Abstract
Hydroxamate groups play key roles in the biological function of diverse natural products. Important examples include trichostatin A, which inhibits histone deacetylases via coordination of the active site zinc(II) ion with a hydroxamate group, and the desferrioxamines, which use three hydroxamate groups to chelate ferric iron. Desferrioxamine biosynthesis in Streptomyces species involves the DesD-catalysed condensation of various N-acylated derivatives of N-hydroxycadaverine with two molecules of N-succinyl-N-hydroxycadaverine to form a range of linear and macrocyclic tris-hydroxamates. However, the mechanism for assembly of the various N-acyl-N-hydroxycadaverine substrates of DesD from N-hydroxycadaverine has until now been unclear. Here we show that the desC gene of Streptomyces coelicolor encodes the acyl transferase responsible for this process. DesC catalyses the N-acylation of N-hydroxycadaverine with acetyl, succinyl and myristoyl-CoA, accounting for the diverse array of desferrioxamines produced by S. coelicolor. The X-ray crystal structure of DesE, the ferrioxamine lipoprotein receptor, in complex with ferrioxamine B (which is derived from two units of N-succinyl-N-hydroxycadaverine and one of N-acetyl-N-hydroxycadaverine) was also determined. This showed that the acetyl group of ferrioxamine B is solvent exposed, suggesting that the corresponding acyl group in longer chain congeners can protrude from the binding pocket, providing insights into their likely function. This article is part of a discussion meeting issue ‘Frontiers in epigenetic chemical biology'.
This article is part of a discussion meeting issue ‘Frontiers in epigenetic chemical biology’.
Keywords: siderophore, hydroxamic acid, hydroxylamine, acyl-coenzyme A, Streptomyces coelicolor
1. Introduction
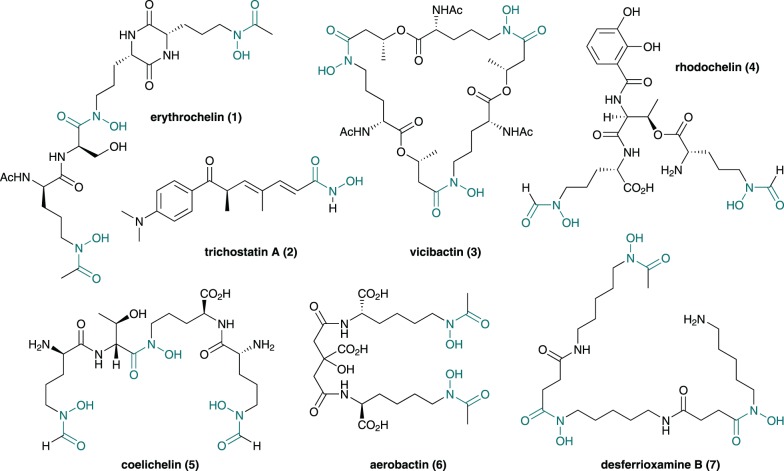
A range of structurally diverse microbial natural products contain one or more hydroxamate functional groups (figure 1), which are employed as metal ion chelators to inhibit or enable important biological processes [1]. Prominent examples include trichostatin A (2), a histone deacetylase (HDAC) inhibitor with anti-cancer activity that is widely used in epigenetics research [2], and desferrioxamine B (7), which is marketed as desferral for the treatment of iron overload and neuroblastoma in humans [3,4]. Trichostatin A blocks histone deacetylation via chelation of its hydroxamate group to the active zinc(II) ion of HDACs [5], whereas desferrioxamine B uses its three hydroxamate groups to sequester ferric iron via formation of a hexadentate complex [6]. Interestingly, desferrioxamine B (and other iron chelators) have been reported to enhance the anti-cancer activity of trichostatin A [7].
Figure 1.
Chemical structures of some bacterial natural products containing hydroxamate groups (in grey). (Online version in colour.)
