure 4.
Relative expression levels of StGA20ox1 in WT and α-4 plants treated with water or 50 μm GA3.
It may be possible that the cycling of StGA20ox1 is due to some input from the circadian clock, although the period of the rhythm is very short for that. When StGA20ox1 transcript levels were analyzed in plants after growing them in constant light for 3 d, no cycling was observed, and they were maintained at a constant high level (data not shown). Therefore, if the cycling is the result of a circadian rhythm, the rhythm has a very short period and damps out fairly quickly.
Reduced Levels of Phytochrome A (PHYA) Does Not Affect the Light Induction of StGA20ox1
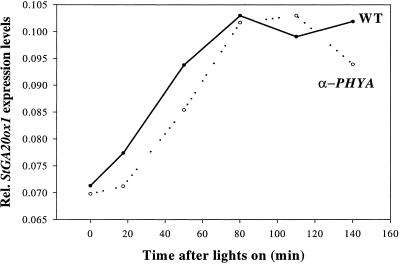
Although PHYB is involved in controlling overall levels of StGA20ox1 transcript, the reduced levels of PHYB in the antisense plants does not appear to affect the diurnal regulation of the levels of this transcript by light, the induction by the light-on signal, and the repression in the dark occurring in a similar manner to WT plants. To assess the potential involvement of PHYA, we used potato plants (cv Désirée) that are antisensed for the PHYA gene (Heyer et al., 1995). We grew these antisense PHYA plants (line Ap9) and WT cv Désirée plants in the same conditions as before and looked at the StGA20ox1 transcript levels in these plants during the first couple of hours after lights on. As is shown in Figure 5, the rate of induction of StGA20ox1 transcript levels is similar in both antisense PHYA and WT plants, suggesting that PHYA is probably also not involved in the induction of this gene by light.
Figure 5.
Relative levels of StGA20ox1 induction by light in WT and antisense PHYA (α-PHYA) plants.
StGA20ox1 Transcript Levels Are Induced by Blue Light
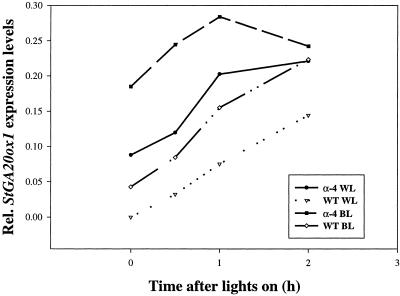
To test whether the levels of StGA20ox1 transcript are affected by blue light we grew potato plants in a cabinet fitted with a blue filter that only transmitted light of wavelengths between 400 and 500 nm. The filters reduced the light levels to 10 μmol m−2 s−1 and so control plants were grown under similar levels of white light. The induction of StGA20ox1 was analyzed for the first couple of hours after lights on in WT and antisense PHYB plants grown under white and blue light. Figure 6 shows that blue light can induce the levels of StGA20ox1 transcript in both WT and antisense PHYB plants. Whereas the levels of StGA20ox1 transcript are higher in the antisense PHYB plants compared with WT plants as has been observed in all previous experiments, it appears that blue light can also result in an increased level of StGA20ox1 transcript in both antisense PHYB and WT plants compared with white light.
Figure 6.
Relative levels of StGA20ox1 in WT and α-4 plants upon induction by white (WL) and blue (BL) light.
In similar experiments using red filters we were not able to obtain a consistent induction of StGA20ox1 by red light, and in some cases a decrease in the level of expression was observed (data not shown).
DISCUSSION
GA 20-oxidase genes have now been isolated from several species, and in both spinach and Arabidopsis the transcript levels of at least one of the GA 20-oxidase genes was shown to be up-regulated in LD (Wu et al., 1996; Xu et al., 1997). These analyses, however, were based on single time point measurements that may be difficult to interpret in the light of these and previous results, showing that the transcript levels of a GA 20-oxidase from potato (StGA20ox1) cycles throughout the day and furthermore that the degree of cycling is variable (Carrera et al., 1999; Figs. 2 and 3). Analysis of this gene in different photoperiods found no difference in transcript levels between LD and SD, a result supported by gas chromatography-mass spectrometry analysis of [14C]GA12 feeding studies of potato plants grown in LD and SD (Van den Berg et al., 1995b). However, the gene was expressed for a longer period in LD than SD and was also up-regulated by a light treatment in the middle of the night (Carrera et al., 1999), indicating that the gene is regulated by light rather than photoperiod in potato plants. The difference between potato and spinach and Arabidopsis may lie in the fact that the latter two are LD plants whereas potato is a SD plant or in the fact that both spinach and Arabidopsis “bolt” in response to photoperiod whereas potato does not.
