Abstract
Within the past two decades, seven epigenetic drugs have received regulatory approval and numerous other candidates are currently in clinical trials. Among the epigenetic targets are the writer and eraser enzymes that are, respectively, responsible for the reversible introduction and removal of structural modifications in the nucleosome. This review discusses the progress achieved in the design and development of inhibitors against the key writer and eraser pairs: DNA methyltransferases and Tet demethylases; lysine/arginine methyltransferases and lysine demethylases; and histone acetyltransferases and histone deacetylases. A common theme for the successful inhibition of these enzymes in a potent and selective manner is the targeting of the cofactors present in the active site, namely zinc and iron cations, S-adenosylmethione, nicotinamide adenine dinucleotide, flavin adenine dinucleotide and acetyl Coenzyme A.
This article is part of a discussion meeting issue ‘Frontiers in epigenetic chemical biology’.
Keywords: epigenetics, enzyme inhibitors, cofactors
1. Readers, writers and erasers
Epigenetics is defined as a change in phenotype without an underlying change in genotype. As succinctly defined in their dictionary Aristotle to Zoos, Medawar and Medawar say it encompasses ‘… all the processes that go into implementation of the genetic instructions contained within the fertilized egg. Genetics proposes; epigenetics disposes' [1, pp. 113–114]. At the molecular level, the epigenetic modulation of gene expression is accomplished through structural modifications of the nucleosome [2,3]. The reversibility of these pathways is of great interest for therapeutic applications, because a disease state can potentially be epigenetically reprogrammed to a healthy one. For convenience, epigenetic targets for drug discovery are grouped into three major families [4]. First, there are the so-called writers, enzymes that catalyse a structural modification of their protein or a nucleic acid macromolecular substrate. Second, there are ‘readers’, recognition units that can discriminate between the native macromolecule and its modified version. Finally, reversibility is ensured by ‘erasers’, enzymes that catalyse the removal of the chemical signal introduced by writers.
Currently, a number of targets from these three families of ‘readers, writers and erasers' have reached different stages of the drug discovery process (table 1). At first sight, the list might not appear particularly impressive, as the approved drugs appear within the narrow confines of haematological cancers and involve only two targets, the DNMTs and HDACs. However, given the long gestation period of drug discovery, the table is only a snapshot in time and neither conveys the tremendous progress achieved within a short period nor the future potential of epigenetic therapy [5]. Even the scope of the approved DNMT and HDAC inhibitors is evolving, as there are ongoing clinical evaluations in non-haematological cancers and non-cancer indications. Concurrently, other clinical trials are exploring these agents in combination therapies with non-epigenetic drugs, not only in cancer but also for other applications such as the activation of latent viral reservoirs in HIV therapy [6]. Apart from the DNMT and HDAC inhibitors, the remaining targets in table 1 that have reached clinical development have moved from bench to bedside at a breathtaking pace. For example, the lysine-specific demethylase 1 (LSD1, KDM1A) was only identified in 2004 and the first clinical trials of LSD1 inhibitors were already announced in 2014. To go from protein discovery to target validation, in vitro and in vivo proof of concept to human clinical trials in 10 years is truly remarkable. Similarly, since the bromodomain was first reported to bind acetyllysine in 1999, multiple small molecule ligands from diverse chemical scaffolds have progressed to clinical trials [7].
Table 1.
The major epigenetic targets and their current drug discovery status.
| epigenetic target | status |
|---|---|
| writers | |
| DNA methyltransferases (DNMTs) | approval for myelodysplastic syndrome, acute myeloid and chronic myelomonocytic leukaemia |
| histone methyltransferases (KMTs) | clinical trials |
| arginine methyltransferases (PRMTs) | clinical trials |
| histone acetyltransferases (KATs) | preclinical |
| erasers | |
| Tet DNA demethylases (Tets) | lead discovery |
| lysine-specific demethylases (KDM1) | clinical trials |
| Jumonji C demethylases (KDM2-7) | preclinical |
| zinc-dependent HDACs | approval for T-cell lymphoma, multiple myeloma |
| sirtuin histone deacetylases (Sirts) | preclinical |
| readers | |
| bromodomains (BRDs) | clinical trials |
| chromodomains (CRDs) | preclinical |
| Tudor domains (TDRs) | preclinical |
Apart from the readers, the other epigenetic players are all enzymes. This augurs well for drug discovery, because many enzymes are successfully targeted in a potent and selective manner by small molecules. However, each epigenetic enzyme has its own idiosyncratic features, as discussed in the next section. A common theme that emerges is the utility of cofactor interference as a platform to inhibit a diverse set of epigenetic enzymes.
2. The organic chemistry of epigenetic enzymes
Within the nucleosome, DNA and histone proteins undergo a bewildering array of chemical modifications, both in type and frequency [8]. As a result, the nucleosome can potentially exist in a vast number of chemically distinct states that is far higher than that of the observable phenotypic states. If there is a ‘histone code’, it is likely to be complex and cooperative, with many redundant states that are biologically equivalent in their phenotype [9]. Despite the seemingly incalculable numbers of possible epigenetic states, at the chemical level, epigenetic writing involves either of two simple organic reactions that are among the first encountered in an undergraduate organic chemistry course, namely the acylation or alkylation of a nucleophile. In the discussion that follows, phosphorylation of serine, threonine and tyrosine residues is deliberately omitted. Although histone proteins undergo phosphorylation, there are a large number of non-epigenetic processes that also involve this post-translational modification. Furthermore, although the protein kinases that perform phosphorylation are highly popular targets for small molecule drug discovery, the inhibitors do not have the exquisite selectivity to distinguish between histone phosphorylation and non-histone substrates.
(a). Lysine acylation
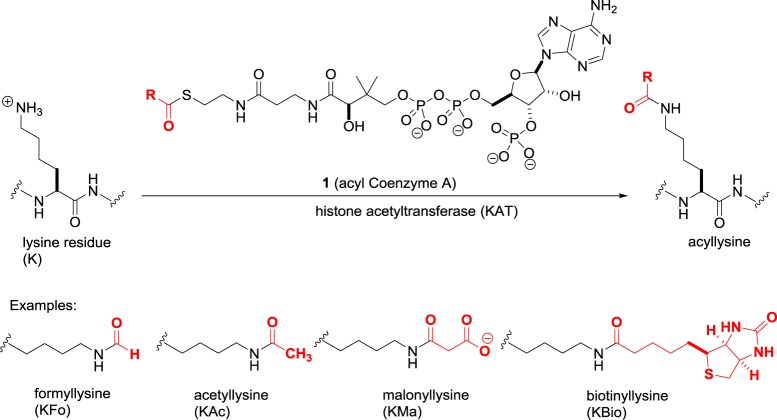
The N-terminal tails of histone proteins contain multiple basic lysine and arginine residues. The histone acetyltransferase (HAT) enzymes employ activated acyl Coenzyme A donors 1 (figure 1) to convert lysine residues to the corresponding acyl-lysine residue. Nearly 20 human HATs are known that vary in nuclear and cytoplasmic distribution [10]. By sequence homology, some HATs are grouped together in the GNAT, MYST and SRC families, while others such as CLOCK are structurally distinct.
Figure 1.
The histone acetyltransferase (KAT)-catalysed acylation of lysine residues.
The predominant reaction of HATs is lysine acetylation, which not only increases the size of the side chain but also alters its net charge from +1 at physiological pH to zero. As these acetylations take place not only in histones but also in a myriad of non-histone proteins in the nucleus as well as other cellular compartments, the traditional ‘histone acetyltransferase’ nomenclature is misleading. The unified nomenclature proposal by Allis et al. [11] suggests the HATs be renamed lysine acetyltransferases (KATs), and this is increasingly adopted in the literature.
While acetylation may be the major reaction catalysed by HATs, at least some of these enzymes accept a variety of other low-molecular-weight acyl donors that differ in size and charge. For example, attachment of dicarboxylic acids such as malonate not only increases the lysine side chain size but also results in a net charge of −1. It is currently unknown whether each acylation has its unique phenotypic response, or merely reflects a stochastic process dependent on the population of acyl donors available to the cell. Meanwhile, acylation is not limited to low-molecular-weight donors, as longer chain carboxylic acids such as biotin and myristic acid can be transferred. To reflect the diversity of acyl donors and the nature of the substrates, the KAT definition should be refined to ‘protein lysine acyltransferases'. In addition to KAT-driven acylation, the inherent reactivity of the thioester bond in acyl Coenzyme A donors enables non-catalysed transfer of acyl groups [12]. The relative importance of enzyme and non-catalysed acylation of lysine residues needs further investigation.
Although it does not involve a small molecule carboxylic acid or proceed through an acyl Coenzyme A donor, it is worth mentioning that a mechanistically similar amide bond formation of the lysine residues is involved in the conjugation of proteins such as SUMO (small ubiquitin-like modifier) and ubiquitin to histones [13,14]. The attachment of these proteins plays a significant role in histone recognition and degradation by the proteasome.
