Abstract
We explored the association of metal levels with subclinical atherosclerosis and epigenetic changes in relevant biological pathways. Whole blood DNA Infinium Methylation 450 K data were obtained from 23 of 73 middle age men without clinically evident cardiovascular disease (CVD) who participated in the Aragon Workers Health Study in 2009 (baseline visit) and had available baseline urinary metals and subclinical atherosclerosis measures obtained in 2010–2013 (follow-up visit). The median metal levels were 7.36 µg g−1, 0.33 µg g−1, 0.11 µg g−1 and 0.07 µg g−1, for arsenic (sum of inorganic and methylated species), cadmium, antimony and tungsten, respectively. Urine cadmium and tungsten were associated with femoral and carotid intima-media thickness, respectively (Pearson's r = 0.27; p = 0.03 in both cases). Among nearest genes to identified differentially methylated regions (DMRs), 46% of metal-DMR genes overlapped with atherosclerosis-DMR genes (p < 0.001). Pathway enrichment analysis of atherosclerosis-DMR genes showed a role in inflammatory, metabolic and transport pathways. In in silico protein-to-protein interaction networks among proteins encoded by 162 and 108 genes attributed to atherosclerosis- and metal-DMRs, respectively, with proteins known to have a role in atherosclerosis pathways, we observed hub proteins in the network associated with both atherosclerosis and metal-DMRs (e.g. SMAD3 and NOP56), and also hub proteins associated with metal-DMRs only but with relevant connections with atherosclerosis effectors (e.g. SSTR5, HDAC4, AP2A2, CXCL12 and SSTR4). Our integrative in silico analysis demonstrates the feasibility of identifying epigenomic regions linked to environmental exposures and potentially involved in relevant pathways for human diseases. While our results support the hypothesis that metal exposures can influence health due to epigenetic changes, larger studies are needed to confirm our pilot results.
This article is part of a discussion meeting issue ‘Frontiers in epigenetic chemical biology’.
Keywords: environmental metals, subclinical atherosclerosis, DNA methylation, cohort study
1. Introduction
Cardiovascular disease (CVD) is a major cause of mortality and morbidity worldwide. However, traditional cardiovascular risk factors do not completely explain the development of atherosclerotic disease [1,2]. Given the elevated burden of CVD, we need to identify novel factors and mechanisms for CVD development. Non-essential environmental metals are emerging as major contributing factors to cardiovascular health in populations exposed to low or moderate concentrations through air, food or water [3–7]. There is mounting evidence to support the idea that metals could be environmental determinants of both epigenetic marks and CVD [8–13], although epigenetic data from epidemiologic samples are scarce. While there is evidence to support a causal association of metals such as cadmium and inorganic arsenic with clinical cardiovascular events [3,5], fewer studies have evaluated the association of metals with subclinical CVD [4].
Our objective was to evaluate the association of metal concentrations with subclinical atherosclerosis and with epigenetic changes in relevant biological pathways for atherosclerosis. We first examined the association of urine metal concentrations (sum of inorganic and methylated arsenic, cadmium, antimony and tungsten) and measures of subclinical atherosclerosis (coronary calcium score, carotid and femoral intima-media thickness (IMT) and atherosclerosis presence overall and in carotid, femoral and coronary territories) in 73 men participating in the Aragon Workers Health Study (AWHS)-metal pilot study. Second, we evaluated the association of subclinical atherosclerosis and urine metal concentrations with DNA methylation markers in approximately 450 k single-nucleotide genomic positions measured in blood samples from 23 study participants. Third, we conducted an in silico analysis to explore biological pathways for the potential role of metal-induced DNA methylation in atherosclerosis. The AWHS is a cohort study of factors associated with metabolic abnormalities and subclinical atherosclerosis in a middle age population from Spain [1,14].
2. Material and methods
(a). Study population
The AWHS is a longitudinal cohort study based on the annual health examinations of 5400 workers of a car assembly plant in Figueruelas (Zaragoza, Spain). The study design and population characterization have been previously described [1,14]. The AWHS included a baseline examination (2009–2010) and a follow-up visit with measures of subclinical atherosclerosis (2011–2013). In 2010, we selected a pilot sample of 174 AWHS baseline participants, mostly men, to evaluate metal exposure in the AWHS and to generate preliminary data to support metal determination in the bigger cohort (AWHS-metal pilot study). After excluding women, 74 of these participants also participated in the follow-up visit and had prospectively collected subclinical atherosclerosis measures. We excluded one participant owing to missing hypertension status. No other participant missed relevant covariates, leaving 73 participants for the analysis. The AWHS study was approved by the Ethics Committee for Clinical Research at the Institutional Review Board of Aragón (CEICA). All study participants provided written informed consent.
(b). Subclinical atherosclerosis
The AWHS design and baseline characteristics from the 2009–2010 visit have been reported elsewhere [1,14]. In 2011–2013, coronary calcium scoring was performed using non-contrast ECG-gated prospective acquisition by a 16 multidetector computed tomography scanner (Mx 8000 IDT 16, Philips Medical Systems, Best, the Netherlands). During a single breath hold, images were acquired from the tracheal bifurcation to below the base of the heart. Scan parameters were 8 × 3 mm collimation, 220 mm field of view, 120 kVp, 55 mA and 3 mm section thickness. Coronary calcium was quantified with calcium scoring software (Workspace CT Viewer, Philips Medical Systems) that follows the Agatston method [15]. We defined a dichotomous variable for the presence of coronary calcium, which was positive if the Agatston score was greater or equal than 1.
Intima-media thickness in the carotid and femoral arteries was determined using the Philips IU22 ultrasound system (Philips Healthcare, Bothell, Washington). Ultrasound images were acquired with linear high-frequency two-dimensional probes (Philips Transducer L9-3, Philips Healthcare) following the Bioimage Study protocol [16]. Examination of the carotid territory included the terminal portion (10 mm) of the common carotid, the bulb and the initial portion (10 mm) of the internal and external carotid arteries. Examination of the femoral territory included the distance between the intima and media layers by ultrasound in a 2 cm segment proximal to the bifurcation. For each participant, average intima-media thickness values were obtained from three determinations at each site in both sides. Carotid and femoral intima-media thickness values were then defined as the maximum average value across all sites and sides for the carotid and femoral territories, respectively. Plaque, both in the carotid and femoral territories, was defined as a focal structure protruding more than 0.5 mm into the lumen or reaching a thickness of more than 50% of the surrounding intima [16]. In all cases, plaques are recorded in both longitudinal and transverse planes to take into consideration circumferential asymmetry. We also defined a combined outcome for the presence of atherosclerosis for participants with coronary calcium or with carotid or femoral plaque.
