ABSTRACT
Although the TEM-1 β-lactamase (BlaTEM-1) hydrolyzes penicillins and narrow-spectrum cephalosporins, organisms expressing this enzyme are typically susceptible to β-lactam/β-lactamase inhibitor combinations such as piperacillin-tazobactam (TZP). However, our previous work led to the discovery of 28 clinical isolates of Escherichia coli resistant to TZP that contained only blaTEM-1. One of these isolates, E. coli 907355, was investigated further in this study. E. coli 907355 exhibited significantly higher β-lactamase activity and BlaTEM-1 protein levels when grown in the presence of subinhibitory concentrations of TZP. A corresponding TZP-dependent increase in blaTEM-1 copy number was also observed, with as many as 113 copies of the gene detected per cell. These results suggest that TZP treatment promotes an increase in blaTEM-1 gene dosage, allowing BlaTEM-1 to reach high enough levels to overcome inactivation by the available tazobactam in the culture. To better understand the nature of the blaTEM-1 copy number proliferation, whole-genome sequence (WGS) analysis was performed on E. coli 907355 in the absence and presence of TZP. The WGS data revealed that the blaTEM-1 gene is located in a 10-kb genomic resistance module (GRM) that contains multiple resistance genes and mobile genetic elements. The GRM was found to be tandemly repeated at least 5 times within a p1ESCUM/p1ECUMN-like plasmid when bacteria were grown in the presence of TZP.
KEYWORDS: Escherichia coli, TEM-1, antibiotic resistance, gene amplification, piperacillin-tazobactam
IMPORTANCE
Understanding how bacteria acquire resistance to antibiotics is essential for treating infected patients effectively, as well as preventing the spread of resistant organisms. In this study, a clinical isolate of E. coli was identified that dedicated more than 15% of its genome toward tandem amplification of a ~10-kb resistance module, allowing it to escape antibiotic-mediated killing. Our research is significant in that it provides one possible explanation for clinical isolates that exhibit discordant behavior when tested for antibiotic resistance by different phenotypic methods. Our research also shows that GRM amplification is difficult to detect by short-read WGS technologies. Analysis of raw long-read sequence data was required to confirm GRM amplification as a mechanism of antibiotic resistance.
INTRODUCTION
β-Lactam antibiotics are an important class of antimicrobial agents that include penicillins, narrow- and extended-spectrum cephalosporins, monobactams, and carbapenems. They act by inhibiting cell wall synthesis and promoting cell lysis upon osmotic shock. There are four main resistance mechanisms that allow bacteria to circumvent the effects of β-lactams: target modification of bacterial penicillin binding proteins (PBPs), upregulation of efflux pumps, alteration of porin expression or function, and β-lactamase enzyme production (1). The combined presence of these mechanisms in individual strains has been documented (2).
β-Lactamases are enzymes that inactivate β-lactam antibiotics by hydrolyzing the β-lactam ring. More than 2,000 unique naturally occurring β-lactamase enzymes have been identified to date (3). The TEM β-lactamases, a group containing more than 200 variants (http://www.lahey.org/Studies/temtable.asp), are all presumably derived from the first identified TEM protein, TEM-1. The BlaTEM-1 enzyme hydrolyzes penicillins and narrow-spectrum cephalosporins, placing it in functional classification group 2b (4, 5). According to the molecular classification system, it is a class A serine β-lactamase (6). Although the blaTEM-1 gene is usually plasmid-borne, it is actively spread via transposition and its insertion into the chromosome has been documented (7–10).
To preserve and extend the utility of β-lactams, some have been used in combination with β-lactmase inhibitors. For example, amoxicillin, ampicillin, and piperacillin are used in combination with the β-lactamase inhibitors clavulanate, sulbactam, and tazobactam, respectively. These classical inhibitors render most organisms that express class A serine β-lactamases susceptible to β-lactams, excluding a few enzymes that are inhibitor resistant, such as Klebsiella pneumoniae carbapenemases (KPCs) (1). However, intensive use of β-lactamase inhibitors has caused the emergence of inhibitor-resistant variants within the TEM and SHV β-lactamase families. In addition, overexpression of inhibitor-sensitive enzymes, such as BlaTEM-1, has led to clavulanate, sulbactam, and/or tazobactam resistance (11–20). In some cases, higher BlaTEM-1 enzyme levels were attributed to a Pa/Pb mutation in the blaTEM-1 promoter that increased transcription (12, 17, 18, 21). BlaTEM-1 hyperproduction resulting from an increase in blaTEM-1 gene dosage has also been documented (12, 13, 15, 22, 23).