Along with several other bis- and tris-hydroxamates, such as coelichelin (5), erythrochelin (1), vicibactin (3), aerobactin (6) and rhodochelin (4) (figure 1), and other members of the desferrioxamine complex (figure 2), desferrioxamine B (7) plays an important role in microbial iron uptake [8–11]. Iron is an essential nutrient for most forms of life because it plays a central role in several key metabolic processes. Although iron is the fourth most abundant element on the Earth, its availability in aqueous environments is extremely low owing to the insolubility of ferric oxide and hydroxide complexes [12,13]. As a consequence, saprophytic microorganisms excrete small molecules known as siderophores that scavenge iron from the environment [14]. The resulting ferric-siderophore complexes are taken up by the microbial cells using ATP-dependent transport mechanisms and the iron is released via reduction or degradation of the ligand [15]. One well-studied example is the model antibiotic-producing actinobacterium Streptomyces coelicolor A3(2), which biosynthesizes and excretes coelichelin (5), desferrioxamine B (7) and desferrioxamine E (8) [11]. Distinct cell surface-associated lipoprotein receptor components of ATP-binding cassette (ABC) transporters (CchF and DesE, respectively) are used by S. coelicolor to import the ferricoelichelin and ferrioxamine complexes [16].
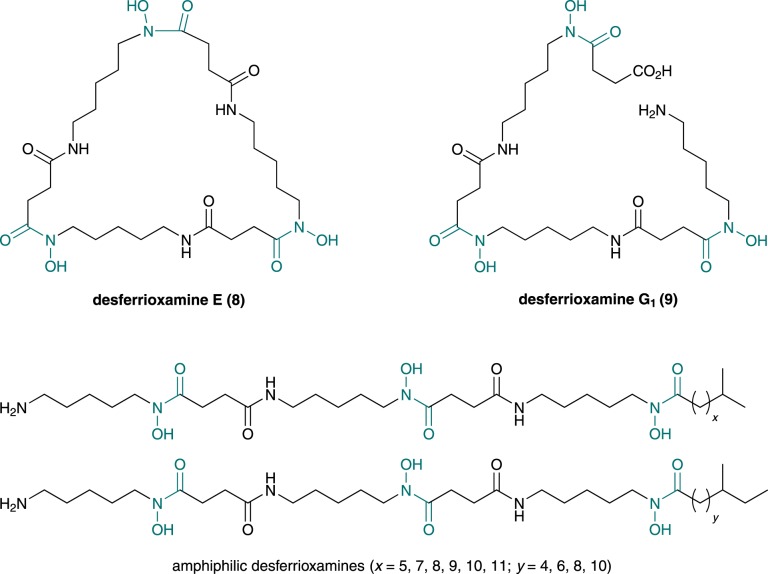
Figure 2.
Chemical structures of desferrioxamine E and G1, and the amphiphilic desferrioxamines reported to be produced, along with desferrioxamine B, by S. coelicolor. Hydroxamate groups are in grey. (Online version in colour.)
Desferrioxamine E (8) and several other siderophores are also used by pathogenic microorganisms to scavenge iron from plant and animal hosts [17–19]. The enzymes involved in the biosynthesis of such molecules thus offer potential as targets for the development of antibacterials. The mechanism for hydroxamate incorporation into several bacterial siderophores has been shown to involve initial oxidation of the amino group in the side chains of ornithine and lysine by flavin-dependent monooxygenases [8–10,20–22]. The resulting hydroxylamines undergo formylation by formyl tetrahydrofolate-dependent formyl transferases (e.g. in rhodochelin (4) [10] and coelichelin (5) [20] biosynthesis), acylation by acyl-CoA-dependent acyl transferases (e.g. in aerobactin (6) [23] and vicibactin (3) [9] biosynthesis), or condensation with amino acyl thioesters catalysed by non-ribosomal peptide synthetase (NRPS) assembly lines [8]. By contrast, trichostatin A (2) biosynthesis involves oxidation of the amido group in glutamine to the corresponding hydroxamate by a non-haem di-iron-dependent monooxygenase [24]. An asparagine synthetase homologue then catalyses the hydrolysis of this hydroxamate, followed by ATP-dependent condensation of the resulting hydroxylamine with trichostatic acid (presumably via an acyl adenylate intermediate).