In biological terms, by altering the properties of the lysine side chain, acylation affects the interactions between the protein substrate and other macromolecules. From an epigenetic perspective, an important consequence of histone acylation is decreased affinity for the negatively charged DNA, leading to DNA unwinding off the nucleosome and becoming transcriptionally active. In addition, acylation serves as a signal for recognition, e.g. acetylation is recognized by the bromodomain and crotonylation by the YEATS domain [15]. Finally, by undergoing acylation, the lysine is ‘locked’ and can no longer undergo other modifications such as methylation.
(b). Acyl-lysine deacylation
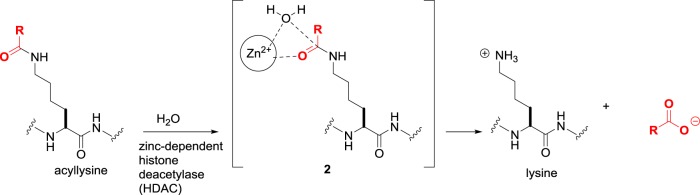
Deacylation is the reverse reaction of lysine acylation and accomplished by two distinct classes of enzymes: the zinc-dependent histone deacetylases (HDACs) and the sirtuins (Sirts) [16]. Biologically, the action of HDACs and sirtuins returns acyl-lysine residues to their native protonated lysine. In the nucleosome, this leads to compaction of chromatin and gene silencing. Much of the interest in inhibiting these enzymes lies in the ensuing reprogamming to reactivate repressed pathways, such as tumour suppression, DNA repair, immunomodulation and apoptosis in cancer cells.
In humans, there are 11 HDAC isoforms that are further subdivided according to sequence homology and localization. Class I constitutes the ubiquitous nuclear HDAC1, HDAC2, HDAC3 and HDAC8, for which histone proteins are likely to be an important substrate. The class IIa HDAC4, HDAC5, HDAC7 and HDAC9 are tissue-specific in their distribution, larger in size than the class I enzymes, and shuttle between the cytoplasm and the nucleus upon activation. Then, there are the class IIb HDAC6 and HDAC10, while HDAC11 is placed in the separate class IV due to similarities to both class I and class II. All these HDACs are metallohydrolases that employ a charge relay mechanism, with the active site Zn(II) cation accelerating hydrolysis through coordination to the carbonyl group of the amide and the water molecule in the intermediate 2 (figure 2).
Figure 2.
Lysine deacylation catalysed by zinc-dependent HDACs.
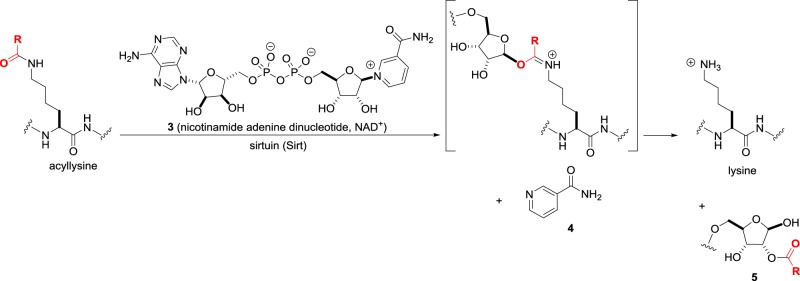
While the catalytic mechanism of HDACs appears straightforward and comparable to other amide hydrolysing enzymes, nature has evolved a second family of sirtuin enzymes that carry out the same conversion in a completely different manner [17]. In the sirtuins, the amide unusually acts as an oxygen nucleophile that attacks the nicotinamide adenine dinucleotide (NAD+) cofactor 3 (figure 3) to eject nicotinamide 4. Intramolecular acyl transfer to the 2′-OH group on ribose to give 5 completes the deacylation. There are seven human sirtuins—Sirt1, Sirt6 and Sirt7 are predominantly nuclear, Sirt2 is in the cytoplasm, and Sirt3, Sirt4 and Sirt5 are in the mitochondria. Both activators and inhibitors of the sirtuins are of potential therapeutic value and the focus of current attention.
Figure 3.
Lysine deacylation catalysed by sirtuins (Sirts).
(c). Lysine and arginine methylation
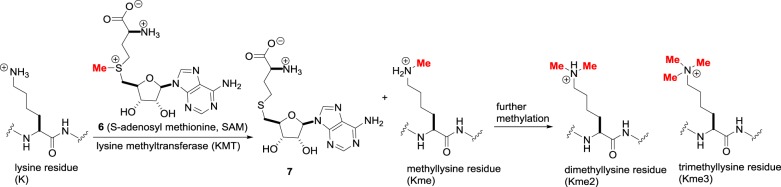
Besides acylation, the other major writing operation in epigenetics is alkylation. Within the active site of lysine methyltransferases, nucleophilic attack of the biological electrophilic methyl donor S-adenosylmethionine 6 (SAM, figure 4) by the lysine tails produces the methylated residues and the S-adenosylhomocysteine 7 (SAH) by-product. In humans, the majority of lysine methyltransferases (KMTs) share a catalytic SET domain named after the Su(var)3–9, Enhancer-of-zeste and Trithorax Drosophila proteins where it was identified [18]. However, although there are nearly 50 human proteins containing SET domains, not all have demonstrable lysine methyltransferase activity. In addition, there are lysine methyltransferases such as DOT1L (disruptor of telomeric silencing 1-like) that do not contain the SET domain.
Figure 4.
Lysine methylation catalysed by lysine methyltransferases (KMTs).
Once the first methyl group is added to give Kme, further methylation by KMT enzymes leads to the dimethylated Kme2 or trimethylated Kme3 residues. Owing to the many proteins within the KMT family, their nomenclature is complex. The KMTs are subdivided into eight subfamilies KMT1–8 and individual members are indicated by additional descriptors. To add to the complications, many proteins were first discovered in Drosophila and have conventional names that are widely used in the literature. For example, KMT1A is the Su(var)3–9 (suppressor of variegation 3–9) homologue, while KMT2H is ASH1 (absent, small or homeotic)-like protein.
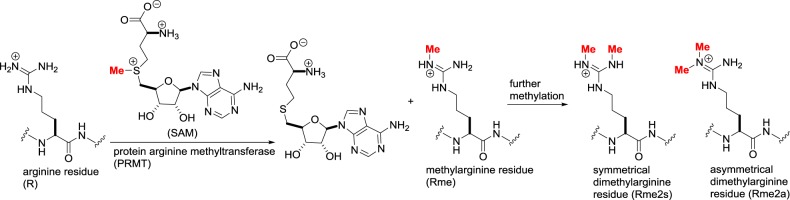
In an analogous manner, the guanidine functionality in arginine side chains undergoes methylation catalysed by protein arginine methyltransferases (PRMTs) [19]. In humans, there are nine PRMTs that share sequence homology and common ‘double E’ and ‘THW’ amino acid motifs. As with lysine methylation, further reaction can introduce a second methyl group, in either a symmetrical or asymmetrical fashion depending upon the specific PRMT (figure 5). There is considerable variation between the KMTs and PRMTs in their substrate tolerance. Some enzymes are highly specific such as DOT1L, the only known human H3K79 trimethylase, while others are relatively promiscuous in their choice of substrate.
Figure 5.
Arginine methylation catalysed by protein arginine methyltransferases (PRMTs).
From a structural point of view, neither lysine nor arginine methylation alters the net charge of the side chain. On the other hand, although methylation states differ from one another by only the addition of a single carbon atom, there is an increase in size and hydrophobicity with each methylation and a corresponding decrease in the number of H-bond donors. Reader domains have evolved to recognize these subtle changes and can specifically differentiate between one methylation state and another. While the previously discussed lysine acylation has primarily an activating effect upon transcription, the biological consequence of methylation is context-dependent [20]. H3K4 trimethylation, for example, recruits the gene-activating Trithorax protein complex, while H3K27 trimethylation recruits the Polycomb repressor complex.
Note that while the definition of ‘A’ as acetylation in KATs and HDACs is too restrictive due to the occurrence of other acylations, the ‘M’ for methyl in KMTs and PRMTs is biologically accurate. In principle, other alkylations are theoretically possible but in cells, it is limited to methylation due to the absence of natural SAM-like donors that can deliver larger alkyl groups.