We have identified a number of clinical E. coli isolates that exhibit discordant behavior and heterogeneous resistance in disk diffusion (DD), agar dilution (AD), and broth microdilution (BMD) antibiotic susceptibility testing (AST) assays (24, 25). A PCR- and sequence-based screen for plasmid-borne β-lactamase genes revealed that many of the isolates harbored only blaTEM-1. In this study, we further characterized one of these clinical isolates, combining phenotypic and genotypic approaches to characterize a unique mechanism of antibiotic resistance.
RESULTS
Previous work revealed that E. coli clinical isolate 907355 is resistant to piperacillin-tazobactam (TZP; MIC for the combination, >128 µg/ml piperacillin and 4 µg/ml tazobactam [expressed here as ≥128/4 µg/ml]) in BMD tests, yet susceptible (MIC, ≤16/4 µg/ml) by the AD method (24, 25). To further investigate the nature of this discordant behavior, BMD assays were performed with two other β-lactam/β-lactamase inhibitor combinations, ampicillin-sulbactam (SAM) and amoxicillin-clavulanate (AMC). According to Clinical and Laboratory Standards Institute (CLSI) breakpoints (26), E. coli 907355 is resistant to both SAM (MIC, >64/32 µg/ml) and AMC (MIC, 32/16 µg/ml) by BMD (Table 1). Thus, the mechanism underlying E. coli 907355 resistance to TZP likely extends to other β-lactam/β-lactamase inhibitor combinations.
TABLE 1 .
Summary of phenotypic and genotypic characteristics of E. coli 907355
| Genotype or phenotype examined | Result |
|---|---|
| SAMa MIC (µg/ml) | >64/32 |
| AMCa MIC (µg/ml) | 32/16 |
| β-Lactamase pIb | 5.4 |
| blaTEM-1 promoter | P3 |
| OmpF expressionc | Yes |
| OmpC expressionc | Yes |
| Increased efflux (PAβN)d | No |
MICs for SAM (ampicillin-sulbactam) and AMC (amoxicillin-clavulanic acid) were determined in broth microdilution assays.
β-Lactamase pI was determined by IEF.
OmpF and OmpC expression was detected by Western immunoblotting.
Increased efflux was measured in an PAβN inhibitor assay. A ≥4-fold decrease in the TZP MIC in the presence of 50 µM PAβN was considered significant.
We next examined whether increasing the concentration of tazobactam within the TZP formulation renders 907355 susceptible to piperacillin in BMD assays. E. coli 907355 exhibited a TZP MIC of ≥128 µg/ml when the tazobactam concentration was between 0 and 8 µg/ml, whereas 16 to 64 µg/ml tazobactam reduced the 907355 TZP MIC to ≤2 µg/ml (Fig. 1A). As a control, a BMD assay was performed with 128 µg/ml tazobactam alone, which did not affect 907355 growth (data not shown). Therefore, the decrease in the 907355 TZP MIC at higher tazobactam concentrations was not due to tazobactam toxicity. The experiment was also performed on E. coli ATCC 35218, a CLSI quality control (QC) organism that expresses BlaTEM-1 and is resistant to piperacillin yet susceptible to TZP. The MICs for ATCC 35218 were ≤2 µg/ml at all tazobactam concentrations tested (Fig. 1B), revealing that 907355 requires a much higher concentration of tazobactam than ATCC 35218 for TZP to be effective.
FIG 1 .
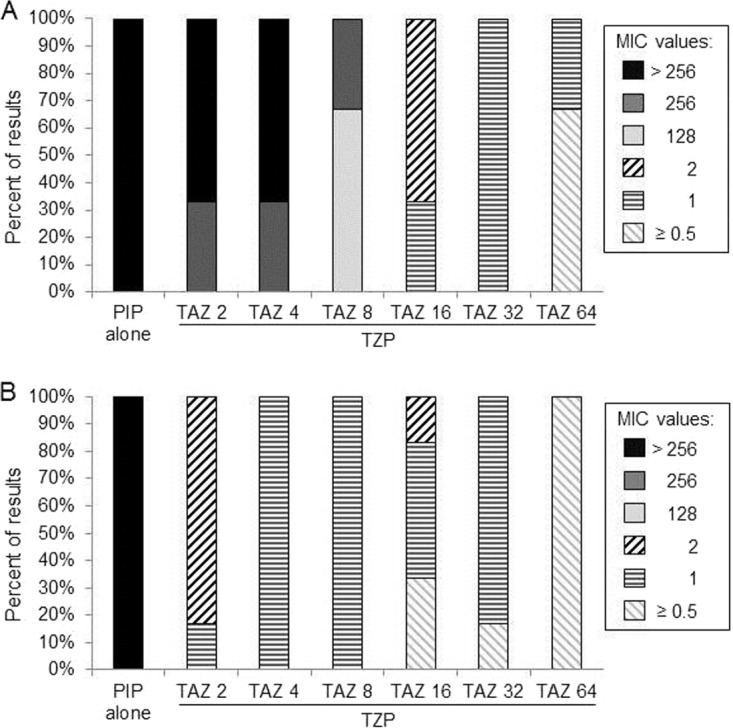
Effect of increasing tazobactam concentrations on E. coli susceptibility to TZP. MICs were determined for 907355 (A) and ATCC 35218 (B) via BMD in the presence of piperacillin (PIP) alone or TZP formulations containing different fixed concentrations (2 to 64 µg/ml) of tazobactam (TAZ). The MIC values in the legend are based on the concentration of piperacillin in the TZP formulation that inhibited growth. The range of MIC values obtained from six experiments is represented within each stacked bar on the graphs.