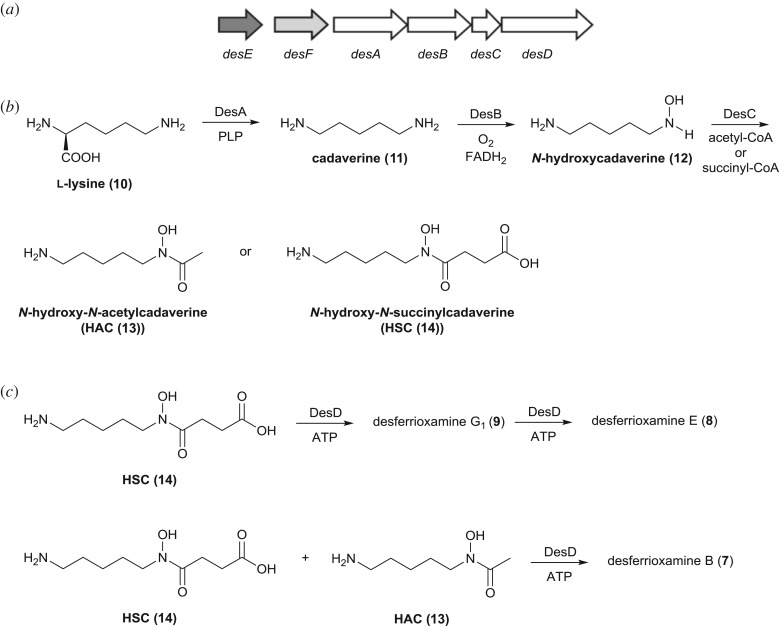
Desferrioxamine E (8) is assembled from three molecules of N-hydroxy-N-succinylcadaverine (HSC (14)) in S. coelicolor by the NRPS-independent siderophore synthetase DesD (figure 3) [21,25,26]. This process involves adenylation of the carboxyl group in one HSC unit followed by condensation with the amino group in another. The resulting homodimeric intermediate is adenylated and condensed with a third molecule of HSC to yield a homotrimer (desferrioxamine G1 (9)). Desferrioxamine E is formed from this homotrimer via a further round of adenylation followed by macrolactamization. DesD assembles desferrioxamine B (7) via condensation of the adenylated HSC homodimer with N-hydroxy-N-acetylcadaverine (HAC (13); figure 3). Although N-hydroxycadaverine (12), arising from DesA-catalysed decarboxylation of l-lysine (10) followed by DesB-catalysed N-hydroxylation of the resulting cadaverine (11), is known to be an intermediate in desferrioxamine biosynthesis (figure 3), the mechanism by which the key iron-chelating hydroxamate groups are incorporated into the desferrioxamines remains to be determined. The desC gene within the desferrioxamine biosynthetic gene cluster (which is highly conserved in many Streptomyces genomes) encodes a putative acyl-CoA-dependent acyl transferase that has been proposed to catalyse the acylation of N-hydroxycadaverine (12) with succinyl- and acetyl-CoA to form HSC (14) and HAC (13), respectively (figure 3) [21]. However, this hypothesis remains experimentally untested and recent reports detailing the production of several desferrioxamine B analogues by S. coelicolor, in which the acetyl group is substituted with various nine- to 15-carbon saturated acyl groups, further complicates the picture [27,28]. Presumably these amphiphilic desferrioxamines arise from the DesD-catalysed condensation of the HSC homodimer with the corresponding N-hydroxy-N-acylcadaverines, suggesting that DesC may be a remarkably substrate-tolerant acyl transferase capable of forming the hydroxamate group in diverse desferrioxamine precursors. Moreover, the reason why S. coelicolor produces so many different desferrioxamine congeners remains unclear. Here we report experimental evidence demonstrating that this is indeed the case. We also report the X-ray crystal structure of DesE (the desferrioxamine lipoprotein receptor) in complex with ferrioxamine B, leading us to propose that the amphiphilic desferrioxamines may function as membrane-bound iron shuttles in desferrioxamine-mediated iron uptake.
Figure 3.