(d). Lysine and arginine demethylation
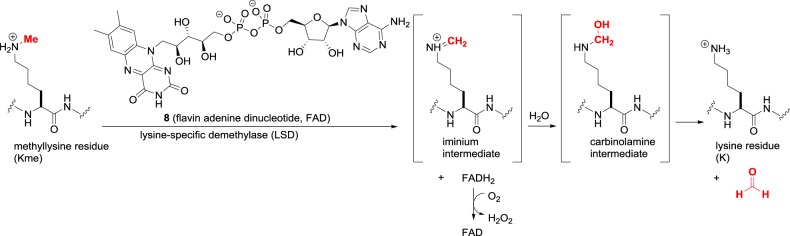
The erasure of methyllysine residues is performed by a family of oxidative enzymes, the lysine demethylases (KDMs). Just as lysine deacylation involves two mechanistically orthogonal routes via HDACs and sirtuins, there are two KDM subfamilies that are distinct in their function: the LSDs and Jumonji C demethylases. Humans have two LSD isoforms, LSD1 (KDM1A) and LSD2 (KDM1B), and are flavin adenine dinucleotide (FAD)-dependent enzymes that promote C–H oxidation of the methyl group by hydride transfer to the FAD cofactor 8 (figure 6) [21]. As this requires an amine with a free lone pair, LSDs act only upon Kme and Kme2 substrates that bind to the enzyme active site in a neutral form. As the permanently charged Kme3 quaternary amine does not have a lone pair, it is not an LSD substrate. The hydride transfer from Kme or Kme2 generates a transient iminium ion that spontaneously undergoes hydrolysis to an unstable carbinolamine that breaks down to native lysine. Meanwhile, the reduced FADH2 cofactor is oxidized by molecular oxygen to regenerate FAD. Overall, each enzyme turnover produces two toxic metabolites, i.e. formaldehyde and hydrogen peroxide.
Figure 6.
Lysine demethylation catalysed by lysine-specific demethylases (LSD, KDM1).
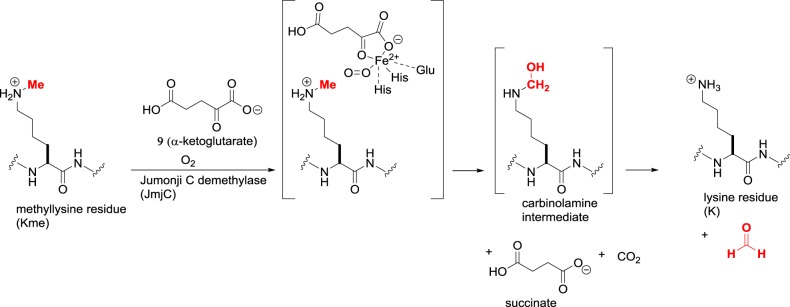
The larger family of lysine demethylases share a catalytic Jumonji C (JmjC) domain and are classified as KDM2–7 [22]. These proteins fall under the Fe(II)/α-ketoglutarate-dependent superfamily of dioxygenases that contain a Fe(II) cation. The active site brings together the methyllysine substrate, the co-substrate α-ketoglutarate 9 and molecular oxygen (figure 7). One oxygen atom effects C–H oxidation of the methyl group to produce a carbinolamine intermediate, and the other oxygen atom oxidizes the co-substrate to succinate and carbon dioxide. Overall, each enzyme turnover produces two toxic metabolites, i.e. formaldehyde and carbon dioxide. As the oxidation does not involve a hydride transfer, Kme, Kme2 or Kme3 can be accepted as substrates. There are approximately 20 human proteins containing JmjC domains with varying degrees of substrate specificity, including some examples for which catalytic activity has not been detectable in laboratory conditions.
Figure 7.
Lysine demethylation catalysed by Jumonji C demethylases (JmjC, KDM2–7).
Although in vitro experiments have shown that oxidation of somewhat larger alkyl groups is possible for certain KDMs [23], there is no evidence of physiological relevance for the enzymatic removal of non-methyl groups. Like methylation, the phenotypic consequences of demethylation are context-dependent. The effect of H3K4 demethylation by LSD1, for example, has a repressive effect important in normal development and haematopoiesis. Meanwhile, the same enzyme when bound to the androgen or estrogen receptor demethylates H3K9 residues with a transcriptional activating effect.
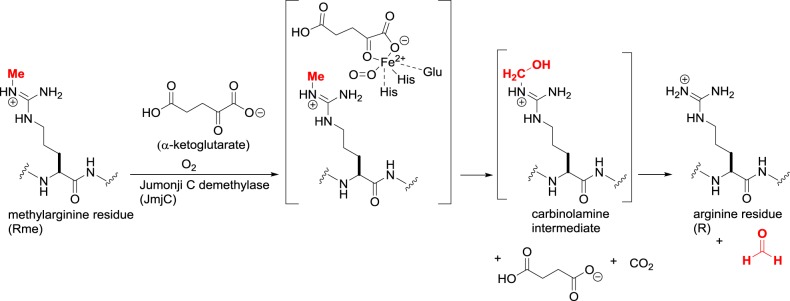
Some of the JmjC demethylases are capable of arginine demethylation by an identical mechanism to lysine demethylation (figure 8) [24]. In addition to KDM-mediated demethylation, methylated arginine residues undergo breakdown to citrulline through the action of calcium-dependent protein arginine deiminases (PADs) [25]. While this is an erasure of the methyl arginine mark, it does not return the side chain to its original state but to the non-canonical amino acid citrulline. Although histones are among PAD substrates, the epigenetic significance of PADs is not clearly defined and they will not be discussed further.
Figure 8.
Arginine demethylation catalysed by Jumonji C demethylases (JmjC, KDM2–7).
(e). Cytosine methylation
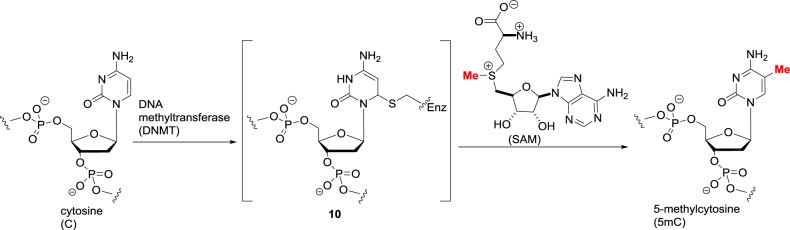
The epigenetic modification of DNA centres around one reaction, the methylation of the C-5 position of cytosine residues using SAM as the cofactor. The cytosine is not normally nucleophilic, and the DNA methyltransferase enzyme activates C-5 by the addition of an active site Cys residue to create an enamine-like electron-rich intermediate 10 (figure 9) [26]. The enamine then attacks SAM, and the methylated intermediate undergoes β-elimination to regenerate the enzyme and 5-methylcytosine. In humans, there are five DNMTs: DNMT1 is ubiquitous and responsible for the replication of methylation marks in daughter strands during DNA replication; DNMT2 is actually an RNA methylase that acts upon tRNA; DNMT3A/B are ‘de novo’ methylases that introduce methylation into both non-methylated and hemi-methylated DNA; and DNMT3 L, which is not catalytically active but enhances the methyltransferase activity of DNMT3A/B. Cytosine methylation is recognized by methyl-binding proteins that have a gene-silencing effect. The inhibition of DNMTs would thus be expected to be gene activating, in a similar manner to HDAC inhibition. Although the recombinant DNMTs can be engineered to accept synthetic higher homologues of SAM [27], their physiological role is presumably limited to methylation only as is the case with lysine methyltransferases.
Figure 9.
Cytosine methylation catalysed by DNA methyltransferases.
(f). Cytosine demethylation
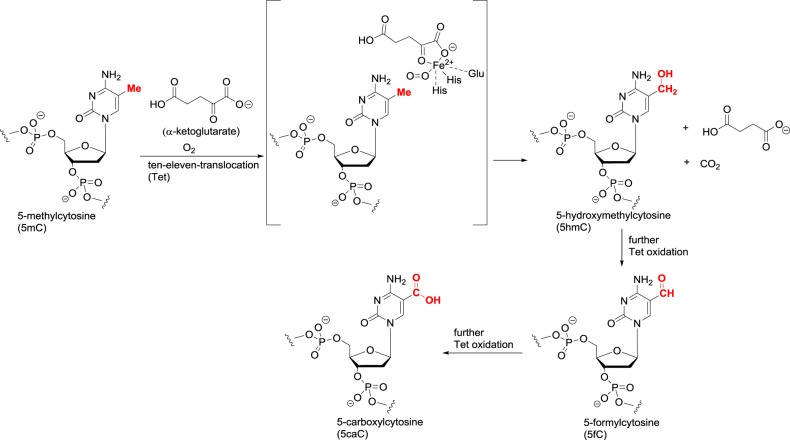
For a long time, there was no evidence for the enzymatic demethylation of 5-methylcytosine. The need for active erasure was considered unnecessary, as during DNA replication the new daughter strand does not contain the methylation pattern present in the parent. The action of DNMT1 reintroduces the methyl groups, and a lack of 100% fidelity in the reproduction would lead to a gradual passive loss of methylation in each replication cycle. However, it was recently shown that the ten–eleven translocation (Tet) enzymes carry out an oxidative demethylation [28]. The three human enzymes Tet1–3, like the JmjC demethylases, are Fe(II)/α-ketoglutarate-dependent dioxygenases and perform a mechanistically similar C–H oxidation of the methyl group in 5-methylcytosine (figure 10). In lysine and arginine demethylation, a single oxidation is sufficient as it produces an unstable N–C–OH carbinolamine that undergoes hydrolysis. In DNA demethylation, the first oxidation gives a stable C–C–OH bond. The resulting 5-hydroxymethylcytosine residue can undergo two further C–H oxidations to give 5-formylcytosine and 5-carboxylcytosine. Both the formyl and carboxyl groups can undergo chemical deformylation and decarboxylation, respectively, to regenerate cytosine. In addition, these modified cytosines are targeted by thymine DNA glycosylase (TDG) for base excision repair that removes the formyl or carboxyl pyrimidine heterocycle. The resulting abasic pair can then be repaired to cytosine, thus achieving an overall cycling of 5-methylcytosine back to cytosine.