We previously performed PCR assays on E. coli 907355 to detect multiple plasmid-borne β-lactamase genes. The product from the only positive reaction was sequenced, revealing that E. coli 907355 contains blaTEM-1 (24). At the deduced amino acid sequence level, E. coli 907355 BlaTEM-1 is 100% identical to the original BlaTEM-1. Therefore, 907355 resistance to TZP, SAM, and AMC is not due to acquisition of inhibitor resistance mutations within the blaTEM-1 gene. Further analysis of 907355 by isoelectric focusing (IEF) confirmed that the isolate expresses only the BlaTEM-1 β-lactamase, which has a pI of 5.4 (Table 1). In addition, the 907355 blaTEM-1 gene has a wild-type P3 promoter (−35 to +1) region (Table 1). In a previous study, the P3 promoter directed relatively low levels of blaTEM expression compared to the Pa/Pb, P4, and P5 promoters (21). Therefore, sequence variations in the RNA polymerase binding site are likely not the primary cause of β-lactam–β-lactamase inhibitor resistance in E. coli 907355.
To better understand why E. coli 907355 is resistant to TZP, BlaTEM-1 levels were determined by Western immunoblotting following growth in liquid media containing no antibiotic, tazobactam (4 µg/ml), piperacillin (4 µg/ml), or TZP (4/4 µg/ml). A subinhibitory concentration of TZP was utilized, as 907355 does not consistently grow in liquid cultures at higher TZP concentrations despite the fact that it is resistant to TZP in BMD assays. E. coli ATCC 25922 was included as a negative control, as this strain lacks the blaTEM-1 gene. As expected, BlaTEM-1 was not detected in the ATCC 25922 protein sample (Fig. 2A). In contrast, a protein with the estimated molecular mass of BlaTEM-1 (31.5 kDa) was detectable in the positive-control strain ATCC 35218 grown in the presence of no antibiotic, tazobactam, or piperacillin.
FIG 2 .

Effects of TZP on E. coli 907355 BlaTEM-1 levels and β-lactamase enzyme activity. (A) Detection of BlaTEM-1 in E. coli ATCC 25922 (lane 2), ATCC 35218 (lanes 3 to 5), and 907355 (lanes 6 to 9) by Western immunoblotting. Strains were cultured in medium containing no antibiotic (lanes 2, 3, and 6), 4 µg/ml tazobactam (lanes 4 and 7), 4 µg/ml piperacillin (lanes 5 and 8), or 4/4 µg/ml TZP (lane 9) prior to protein sample collection. Molecular mass standards are shown in lane 1 (kilodaltons are indicated to the left of the blot). The estimated molecular mass of BlaTEM-1 is 31.5 kDa. This experiment was repeated on independently collected samples and yielded similar results. (B) Determination of BlaTEM-1 activity in E. coli 907355 via a nitrocefin hydrolysis assay after growth in the presence or absence of 4/4 µg/ml TZP. ATCC 35218 BlaTEM-1 activity is included as a comparison but was not determined (ND) in the presence of TZP due to lack of growth. Units on the y axis denote the micromoles of nitrocefin hydrolyzed per milligram of total protein per minute. The values above each bar on the graph represent the means of 3 experiments, and the error bars indicate the standard deviations of the means.
The Western immunoblotting results showed that E. coli 907355 BlaTEM-1 levels varied in response to the antibiotic treatment. In the presence of tazobactam alone, BlaTEM-1 levels were lower than under the other growth conditions (Fig. 2A). Quantitation of the 907355 BlaTEM-1 band intensities in each lane, which were normalized to total protein levels (data not shown), revealed that tazobactam reduced BlaTEM-1 levels by approximately 2.7-fold below those for cells grown without supplementation. This phenomenon was not specific to 907355 BlaTEM-1, as it also occurred in ATCC 35218 (Fig. 2A). One possible explanation for this finding is that tazobactam binding destabilizes BlaTEM-1, leading to increased degradation of the protein. Alternatively, BlaTEM-1 levels may be equal in untreated and tazobactam-treated cells, but tazobactam may partially block the primary antibody binding site during the immunoblotting procedure. We did not investigate this observation further, as the primary goal of this study was to determine the mechanism of BlaTEM-1-mediated TZP resistance in E. coli 907355.