(c). Urine metals
Spot urine samples were collected in polypropylene tubes, frozen within 1–2 h of collection and stored at −70°C in the Occupational Medicine Service Unit, Opel Factory, Figueruelas (Spain). In 2010, up to 1.0 ml of urine from each participant was transported on dry ice to the Trace Element Laboratory of the Institute of Chemistry, University of Graz, Austria. Urine samples were thawed, and arsenic, cadmium, antimony and tungsten were measured using inductively coupled plasma-mass spectrometry (ICPMS), as previously described by Scheer et al. [17]. Arsenic species (inorganic arsenic (iAs; arsenite, arsenate), methylarsonate (MMA) and dimethylarsinate (DMA)) were measured in the same urine samples using anion exchange high-performance liquid chromatography coupled with ICPMS. The limits of detection were 0.1 µg l−1 for arsenic species and 0.05 µg l−1 for cadmium, antimony and tungsten. None of the samples included in this study was below the limit of detection. The interassay coefficients of variation for arsenite, arsenate, MMA, DMA, cadmium, antimony and tungsten were 14.7%, 6.9%, 6.4%, 6.0%, 8.7%, 30.0% and 14.5%, respectively. For every batch of 79 samples, 10 of the samples were analysed in duplicate. The mean intra-assay coefficients of variation for arsenite, arsenate, MMA, DMA, cadmium, antimony and tungsten were 4.46%, 4.23%, 3.02%, 1.49%, 1.34%, 3.29%, and 0.57%, respectively. The estimated intra-assay intraclass correlations (ICCs) in these samples for arsenite, arsenate, MMA, DMA, cadmium, antimony and tungsten were 0.990, 0.996, 0.992, 0.987, 0.994, 0.966 and 0.990, respectively. To account for urine dilution, urine metal concentrations (micrograms per litre) were divided by urine creatinine concentrations (grams per litre) and reported in micrograms per gram creatinine.
(d). Microarray epigenomic determinations
Each participant also provided a sample of blood after overnight fasting (longer than 8 h) for laboratory analyses and for biobanking. Genomic DNA was extracted from peripheral blood cells by use of commercial reagents (FlexiGene DNA Kit, Qiagen, USA). DNA purity and concentration were determined by absorbance at 260 nm (A260) and 280 nm (A280) using Nanovue (GE Healthcare, Munich, Germany). DNA was stored in 0.3 ml vials at −80°C. gDNA (500 ng) was bisulfite converted using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA) according to the manufacturer's instructions. Bisulfite-converted DNA methylation was measured using a microarray-based analysis with the Infinium HumanMethylation450 BeadChip (450 k array). DNA quality checks, bisulfite modification, hybridization, data normalization, statistical preprocessing and β-value calculation, which represents the proportion of cytosines (Cs) in bisulfite-converted DNA, at specific locations, were performed using the R package minfi [18]. CG dinucleotide (CpG) sites with a detection p-value higher than 0.01 were removed. Intra-array normalization was done using quantile normalization. In exploratory analysis, we detected and corrected a potential batch effect by slide using the function ‘combat’ from the ‘sva’ R package [19].
(e). Other variables
Participants were interviewed and physically examined by trained and certified staff following a standard protocol [14]. Baseline information on sociodemographic data (age, sex and education), smoking status (never, former and current), completed education level (less than high school and high school or beyond) and medication use was obtained by questionnaire during the interview. The examination included anthropometry (height, weight and waist circumference) and blood pressure measurements. Blood pressure was measured three consecutive times using an automatic oscillometric sphygmomanometer OMRON M10-IT (OMRON Healthcare Co., Ltd, Japan) with the participant sitting after a 5-min rest. Hypertension was defined as a measured blood pressure ≥140/90 mmHg, a self-reported diagnosis or current use of antihypertensive medication.
Hypercholesterolaemia was defined as a total cholesterol level of greater than or equal to 200 mg dl−1, a self-reported diagnosis or current use of lipid-lowering medication. Diabetes was defined as a diagnosis of diabetes in the clinical record, current use of antidiabetic medication, a fasting serum glucose greater than or equal to 126 mg dl−1 or a haemoglobin A1c (HbA1c) of greater than or equal to 6.5%.
(f). Statistical methods
(i). Summary statistics for the association of metals and subclinical atherosclerosis
We descriptively report median and interquartile range of urine biomarkers, and the proportion of individuals with subclinical atherosclerosis binary endpoints, overall and by subgroups. To estimate multi-adjusted correlations of atherosclerosis with urine metal measures, we first run separate regression models of standardized atherosclerosis measures (z scores of coronary artery calcium, and carotid and femoral intima-media thickness) and metal-level measurements (arsenic, cadmium, antimony and tungsten) (continuous dependent variables) to obtain residuals adjusted by age, smoking status, body mass index, hypertension, dyslipidaemia and diabetes (independent variables). Second, we estimated Pearson correlation coefficients on the adjusted residuals recalibrated to the corresponding overall mean.
(ii). Differentially methylated regions
Methylation proportions were represented as β-values ranging from 0 to 1. β-values were converted into M-values (logit transformation) before the statistical analysis. We identified differentially methylated regions (DMRs) with respect to subclinical atherosclerosis (i.e. ‘atherosclerosis-DMRs') in coronary, carotid and femoral territories (both continuous and binary endpoints) and metal concentrations (i.e. ‘metal-DMRs’) by using the ‘bumphunter’ function in R package ‘minfi’ [18] with the following adjustment variables: age, smoking status (never, former and current smoking), body mass index and empirical cell estimates (B cell, CD4+ and CD8+ T cells, granulocytes, monocytes and natural killer cells) [20]. We then ranked DMRs by calculating an exact p-value (i.e. ‘p-value area’ in the ‘bumphunter’ output) for each region by creating 1000 bootstrap samples. The p-value was defined as the per cent of regions obtained from the bootstrap samples that showed an area as extreme as in the observed region. We conducted annotation of DMRs to the nearest gene according to the Homo sapiens data from UCSC build hg19 as implemented by the default behaviour of the ‘matchGenes’ function in ‘minfi’ R package [18]. Among DMRs with a p-value of <0.05, we created a list of unique genes attributed the union of the identified atherosclerosis-DMRs, and a corresponding list of unique genes attributed to the union of the identified metal-DMRs. Finally, we evaluated the overlap of genes from atherosclerosis-DMRs and metal-DMRs by using a hypergeometric test [21].
(iii). Molecular characterization of atherosclerosis and in silico analysis of potential metal-related epigenetic changes underlying mechanisms in atherosclerosis
The molecular characterization of atherosclerosis has been performed through manual review of the literature to identify key effector proteins in atherosclerosis (Biological Effectors Database (BED), Anaxomics Biotech). Subsequently, we first evaluated the relatedness of proteins encoded by the identified atherosclerosis-DMR genes with respect to the atherosclerosis protein effectors from the BED database (protein-to-protein interactions) by applying an artificial neuronal network (ANN) algorithm (Anaxomics Biotech) [22,23]. The ANN algorithm provides a predictive score (from 0 to 100%) that quantifies the amount and strength of relationships between the evaluated proteins. Each score is associated with a p-value that describes the probability of the results being a true positive result, wherein a score greater than 91% indicates a very strong relationship with a p-value <0.01; a score between 76–91% indicates a strong relationship with a p-value of 0.01–0.05; a score between 40–76% indicates a medium–strong relationship with a p-value of 0.05–0.25; and a score less than 40% indicates a weak relationship with a p-value >0.25. Second, we conducted a hypergeometric enrichment analysis [21] to identify the enriched protein sets in the data using KEGG [24], Gene Ontology [25], PHARMGKB [26], SMPDB and TRRUST [27]. The enrichment analysis aims to provide a global view of differential data, as it takes into account the accumulated biological knowledge of how genes work together, allowing the identification of predominant or enriched pathways and clinical conditions. Finally, we displayed protein interaction networks with Cytoscape (v. 3.5.1). The protein interactions were obtained from public datasets including KEGG [24], Reactome [28], IntAct [29], BIOGRID [30], HDPR [31] and MINT [32]. Only proteins encoded by identified atherosclerosis- and metal-DMRs that interacted with well-established atherosclerosis-effector protein from BED were included in the protein-to-protein interaction networks (figure 1).