(a) Organization of the desferrioxamine biosynthetic gene cluster in S. coelicolor. Genes encoding the four desferrioxamine biosynthetic enzymes (DesA–D) are white. The desE gene (dark grey) encodes a cell surface-associated lipoprotein receptor component of an ABC transporter involved in ferrioxamine uptake and desF (light grey) encodes a putative ferrioxamine reductase. (b) Previously proposed pathway for the assembly of HSC and HAC by the DesA, DesB and DesC enzymes. (c) DesD-catalysed assembly of desferrioxamine E, via desferrioxamine G1, from three molecules of HSC, and the assembly of desferrioxamine B from two units of HSC and one of HAC.
2. Results and discussion
(a). DesC catalyses N-acylation of N-hydroxycadaverine with diverse acyl-CoA thioesters
To investigate the catalytic properties of DesC, it was overproduced in Escherichia coli as a soluble N-terminal hexahistidine fusion and purified to homogeneity using nickel affinity chromatography (see electronic supplementary material for experimental details). The identity of the purified protein was confirmed by peptide mass fingerprinting and gel filtration chromatography showed that it was monomeric (see electronic supplementary material).
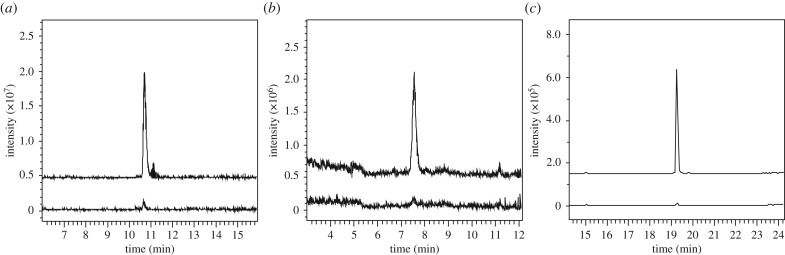
N-hydroxycadaverine (12) was synthesized from N-benzyloxy-N-BOC-cadaverine (15) [26] via hydrogenolytic cleavage of the benzyl ether, followed by removal of the BOC group under acidic conditions (scheme 1; see electronic supplementary material for experimental details). To establish whether DesC is able to catalyse the assembly of HSC (14), the core hydroxamate building block common to all desferrioxamines, the purified recombinant enzyme was incubated with N-hydroxycadaverine (12) and succinyl-CoA for 10 min at 37°C. The reaction was stopped by addition of trichloracetic acid and the mixture was analysed by liquid chromatography–mass spectrometry (LC-MS). A prominent species with m/z = 219, corresponding to the [M + H]+ ion for HSC (14), was observed in these analyses (figure 4). This product was confirmed as HSC by LC-MS/MS comparisons with an authentic synthetic standard, as described previously [26]. The reaction was monitored using a continuous spectroscopic assay employing 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) to detect the production of coenzyme A (see electronic supplementary material). A small quantity of HSC was observed in control reactions from which the enzyme had been omitted (figure 4). This is unsurprising given the high electrophilicity and nucleophilicity of the thioester group in succinyl-CoA and the hydroxylamino group in N-hydroxycadaverine, respectively. Negligible production of coenzyme A was observed in the control reaction using the DTNB assay (see electronic supplementary material). Incubation of DesC with N-hydroxycadaverine (12) and acetyl-CoA resulted in a product with m/z = 161, corresponding to the [M + H]+ ion for HAC (13), in LC-MS analyses (figure 4). As before, this was confirmed as HAC by comparison with an authentic standard [26], coenzyme A was confirmed as the by-product of the reaction using the DTNB assay (see electronic supplementary material) and small quantities of HAC were observed in the no-enzyme control.
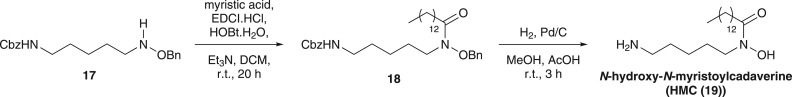
Scheme 1.
Synthesis of N-hydroxycadaverine from N-benzyloxy-N-BOC-cadaverine.
Figure 4.