The Western immunoblotting also revealed that E. coli BlaTEM-1 levels are significantly higher in the presence of TZP in comparison to no antibiotic, tazobactam, or piperacillin (Fig. 2A). Quantitation of the BlaTEM-1 band intensities indicated that BlaTEM-1 levels were 3.7-fold higher in the presence of TZP versus no antibiotic and 9.9-fold higher in the presence of TZP than with tazobactam alone. Thus, TZP selection considerably increased BlaTEM-1 protein levels in E. coli 907355.
To confirm that TZP elevates the amount of BlaTEM-1 in E. coli 907355, we measured β-lactamase enzyme activity via a nitrocefin hydrolysis assay after growth in the presence or absence of 4/4 µg/ml TZP. E. coli 907355 BlaTEM-1 activity was approximately 2.2-fold higher than that of ATCC 35218 BlaTEM-1 in the absence of antibiotic (Fig. 2B). In addition, 907355 BlaTEM-1 activity increased by ~3.5-fold in the presence of TZP in comparison to no antibiotic (Fig. 2B). As controls, the experiment was also performed on E. coli 907355 grown in 4 µg/ml piperacillin or tazobactam alone. Piperacillin did not significantly affect BlaTEM-1 activity in comparison to no additive, whereas tazobactam inhibited β-lactamase activity, as expected (data not shown). These results are consistent with the finding that TZP increases E. coli 907355 BlaTEM-1 production.
Based on previous studies, we suspected that the TZP-induced increase in 907355 BlaTEM-1 activity might be due to an increase in blaTEM-1 gene dosage (13, 15, 22, 23). The copy numbers per cell for E. coli 907355 and ATCC 35218 blaTEM-1 genes were determined using a SYBR green quantitative PCR (qPCR) assay after bacterial growth in cation-adjusted Mueller-Hinton broth (CAMHB) containing or lacking subinhibitory concentrations of TZP (4/4 µg/ml). As a reference for cell number, qPCR was also performed on the dxs gene, which is present in single copy on the E. coli chromosome. The concentration of each blaTEM-1 product was divided by the concentration of dxs in the same sample to determine the relative number of copies of blaTEM-1 per cell.
The blaTEM-1 gene was found to be present at an average copy number of only 2.3 per cell in ATCC 35218 (Fig. 3). In contrast, the 907355 blaTEM-1 gene copy number per cell rose from 8.4 in the absence of TZP to 33.8 in the presence of TZP. These results suggest that 4/4 µg/ml TZP induces a ~4-fold increase in blaTEM-1 gene dosage, which is responsible for the elevated BlaTEM-1 activity. As controls, piperacillin and tazobactam were tested individually for effects on blaTEM-1 gene copy number. Neither piperacillin nor tazobactam alone influenced blaTEM-1 gene dosage (data not shown), indicating that the increase in copy number per cell is due to treatment with a combination of antibiotic and inhibitor.
FIG 3 .

Estimation of blaTEM-1 copy number by qPCR. blaTEM-1 copy number per cell was measured using a real-time SYBR green qPCR assay after bacterial growth in the absence or presence of 4/4 µg/ml TZP. The copy number was not determined (ND) for ATCC 35218 in the presence of TZP due to lack of growth. The values above each bar on the graph represent the mean results of 3 experiments, and the error bars indicate the standard deviations of the means.
To further investigate the mechanism of blaTEM-1 copy number proliferation and genomic changes in E. coli 907355, bacteria grown in absence or presence of TZP were sequenced using Illumina technology. After assembly and annotation of the WGS data, the contig containing blaTEM-1 was only ~ 2 kb. Therefore, the same samples were sequenced again using the PacBio and MinION platforms, which allow longer raw sequence reads. A draft hybrid assembly constructed from the Illumina and PacBio data, available under BioProject PRJNA431448 on the NCBI website (https://www.ncbi.nlm.nih.gov), revealed that the genome is approximately 5.38 Mb. The blaTEM-1 gene resides within an approximately 10-kb region, similar to the genomic resistance module (GRM) found in the chromosome of E. coli UMN026 (Fig. 4) (10). Unlike the chromosomal GRM in E. coli UMN026, the 907355 GRM is located on a large plasmid similar to p1ESCUM (additionally called p1ECUMN), which was also first identified in UMN026 (10, 27). MinION sequence data further illuminated the genomic structure and location of the GRM. Examination of these reads confirmed that the GRM in 907355 p1ESCUM interrupts traI, which encodes a DNA helicase/relaxase normally required for plasmid conjugative transfer (28).