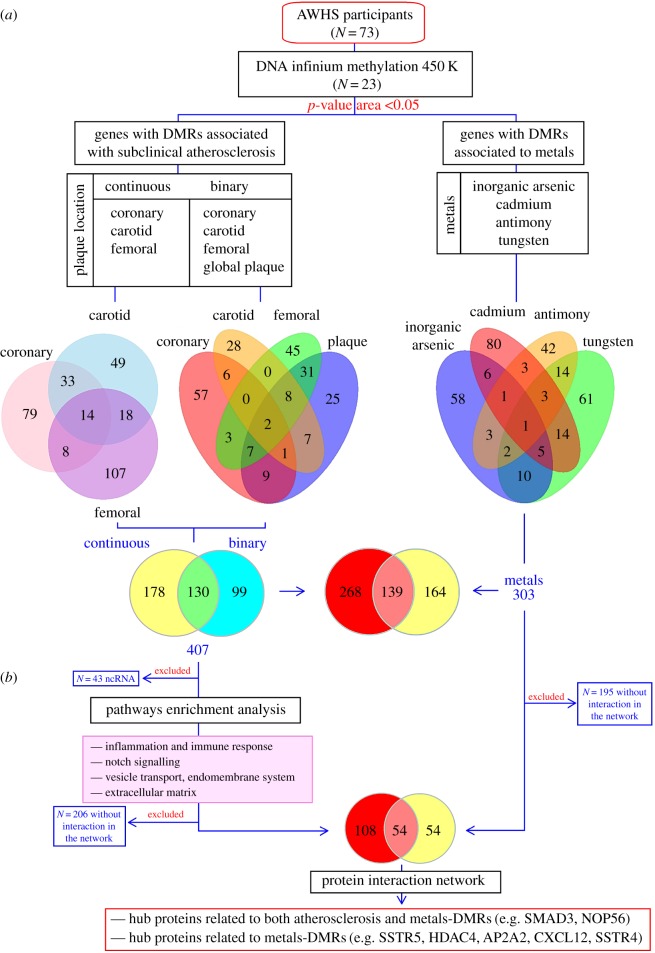
Figure 1.
Summary of the in silico analysis and results obtained from DMRs identified in the AWHS-metal pilot study. (a) A subset of 73 AWHS participants had available preliminary metal biomarker and subclinical atherosclerosis data. Among these, 23 participants additionally had whole blood DNA Infinium Methylation 450 K data. We identified 407 and 303 DMRs with respect to subclinical atherosclerosis measures and metals, respectively (p-value area less than 0.05 as estimated by the bumphunter function in the R package ‘minfi’ [18]). Among these regions, 139 were in common for both DMR lists. The exact number of differentially methylated genes for each condition (subclinical atherosclerosis and metals) is shown in the Venn diagrams. (b) After excluding non-protein-coding genomic regions (e.g. non-coding RNAs (ncRNAs)), pathway enrichment analysis showed enriched categories related to inflammation and cellular transport. We subsequently evaluated protein interaction networks among genes encoding proteins with at least one connection with known atherosclerosis-effector proteins, first by ANNs and second by graphically displaying the global protein network. We observed hub proteins, which were directly or indirectly related to metals, pointing to the potential role of epigenetic mechanisms in explaining metal-related cardiovascular risk.
3. Results
(a). Metal biomarkers and subclinical atherosclerosis measures
The median urinary metal levels were 7.36 µg g−1, 0.33 µg g−1, 0.11 µg g−1 and 0.07 µg g−1, for the sum of inorganic arsenic, cadmium, antimony and tungsten, respectively (electronic supplementary material 1 (ESM1), supplementary table 1). Older, more educated, obese participants, participants with diabetes and participants without dyslipidaemia showed higher arsenic concentrations. Older, less educated and non-obese participants had higher cadmium concentrations. For antimony and tungsten, the metal concentrations did not substantially differ by participant characteristics, except for smoking. Ever smokers displayed higher arsenic, cadmium, antimony and tungsten concentrations compared with never smokers (ESM1, supplementary table 1). The adjusted Pearson correlation coefficients for metal levels ranged from −0.08 for the correlation between arsenic and cadmium to 0.24 for the correlation between tungsten with arsenic and cadmium (ESM1, supplementary figure 1).
The prevalence of atherosclerosis endpoints was 74.0%, 38.4%, 39.7%, and 54.8%, respectively, for combined atherosclerosis, presence of coronary calcium and presence of carotid and femoral plaque (table 1). Older, less educated, obese participants, participants with hypertension and participants with diabetes showed a higher prevalence of subclinical atherosclerosis, generally for all atherosclerosis endpoints. Participants with dyslipidaemia did not show differences in the proportion of atherosclerosis endpoints, except for a higher proportion with the presence of coronary calcium, compared with participants without dyslipidaemia. In analyses adjusted for age, obesity and ever smoking status, there was a positive correlation of cadmium with femoral intima-media thickness (IMT) and tungsten with carotid IMT (data not shown), which remained after additional adjustment for hypertension, diabetes and dyslipidaemia (ESM1, supplementary figure 1).
Table 1.
Number of individuals (%) with subclinical atherosclerosis endpoints by participant characteristics.
| N | combined atherosclerosis | presence of coronary calcium | presence of carotid plaque | presence of femoral plaque | |
|---|---|---|---|---|---|
| overall | 73 | 54 (74.0) | 28 (38.4) | 29 (39.7) | 40 (54.8) |
| age (years) | |||||
| ≤50 | 43 | 28 (65.1) | 11 (25.5) | 14 (32.6) | 20 (46.5) |
| >50 | 30 | 26 (86.7) | 17 (56.7) | 15 (50.0) | 20 (66.7) |
| completed education | |||||
| ≥high-school | 32 | 21 (65.6) | 11 (34.4) | 11 (34.4) | 17 (53.1) |
| <high-school | 41 | 33 (80.4) | 17 (41.5) | 18 (43.9) | 23 (56.1) |
| body mass index (kg m−2) | |||||
| <30 | 54 | 38 (70.4) | 21 (38.9) | 20 (37.0) | 27 (50.0) |
| ≥30 | 19 | 16 (84.2) | 7 (36.8) | 9 (47.4) | 13 (68.4) |
| smoking status | |||||
| never | 16 | 9 (56.3) | 5 (31.3) | 4 (25.0) | 4 (25.0) |
| ever | 57 | 45 (79.0) | 23 (40.4) | 25 (43.9) | 36 (63.2) |
| hypertension | |||||
| no | 41 | 28 (68.3) | 14 (34.1) | 10 (24.4) | 21 (21.2) |
| yes | 32 | 26 (81.3) | 14 (43.8) | 19 (59.4) | 19 (59.3) |
| diabetes | |||||
| no | 67 | 49 (73.1) | 26 (38.8) | 26 (38.8) | 36 (53.7) |
| yes | 6 | 5 (83.3) | 2 (33.3) | 3 (50.0) | 4 (66.7) |
| dyslipidaemia | |||||
| no | 27 | 20 (74.0) | 9 (33.3) | 11 (40.7) | 15 (55.5) |
| yes | 46 | 34 (73.9) | 19 (41.3) | 18 (39.1) | 25 (54.3) |
| inorganic arsenic (µg g−1) | |||||
| ≤7.36 | 36 | 28 (77.8) | 15 (41.7) | 17 (47.2) | 18 (50.0) |
| >7.36 | 37 | 26 (70.3) | 13 (35.1) | 12 (32.4) | 22 (59.5) |
| cadmium (µg g−1) | |||||
| ≤0.33 | 37 | 25 (67.6) | 15 (40.5) | 12 (32.4) | 16 (43.2) |
| >0.33 | 36 | 29 (80.6) | 13 (36.1) | 17 (47.2) | 24 (66.7) |
| antimony (µg g−1) | |||||
| ≤0.11 | 36 | 24 (66.7) | 13 (36.1) | 14 (38.9) | 17 (47.2) |
| >0.11 | 37 | 30 (81.1) | 15 (40.5) | 15 (40.5) | 23 (62.2) |
| tungsten (µg g−1) | |||||
| ≤0.07 | 37 | 29 (78.4) | 14 (37.8) | 16 (43.2) | 23 (62.1) |
| >0.07 | 36 | 25 (69.4) | 14 (38.9) | 13 (36.1) | 17 (47.2) |
(b). Genes annotated to atherosclerosis-differentially methylated regions and the interaction of their encoded proteins with atherosclerosis-effector proteins
We identified a total of 308 and 229 genes nearest to the identified DMRs with respect to continuous (coronary calcium score, and carotid and femoral intima-media thickness) and dichotomous (combined atherosclerosis, positive coronary calcium and carotid and femoral plaque) subclinical atherosclerosis measures, respectively (figure 1). Among these, 130 genes were common for both continuous and dichotomous atherosclerosis measures (figure 1a, right). Overall, there were 407 unique genes associated with atherosclerosis-DMRs (electronic supplementary material 2 (ESM2)).