Extracted ion chromatograms (EICs) from LC-MS analyses of DesC-catalysed reactions (top traces) and control reactions from which the enzyme was omitted (bottom traces). (a) EICs at m/z = 219 (corresponding to [M + H]+ for HSC (14)); (b) EICs at m/z = 161 (corresponding to [M + H]+ for HAC (13)); (c) EICs at m/z = 329 (corresponding to [M + H]+ for HMC (19)).
Having established that DesC is able to biosynthesize not only the HSC (14) building block used by DesD for the assembly of desferrioxamine E (8), but also the HAC (13) unit used, along with HSC, for the assembly of desferrioxamine B (7) [26], we next investigated whether DesC is also able to produce medium chain acyl hydroxamates similar to those incorporated into the amphiphilic desferrioxamines. Thus, DesC was incubated with N-hydroxycadaverine (12) and myristoyl-CoA and the reaction mixture was analysed by LC-MS. A prominent species with m/z = 329, corresponding to the [M + H]+ ion for N-hydroxy-N-myristoylcadaverine (HMC (19)), was detected (figure 4). An authentic standard of HMC was synthesized via EDCI-mediated coupling of N-benzyloxy-N'-(benzyloxycarbonyl)cadaverine (17) [26] with myristic acid to yield 18, followed by deprotection with H2 over Pd/C in MeOH/MeCO2H (scheme 2; see electronic supplementary material for experimental details). The product of the enzymatic reaction and the authentic standard of HMC (19) had identical retention times and fragmentation patterns in LC-MS/MS analyses (see electronic supplementary material). As observed with acetyl- and succinyl-CoA (see above), a small amount of spontaneously generated HMC was present in the negative control (figure 4).
Scheme 2.
Synthesis of an authentic standard of HMC from N-benzyloxy-N'-(benzyloxycarbonyl)cadaverine. CbzHN stands for carboxybenzyl.
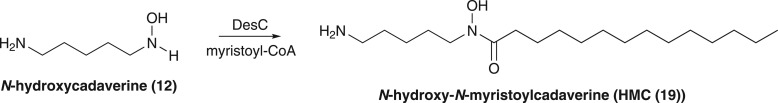
Our data show that DesC is the key hydroxamate-forming enzyme in the desferrioxamine biosynthetic pathway. It catalyses not only the acylation of N-hydroxycadaverine with succinyl-CoA to form HSC, the precursor of desferrioxamines G1, and E, but also the acylation of N-hydroxycadaverine with acetyl-CoA to form HAC, which is used along with HSC for the assembly of desferrioxamine B. Remarkably, DesC is also able to catalyse the acylation of N-hydroxycadaverine with myristoyl-CoA (figure 5), suggesting that it assembles the various N-hydroxy-N-acylcadaverines incorporated into the amphiphilic desferrioxamines. The broad substrate tolerance exhibited by DesC is atypical for acyl transferases involved in hydroxamate biosynthesis (e.g. IucB, which shows a high degree of specificity towards acetyl-CoA) [23].
Figure 5.
DesC-catalysed acylation of N-hydroxycadaverine with myristoyl-CoA to form HMC, suggesting that DesC can assemble the various N-hydroxy-N-acylcadaverines incorporated into the amphiphilic desferrioxamines.