Figure 10.
Cytosine demethylation catalysed by Tet dioxygenases.
3. The organic chemistry of epigenetic enzyme inhibition
Many drugs exert their action by the inhibition of an enzyme target and fall into one of five broad categories based on their mechanism of interaction:
(1) Active site binding through analogy to the enzyme substrate.
(2) Active site binding through analogy to the transition state of the enzyme-catalysed reaction.
(3) Active site binding through analogy to the enzyme product.
(4) Active site binding through analogy to the enzyme cofactor or its inactivation.
(5) Allosteric binding outside the enzyme active site.
As an illustrative example of how these strategies can be applied to an epigenetic enzyme, consider the LSD1 lysine demethylase. The best-characterized substrates of LSD1 are histone H3K4me and H3K4me2, and it is possible to design substrate or product analogues based on the histone H3 sequence. The product of demethylation, histone H3K4, is itself an LSD1 inhibitor, although the level is strongly dependent on the size (table 2). While the full-length H3 protein inhibits LSD1 with an impressive Ki of 20 nM [29], truncation to the H3 21-mer peptide leads to a significant drop in Ki to 2 µM, and the shorter H3 12-mer sequence has only a weak Ki of 200 µM [30]. A second series of LSD1 peptide inhibitors is based on the N-terminal sequence of Snail transcription factors that contain an F4 residue instead of K4. X-ray crystallographic studies confirm that the phenylalanine acts as a hydrophobic analogue of the methyllysine substrate that binds to LSD1 more tightly than the histone H3 sequence [31]. Nevertheless, for both the histone H3 and Snail peptides, achieving reasonable target affinity requires dimensions that are beyond small molecule chemical space. This is an inherent issue with all epigenetic enzyme targets, as the substrate, transition state and product are macromolecular proteins or nucleic acids that are difficult to mimic within the constraints of a small molecule suitable for cell penetration and oral administration. That leaves two other options for enzyme inhibition—cofactor modulation or allosteric binding. Taking LSD1 again as an example, computational modelling identified several small-molecule binding pockets outside the enzyme active site [32], although there are no known LSD1 inhibitors confirmed to bind through interactions with these allosteric sites. Indeed, although LSD1 and other epigenetic enzymes have undoubtedly been the focus of high-throughput screening with large compound libraries, few examples of allosteric inhibitors have been reported for these targets.
Table 2.
Examples of LSD1 inhibitors based on substrate or product analogues. The K4 site of enzyme action or the mimetic F4 in the Snail peptides is highlighted in bold.
| peptide | Ki (µM) |
|---|---|
| full-length histone H3 | 0.02 |
| histone H3 1–21 ARTKQTARKSTGGKAPRKQLA | 2 |
| histone H3 1–12 ARTKQTARKSTG | 200 |
| Snail1 1–20 PRSFLVRKPSDPNRKPNYSE | 0.2 |
| Snail1 1–6 PRSFLV | 28 |
4. Cofactor interference as a strategy for epigenetic enzyme inhibition
Given the challenges in identifying small molecule epigenetic enzyme inhibitors based on substrate, transition state, product or allosteric binding, cofactor interference has emerged as the drug discovery platform that is not only the most general in addressing multiple targets but also the most powerful in leading to clinical candidates. There are three basic mechanisms by which interference with an active site cofactor is achieved:
(1) Competitive displacement of the cofactor from its binding pocket.
(2) Reversible binding with the cofactor that prevents substrate occupancy or productive catalysis.
(3) Irreversible inactivation of the cofactor.
All three of these approaches have made significant progress against epigenetic targets (table 3). In many cases, they have overtaken non-cofactor-based inhibitor design particularly for the zinc-dependent HDACs and the SAM cofactor utilizing KMTs. We will now discuss each of these cofactors in order of increasing size, as size does matter—the larger the cofactor, the greater its interactions within the enzyme-binding pocket and the less likely it can be competitively displaced by a small molecule. While there are now good examples of cofactor displacement for ATP and SAM, achieving this for larger cofactors with significant polar or electrostatic non-covalent interactions with the enzyme is a challenge for drug discovery.
Table 3.
The potency of epigenetic enzyme inhibitors based on cofactors versus alternative approaches, and their current status in drug discovery.
| cofactor, target | cofactor-based potency | alternative approach potency |
|---|---|---|
| zinc (II), histone deacetylases | nM, approvals | µM, lead discovery |
| iron (II), JmjC demethylases | nM, preclinical | µM, preclinical |
| iron (II), Tet demethylases | µM, lead discovery | none |
| SAM, lysine/arginine methyltransferases | nM, clinical trials | nM, clinical trials |
| SAM, DNA methyltransferases | µM, lead discovery | µM, approvals |
| NAD+, sirtuins | µM, preclinical | nM, preclinical |
| FAD, lysine-specific demethylases | nM, clinical trials | nM, clinical trials |
| acetyl CoA, histone acetyltransferases | nM, preclinical | µM, preclinical |
(a). Zinc (II)
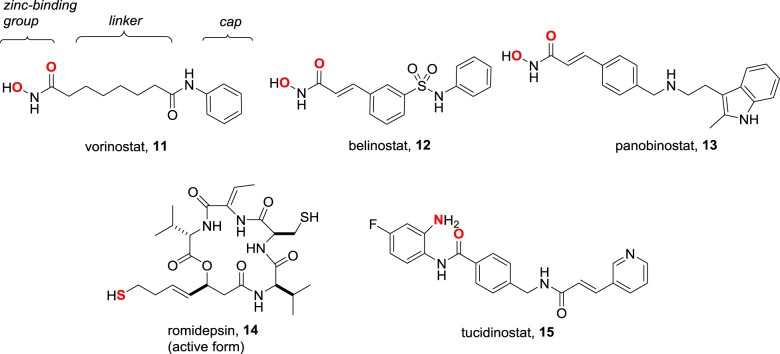
Metal coordination is a popular design strategy for the inhibition of metalloenzymes [33]. Although off-target indiscriminate binding to a host of metalloenzymes might be a concern, in practice it is often not significant [34]. This is because efficient metal coordination also requires the drug to maximize other interactions within the substrate-binding pocket. As the metalloenzymes vary in the topology of their active site, it is usually possible to achieve high selectivity between them. Among the epigenetic targets, metal coordination is most successful in the inhibition of the zinc-dependent HDACs (figure 11). The five approved drugs conform to the common pharmacophore illustrated for vorinostat and contain a zinc-binding group that plays a critical role in the reversible binding of these inhibitors to HDACs. Vorinostat 11, the first to be approved, contains a bidentate hydroxamic acid as a zinc-binding group and arose from Breslow's remarkable lead optimization of the phenotypic differentiation observed with the solvent dimethyl sulfoxide [35]. Without knowing the molecular target, Breslow's keen insight suggested that metal binding might be involved, eventually leading to the exploration of the hydroxamic acid motif. Belinostat 12 and panobinostat 13, two second-generation hydroxamic acid HDAC inhibitors, are now approved, while others are in clinical trials.
Figure 11.
The five approved HDAC inhibitors, with the common pharmacophore indicated for vorinostat and zinc-binding atoms shown in red.
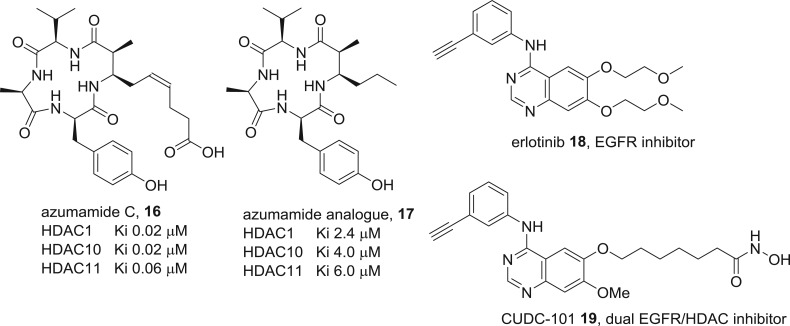
While the zinc-binding group is critical, the rest of the molecule in HDAC inhibitors is important for additional binding interactions that augment potency and influence selectivity between the 11 human isoforms [36,37]. Compared with the minimal structure of vorinostat, which is a pan-HDAC inhibitor, the more elaborate scaffolds present in romidepsin 14 and tucidinostat 15 are more potent and display selectivity levels of greater than 1000-fold between isoforms that are potentially useful in reducing on-target toxicity [38,39]. Meanwhile, the interesting question arises whether zinc-binding is obligatory if the additional interactions are sufficiently strong to impose tight binding. The experiments by Olsen and colleagues based on macrocyclic natural product HDAC inhibitors (figure 12) indicate a 100-fold loss of activity when the zinc-binding group in the natural product azumamide C 16 is excised [40]. Nevertheless, the analogue 17 is still a micromolar inhibitor without the pharmockinetic liabilities of zinc-binding groups, and further fine-tuning may produce compounds with sufficient potency for in vivo studies. At the other extreme are inhibitors where zinc coordination is of paramount importance and the ‘cap’ region essentially acts as a stopper that plugs the substrate-binding channel. If this is true, the nature of the cap should be highly variable without losing potency. One way to take advantage of this flexibility is the design of hybrid drugs where the cap incorporates the pharmacophore for a second target [41]. Two examples of this strategy from Curis that are dual kinase–HDAC inhibitors are in clinical trials. One of these is inspired by the epidermal growth factor receptor (EGFR) inhibitor 18, with the hybrid CUDC-101 19 being a potent EGFR and HDAC inhibitor [42].