Among the 407 genes, we identified 364 genes encoding proteins that were included in the protein interaction networks and ANN analysis. Two hundred and seventy protein-coding genes (of 364) were linked with well-known atherosclerosis effectors (figure 1b; electronic supplementary material 3 (ESM3)). In ANN analysis, a total of 58 protein-coding genes were predicted to be linked to atherosclerosis or its related factors with a medium–strong relationship (ANN score greater than 40%). Among these, the proteins encoded by IRF1, MSR1 and PF4 genes had been previously identified as protein effectors for atherosclerosis. The genes showing the top 5 ANN scores were HLA-DPA1, MSR1, PF4, IRF1 and SNTB1 (ESM3). In protein interaction networks, which display biological connections between proteins with a known role in atherosclerosis and identified proteins from our results, we identified relevant hub proteins, such as proteins encoded by APP, SMAD3, MAPK10, GNAI1 and HLA-DPA1 (the numbers of connections in the network were 72, 24, 18, 14 and 13, respectively) (figure 2).
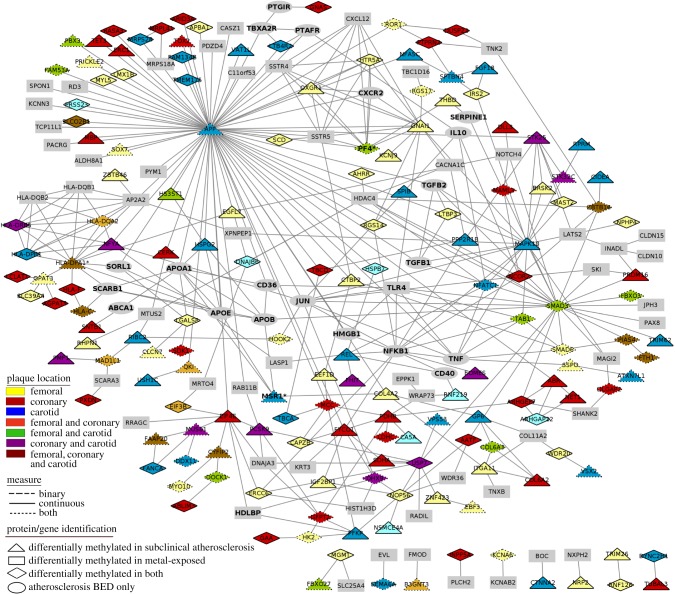
Figure 2.
Protein interaction network between protein attributed to metal and subclinical atherosclerosis-DMRs in the AWHS-pilot study and atherosclerosis-effector protein from public databases included in the BED.
(c). Enrichment analysis: relationship between methylation data with biological pathways and clinical conditions
The pathways and biological processes enriched within the genes related to atherosclerosis-DMRs were assessed through a hypergeometric enrichment analysis approach [19] (ESM1, supplementary table 2). Twenty-one protein sets were found enriched among the 364 coding genes attributed to atherosclerosis-DMRs (figure 1 and ESM1, supplementary table 2). The reported enriched protein sets were mainly related to immune response and inflammation (including HLA-DPA1, NFYA, HLA-DPB1, HLA-C and HLA-DQA2 genes with strong ANN scores from previous ANN analysis); extracellular matrix (including LTBP3 and COL4A2 genes with medium–strong ANN scores); altered metabolism (including PDHB and PCSK9 genes associated with sterol regulatory element-binding transcription factor-1 (SREBF1) with strong ANN scores) and endomembrane system and vesicle transport (including the PTPRN2 gene with a medium–strong ANN score) (ESM1). Notch signalling categories were also enriched, although the genes included in these categories showed weak evidence of a link with atherosclerosis based on the ANN scores.
(d). Metal-related differentially methylated regions and overlap with atherosclerosis-differentially methylated regions
In this study, we identified a total of 303 unique genes annotated to metal-DMRs. Among these genes, approximately 50% were associated with more than one metal exposure (figure 1a, right). However, there were also 58, 80, 42 and 61 genes that were uniquely associated with arsenic, cadmium, antimony and tungsten, respectively. To explore the potential role of metals in explaining atherosclerosis-related DNA methylation changes, we identified metal-DMRs and evaluated their overlap with atherosclerosis-DMRs. We observed that approximately 46% of the differentially methylated genes in metals (139 genes) were also differentially methylated in subclinical atherosclerosis (figure 1b; ESM1, supplementary table 3). (The estimated probability of finding these results by chance using a hypergeometric test was less than 0.001.)
We also explored whether proteins encoded by metal-DMR genes directly interacted with known atherosclerosis effectors or proteins encoded by atherosclerosis-DMR genes. In protein interaction analysis, 108 proteins attributed to metal-DMR genes showed any degree of relatedness to BED effectors (top 5: PF4, ANN 83%; IRF1, ANN 75%; PCSK9, ANN 72%; LTBP3, ANN 71% and HLA-DPB1, ANN 70%) (figure 2; ESM3). Moreover, 54 proteins attributed exclusively to metal-DMR genes directly interacted with at least one of the proteins attributed to atherosclerosis-DMR genes or the BED effectors (figure 1b; electronic supplementary material 4 (ESM4)). Figure 2 summarizes the protein-to-protein interactions of protein encoded by identified atherosclerosis and metal-DMR genes with known atherosclerosis effectors. We observed hub proteins in the network, such as the ones encoded by SMAD3 and NOP56 (12 and 6 connections, respectively), which were associated with both atherosclerosis- and metal-DMR genes. Other hub proteins, such as the ones encoded by SSTR5, HDAC4, AP2A2, CXCL12 and SSTR4 (7, 7, 6, 5 and 5, respectively), were only associated with metal-DMR genes, but did have connections with atherosclerosis effectors (figure 2; ESM4).
4. Discussion
We identified a total of 364 unique genes annotated to atherosclerosis-DMRs, which encoded proteins with existing interactions with known atherosclerosis-effector proteins in protein-to-protein interaction networks. Moreover, we identified 108 genes attributed to metal-DMRs directly related to known atherosclerosis effectors, which largely overlapped with atherosclerosis-DMR genes. While our pilot study cannot be used to establish firm conclusions, our analysis indicates the feasibility of identifying associations between metal levels, epigenetic markers and measures of subclinical atherosclerosis that may correspond to epigenetic mechanisms induced by metal exposures in relevant biological pathways.