(b). X-ray crystal structure of DesE-ferrioxamine B complex
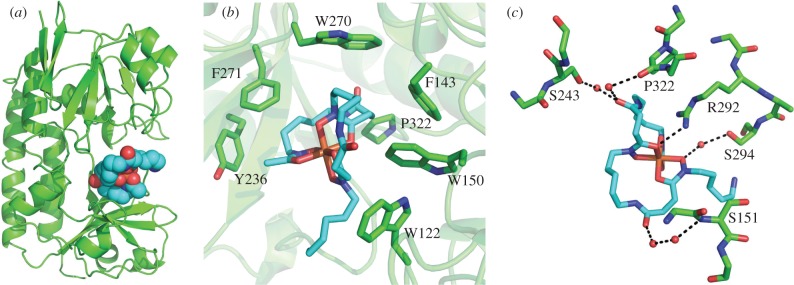
We previously reported the X-ray crystal structure of S. coelicolor DesE [29], an extracellular lipoprotein that binds ferrioxamines E/B (the ferric complexes of the corresponding desferrioxamines) [16], initiating the uptake of these ferric-siderophore complexes via an ABC transporter [11]. To gain further insight into the mechanism of ferrioxamine recognition by DesE, we determined the crystal structure of the DesE-ferrioxamine B complex to 1.96 Å resolution (figure 6; see electronic supplementary material for experimental details). The conformation of DesE in the apo and ferrioxamine B-bound forms is essentially identical (see electronic supplementary material). Visual inspection of the structure shows that the ferric ion is coordinated by the six oxygen atoms of the three hydroxamate groups in desferrioxamine B, as expected, with Fe–O distances between 1.9 and 2.0 Å. The substrate binding site of DesE is lined with the side chains of Trp122, Trp150, Trp270, Phe143, Phe271, Pro322 and Tyr236, which make multiple van der Waals contacts with hydrophobic regions of ferrioxamine B. The only direct polar contact between DesE and ferrioxamine B is mediated by the guanidinium group of Arg292, which is positioned within hydrogen bonding distance of one of the hydroxamate O atoms. Five ordered water molecules mediate indirect polar contacts between Ser243, Ser294, Ser151 and Pro322 of DesE and three of the oxygen atoms in ferrioxamine B. The N-acetyl group of ferrioxamine B points out of the binding cleft. This indicates that DesE can also bind ferric complexes of the amphiphilic desferrioxamines, suggesting they may act as membrane-embedded shuttles that receive ferric ions scavenged from the environment by soluble siderophores, such as desferrioxamines B/E (7/8) and coelichelin (5).
Figure 6.
X-ray crystal structure of DesE (green) bound to ferrioxamine B (cyan). (a) Overall structure of the complex; (b) the ferrioxamine binding site, highlighting the hydrophobic residues lining it; (c) polar contacts between DesE and ferrioxamine B.
3. Conclusion
In this work, we have shown that DesC is a remarkably substrate-tolerant acyl transferase that catalyses the key hydroxamate-forming step in desferrioxamine biosynthesis. DesC is able to catalyse the acylation of N-hydroxycadaverine not only with succinyl and acetyl-CoA to form HSC and HAC, known intermediates in the biosynthesis of desferrioxamines E and B, but also with myristoyl-CoA to form HMC. This provides a rationale for the recently reported production of amphiphilic desferrioxamines by S. coelicolor. While the biological function of these unusual tris-hydroxamates remains unclear, the X-ray crystal structure of the DesE-ferrioxamine B complex suggests that ferric complexes of the amphiphilic desferrioxamines are likely to be recognized and imported by the S. coelicolor ferrioxamine uptake system. The nine to 15-carbon acyl chains in these molecules likely targets them to the extracytoplasmic membrane. It is, therefore, conceivable that ferric iron scavenged from the environment by desferrioxamines E/B and other soluble siderophores is transferred to the membrane-associated ferrioxamines prior to DesE-initiated uptake. Further experiments will be required to test this hypothesis.
Desferrioxamine B is used in the clinic as a treatment for iron overload. It will, therefore, be interesting to investigate whether the broad substrate tolerance of DesC can be harnessed for chemoenzymatic synthesis of novel desferrioxamine B analogues with improved therapeutic properties. DesC may also prove to be a useful tool in the development of biocatalytic approaches for the synthesis of hydroxamate-based HDAC inhibitors.
Supplementary Material
Acknowledgements
Dr Lijiang Song, Dr Ivan Prokes and Mr Philip Aston are thanked for assistance in obtaining spectroscopic data.
Data accessibility
Supporting data and experimental procedures are included in the electronic supplementary material. The coordinates of the DesE-ferrioxamine B complex have been deposited in the Protein Data Bank (accession no. 6ENK).
Authors' contributions
G.L.C. conceived the study and G.L.C., N.K., J.L.R., S.A.M. and J.H.N. designed the experiments. N.K., J.L.R. and S.A.M. acquired the data, and all authors contributed to its analysis and interpretation. G.L.C., L.M.A, N.K., J.L.R. and S.A.M. drafted the article and all authors approved the content.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by grants from the BBSRC (BB/S/B14450 to J.H.N. and G.L.C., and BB/L502017/1 to G.L.C.) and a Chancellor's Scholarship from the University of Warwick (to J.L.R.). G.L.C. is the recipient of a Wolfson Research Merit Award (WM130033) from the Royal Society.