Figure 12.
Examples of ‘extreme’ HDAC inhibitors. Azumamide is a macrocyclic natural product, and an analogue without a zinc-binding group still inhibits the enzyme. CUDC-101 is a dual target inhibitor designed by grafting a zinc-binding inhibitor onto the approved drug erlotinib.
(b). Iron (II)
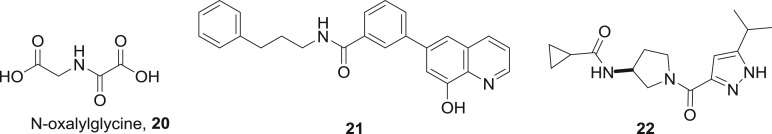
Iron (II) is the common cofactor in the Fe(II)/α-ketoglutarate-dependent superfamily of dioxygenases [43]. The stable α-ketoglutarate co-substrate mimic N-oxalylglycine 20 (figure 13) is a generic micromolar inhibitor of this enzyme class including the epigenetic JmjC lysine demethylases. Other metal-binding compounds including the bidentate hydroxamic acids that inhibit the zinc-dependent HDACs are also capable of inhibiting JmjC lysine demethylases. The approved drug vorinostat 11, for example, inhibits KDM4E with a Ki of 14 µM, although this modest level of activity is unlikely to be pharmacologically important in its therapeutic use [44]. In terms of our classification, it is a moot point whether such inhibitors are best considered as substrate analogues of α-ketoglutarate or as cofactor interfering compounds that bind to the iron (II) centre as the two roles are indistinguishable for this target. Over the years, more selective iron-binding JmjC inhibitors have been discovered [45,46]. The 8-hydroxyquinoline 21 is a selective nanomolar KDM4B inhibitor with in vivo efficacy in cancer models that is likely to be metal binding in its action [47]. A recent publication from Genentech and Constellation described the lead optimization of a screening hit to the pyrazole 22, which has an IC50 of 45 nM against KDM5A and an oral bioavailability of F34% in mice [48]. Bidentate metal coordination through the pyrazole nitrogen and the carbonyl was confirmed by X-ray crystallography. These examples and others in the journal and patent literature demonstrate that potent and selective JmjC inhibitors can be discovered through metal-coordinating small molecules. Besides the JmjC enzymes, the Tet DNA demethylases are the other epigenetic target within the Fe(II)/α-ketoglutarate-dependent superfamily of dioxygenases. At the present time, the discovery of potent inhibitors for these newly identified epigenetic Tet enzyme functions is in its infancy. Although not relevant for drug discovery, nickel (II) was reported to inhibit the Tet enzymes by displacement of the iron (II) cofactor with an IC50 of 1.2 µM [49].
Figure 13.
Examples of iron (II)-binding inhibitors of Jumonji C lysine demethylases.
(c). S-Adenosylmethionine
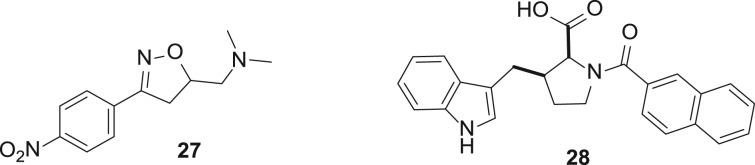
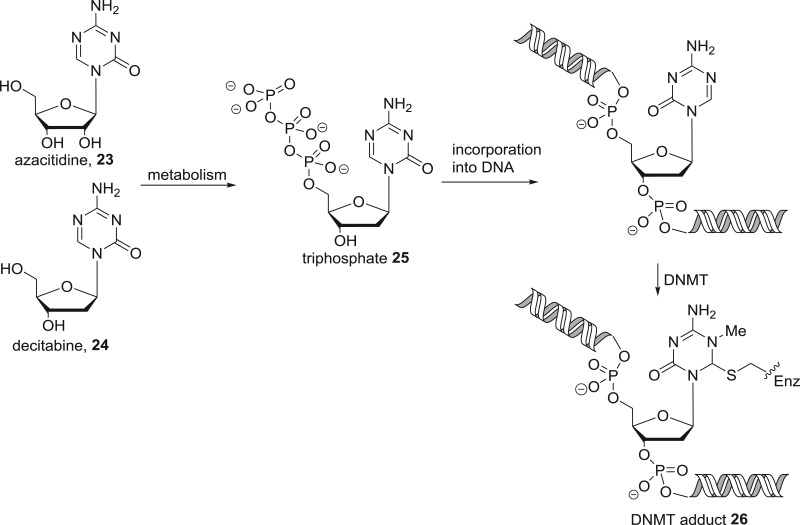
SAM is the biological methyl transfer agent employed by the DNA methyltransferases and the lysine/arginine methyltransferases. DNA methyltransferases are the only epigenetic enzyme target for which alternative approaches to cofactor interference are more successful for inhibitor design. Through phenotypic screening, the cytidine analogues azacitidine 23 and decitabine 24 (figure 14) were identified as potent cytotoxic agents. On this basis, azacitidine entered clinical trials 50 years ago, but did not receive approval due to toxicity. Later, interest was rekindled in azacitidine and decitabine when their mechanism of action as DNA methyltransferase inhibitors was discovered. These nucleosides undergo metabolic activation to the nucleotide 25, which is incorporated into DNA and irreversibly inhibits DNMTs by formation of the covalent adduct 26. Although both compounds are approved for the treatment of all five stages of myelodysplastic syndrome, their pharmacodynamic and pharmacokinetic liabilities have sparked interest in the discovery of second-generation nucleoside DNMT inhibitors [50]. In principle, cofactor interference would be an attractive alternative strategy. Although a number of non-nucleoside scaffolds are reported to modestly inhibit DNMTs in the micromolar range, cofactor interference was clearly identified as the mechanism of action only in rare cases such as compounds 27 and 28 [51–53].
Figure 15.
Examples of SAM competitive DNA methyltransferase inhibitors.
Figure 14.
Mechanism of action of the DNA methyltransferase inhibitors azacitidine and decitabine.
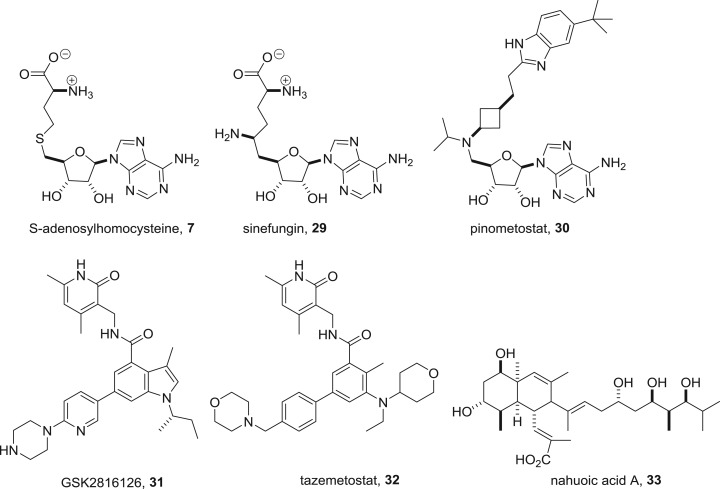
The relative lack of success in identifying potent DNMT inhibitors that are SAM antagonists and the need for selectivity between the many enzymes that use this cofactor suggest that SAM competition is not a viable approach for drug discovery. Examples of known SAM mimics such as the by-product of methyl group transfer S-adenosylhomocysteine 7 or the natural product sinefungin 29 (figure 16) are relatively modest methyltransferase inhibitors and non-selective in their action. Nevertheless, these hurdles have been spectacularly overcome for another SAM utilizing epigenetic target, the lysine methyltransferases [54,55]. Pinometostat 30 discovered by Epizyme was the first SAM-mimetic KMT inhibitor to enter clinical trials. The compound is a DOT1 L inhibitor with an IC50 of 80 pM and 37 000-fold selectivity over all other methyltransferases tested [56]. Although the structural resemblance of pinometostat to the cofactor is clear, the homology between other recently developed mimetics and SAM is less obvious. GlaxoSmithKline and Epizyme, for example, have launched clinical trials with SAM mimetics GSK2816126 31 and tazemetostat 32 with a pyridone scaffold [57,58]. These compounds are potent nanomolar EZH2 inhibitors with high selectivity over the isoform EZH1 as well as other methyltransferase enzymes. An unusual example lacking any nitrogen or a heterocycle is nahuoic acid 33, a Streptomyces natural product that is a SAM competitive inhibitor of KMT5A with an IC50 of 6.5 µM [59].