Altered DNA methylation is a novel factor potentially involved in the development and progression of CVDs. In our data, some genes were differentially methylated for more than one atherosclerosis location and endpoint. Many of these genes have been consistently reported as DMRs in relation to atherosclerosis in genome-wide studies [11,13,33–35]. For instance, KCTD5 (coronary and femoral), GDAP2 (combined atherosclerosis and femoral) and CHST8 (combined atherosclerosis and femoral) were differentially methylated in previous studies comparing human carotid plaques of patients with cerebrovascular events (n = 19) versus asymptomatic counterparts (n = 19) [35]. MYO10 (combined atherosclerosis and femoral) and THSD4 (coronary and femoral) were differentially methylated comparing 15 post-mortem donor-matched atherosclerotic versus normal aortic sample pairs obtained from a Spanish population [11]. THSD4 was also differentially methylated in coronary arteries (n = 6) compared with internal mammary arteries (n = 6) and great saphenous vein (n = 6) in Russian men who underwent surgery for severe coronary artery stenosis [34]. This study also reported DMRs in C1QTNF7 (combined atherosclerosis and coronary), MS4A4A (combined atherosclerosis and coronary) and ANXA13 (combined atherosclerosis and femoral). Conversely, DMRs in two protein-encoding genes, the adhesion G protein-coupled receptor A1 (ADGRA1, also known as GPR123) and the fibrosin-like 1 (FBRSL1), were differentially methylated with respect to all atherosclerosis measures (data not shown), but these genes have not been related to atherosclerosis before. Thus, future studies to confirm the findings are needed. Other genes that in our study were associated with DMRs with respect to only one vascular territory (see ESM2, supplementary table 2) have also been reported before, including PXDN [13], ABR [11,33], PLEKHA4 [34], C6orf10 [35], PRDM16, INPP5A [11], MSR1 [34], ABHD8 [35], FAM134B [11], IRS2, COL4A2, MSRA and IGF2BP1 [11] (ESM3). Overall previous studies evaluated clinical cardiovascular outcomes. However, our data suggest that methylation changes may occur early before the cardiovascular event is produced.
Importantly, the results of enrichment pathway analyses reasonably reflected the well-known involvement of inflammation in atherosclerosis [36,37], because most of the significant differentially methylated genes were related to chronic inflammation and associated pathways such as the RFX transcription factor complex (RFXAP, RFXANK and RFX5), which activates MCH-II transcription [38], or Notch receptors and their ligands, which play a major role in the modulation of numerous cellular functions in macrophages, endothelial and vascular smooth muscle cells, particularly under inflammatory conditions [39–41]. Genes related to inflammation also were the most important hubs in networks of proteins related to atherosclerosis in our study. For instance, the amyloid beta A4 protein (encoded by APP), widely known for its role in Alzheimer's disease, has also been associated with inflammatory vascular dysfunction [42,43]. The interferon regulatory factor-1 (a transcription factor encoded by IRF1) is involved in inflammatory processes, by regulating the transcription of a set of target genes that play essential roles in various physiological and pathological processes [44]. Other hub proteins in our data have also been related to inflammatory responses in the literature, including SMAD3 [45,46], the NF-κB protein REL [47], the serotonin receptor HTR5A [48], the leukotriene B4 receptor 2 (LTB4R2) [49], the nuclear receptor corepressor 2 (NCOR2) [50], the nuclear factor of activated T-cells 1 (NFATC1) [51], the DExH-box helicase 9 (DHX9) [52] and the TGF-β activated kinase 1 binding protein 1 (TAB1) [53].
The Gene Ontology term ‘extracellular matrix’ was additionally enriched in our list of atherosclerosis-DMR genes. Extracellular matrix not only provides the structural integrity of the plaques, but participates in several key events such as cell migration and proliferation, lipoprotein retention and thrombosis [54]. In protein-to-protein interaction networks, we identified hub proteins also involved in plaque stability. For instance, MAPK10 has been linked to foam cell formation by inducing scavenger receptor expression [55]; NFATC1 [56] and HSPG2 [57] have been shown to promote excessive angiogenesis, a key feature of vulnerable atherosclerotic plaques; and the platelet factor 4 (PF4) has been involved in atherosclerosis cell recruitment and homeostasis [58]. Other pathways related to cardiometabolic factors appear to be also enriched, including a protein set related to the SREBF1. SREBFs are a family of transcription factors that mediate cholesterol metabolism by increasing the transcription of their downstream target LDL receptor. The increase in LDL receptors could lead to increased LDL transport into the lesions, contributing to foam cell formation [59]. Overall, protein-encoding genes from differential methylation data analysis show a strong association with atherosclerosis' pathophysiology.
Pathways associated with the endomembrane system, a complex network of membrane-bound compartments (such as the nuclear envelope, the endoplasmic reticulum, the Golgi apparatus, the plasma membrane and dynamic vesicles) also showed enrichment among atherosclerosis-DMR genes. The endomembrane system plays a key role in metal-ion homeostasis within the cell. Specific sets of transporters function in each cellular compartment to maintain cell homeostasis, as essential metal ions are vital for several life processes. However, non-essential metals have no known physiological role and can compete for transporter and binding sites for essential metals as a mechanism for toxicity [60]. In experimental studies, exposure to non-essential metals has been related to pro-oxidant properties [61], altered signalling pathways [62,63], endocrine disruption functions [64] and direct DNA and protein damage [65]. In recent years, there has been an increasing body of evidence pointing to the possibility that metals can alter epigenetic factors and cardiovascular risk [4]. Among the metal-DMR genes, we identified three genes encoding atherosclerosis-effector proteins (IRF1, PF4 and TLR4), which are either involved in unbalanced lipid metabolism or chronic inflammation of the arterial wall [66–68]. Others genes, including SMAD3, HLA-C, GNAS or SDK1, have been previously reported as differentially methylated in CVDs [11,13,34,35], and some of these had a strong and medium–strong score of relationship with atherosclerosis in our ANN analysis. In addition to the large overlap of atherosclerosis- and metal-DMRs in our data, there were 54 genes exclusively differentially methylated with respect to metal exposure that encoded protein interacting directly with at least one known effector protein or the differentially methylated genes in atherosclerosis and could, thus, also influence atherosclerosis pathways.
Our study is not free from limitations. First, we cannot ignore the possibility of having obtained spurious results by chance and we could not protect our results from a potentially high false-negative rate. Second, we only explored a subset of the genome, which may yield a partial view of the biologically relevant methylation sites in the genome. Third, smoking is a source of metals and an established cardiovascular risk factor. Thus, even if we adjusted statistical models for active smoking, we cannot discard residual confounding by smoking. Accordingly, we conducted post hoc exploratory analysis to evaluate the overlap of genes attributed to metal-DMRs with genes attributed to active smoking DMRs. Among 107 genes related to smoking DMRs, only 39 were also differentially methylated with both metals and atherosclerosis (data not shown), thus suggesting that specific metals may potentially have specific epigenetic signatures when compared with smoking, which is a mixture of compounds, including not only metals but also other compounds.