References
- 1.Bertrand S, Helesbeux JJ, Larcher G, Duval O. 2013. Hydroxamate, a key pharmacophore exhibiting a wide range of biological activities. Mini-Rev. Med. Chem. 13, 1311–1326. ( 10.2174/13895575113139990007) [DOI] [PubMed] [Google Scholar]
- 2.Yoshida M, Kijima M, Akita M, Beppu T. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17 174–17 179. [PubMed] [Google Scholar]
- 3.Blatt J, Stitely S. 1987. Antineuroblastoma activity of desferoxamine in human cell lines. Cancer Res. 47, 1749–1750. [PubMed] [Google Scholar]
- 4.Norman CS. 1964. The treatment of iron overload with desferrioxamine B. Ir. J. Med. Sci. 134, 13–18. [DOI] [PubMed] [Google Scholar]
- 5.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401, 188–193. ( 10.1038/43710) [DOI] [PubMed] [Google Scholar]
- 6.Dhungana S, White PS, Crumbliss AL. 2001. Crystal structure of ferrioxamine B: a comparative analysis and implications for molecular recognition . J. Biol. Inorg. Chem. 6, 810–818. ( 10.1007/s007750100259) [DOI] [PubMed] [Google Scholar]
- 7.Kilinc V, Bedir A, Okuyucu A, Salis O, Alacam H, Gulten S. 2014. Do iron chelators increase the antiproliferative effect of trichostatin A through a glucose-regulated protein 78 mediated mechanism? Tumour Biol. 35, 5945–5951. ( 10.1007/s13277-014-1788-1) [DOI] [PubMed] [Google Scholar]
- 8.Robbel L, Helmetag V, Knappe TA, Marahiel M. 2001. Consecutive enzymatic modification of ornithine generates the hydroxamate moieties of the siderophore erythrochelin. Biochemistry 50, 6073–6080. ( 10.1021/bi200699x) [DOI] [PubMed] [Google Scholar]
- 9.Heemstra JR, Walsh CT, Sattely ES. 2009. Enzymatic tailoring of ornithine in the biosynthesis of the rhizobium cyclic trihydroxamate siderophore vicibactin. J. Am. Chem. Soc. 131, 15 317–15 329. ( 10.1021/ja9056008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosello M, Mielacarek A, Giessen TW, Marahiel MA. 2012. An enzymatic pathway for the biosynthesis of the formylhydroxyornithine required for rhodochelin iron coordination. Biochemistry 51, 3059–3066. ( 10.1021/bi201837f) [DOI] [PubMed] [Google Scholar]
- 11.Barona-Gomez F, Lautru S, Francou F-X, Pernodet J-L, Leblond P, Challis GL. 2006. Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor and Streptomyces ambofaciens. Microbiology 152, 3355–3366. ( 10.1099/mic.0.29161-0) [DOI] [PubMed] [Google Scholar]
- 12.Taylor SR. 1964. Abundance of chemical elements in the continental crust: a new table. Geochim. Cosmochim. Acta 28, 1273–1285. ( 10.1016/0016-7037(64)90129-2) [DOI] [Google Scholar]
- 13.Carrano CJ, Drechsel H, Kaiser D, Jung G, Matzanke B, Winkelmann G, Rochel N, Albrecht-Gary AM. 1996. Coordination chemistry of the carboxylate type siderophore rhizoferrin: the iron(III) complex and its metal analogs. Inorg. Chem. 35, 6429–6436. ( 10.1021/ic960526d) [DOI] [PubMed] [Google Scholar]
- 14.Neilands JB. 1993. Siderophores. Arch. Biochem. Biophys. 302, 1–3. ( 10.1006/abbi.1993.1172) [DOI] [PubMed] [Google Scholar]
- 15.Braun V, Braun M. 2002. Active transport of iron and siderophore antibiotics. Curr. Opin. Microbiol. 5, 194–201. ( 10.1016/S1369-5274(02)00298-9) [DOI] [PubMed] [Google Scholar]
- 16.Patel P, Song L, Challis GL. 2010. Distinct extracytoplasmic siderophore binding proteins recognize ferrioxamines and ferri-coelichelin in Streptomyces coelicolor A3(2). Biochemistry 49, 8033–8042. ( 10.1021/bi100451k) [DOI] [PubMed] [Google Scholar]