PHYB is involved in controlling the level of StGA20ox1 gene transcript in potato. Increased levels of expression are observed in PHYB-deficient antisense transgenic plants in all leaves of the plant and at all times of the day and night. The increased levels of StGA20ox1 mRNA could possibly lead to higher levels of GA20 and GA1, and this may explain some of the observed phenotypes of the PHYB antisense plants such as increased internode elongation and reduced chlorophyll levels (Jackson et al., 1996), both of which can be caused by increased GA levels. Whether the increased levels of StGA20ox1 transcript are responsible for the reduced photoperiodic sensitivity of the PHYB antisense plants enabling them to tuberize in LD is still to be determined. It is known, however, that a PHYB mutant of sorghum, the S. bicolor ma3R mutant, which also exhibits a reduced sensitivity to photoperiod (Pao and Morgan, 1986), has higher levels of GA20 and reduced levels of GA53 than WT plants (Foster and Morgan, 1995; Lee et al., 1998). These authors also showed that the levels of all GAs, from GA12 to GA1, cycle throughout the day and night in sorghum and furthermore that a shift in the rhythm of GA20 levels is reported in the PHYB-deficient ma3R mutant compared with WT sorghum plants. This reflects our observations of the longer period of StGA20ox1 cycling in the PHYB antisense potato plants compared with WT. Thus there are striking similarities in the effects of PHYB deficiency on StGA20ox1 transcript levels in potato and on GA levels in S. bicolor, both potato and S. bicolor being SD plants.
It is known that GA 20-oxidase genes are subject to feedback inhibition by active GAs further down the pathway such as GA1 and are accordingly expressed at higher levels in GA-deficient mutants (Phillips et al., 1995; Xu et al., 1997; Carrera et al., 1999). It is possible that the cycling of StGA20ox1 transcript levels are caused by a build-up of GA1 that inhibits expression of StGA20ox1. This in turn would lead to reduced levels of GA1 and a resulting increase of StGA20ox1 transcript. It should be noted that whereas the cycling of StGA20ox1 transcript levels has been a consistent observation, the degree of cycling does vary between experiments (compare Figs. 2 and 3). This may reflect different GA statuses of the plants in the different experiments, perhaps caused by different ages of the plants at the time of the experiments.
Whereas the feedback inhibition of GA 20-oxidase expression is still present in the antisense PHYB plants, the increased transcript levels in these plants may be explained by a reduced level of feedback inhibition by GA1, resulting in overall higher levels of StGA20ox1 mRNA. Such a reduced feedback mechanism in the PHYB antisense plants and phyB mutants may explain why they appear to have a greater response to GAs (Weller et al., 1994; Lopez-Juez et al., 1995; Reed et al., 1996). If the oscillations in StGA20ox1 transcript levels are due to feedback inhibition, a weaker feedback mechanism may also explain why the cycling of StGA20ox1 is different in the antisense PHYB plants compared with WT. Reduced levels of PHYB do not affect other aspects of the control of StGA20ox1 mRNA levels such as the induction by light or the down-regulation in the dark. Likewise, reduced levels of PHYA also do not affect the induction of StGA20ox1 transcript by light, which is consistent with the fact that PHYA antisense potato plants do not exhibit elongated internodes and reduced chlorophyll levels in white light as are observed in PHYB antisense plants. A photoreceptor other than PHYA or PHYB is probably responsible for the light induction of this GA 20-oxidase.
As we observed no induction by red light, an observation also reported for the two GA 20-oxidase genes from lettuce Ls20ox1 and Ls20ox2 (Toyomasu et al., 1998), it suggests that none of the phytochromes are involved in the induction of GA 20-oxidase by light. This is in contrast to the findings of Ait-Ali et al. (1999) who observed induction of a GA 20-oxidase in pea under their red light conditions. This induction is also observed in PHYA-deficient and PHYB-deficient mutants of pea, indicating that as in potato these phytochromes are unlikely to be involved in the induction of the GA 20-oxidase by light and leading the authors to propose that other phytochromes may be involved. That blue light alone can induce the levels of StGA20ox1 transcript to the same, if not greater, extent supports this conclusion and implies that a blue light receptor may be involved. If a blue light receptor does play a role in the control of StGA20ox1 transcript levels this may explain some of the differences in expression pattern that have been observed. In previous experiments where incandescent lights were used, a different time course with a much slower induction by light was observed (Carrera et al., 1999). This weaker induction may be explained by the lower levels of blue light present in the incandescent lights compared with the fluorescent lights we have used in our experiments, which have greater amounts of blue light and induce StGA20ox1 much more rapidly. The higher levels of expression in blue light compared with white light can either be explained by a reduced level of feedback inhibition in blue light or alternatively by some degree of inhibition by a component of white light other than the 400- to 500- nm wavelengths transmitted through the blue filter.