Figure 16.
Examples of SAM-mimetic reversible inhibitors of lysine methyltransferases.
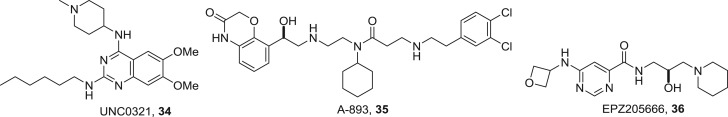
While SAM competition is a successful strategy for histone methyltransferase inhibition, other approaches have also given rise to inhibitors of these enzymes. Two examples from optimization of high-throughput screening against lysine methyltransferases are UNC0321 34 (figure 17) that selectively inhibits the G9a/GLP heterotrimer with an IC50 of 7 nM against GLP and 992 nM against the closely related G9a, and A-893 35, a selective SMYD2 inhibitor with an IC50 of 2.8 nM [60,61]. Similarly, screening against the arginine methyltransferase PRMT5 and lead optimization culminated in the potent inhibitor EPZ015666 (GSK3235025) 36 with an IC50 of 22 nM [62]. From crystallographic studies, all three compounds 32–34 occupy the substrate-binding pocket and are not competitive with SAM.
Figure 17.
Examples of substrate mimetic inhibitors of protein methyltransferases.
(d). Nicotinamide adenine dinucleotide
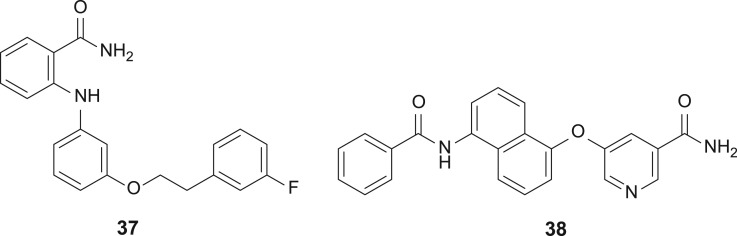
The erasure of acyl-lysine marks catalysed by the sirtuin enzymes involves transfer of the acyl group to the NAD+ cofactor (figure 3). The size and multiply charged nature of NAD+ suggests that, for this target, cofactor interference will be less attractive than other strategies for enzyme inhibition. Indeed, potent and selective small inhibitors of sirtuins tend to operate by other mechanisms of action [63]. One example that is cofactor-related involves the nicotinamide by-product of sirtuin catalysis. Nicotinamide itself is a micromolar inhibitor of the sirtuins and further optimization led Suzuki to anilinobenzamide 37 (figure 18), a submicromolar Sirt2 inhibitor suggested by molecular docking to bind to the NAD+-binding pocket [64]. Meanwhile, starting from a fragment-based approach, the nicotinamide analogue 36 was identified as a submicromolar selective Sirt2 inhibitor that was competitive against the peptide substrate and likely to be a non-competitive inhibitor of NAD+ [65].
Figure 18.
Examples of nicotinamide-based inhibitors of sirtuin deacylases.
(e). Flavin adenine dinucleotide
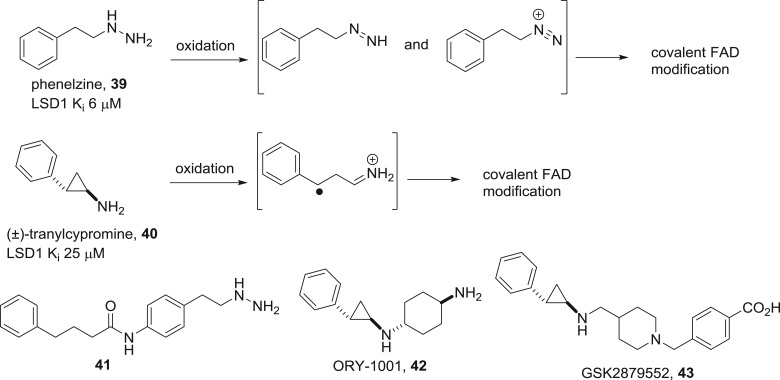
FAD is the oxidizing cofactor present in the LSDs. The cofactor is tightly bound by non-covalent interactions to the enzyme, and there are no known LSD inhibitors that work by ejection of FAD from its binding pocket. However, irreversible inactivation of the cofactor has proven to be a successful means for enzyme inhibition [66,67]. The LSDs are mechanistically homologous to the monoamine oxidases (MAOs), well-established targets for CNS drug discovery with approved drugs including phenelzine 39 (figure 19) and tranylcypromine 40. These drugs are oxidized by FAD to produce reactive intermediates that covalently modify the cofactor, leading to irreversible enzyme inactivation. Based on this discovery, the repurposing of tranylcypromine as an LSD1 inhibitor for the treatment of leukaemia is under clinical investigation. Meanwhile, lead optimization has uncovered the more potent nanomolar phenelzine analogue 41 [68] and the clinical candidate tranylcypromine analogues ORY-1001 42 and GSK2879552 43 that are highly selective for LSD1 inhibition over the isoform LSD2 or other amine oxidases [69,70]. Non-cofactor-based approaches have also had some success, although the exact binding mode has usually not been corroborated by X-ray crystallography. One recent exception reports the optimization of a bicyclic heterocyclic scaffold to provide nanomolar LSD1 inhibitors that occupy the substrate-binding pocket [71].
Figure 19.
Mechanism of action of phenelzine and tranylcypromine, irreversible MAO/LSD1 inhibitors and structures of second-generation analogues.
(f). Acetyl Coenzyme A
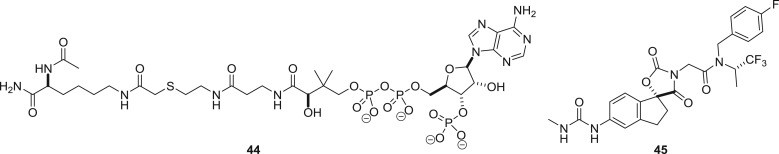
Acetyl Coenzyme A (and its congeners for the transfer of other acyl groups) is the largest of the epigenetic cofactors. The size and the need for selectivity against the multitude of other enzymes that employ acyl Coenzyme A suggest that cofactor interference will be at its most challenging when applied to histone acetyltransferases (KATs). Furthermore, regardless of whether cofactor interference or other strategies are employed, the KATs have a reputation for difficulty in the discovery of selective cell-permeable submicromolar inhibitors [72,73]. An example of successful cofactor mimicry from Cole and colleagues features the bisubstrate mimic Lys-CoA 44 (figure 20) [74]. The compound was tested against two KATs, p300 and P300/CBP-associated factor (PCAF), and selectively inhibited p300 with an approximate IC50 of 500 nM. Extending the lysine to the 20-mer histone H3 sequence gave a peptide that instead selectively inhibited PCAF. By virtual screening of 800 000 compounds to the binding pocket for 44, a hydantoin hit was identified and optimized to 45 [75]. Impressively, compound 45 was a nanomolar inhibitor of p300/CBP without inhibiting other tested KATs at 10 µM. Competition with acetyl CoA was confirmed through X-ray crystallography, and 45 was active in a xenograft model of prostate cancer.
Figure 20.
Structures of p300 histone acetyltransferase inhibitors competitive with acetyl Coenzyme A.
5. Conclusion
There are many possible approaches to small molecule drug discovery and each of these needs to be evaluated for its appropriateness for a given target. Within the field of epigenetic enzyme inhibition, cofactor interference has become the solution of broadest applicability. The ability to design metal cation coordinating compounds with target selectivity has produced multiple approvals and clinical candidates for zinc-dependent HDACs and preclinical chemical probes for individual JmjC demethylases. The reversible displacement of SAM from its binding pocket with high selectivity between SAM-utilizing enzymes was believed to be chemically demanding, but is now successfully achieved for several lysine methyltransferases with examples undergoing clinical trials. The larger cofactors—NAD+, FAD and acetyl Coenzyme A—are less likely to be targeted in the same way by competitive displacement. Nevertheless, for the lysine-specific demethylases, irreversible inactivation of the FAD cofactor in a potent and selective manner has led to several clinical candidates, while a nanomolar small molecule p300 inhibitor that competes with acetyl Coenzyme A was recently described. Further developments can be anticipated in the field of cofactor interference applied to epigenetic therapy.
Data accessibility
This article has no additional data.
Competing interests
I have no competing interests.
Funding
I thank the COST Action CM1406 Epigenetic Chemical Biology for financial support and the Freiburg Institute of Advanced Studies (FRIAS) for a fellowship.