Fourth, the functional implication of our findings is unclear. We assumed that the identified DMRs control the protein coding of the nearest gene. Some DMRs, however, may be regulatory of other genomic regions in trans. In our data, the identified DMRs were mostly located inside intron and exon regions (data not shown). Thus, it is also possible that methylation may have a role in the regulation of splicing inducing a change in the isoform expression [69]. Moreover, genes such as SDK1, NINJ2, DIP2C, SLC25A21, PRDM16, GNAI1, RHPN1 and HS3ST1 that are associated with DMRs in our data were reported as differentially expressed in human atherosclerotic plaque [70]. Functional studies will be needed to support the causality of the reported pathways and the mechanisms underlying epigenetic changes for the cardiovascular toxicity of metals. Fifth, we measured DNA methylation from bisulfite-converted DNA, which does not distinguish methyl cytosine from hydroxymethyl cytosine. A previous study reported a statistically significant correlation between global DNA methylation and hydroxymethylation at the population level [71]. Future studies evaluating the role of site-specific DNA hydroxymethylation in atherosclerosis development are needed. Sixth, IMT in the distal common carotid is not a well-established measure of atherosclerosis [72], although it has been used in several epidemiological studies [73,74]. In our statistical framework, we attempted to summarize the individual likelihood of having atherosclerosis in the whole carotid territory by using the maximum average IMT value at both carotid territories including multiple sites, not only the terminal portion of the common carotid. Finally, while we had prospectively collected subclinical atherosclerosis measures, we did not have available information on baseline atherosclerosis. Repeated subclinical atherosclerosis measures would be needed to give a view of the potential role of epigenetics of metals in the change in atherosclerosis over time.
5. Conclusion
Our results showed a large overlap for metal- and atherosclerosis-DMRs, consistent with the hypothesis that metals can affect methylation profiles, which in turn may be involved in the CVD development. In addition, in protein interaction networks, proteins attributed to DMRs exclusively in association with metal exposure could influence the effect of proteins related to atherosclerosis. Our integrative in silico analysis demonstrates the feasibility of identifying epigenomic regions linked to environmental exposures and potentially involved in relevant pathways for human diseases. While our results support the possibility that metal exposures can influence health due to epigenetic changes, larger studies are needed to confirm our pilot results.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank the Bioinformatics Unit at the Institute for Biomedical Research Hospital Clinic of Valencia INCLIVA (especially Alejandro Dominguez-Lucas and Miguel Herreros) for their kind support and troubleshooting when using the computation server at INCLIVA for the statistical analysis in this study.
Data accessibility
The computing code is available upon request to the corresponding author. The data are not available as the steering committee and the participants did not approve unrestricted data sharing at the time of ethical approval of the study and data sharing was not included in the consent form. The lists of genes attributed to DMRs supporting protein interaction networks in this article have been uploaded as part of the electronic supplementary material, files ESM2, ESM3 and ESM4.
Authors' contributions
A.L.R.-C. and M.T.-P. conceived and designed the study. W.Y.T., Z.S., G.D.M., P.R.-G., K.F., W.G., M.L.-L., J.A.C., E.G. and M.T.-P. performed acquisition of data. A.L.R.-C., A.F.-T., V.A.-F., V.L.-T., C.L.-A., F.J.C., A.N.-A., E.G. and M.T.-P. analysed and interpreted the data. A.L.R.-C., A.F.-T., W.Y.T., Z.S., G.D.M., P.R.-G., V.A.-F., V.L.-T., K.F., W.G., C.L.-A., M.L.-L., J.A.C., F.J.C., A.N.-A., E.G. and M.T.-P. drafted the article or revised it critically for important intellectual content. A.L.R.-C., A.F.-T., W.Y.T., Z.S., G.D.M., P.R.-G., V.A.-F., V.L.-T., K.F., W.G., C.L.-A., M.L.-L., J.A.C., F.J.C., A.N.-A., E.G. and M.T.-P. finally approved the version to be published.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the Strategic Action for Research in Health Sciences (CP12/03080, PI10/0082, PI13/ 01848, PI15/00071 and PI11/00726), GRUPOS 03/101; PROMETEO/ 2009/029 and ACOMP/2013/039 from the Valencia Government; GRS/279/A/08 from Castilla-Leon Government; the European Network of Excellence Ingenious Hypercare (EPSS-037093) from the European Commission; CIBER Fisiopatologia Obesidad y Nutricion (CIBERobn) (CIBER-02-08-2009, CB06/03 and CB12/03/30016) and CIBER de Diabetes y Enfermedades Metabolicas Relacionadas (CIBERDEM). The Strategic Action for Research in Health Sciences, CIBEROBN and CIBERDEM are initiatives from Carlos III Health Institute Madrid and the Spanish Ministry of Economy and Competitiveness and are co-funded with European Funds for Regional Development (FEDER). A.N.-A. was supported by National Institute of Environmental Health Sciences (R01ES025216). The AWHS was co-funded by the local Government of Aragon (Spain) through the Institute for Health Sciences of Aragon and the National Center for Cardiovascular Research at the Carlos III Health Institute in Madrid. The authors are participants of the EU-funded COST Action CMST COST Action CM1406 EPICHEM (Epigenetic Chemical Biology).
References
- 1.Laclaustra M, et al. 2016. Femoral and carotid subclinical atherosclerosis association with risk factors and coronary calcium. J. Am. Coll. Cardiol. 67, 1263–1274. ( 10.1016/j.jacc.2015.12.056) [DOI] [PubMed] [Google Scholar]
- 2.Patel MR, Peterson ED, Dai D, Brennan JM, Redberg RF, Anderson HV, Brindis RG, Douglas PS. 2010. Low diagnostic yield of elective coronary angiography. N. Engl. J. Med. 362, 886–895. ( 10.1056/NEJMoa0907272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tellez-Plaza M, Jones MR, Dominguez-Lucas A, Guallar E, Navas-Acien A. 2013. Cadmium exposure and clinical cardiovascular disease: a systematic review. Curr. Atheroscler. Rep. 15, 162 ( 10.1007/s11883-013-0356-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigra AE, Ruiz-Hernandez A, Redon J, Navas-Acien A, Tellez-Plaza M. 2016. Environmental metals and cardiovascular disease in adults: a systematic review beyond lead and cadmium. Curr. Environ. Health Rep. 3, 416–433. ( 10.1007/s40572-016-0117-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon K, Guallar E, Navas-Acien A. 2012. Arsenic exposure and cardiovascular disease: an updated systematic review. Curr. Atheroscler. Rep. 14, 542–555. ( 10.1007/s11883-012-0280-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navas-Acien A, Guallar E, Silbergeld EK, Rothenberg SJ. 2006. Lead exposure and cardiovascular disease—a systematic review. Environ. Health Perspect. 115, 472–482. ( 10.1289/ehp.9785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamas GA, Navas-Acien A, Mark DB, Lee KL. 2016. Heavy metals, cardiovascular disease, and the unexpected benefits of chelation therapy. J. Am. Coll. Cardiol. 67, 2411–2418. ( 10.1016/j.jacc.2016.02.066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz-Hernandez A, Kuo C-C, Rentero-Garrido P, Tang W-Y, Redon J, Ordovas JM, Navas-Acien A, Tellez-Plaza M. 2015. Environmental chemicals and DNA methylation in adults: a systematic review of the epidemiologic evidence. Clin. Epigenet. 7, 9 ( 10.1186/s13148-015-0055-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, Sparrow D, Vokonas P, Schwartz J. 2010. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology 21, 819–828. ( 10.1097/EDE.0b013e3181f20457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Istas G, et al. 2017. Identification of differentially methylated BRCA1 and CRISP2 DNA regions as blood surrogate markers for cardiovascular disease. Sci. Rep. 7, 2919 ( 10.1038/s41598-017-03434-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valencia-Morales MP, et al. 2015. The DNA methylation drift of the atherosclerotic aorta increases with lesion progression. BMC Med. Genet. 8, 425 ( 10.1186/s12920-015-0085-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, Ballestar E, Esteller M, Zaina S. 2004. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J. Biol. Chem. 279, 29 147–29 154. ( 10.1074/jbc.M403618200) [DOI] [PubMed] [Google Scholar]