We have demonstrated that PHYB levels can affect StGA20ox1 transcript levels, however, it is clear that other photoreceptors are also likely to be involved in the control of this gene and of other genes involved in GA biosynthesis, which would at least in part explain how light quality can have such diverse and dramatic effects on plant development.
MATERIALS AND METHODS
Plant Material and Growth Conditions
WT potato (Solanum tuberosum cv Désirée), and the photoperiodic S. tuberosum L. subsp. andigena WT line 7540 were obtained from the Institute für Pflanzenbau und Pflanzenzüchtung Bundesforschungsanstalt für Landwirtschaft Braunschweig-Volkenrode (Braunschweig, Germany). The antisense PHYB transgenic lines were produced as described by Jackson et al. (1996). These antisense lines are deficient in PHYB1, however they may also have reduced levels of PHYB2 and therefore are referred to as antisense PHYB lines. The antisense PHYA transgenic lines were produced as described in Heyer et al. (1995). Plants were derived from in vitro grown plants that had been planted out into soil and then subsequently propagated through stem cuttings. The plants were grown in soil in growth cabinets (Sanyo, Gallenkamp PLC, Loughborough, UK) under cool-white Pluslux 3,500 fluorescent tubes (Thorn, Borehamwood, Herts, UK) at light levels of 100 μmol m−2 s−1 and at constant 70% humidity and 22°C. The blue filter (no. 119, Dark Blue) was obtained from LEE Filters Ltd. (Andover, UK).
For each time point or leaf position, two samples were taken from duplicate plants (i.e. four leaves, two per plant) and combined before RNA extraction in an attempt to average out differences between leaves and plants and to obtain a more representative picture of the effects of the light environment on StGA20ox1 transcript levels. Each data point therefore represents the average transcript level from four separate leaves and two different plants.
Northern-Blot Analysis
RNA was extracted from leaves as described (Logemann et al., 1987). Thirty micrograms of total RNA was loaded per track and run out on agarose/formaldehyde gels. The RNA was blotted onto a nylon membrane and hybridized with a radioactively labeled (Rediprime kit, Amersham Life Science, Buckinghamshire, UK) 180-bp StGA20ox1 probe fragment that specifically recognizes StGA20ox1 and no other potato GA 20 oxidase (fragment 7 in Carrera et al., 1999). Hybridization conditions were as described by Amasino (1986), filters were washed twice in 3× SSC, 0.5% (w/v) SDS, at 60°C. The strength of the signal on the filters were analyzed with a phosphor imager using ImageQuant software (version 5.0, Molecular Dynamics, Sunnyvale, CA).
To correct for any differences in the amounts of RNA loaded in each sample, the northern blot was subsequently hybridized with a probe to the constitutively expressed S4 ribosomal protein gene of potato (Braun et al., 1994), and the relative levels of StGA20ox1 transcript were calculated. Due to the uniformity of the tissue analyzed (only leaf tissue and in most experiments just the first three fully expanded leaves were sampled), and the spectrophotometric quantitation of the amount of RNA loaded, we observed little variation in S4 gene transcript levels in our experiments (Fig. 2). The S4 signal levels obtained from the northern blots correlated well with the levels of RNA in the ethidium bromide-stained gels.
ACKNOWLEDGMENT
We are very grateful to Christine Richardson for her maintenance of the plants in tissue culture.
Footnotes
This work was supported by the Biotechnology and Biological Science Research Council and the Comision Interministerial de Ciencia y Tecnologia Plan Nacional (grant no. BIO96–0532–C02–02).