FIG 4 .
Resistance island containing blaTEM-1 gene in E. coli 907355. Annotation is based upon the E. coli UMN026 complete genome sequence (10, 27). Genes are shaded according to function: gray, transposon elements and integrase; blue, pseudogenes; orange, antibiotic resistance; green, heavy metal or toxic compound efflux; purple, hypothetical protein.
Previous studies in E. coli have attributed β-lactam/β-lactamase inhibitor or broad-spectrum cephalosporin resistance to the localization of blaTEM-1 on high-copy-number plasmids (15, 22, 23). To determine whether the observed increase in E. coli 907355 blaTEM-1 gene dosage is due to p1ESCUM accumulation, we analyzed raw Illumina and PacBio WGS data and calculated approximate copy numbers for blaTEM-1, ebr, and three tra genes (traI, traJ, and traK) averaged together (Table 2). Copy numbers were estimated by dividing the sequence coverage for each p1ESCUM gene by the mean coverage for seven single-copy E. coli multilocus sequence type (MLST) genes (adk, fumC, gyrB, icd, mdh, purA, and recA). Independent calculations from the Illumina and PacBio WGS data indicated that blaTEM-1 copy number increased ~4- to 5-fold in the presence of 4/4 µg/ml TZP (Table 2), similar to the results obtained with qPCR (Fig. 3). Raising the TZP concentration to 8/8 µg/ml further augmented the blaTEM-1 copy number to more than 90 per cell (Table 2). Similarly, the estimated copy number for ebr, another gene on the GRM (Fig. 4), increased 4- to 5-fold upon exposure to 4/4 µg/ml TZP and reached 89 copies or more per cell in 8/8 µg/ml TZP (Table 2). In contrast, the average copy number for the three tra genes, which are not on the GRM, was ~1 in the absence of antibiotic selection and remained stable upon exposure to TZP (Table 2). Thus, the increase in E. coli 907355 blaTEM-1 copy number in response to TZP is not simply due to accumulation of the entire p1ESCUM plasmid.
TABLE 2 .
Estimation of blaTEM-1 copy number based on two WGS technologies
| NGS technology | TZP concn (µg/ml) | Mean sequence coveragea |
Estimated gene copy no. |
|||||
|---|---|---|---|---|---|---|---|---|
| MLST genes | blaTEM-1 gene | tra genes | ebr gene | blaTEM-1 gene | tra genes | ebr gene | ||
| Illumina | None | 52.3 | 554.8 | 53.0 | 556.1 | 10.6 | 1.0 | 10.6 |
| 4/4 | 62.3 | 3,358.0 | 94.5 | 3,218.8 | 53.9 | 1.5 | 51.7 | |
| 8/8 | 30.5 | 3,450.0 | 38.9 | 2,995.7 | 113.3 | 1.3 | 98.3 | |
| PacBio | None | 113.9 | 1,363.5 | 104.2 | 1,094.0 | 12.0 | 0.9 | 9.6 |
| 4/4 | 163.4 | 7,537.5 | 264.7 | 6,825.8 | 46.1 | 1.6 | 41.8 | |
| 8/8 | 83.2 | 7,523.3 | 160.5 | 7,410.4 | 90.4 | 1.9 | 89.1 | |
See text for calculation method.
Based on the copy number analysis of p1ESCUM genes, we hypothesized that the ~10-kb GRM is amplified within the plasmid as a result of TZP selection. In support of this theory, five PacBio reads were identified that contained one full GRM and two partial GRMs on either side (suggesting up to three adjacent GRMs) when 907355 was grown in 8/8 µg/ml TZP. Furthermore, one MinION read was identified that contained three full adjacent GRMs with two partial GRMs on either side (suggesting up to five adjacent GRMs). In both cases, the number of repeats detected was limited by the read lengths provided by the sequencing platform. Reads containing the GRM bordered on both sides by the interrupted traI gene were not observed (data not shown). These findings suggest that at higher TZP levels, we were unable to detect the entire amplified region and that the 907355 p1ESCUM plasmid likely contains more than five tandem copies of the GRM.