References
- 1.Medawar PB, Medawar JS. 1983. Aristotle to zoos. A philosophical dictionary of biology, pp. 113–114. Cambridge, MA: Harvard University Press. [Google Scholar]
- 2.Chen Y, Hong T, Wang S, Mo J, Tian T, Zhou X. 2017. Epigenetic modification of nucleic acids: from basic studies to medical applications. Chem. Soc. Rev. 46, 2844–2872. ( 10.1039/C6CS00599C) [DOI] [PubMed] [Google Scholar]
- 3.Andrews FH, Strahl BD, Kutateladze TG. 2016. Insights into newly discovered marks and readers of epigenetic information. Nat. Chem. Biol. 12, 662–668. ( 10.1038/nchembio.2149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang GG, Allis CD, Chi P. 2007. Chromatin remodeling and cancer, part I: covalent histone modifications. Trends Mol. Med. 13, 363–372. ( 10.1016/j.molmed.2007.07.003) [DOI] [PubMed] [Google Scholar]
- 5.Pister SX, Ashworth A. 2017. Marked for death: targeting epigenetic changes in cancer. Nat. Rev. Drug Discov. 16, 241–263. ( 10.1038/nrd.2016.256) [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Issa JPJ, Baylinn S. 2016. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 17, 630–641. ( 10.1038/nrg.2016.93) [DOI] [PubMed] [Google Scholar]
- 7.Pérez-Salvia M, Esteller M. 2017. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics 12, 323–339. ( 10.1080/15592294.2016.1265710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Zhou J, Lin S, Deng W, Zhang Y, Xue Y. 2017. PLMD: an updated data resource of protein lysine modifications. J. Genet. Genomics 44, 243–250. ( 10.1016/j.jgg.2017.03.007) [DOI] [PubMed] [Google Scholar]
- 9.Su Z, Denu JM. 2016. Reading the combinatorial histone language. ACS Chem. Biol. 11, 564–574. ( 10.1021/acschembio.5b00864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marmorstein R, Zhou MM. 2014. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762 ( 10.1101/cshperspect.a018762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allis CD, et al. 2007. New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636. ( 10.1016/j.cell.2007.10.039) [DOI] [PubMed] [Google Scholar]
- 12.Wagner GR, Hirschey MD. 2014. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases . Mol. Cell 54, 5–16. ( 10.1016/j.molcel.2014.03.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dwane L, Gallagher WM, Ní CT, O'Connor DP. 2017. The emerging role of non-traditional ubiquitination in oncogenic pathways. J. Biol. Chem. 292, 3543–3551. ( 10.1074/jbc.R116.755694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendriks IA, Vertegaal AC. 2016. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581–595. ( 10.1038/nrm.2016.81) [DOI] [PubMed] [Google Scholar]
- 15.Zhao D, Li Y, Xiong X, Chen Z, Li H. 2017. YEATS domain—A histone acylation reader in health and disease. J. Mol. Biol. 429, 1994–2002. ( 10.1016/j.jmb.2017.03.010) [DOI] [PubMed] [Google Scholar]
- 16.Yoshida M, Kudo N, Kosono S, Ito A. 2017. Chemical and structural biology of protein lysine deacetylases . Proc. Jpn. Acad. Ser. B 93, 297–321. ( 10.2183/pjab.93.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bheda P, Jing H, Wolberger C, Lin H. 2016. The substrate specificity of sirtuins. Annu. Rev. Biochem. 85, 405–429. ( 10.1146/annurev-biochem-060815-014537) [DOI] [PubMed] [Google Scholar]
- 18.Boriack-Sjodin PA, Swinger KK. 2016. Protein methyltransferases: a distinct, diverse, and dynamic family of enzymes. Biochemistry 55, 1557–1569. ( 10.1021/acs.biochem.5b01129) [DOI] [PubMed] [Google Scholar]
- 19.Morales Y, Cáceres T, May K, Hevel JM. 2016. Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch. Biochem. Biophys. 590, 138–152. ( 10.1016/j.abb.2015.11.030) [DOI] [PubMed] [Google Scholar]
- 20.Hyun K, Jeon J, Park P, Kim J. 2017. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49, e324 ( 10.1038/emm.2017.11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosseini A, Minucci S. 2017. A comprehensive review of lysine-specific demethylase 1 and its roles in cancer. Epigenomics 9, 1123–1142. ( 10.2217/epi-2017-0022) [DOI] [PubMed] [Google Scholar]
- 22.Markolovic S, Leissing TM, Chowdhury R, Wilkins SE, Lu X, Schofield CJ. 2016. Structure–function relationships of human JmjC oxygenases — demethylases versus hydroxylases. Curr. Opin. Struct. Biol. 41, 62–72. ( 10.1016/j.sbi.2016.05.013) [DOI] [PubMed] [Google Scholar]
- 23.Hopkinson RJ, Walport LJ, Münzel M, Rose NR, Smart TJ, Kawamura A, Claridge TD, Schofield CJ. 2013. Is JmjC oxygenase catalysis limited to demethylation? Angew. Chem. Int. Ed. 52, 7709–7713. ( 10.1002/anie.201303282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walport LJ, Hopkinson RJ, Chowdhury R, Schiller R, Ge W, Kawamura A, Schofield CJ. 2016. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974 ( 10.1038/ncomms11974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witalison EE, Thompson PR, Hofseth LJ. 2015. Protein arginine deiminases and associated citrullination: physiological functions and diseases associated with dysregulation. Curr. Drug Targets 16, 700–710. ( 10.2174/1389450116666150202160954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyko F. 2018. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92. ( 10.1038/nrg.2017.80) [DOI] [PubMed] [Google Scholar]
- 27.Lukinavičius G, Lapinaitė A, Urbanavičiūtė G, Gerasimaitė R, Klimašauskas S. 2012. Engineering the DNA cytosine-5 methyltransferase reaction for sequence-specific labeling of DNA. Nucleic Acids Res. 40, 11 594–11 602. ( 10.1093/nar/gks914) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Zhang Y. 2017. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534. ( 10.1038/nrg.2017.33) [DOI] [PubMed] [Google Scholar]
- 29.Burg JM, Gonzalez JJ, Maksimchuk KR, McCafferty DG. 2016. Lysine-specific demethylase 1A (KDM1A/LSD1): product recognition and kinetic analysis of full-length histones. Biochemistry 55, 1652–1662. ( 10.1021/acs.biochem.5b01135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. 2005. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 280, 41 360–41 365. ( 10.1074/jbc.M509549200) [DOI] [PubMed] [Google Scholar]
- 31.Tortorici M, et al. 2013. Protein recognition by short peptide reversible inhibitors of the chromatin-modifying LSD1/CoREST lysine demethylase. ACS Chem. Biol. 8, 1677–1682. ( 10.1021/cb4001926) [DOI] [PubMed] [Google Scholar]
- 32.Robertson JC, Hurley NC, Tortorici M, Ciossani G, Borrello MT, Vellore NA, Ganesan A, Mattevi A, Baron R. 2013. Expanding the druggable space of the LSD1/CoREST epigenetic target: new potential binding regions for drug-like molecules, peptides, protein partners, and chromatin. PLoS Comput. Biol. 9, e1003158 ( 10.1371/journal.pcbi.1003158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Hu XQ, Li QS, Zhang XX, Ruan BF, Xu J, Liao C. 2016. Metalloprotein inhibitors for the treatment of human diseases. Curr. Top. Med. Chem. 16, 384–396. ( 10.2174/1568026615666150813145218) [DOI] [PubMed] [Google Scholar]
- 34.Day JA, Cohen SM. 2013. Investigating the selectivity of metalloenzyme inhibitors. J. Med. Chem. 56, 7997–8007. ( 10.1021/jm401053m) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marks PA, Breslow R. 2007. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 25, 84–90. ( 10.1038/nbt1272) [DOI] [PubMed] [Google Scholar]
- 36.Roche J, Bertrand P. 2016. Inside HDACs with more selective HDAC inhibitors. Eur. J. Med. Chem. 121, 451–483. ( 10.1016/j.ejmech.2016.05.047) [DOI] [PubMed] [Google Scholar]
- 37.Maolanon AR, Madsen AS, Olsen CA. 2016. Innovative strategies for selective inhibition of histone deacetylases. Cell Chem. Biol. 23, 759–768. ( 10.1016/j.chembiol.2016.06.011) [DOI] [PubMed] [Google Scholar]
- 38.Ganesan A. 2016. Romidepsin and the zinc-binding thiol family of natural product HDAC inhibitors. In Successful drug discovery, vol. 2 (eds Fischer J, Childers WE), pp. 13–30. Weinheim, Germany: Wiley. [Google Scholar]
- 39.Lu XP, Ning ZQ, Li ZB, Pan DS, Shan S, Guo X, Cao HX, Yu JD, Yang QJ. 2016. Discovery and development of HDAC subtype selective inhibitor chidamide: potential immunomodulatory activity against cancers. In Successful drug discovery, Vol. 2 (eds Fischer J, Childers WE), pp. 89–114. Weinheim, Germany: Wiley. [Google Scholar]