- 13.Zaina S, et al. 2014. DNA methylation map of human atherosclerosis. Circ. Cardiovasc. Genet. 7, 692–700. ( 10.1161/CIRCGENETICS.113.000441) [DOI] [PubMed] [Google Scholar]
- 14.Casasnovas JA, et al. 2012. Aragon workers' health study – design and cohort description. BMC Cardiovasc. Disord. 12, 675 ( 10.1186/1471-2261-12-45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budoff MJ, Nasir K, McClelland RL, Detrano R, Wong N, Blumenthal RS, Kondos G, Kronmal RA. 2009. Coronary calcium predicts events better with absolute calcium scores than age-sex-race/ethnicity percentiles. J. Am. Coll. Cardiol. 53, 345–352. ( 10.1016/j.jacc.2008.07.072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muntendam P, McCall C, Sanz J, Falk E, Fuster V. 2010. The BioImage study: novel approaches to risk assessment in the primary prevention of atherosclerotic cardiovascular disease—study design and objectives. Am. Heart J. 160, 49–57.e1. ( 10.1016/j.ahj.2010.02.021) [DOI] [PubMed] [Google Scholar]
- 17.Scheer J, et al. 2012. Arsenic species and selected metals in human urine: validation of HPLC/ICPMS and ICPMS procedures for a long-term population-based epidemiological study. Anal. Methods. 4, 406 ( 10.1039/c2ay05638k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. 2014. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369. ( 10.1093/bioinformatics/btu049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. ( 10.1093/bioinformatics/bts034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houseman E, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86 ( 10.1186/1471-2105-13-86) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivals I, Personnaz L, Taing L, Potier M-C. 2007. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23, 401–407. ( 10.1093/bioinformatics/btl633) [DOI] [PubMed] [Google Scholar]
- 22.Valls R, Pujol A, Farrés J, Artigas L, Mas JM.2017. Anaxomics’ methodologies—understanding the complexity of biological processes. Available from: http://www.anaxomics.com/pdf/TPMS_General.pdf. [Accessed 12/03/2018].
- 23.Bishop CM. 2006. Pattern recognition and machine learning. Berlin, Germany: Springer. [Google Scholar]
- 24.Kanehisa M, Goto S, Kawashima S, Nakaya A. 2002. The KEGG databases at GenomeNet. Nucleic Acids Res. 30, 42–46. ( 10.1093/nar/30.1.42) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huntley RP, et al. 2015. The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res. 43(Database issue), D1057–D1063. ( 10.1093/nar/gku1113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. 2012. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Therapeut. 92, 414–417. ( 10.1038/clpt.2012.96) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han H, et al. 2015. TRRUST: a reference database of human transcriptional regulatory interactions. Sci. Rep. 5, 252 ( 10.1038/srep11432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croft D, et al. 2014. The reactome pathway knowledgebase. Nucleic Acids Res. 42(D1), D472–D477. ( 10.1093/nar/gkt1102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerrien S, et al. 2012. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40(D1), D841–D846. ( 10.1093/nar/gkr1088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salwinski L, et al. 2009. Recurated protein interaction datasets. Nat. Methods 6, 860–861. ( 10.1038/nmeth1209-860) [DOI] [PubMed] [Google Scholar]
- 31.Keshava PTS, et al. 2009. Human protein reference database--2009 update. Nucleic Acids Res. 37(Database), D767–D772. ( 10.1093/nar/gkn892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Licata L, et al. 2012. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 40(D1), D857–D861. ( 10.1093/nar/gkr930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada Y, Nishida T, Horibe H. 2014. Identification of hypo- and hypermethylated genes related to atherosclerosis by a genome-wide analysis of DNA methylation. Int. J. Mol. Med. 33, 1355–1363. ( 10.3892/ijmm.2014.1692) [DOI] [PubMed] [Google Scholar]
- 34.Nazarenko MS, Markov A V, Lebedev IN, Freidin MB. 2015. A comparison of genome-wide DNA methylation patterns between different vascular tissues from patients with coronary heart disease. PLoS ONE 10, e0122601 ( 10.1371/journal.pone.0122601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaina S, et al. 2015. DNA methylation dynamics in human carotid plaques after cerebrovascular events. Arterioscler. Thromb. Vasc. Biol. 35, 1835–1842. ( 10.1161/ATVBAHA.115.305630) [DOI] [PubMed] [Google Scholar]
- 36.Hansson GK. 2005. Inflammation, atherosclerosis, and coronary artery disease. New England J. Med. 352, 1685–1695. ( 10.1056/NEJMra043430) [DOI] [PubMed] [Google Scholar]
- 37.Pant S, et al. 2014. Inflammation and atherosclerosis—revisited. J. Cardiovasc. Pharmacol. Ther. 19, 170–178. ( 10.1177/1074248413504994) [DOI] [PubMed] [Google Scholar]
- 38.van Eggermond MCJA, Tezcan I, Heemskerk MHM, van den Elsen PJ. 2008. Transcriptional silencing of RFXAP in MHC class II-deficiency. Mol. Immunol. 45, 2920–2928. ( 10.1016/j.molimm.2008.01.026) [DOI] [PubMed] [Google Scholar]
- 39.Briot A, Bouloumié A, Iruela-Arispe ML. 2016. Notch, lipids, and endothelial cells. Curr. Opin Lipidol 27, 513–520. ( 10.1097/MOL.0000000000000337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rusanescu G, Weissleder R, Aikawa E. 2008. Notch signaling in cardiovascular disease and calcification. Curr. Cardiol. Rev. 4, 148–156. ( 10.2174/157340308785160552) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quillard T, Charreau B. 2013. Impact of notch signaling on inflammatory responses in cardiovascular disorders. Int. J. Mol. Sci. 14, 6863–6888. ( 10.3390/ijms14046863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kokjohn TA, et al. 2011. Chemical characterization of pro-inflammatory amyloid-beta peptides in human atherosclerotic lesions and platelets. Biochim. Biophys. Acta 1812, 1508–1514. ( 10.1016/j.bbadis.2011.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lathe R, Sapronova A, Kotelevtsev Y. 2014. Atherosclerosis and Alzheimer diseases with a common cause? Inflammation, oxysterols, vasculature. BMC Geriatr. 14, 7 ( 10.1186/1471-2318-14-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dou L, Liang H-F, Geller DA, Chen Y-F, Chen X-P. 2014. The regulation role of interferon regulatory factor-1 gene and clinical relevance. Hum. Immunol. 75, 1110–1114. ( 10.1016/j.humimm.2014.09.015) [DOI] [PubMed] [Google Scholar]