In some bacteria, inactivation of major porin genes plays a role in blaTEM-1-mediated β-lactam/β-lactamase inhibitor or cephalosporin resistance (29–32). To determine whether E. coli 907355 major porin genes are intact, we identified homologues of the E. coli K-12 MG1655 ompF and ompC genes within the 907355 assembled WGS data by using the NCBI BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The E. coli 907355 ompF and ompC genes are predicted to encode full-length proteins that are 99% and 95% identical to their respective homologues in E. coli K-12 MG1655. In addition, the levels of OmpF and OmpC were examined in E. coli 907355 and ATCC 35218 by Western immunoblotting after bacterial growth in CAMHB, the medium used for BMD assays. OmpF expression was barely detectable in both strains, likely due to the relatively high osmolarity of the medium. However, a protein with the predicted molecular mass of OmpF was apparent in both 907355 and ATCC 35218 upon overexposure of the blot (see Fig. S1 in the supplemental material). OmpC, which is highly expressed in CAMHB, was also present at similar levels in 907355 and ATCC 35218 (data not shown). These results show that reduction of OmpF and OmpC expression does not account for 907355 TZP resistance in BMD assays.
Detection of major porins OmpF and OmpC in E. coli. Download FIG S1, DOC file, 0.1 MB (53KB, doc) .
Copyright © 2018 Schechter et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Increased antibiotic efflux might also influence β-lactam/β-lactamase inhibitor resistance in 907355. To examine this possibility, we utilized Phe–Arg–β-naphthylamide (PAβN), a known efflux inhibitor (33, 34). The presence of 50 µM PAβN did not reduce the 907355 TZP MIC in a BMD assay, suggesting that increased efflux may not play a significant role in 907355 TZP resistance (Table 1).
DISCUSSION
In this study, the mechanism of β-lactam/β-lactamase inhibitor resistance was investigated in E. coli 907355, a clinical isolate containing the blaTEM-1 gene. Experimental and genomic data support a role for blaTEM-1 gene amplification, leading to BlaTEM-1 hyperproduction, as the primary basis for β-lactam/β-lactamase inhibitor resistance in this isolate. Notably, we found that blaTEM-1 is located within a ~10-kb region that includes other resistance genes, as well as transposon- and integron-related sequences. Others have termed this element a genomic resistance module, or GRM (10).
In the absence of antibiotic selection, our qPCR and WGS raw read data suggest that the 907355 GRM is present at ~8 to 12 copies per cell (Fig. 3; Table 2). In the presence of 8/8 µg/ml TZP, the GRM copy number per cell increased approximately 10-fold to >90. These results translate to at least an ~800-kb increase in 907355 genomic DNA during TZP treatment and dedication of more than 15% of the E. coli 907355 bacterial genome to antibiotic resistance. Because 907355 TZP resistance is heterogeneous (25), our estimation of GRM levels is likely an average of the various copy numbers present within the population. Therefore, GRM amplification may actually be higher than 10-fold within a proportion of the population.
Multiple examples in the literature document the contributions of tandem gene duplication and amplification to the development of antibiotic resistance (35–38). In fact, two studies have reported that BlaTEM-1 hyperproduction may result from tandem amplification of the blaTEM-1 gene. In one report, Sun et al. engineered a laboratory strain of Salmonella enterica serovar Typhimurium to carry the blaTEM-1 gene on an F′ plasmid, and then they exposed it to increasing levels of cephalothin or cefaclor by serial passage (32). Several clones with higher MICs contained large regions (36 to 134 kb) of tandemly amplified DNA that included the blaTEM-1 gene. In another study on a clinical isolate of E. coli, genetic rearrangements and an increase in blaTEM-1 copy number were observed within a single large, low-copy-number plasmid (13). However, the nature of the amplification was not precisely determined. Thus, our work is significant in that it provides direct evidence for tandem gene amplification of blaTEM-1 in a clinical isolate.
Our findings lend support to multiple aspects of the model proposed by Sandegren and Andersson, which details a stepwise process for the contribution of gene amplification to the development of antibiotic resistance (35). In the first step, gene duplication and amplification provide initial increased tolerance against an antibiotic (35). Accordingly, our data show that amplification of blaTEM-1 increases E. coli 907355 tolerance to TZP. Second, tandemly amplified regions are usually genetically unstable and lost in the absence of selection. This theory is supported by the fact that E. coli 907355 blaTEM-1 copy number and BlaTEM-1 levels were much lower when cultures were grown without TZP (Fig. 2 and 3). Third, mutations within the amplified genes or other single-copy genes may allow cells to develop more durable resistance that does not require tandem gene amplification. In fact, several studies have identified decreased membrane permeability as a contributor to β-lactam/β-lactamase inhibitor or cephalosporin resistance in bacteria containing blaTEM-1 (29–32). Our initial examination of E. coli 907355 membrane permeability revealed that OmpF and OmpC are expressed (see Fig. S1 in the supplemental material) and that PAβN did not significantly reduce the TZP MIC. These results may explain why E. coli 907355 amplifies blaTEM-1 to such high levels in the presence of TZP. However, it is important to note that E. coli 907355 membrane permeability was not extensively characterized and the isolate may have a deficiency that we did not detect.