LITERATURE CITED
- Ait-Ali T, Frances S, Weller JL, Reid JB, Kendrick RE, Kamiya Y. Regulation of gibberellin 20-oxidase and gibberellin 3b-hydroxylase transcript accumulation during de-etiolation of pea seedlings. Plant Physiol. 1999;121:783–791. doi: 10.1104/pp.121.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino RM. Acceleration of nucleic acid hybridization rate by polyethylene glycol. Ann Biochem. 1986;152:304–307. doi: 10.1016/0003-2697(86)90413-6. [DOI] [PubMed] [Google Scholar]
- Braun HP, Emmerman H, Mentzel H, Schmitz UK. Primary structure and expression of a gene encoding the cytosolic ribosomal protein S4 from potato. Biochim Biophys Acta. 1994;1218:435–438. doi: 10.1016/0167-4781(94)90200-3. [DOI] [PubMed] [Google Scholar]
- Carrera E, Jackson SD, Prat S. Potato GA 20-oxidases show a diurnal rythmicity that is altered by a short interruption of the dark period. Plant Physiol. 1999;119:765–773. doi: 10.1104/pp.119.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KR, Miller FR, Childs KL, Morgan P. Genetic regulation of development in Sorghum bicolor: VIII. Shootgrowth, tillering, flowering, gibberellin biosynthesis and phytochrome levels are differentially affected by dosage of the ma3R allele. Plant Physiol. 1994;105:941–948. doi: 10.1104/pp.105.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KR, Morgan PW. Genetic regulation of development in Sorghum bicolor: IX. The ma3R allele disrupts diurnal control of gibberellin biosynthesis. Plant Physiol. 1995;108:337–343. doi: 10.1104/pp.108.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Lopez-Diaz I, Sanchez-Beltran MJ, Phillips AL, Ward DA, Gaskin P, Hedden P. Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol. 1997;33:1073–1084. doi: 10.1023/a:1005715722193. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Bleecker AB, Zeevaart JAD. Partial purification of gibberellin oxidases from spinach leaves. Plant Physiol. 1987;85:87–90. doi: 10.1104/pp.85.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zeevaart JAD, Schwenen L, Graebe JE. Gibberellin metabolism in cell-free extracts from spinach leaves in relation to photoperiod. Plant Physiol. 1986;82:190–195. doi: 10.1104/pp.82.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Croker SJ. Regulation of gibberellin biosynthesis in maize seedlings. In: Karssen CM, van Loon LC, Vreugdenhil D, editors. Progress in Plant Growth Regulation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 534–544. [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Heyer AG, Mozley D, Landschutze V, Thomas B, Gatz C. Function of phytochrome A in potato plants as revealed through the study of transgenic plants. Plant Physiol. 1995;109:53–61. doi: 10.1104/pp.109.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SD, Heyer A, Dietze J, Prat S. Phytochrome-B mediates the photoperiodic control of tuber formation in potato. Plant J. 1996;9:159–166. [Google Scholar]
- Jackson SD, Prat S. Control of tuberization in potato by gibberellins and phytochrome B. Physiol Plant. 1996;98:407–412. [Google Scholar]
- Kamiya Y, Garcia-Martinez JL. Regulation of gibberellin biosynthesis by light. Curr Opin Plant Biol. 1999;2:398–403. doi: 10.1016/s1369-5266(99)00012-6. [DOI] [PubMed] [Google Scholar]
- Lee I-J, Foster KR, Morgan PW. Photoperiod control of gibberellin levels and flowering in sorghum. Plant Physiol. 1998;116:1003–1011. doi: 10.1104/pp.116.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Ann Biochem. 1987;163:21–26. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Kobayashi M, Sakurai A, Kamiya Y, Kendrick RE. Phytochrome, gibberellins and hypocotyl growth. Plant Physiol. 1995;107:131–140. doi: 10.1104/pp.107.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao C-I, Morgan PW. Genetic regulation of development in Sorghum bicolor. Plant Physiol. 1986;82:581–584. doi: 10.1104/pp.82.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Railton ID, Wareing PF. Effects of daylength on endogenous gibberellin levels in leaves of Solanum andigena. Physiol Plant. 1973;28:88–94. doi: 10.1007/BF00386032. [DOI] [PubMed] [Google Scholar]
- Reed JW, Foster KR, Morgan PW, Chory J. Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol. 1996;112:337–342. doi: 10.1104/pp.112.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood SB, Williams PH, Pearce D, Murofushi N, Pharis P. A mutant gene that increases gibberellin production in Brassica rapa. Plant Physiol. 1990;93:1168–1174. doi: 10.1104/pp.93.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M, Zeevaart JAD, Gage DA. Identification of gibberellins in spinach and effects of light and darkness on their levels. Plant Physiol. 1991;97:1521–1526. doi: 10.1104/pp.97.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y. Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 1998;118:1517–1523. doi: 10.1104/pp.118.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg JH, Davies PJ, Ewing EE, Halinska A. Metabolism of gibberellin A12 and A12-aldehyde and the identification of endogenous gibberellins in potato (Solanum tuberosum ssp. andigena) shoots. J Plant Physiol. 1995a;146:459–466. [Google Scholar]
- Van den Berg JH, Simko I, Davies PJ, Ewing EE, Halinska A. Morphology and [14C]-gibberellin A12 metabolism in wild-type and dwarf Solanum tuberosum ssp. andigena grown under long and short photoperiods. J Plant Physiol. 1995b;146:467–473. [Google Scholar]
- Van Huizen R, Ozga JA, Reinecke DM. Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiol. 1997;115:123–128. doi: 10.1104/pp.115.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Ross JJ, Reid JB. Gibberellins and phytochrome regulation of stem elongation in pea. Planta. 1994;192:489–496. [Google Scholar]
- Wu K, Li L, Gage DA, Zeevaart JAD. Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach. Plant Physiol. 1996;110:547–554. doi: 10.1104/pp.110.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gage DA, Zeevaart JAD. Gibberellins and stem growth in Arabidopsis thaliana. Plant Physiol. 1997;114:1471–1476. doi: 10.1104/pp.114.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun T-P. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1998;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]