- 40.Villadsen JS, Kitir B, Wich K, Friis T, Madsen AS, Olsen CA. 2014. An azumamide C analogue without the zinc-binding functionality. Med. Chem. Commun. 5, 1849–1855. ( 10.1039/C4MD00252K) [DOI] [Google Scholar]
- 41.Ganesan A. 2016. Multitarget drugs: an epigenetic epiphany . ChemMedChem 11, 1227–1241. ( 10.1002/cmdc.201500394) [DOI] [PubMed] [Google Scholar]
- 42.Cai X, Zhai HX, Wang J, Forrester J, Qu H, Yin L, Lai CJ, Bao R, Qian C. 2010. Discovery of 7-(4-(3-ethynylphenylamino)-7-methoxyquinazolin-6-yloxy)-N-hydroxyheptanamide (CUDC-101) as a potent multi-acting HDAC, EGFR, and HER2 inhibitor for the treatment of cancer. J. Med. Chem. 53, 2000–2009. ( 10.1021/jm901453q) [DOI] [PubMed] [Google Scholar]
- 43.Martinez S, Hausinger RP. 2015. Catalytic mechanisms of Fe(II)- and 2-oxoglutarate-dependent oxygenases. J. Biol. Chem. 290, 20 702–20 711. ( 10.1074/jbc.R115.648691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose NR, Ng SS, Mecinović J, Liénard BM, Bello SH, Sun Z, McDonough MA, Oppermann U, Schofield CJ. 2008. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. J. Med. Chem. 51, 7053–7056. ( 10.1021/jm800936s) [DOI] [PubMed] [Google Scholar]
- 45.Thinnes CC, England KS, Kawamura A, Chowdhury R, Schofield CJ, Hopkinson RJ. 2014. Targeting histone lysine demethylases — progress, challenges, and the future. Biochim. Biophys. Acta 1839, 1416–1432. ( 10.1016/j.bbagrm.2014.05.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McAllister TE, England KS, Hopkinson RJ, Brennan PE, Kawamura A, Schofield CJ. 2016. Recent progress in histone demethylase inhibitors. J. Med. Chem. 59, 1308–1329. ( 10.1021/acs.jmedchem.5b01758) [DOI] [PubMed] [Google Scholar]
- 47.Duan L, et al. 2015. KDM4/JMJD2 histone demethylase inhibitors block prostate tumor growth by suppressing the expression of AR and BMYB-regulated genes. Chem. Biol. 22, 1185–1196. ( 10.1016/j.chembiol.2015.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang J, et al. 2017. From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors. Bioorg. Med. Chem. Lett. 27, 2974–2981. ( 10.1016/j.bmcl.2017.05.016) [DOI] [PubMed] [Google Scholar]
- 49.Yin R, Mo J, Dai J, Wang H. 2017. Nickel(II) inhibits Tet-mediated 5-methylcytosine oxidation by high affinity displacement of the cofactor iron(II). ACS Chem. Biol. 12, 1494–1498. ( 10.1021/acschembio.7b00261) [DOI] [PubMed] [Google Scholar]
- 50.Erdmann A, Halby L, Fahy J, Arimondo PB. 2015. Targeting DNA methylation with small molecules: what's next? J. Med. Chem. 58, 2569–2583. ( 10.1021/jm500843d) [DOI] [PubMed] [Google Scholar]
- 51.Castillo-Aguilera O, Depreux P, Halby L, Arimondo PB, Goossens L. 2017. DNA methylation targeting: the DNMT/HMT crosstalk challenge. Biomolecules 7, 3 ( 10.3390/biom7010003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castellano S, Kuck D, Viviano M, Yoo J, López-Vallejo F, Conti P, Tamborini L, Pinto A, Medina-Franco JL, Sbardella G. 2011. Synthesis and biochemical evaluation of δ(2)-isoxazoline derivatives as DNA methyltransferase 1 inhibitors. J. Med. Chem. 54, 7663–7677. ( 10.1021/jm2010404) [DOI] [PubMed] [Google Scholar]
- 53.Asgatay S, Champion C, Marloie G, Drujon T, Senamaud-Beaufort C, Ceccaldi A, Erdmann A, Rajavelu A, Schambel P, Jeltsch A, Lequin O, Karoyan P, Arimondo PB, Guianvarc'h D. 2014. Synthesis and evaluation of analogues of N-phthaloyl-l-tryptophan (RG108) as inhibitors of DNA methyltransferase 1. J. Med. Chem. 57, 421–434. ( 10.1021/jm401419p) [DOI] [PubMed] [Google Scholar]
- 54.Morera L, Lübbert M, Jung M. 2016. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenet. 8, 57 ( 10.1186/s13148-016-0223-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaniskan HU, Martini ML, Jin J. 2018. Inhibitors of protein methyltransferases and demethylases. Chem. Rev. 118, 989–1068. ( 10.1021/acs.chemrev.6b00801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daigle SR, et al. 2013. Potent inhibition of DOT1 L as treatment of MLL-fusion leukemia. Blood 122, 1017–1025. ( 10.1182/blood-2013-04-497644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCabe MT, et al. 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations . Nature 492, 108–112. ( 10.1038/nature11606) [DOI] [PubMed] [Google Scholar]
- 58.Knutson SK, et al. 2014. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 13, 842–854. ( 10.1158/1535-7163.MCT-13-0773) [DOI] [PubMed] [Google Scholar]
- 59.Williams DE, et al. 2013. Nahuoic acid A produced by a Streptomyces sp. isolated from a marine sediment is a selective SAM-competitive inhibitor of the histone methyltransferase SETD8. Org. Lett. 15, 414–417. ( 10.1021/ol303416k) [DOI] [PubMed] [Google Scholar]
- 60.Xiong Y, et al. 2017. Discovery of potent and selective inhibitors for G9a-like protein (GLP) lysine methyltransferase. J. Med. Chem. 60, 1876–1891. ( 10.1021/acs.jmedchem.6b01645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sweis RF, et al. 2015. Discovery of A-893, a new cell-active benzoxazinone inhibitor of lysine methyltransferase SMYD2. ACS Med. Chem. Lett. 6, 695–700. ( 10.1021/acsmedchemlett.5b00124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan-Penebre E, et al. 2015. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 11, 432–437. ( 10.1038/nchembio.1810) [DOI] [PubMed] [Google Scholar]
- 63.Schiedel M, Robaa D, Rumpf T, Sippl W, Jung M. 2018. The current state of NAD+-dependent histone deacetylases (sirtuins) as novel therapeutic targets. Med. Res. Rev. 38, 147–200. ( 10.1002/med.21436) [DOI] [PubMed] [Google Scholar]
- 64.Suzuki T, et al. 2012. Design, synthesis, and biological activity of a novel series of human sirtuin-2-selective inhibitors. J. Med. Chem. 55, 5760–5773. ( 10.1021/jm3002108) [DOI] [PubMed] [Google Scholar]
- 65.Cui H, Kamal Z, Ai T, Xu Y, More SS, Wilson DJ, Chen L. 2014. Discovery of potent and selective sirtuin 2 (SIRT2) inhibitors using a fragment-based approach. J. Med. Chem. 57, 8340–8357. ( 10.1021/jm500777s) [DOI] [PubMed] [Google Scholar]
- 66.Wang X, Huang B, Suzuki T, Liu X, Zhan P. 2015. Medicinal chemistry insights in the discovery of novel LSD1 inhibitors. Epigenomics 7, 1379–1396. ( 10.2217/epi.15.86) [DOI] [PubMed] [Google Scholar]
- 67.Niwa H, Umehara T. 2017. Structural insight into inhibitors of flavin adenine dinucleotide-dependent lysine demethylases. Epigenetics 12, 340–352. ( 10.1080/15592294.2017.1290032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prusevich P, et al. 2014. A selective phenelzine analogue inhibitor of histone demethylase LSD1. ACS Chem. Biol. 9, 1284–1293. ( 10.1021/cb500018s) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maes T, et al. 2018. ORY-1001, a potent and selective covalent KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell 33, 495–511. ( 10.1016/j.ccell.2018.02.002) [DOI] [PubMed] [Google Scholar]
- 70.Mohammad HP, et al. 2015. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 28, 57–69. ( 10.1016/j.ccell.2015.06.002) [DOI] [PubMed] [Google Scholar]
- 71.Vianello P, et al. 2017. Thieno[3,2-b]pyrrole-5-carboxamides as new reversible inhibitors of histone lysine demethylase KDM1A/LSD1. Part 2: Structure-based drug design and structure–activity relationship. J. Med. Chem. 60, 1693–1715. ( 10.1021/acs.jmedchem.6b01019) [DOI] [PubMed] [Google Scholar]
- 72.Wapenaar H, Dekker FJ. 2016. Histone acetyltransferases: challenges in targeting bi-substrate enzymes. Clin. Epigenet. 8, 59 ( 10.1186/s13148-016-0225-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dahlin JL, et al. 2017. Assay interference and off-target liabilities of reported histone acetyltransferase inhibitors. Nat. Commun. 8, 1527 ( 10.1038/s41467-017-01657-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lau OD, et al. 2010. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell 5, 589–595. ( 10.1016/S1097-2765(00)80452-9) [DOI] [PubMed] [Google Scholar]
- 75.Lasko LM, et al. 2017. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.