In addition to supporting a role for tandem blaTEM-1 amplification in the development of β-lactam/β-lactamase inhibitor resistance, our work underscores two important implications for clinical microbiology laboratories. First, our data may explain why E. coli isolates that contain only blaTEM-1 frequently exhibit discordant or heterogeneous behavior when tested by different AST methods for TZP (24, 25). Because the BMD reference method uses a fixed concentration of 4 µg/ml of tazobactam, which binds irreversibly to β-lactamase enzymes, a threshold level of blaTEM-1 may overcome the available inhibitor. E. coli 907355 may be resistant to TZP in BMD assays, because a cell that amplifies blaTEM-1 to high enough levels outgrows other cells with fewer copies of blaTEM-1. In contrast, 907355 may be susceptible to TZP when tested via agar-based methods because cells that express high enough BlaTEM-1 levels appear as isolated colonies. Accordingly, increasing the concentration of tazobactam overcame 907355 resistance to TZP in BMD assays (Fig. 1). A similar observation was described previously for other BlaTEM-1 hyperproducing strains (39). Taken together, our findings suggest that the in vitro resistance of 907355 may represent an artifact of the BMD testing system, which uses a fixed concentration of tazobactam in combination with increasing concentrations of piperacillin. TZP resistance in this case may not translate into clinical resistance of the organism in a patient therapeutic setting, where higher concentrations of tazobactam could be obtained. In support of this theory, neutropenic mice infected with bacteria identified as TZP resistant by BMD, but susceptible to other classes of β-lactams, were successfully treated with humanized exposures of TZP (40, 41).
A second important clinical implication of our work is that blaTEM-1 gene amplification is difficult to detect by current genotypic antibiotic susceptibility prediction methods. First, clinical isolates are typically subcultured on media without antibiotic selection prior to carrying out both genotypic and phenotypic assays. Because tandemly amplified regions are inherently unstable, they may be lost prior to testing. Second, blaTEM-1 amplification in E. coli 907355 was not detectable by Illumina sequencing alone, as this method generates short sequencing reads (151 bp) that are not easily assembled in repetitive regions of the genome. Similarly, a recent study employing Illumina sequencing to generate WGS data was unable to accurately predict TZP susceptibility in 13 of the isolates tested (42). Combining Illumina data with longer PacBio reads, which were an average length of ~1.7 kb in our study, allowed a more complete assembly of the E. coli 907355 blaTEM-1 region. In addition, MinION technology was useful because it produced even longer reads than PacBio, allowing us to detect at least five adjacent copies of the GRM within the genome of the culture treated with 8/8 µg/ml TZP. Finally, classical PCR detection assays for blaTEM only reveal that the gene is present and not whether it is tandemly amplified. Comparison of blaTEM regions in a variety of clinical isolates resistant to TZP may reveal common sequences that can be utilized for development of PCR assays that detect tandem gene amplification.
Our finding that a clinical isolate of E. coli significantly amplifies blaTEM-1 in response to TZP raises important questions. For example, how commonly are isolates similar to E. coli 907355 found in clinical settings? Although we cannot answer this question definitively, a preliminary screening of nine other E. coli isolates from our previously published studies revealed that at least four other independent clinical isolates have the ability to substantially amplify blaTEM-1 (data not shown). We are currently working on characterizing these isolates. Further studies will be required to determine the frequency of similar strains in natural populations.
Another question is whether blaTEM-1 gene amplification occurs in infected patients during therapy with β-lactam antibiotics or β-lactam/β-lactamase inhibitor combinations. Unfortunately, the antibiotic treatment regimen of the patient from which E. coli 907355 was isolated was not available to us. Furthermore, E. coli 907355 isolates were not collected prior to and over the course of the antibiotic therapy. Future studies in mice might address whether blaTEM-1 amplification occurs in E. coli 907355 during host infection (40, 41). Recently, McGann et al. reported that aph1 amplification occurred in Acinetobacter baumanii during tobramycin treatment of an infected patient, leading to antibiotic therapy failure (36). Clinical studies will be required to determine whether extensive blaTEM-1 amplification contributes to antibiotic therapy failure for β-lactam antibiotics or β-lactam/β-lactamase inhibitor combinations.
MATERIALS AND METHODS
Bacterial isolates.
E. coli 907355, a clinical isolate received from Austria, has been described previously (24, 25). E. coli ATCC 25922 (blaTEM-1 negative) and/or ATCC 35218 (blaTEM-1 positive) quality control organisms recommended by CLSI for AST were used as comparators in experiments. Although ATCC 35218 produces BlaTEM-1, it is susceptible to β-lactam/inhibitor combinations, including TZP.
AST.