- 17.Schaible UE, Kaufmann SHE. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2, 946–953. ( 10.1038/nrmicro1046) [DOI] [PubMed] [Google Scholar]
- 18.Miranda-CasoLuengo R, Coulson GB, Miranda-CasoLuengo A, Vazquez-Boland JA, Hondalus MK, Meijer WG. 2012. The hydroxamate siderophore rhequichelin is required for virulence of the pathogenic actinomycete Rhodococcus equi. Infect. Immun. 80, 4106–4114. ( 10.1128/IAI.00678-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. 2012. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 8, 731–736. ( 10.1038/nchembio.1020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lautru S, Deeth RJ, Bailey LM, Challis GL. 2005. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat. Chem. Biol. 1, 265–269. ( 10.1038/nchembio731) [DOI] [PubMed] [Google Scholar]
- 21.Barona-Gomez F, Wong U, Giannakopulos A, Derrick PJ, Challis GL. 2004. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 126, 16 282–16 283. ( 10.1021/ja045774k) [DOI] [PubMed] [Google Scholar]
- 22.Thariath A, Socha D, Valvano MA, Viswanatha T. 1993. Construction and biochemical characterization of recombinant cytoplasmic forms of the IucD protein (lysine:N6-hydroxylase) encoded by the pColV-K30 aerobactin gene cluster. J. Bacteriol. 175, 589–596. ( 10.1128/jb.175.3.589-596.1993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coy M, Paw BH, Bindereif A, Neilands JB. 1986. Isolation and properties of N.epsilon.-hydroxylysine:acetyl coenzyme A N.epsilon.-transacetylase from Escherichia coli pABN11. Biochemistry 25, 2485–2489. ( 10.1021/bi00357a030) [DOI] [PubMed] [Google Scholar]
- 24.Kudo K, Ozaki T, Shin-ya K, Nishiyama M, Kuzuyama T. 2017. Biosynthetic origin of the hydroxamic acid moiety of trichostatin A: identification of unprecedented enzymatic machinery involved in hydroxylaminetransfer. J. Am. Chem. Soc. 139, 6799–6802. ( 10.1021/jacs.7b02071) [DOI] [PubMed] [Google Scholar]
- 25.Oves-Costales D, Kadi N, Challis GL. 2009. The long-overlooked enzymology of a nonribosomal-peptide synthetase independent pathway for virulence-conferring siderophore biosynthesis. Chem. Commun. 2009, 6530–6541. ( 10.1039/b913092f) [DOI] [PubMed] [Google Scholar]
- 26.Kadi N, Oves-Costales D, Barona-Gomez F, Challis GL. 2007. A new family of ATP-dependent oligomerization-macrocyclization biocatalysts. Nat. Chem. Biol. 3, 652–656. ( 10.1038/nchembio.2007.23) [DOI] [PubMed] [Google Scholar]
- 27.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. 2013. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 4, e00459-13 ( 10.1128/mBio.00459-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidebottom AM, Johnson AR, Karty JA, Trader DJ, Carlson EE. 2013. Integrated metabolomics approach facilitates discovery of an unpredicted natural product suite from Streptomyces coelicolor M145. ACS Chem. Biol. 8, 2009–2016. ( 10.1021/cb4002798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oke M, et al. 2010. The Scottish Structural Proteomics Facility: targets, methods and outputs. J. Struct. Funct. Genomics 11, 167–180. ( 10.1007/s10969-010-9090-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supporting data and experimental procedures are included in the electronic supplementary material. The coordinates of the DesE-ferrioxamine B complex have been deposited in the Protein Data Bank (accession no. 6ENK).