- 45.Itoh F, et al. 2012. Smad2/Smad3 in endothelium is indispensable for vascular stability via S1PR1 and N-cadherin expressions. Blood 119, 5320–5328. ( 10.1182/blood-2011-12-395772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai X, et al. 2015. SMAD3 deficiency promotes vessel wall remodeling, collagen fiber reorganization and leukocyte infiltration in an inflammatory abdominal aortic aneurysm mouse model. Sci. Rep. 5, 659 ( 10.1038/srep10180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pamukcu B, Lip GYH, Shantsila E. 2011. The nuclear factor kappa B pathway in atherosclerosis: a potential therapeutic target for atherothrombotic vascular disease. Thromb. Res. 128, 117–123. ( 10.1016/j.thromres.2011.03.025) [DOI] [PubMed] [Google Scholar]
- 48.Junhua L, Feng L. 2017. The role of serotonin beyond the central nervous system during embryogenesis. Front. Cell Neurosci. 11, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sánchez-Galán E, et al. 2009. Leukotriene B4 enhances the activity of nuclear factor-κB pathway through BLT1 and BLT2 receptors in atherosclerosis. Cardiovasc. Res. 81, 216–225. ( 10.1093/cvr/cvn277) [DOI] [PubMed] [Google Scholar]
- 50.Barish GD, et al. 2012. The Bcl6–SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab. 15, 554–562. ( 10.1016/j.cmet.2012.02.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zetterqvist AV, et al. 2013. Inhibition of nuclear factor of activated T-cells (NFAT) suppresses accelerated atherosclerosis in diabetic mice. PLoS ONE 8, e65020 ( 10.1371/journal.pone.0065020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee T, Pelletier J. 2016. The biology of DHX9 and its potential as a therapeutic target. Oncotarget 7, 42 716–42 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Y, Zhang Y, Li L, Feng X, Ding S, Zheng W, Li J, Shen P. 2014. TAB1: a target of triptolide in macrophages. Chem. Biol. 21, 246–256. ( 10.1016/j.chembiol.2013.12.009) [DOI] [PubMed] [Google Scholar]
- 54.Katsuda S, Kaji T. 2003. Atherosclerosis and extracellular matrix. J. Atheroscler. Thromb. 10, 267–274. ( 10.5551/jat.10.267) [DOI] [PubMed] [Google Scholar]
- 55.Sumara G, Belwal M, Ricci R. 2005. ‘Jnking’ atherosclerosis. Cell. Mol. Life Sci. 62, 2487–2494. ( 10.1007/s00018-005-5253-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weng J, Wang C, Zhong W, Li B, Wang Z, Shao C, Yan J. 2017. Activation of CD137 signaling promotes angiogenesis in atherosclerosis via modulating endothelial Smad1/5-NFATc1 pathway. J. Am. Heart Assoc. 6, e004756 ( 10.1161/JAHA.116.004756) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Mongiat M, Sweeney SM, San Antonio JD, Fu J, Iozzo RV. 2003. Endorepellin, a novel inhibitor of angiogenesis derived from the C terminus of Perlecan. J. Biol. Chem. 278, 4238–4249. ( 10.1074/jbc.M210445200) [DOI] [PubMed] [Google Scholar]
- 58.Weber C, Noels H. 2011. Atherosclerosis: current pathogenesis and therapeutic options. Nat. Med. 17, 1410–1422. ( 10.1038/nm.2538) [DOI] [PubMed] [Google Scholar]
- 59.Yeh M, et al. 2004. Role for sterol regulatory element-binding protein in activation of endothelial cells by phospholipid oxidation products. Circ. Res. 95, 780–788. ( 10.1161/01.RES.0000146030.53089.18) [DOI] [PubMed] [Google Scholar]
- 60.Nordberg GF, Gerhardsson L, Broberg K, Mumtaz M, Ruiz P, Fowler BA. 2007. Chapter 7 — Interactions in Metal Toxicology. In Handbook on the toxicology of metals, pp. 117–145. Academic Press. [Google Scholar]
- 61.Valko M, Morris H, Cronin MTD. 2005. Metals, toxicity and oxidative stress. Curr. Med. Chem. 12, 1161–1208. ( 10.2174/0929867053764635) [DOI] [PubMed] [Google Scholar]
- 62.McCarty MF. 1996. Up-regulation of intracellular signalling pathways may play a central pathogenic role in hypertension, atherogenesis, insulin resistance, and cancer program — the ‘PKC syndrome’. Med. Hypotheses 46, 191–221. ( 10.1016/S0306-9877(96)90243-1) [DOI] [PubMed] [Google Scholar]
- 63.Messner B, Bernhard D. 2010. Cadmium and cardiovascular diseases: cell biology, pathophysiology, and epidemiological relevance. Biometals 23, 811–822. ( 10.1007/s10534-010-9314-4) [DOI] [PubMed] [Google Scholar]
- 64.Iavicoli I, Fontana L, Bergamaschi A. 2009. The effects of metals as endocrine disruptors. J. Toxicol. Environ. Health Part B 12, 206–223. ( 10.1080/10937400902902062) [DOI] [PubMed] [Google Scholar]
- 65.Blaby-Haas CE, Merchant SS. 2014. Lysosome-related organelles as mediators of metal homeostasis. J. Biol. Chem. 289, 28 129–28 136. ( 10.1074/jbc.R114.592618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiu DS, Oram JF, LeBoeuf RC, Alpers CE, O'Brien KD. 1997. High-density lipoprotein-binding protein (HBP)/vigilin is expressed in human atherosclerotic lesions and colocalizes with apolipoprotein E. Arterioscler. Thromb. Vasc. Biol. 17, 2350–2358. ( 10.1161/01.ATV.17.11.2350) [DOI] [PubMed] [Google Scholar]
- 67.Szelag M, Piaszyk-Borychowska A, Plens-Galaska M, Wesoly J, Bluyssen HAR. 2016. Targeted inhibition of STATs and IRFs as a potential treatment strategy in cardiovascular disease. Oncotarget 7, 48 788–48 812. ( 10.18632/oncotarget.9195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao X, et al. 2017. The critical role of ABCG1 and PPARγ/LXRα signaling in TLR4 mediates inflammatory responses and lipid accumulation in vascular smooth muscle cells. Cell Tissue Res. 368, 145–157. ( 10.1007/s00441-016-2518-3) [DOI] [PubMed] [Google Scholar]
- 69.Maunakea AK, Chepelev I, Cui K, Zhao K. 2013. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 23, 1256–1269. ( 10.1038/cr.2013.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Puig O, et al. 2011. A gene expression signature that classifies human atherosclerotic plaque by relative inflammation status. Circ. Cardiovasc. Genet. 4, 595–604. ( 10.1161/CIRCGENETICS.111.960773) [DOI] [PubMed] [Google Scholar]
- 71.Tellez-plaza M, et al. 2014. Association of global DNA methylation and global DNA hydroxymethylation with metals and other exposures in human blood DNA samples. Environ. Health Perspect. 122, 946–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Finn AV, Kolodgie FD, Virmani R. 2010. Correlation between carotid intimal/medial thickness and atherosclerosis: a point of view from pathology. Arterioscler. Thromb. Vasc. Biol. 30, 177–181. ( 10.1161/ATVBAHA.108.173609) [DOI] [PubMed] [Google Scholar]
- 73.Mateen FJ, et al. 2017. Chronic arsenic exposure and risk of carotid artery disease: the strong heart study. Environ. Res. 157, 127–134. ( 10.1016/j.envres.2017.05.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaufman JD, et al. 2012. Prospective study of particulate air pollution exposures, subclinical atherosclerosis, and clinical cardiovascular disease: the multi-ethnic study of atherosclerosis and air pollution (MESA Air). Am. J. Epidemiol. 176, 825–837. ( 10.1093/aje/kws169) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The computing code is available upon request to the corresponding author. The data are not available as the steering committee and the participants did not approve unrestricted data sharing at the time of ethical approval of the study and data sharing was not included in the consent form. The lists of genes attributed to DMRs supporting protein interaction networks in this article have been uploaded as part of the electronic supplementary material, files ESM2, ESM3 and ESM4.