MICs were determined by BMD according to CLSI standard methods, using CAMHB supplemented with appropriate ranges of β-lactam antibiotics (26, 43). β-Lactamase inhibitors were included at the recommended fixed concentrations, except that the concentration of tazobactam varied between 2 and 64 µg/ml for the experiment in Fig. 1 (26). Resistance or susceptibility was interpreted using CLSI breakpoints (26). When skipped wells were encountered during reading of the MIC, CLSI recommendations were followed for interpretation.
Bacterial growth and sample collection.
For all BlaTEM-1 protein characterization assays and blaTEM-1 qPCR studies, bacteria were cultured in CAMHB with no antibiotic, 4 µg/ml tazobactam, 4 µg/ml piperacillin, and/or 4/4 µg/ml TZP. Cultures were inoculated with a 1:300 dilution of a 0.5 McFarland suspension and incubated overnight at 35°C without agitation. After 18 to 20 h, cells were harvested by centrifugation. For Western immunoblotting, equal amounts of cells (~1.5 ml of each culture) were pelleted based on readings of the optical density at 600 nm. Pellets were then suspended in 100 µl 2× Laemmli sample buffer (Bio-Rad) and frozen at −20°C. For enzyme activity and PCR experiments, pellets (from 1-ml culture aliquots) were suspended in 0.1 M sodium phosphate buffer, pH 7.0.
For WGS analysis, E. coli 907355 bacteria were cultured overnight on Mueller-Hinton agar (MHA) plates supplemented with 4/4 µg/ml, 8/8 µg/ml, or no TZP. Approximately 10 isolated colonies were then inoculated into CAMHB containing the same concentrations of antibiotics as the MHA plates. Cultures were grown overnight at 35°C with agitation and pelleted by centrifugation. Pellets were stored at −20°C prior to DNA isolation.
Isoelectric focusing and Western immunoblotting.
IEF was performed using pH 3 to 10 precast IEF protein gels (Novex) and the XCell SureLock Mini-Cell (Life Technologies, Inc.) per the manufacturer’s instructions. β-Lactamase proteins were detected with 1 mM nitrocefin (Remel), using known enzymes as standards.
For Western immunoblotting, samples were boiled for 5 min and loaded (12 µl) onto a Mini-Protean TGX Stain-Free gel (Bio-Rad). Precision Plus protein standards (Bio-Rad) were run simultaneously to estimate molecular weight. Following separation by electrophoresis in a Protean Tetra cell (Bio-Rad), proteins were transferred onto a polyvinylidene difluoride membrane by using the Trans-Blot Turbo system (Bio-Rad). BlaTEM-1 was detected using a standard Western immunoblotting procedure with the Clarity Western ECL substrate (Bio-Rad), as instructed by the manufacturer. Primary anti-β-lactamase 8A5.A10 mouse monoclonal antibodies (Santa Cruz Biotechnology) were used at a 1:200 dilution, and secondary goat anti-mouse IgG–horseradish peroxidase conjugate antibodies (Bio-Rad) were used at a 1∶3,000 dilution. Blots were developed on a ChemiDoc MP imager with Image Lab v5.2 software (Bio-Rad). Total protein on the blot was visualized using the Stain-Free protocol, whereas chemiluminescent signals were captured using the Chemi-Hi resolution protocol. Relative levels of BlaTEM-1 in E. coli 907355 samples were calculated by the Image Lab software, which quantitated the intensities of the chemiluminescent BlaTEM-1 bands and normalized them to total protein.
β-Lactamase enzyme activity assays.
Lysates for β-lactamase enzyme studies were prepared using glass beads as previously described (44). Briefly, cell suspensions in phosphate buffer were transferred to microcentrifuge tubes containing about 0.25 g of 100-µm glass beads. The tubes were vortexed at full speed with a bead beater attachment at 4°C for 10 min and then centrifuged at 16,000 × g at 4°C for 15 min. Supernatants were transferred to clean tubes and stored at −20°C until testing. β-Lactamase activity assays were carried out in triplicate in an Infinite M200 Pro microplate reader (Tecan) at 25°C using 100 µM nitrocefin and 5 µl of each crude glass bead lysate, as previously described (11, 45). Enzyme rates were normalized to the total protein present in each lysate, which was determined with a bicinchoninic acid assay kit (Thermo Scientific) per the manufacturer’s instructions.
Determination of blaTEM-1 copy number by PCR.
Lysates for blaTEM-1 gene copy number analysis were prepared by boiling bacterial suspensions for 10 min, followed by storage at −20°C prior to testing (46). Real-time qPCR was performed in a LightCycler 1.5 system (Roche) with SYBR green I (Roche) and primers that hybridize to the target gene blaTEM-1 or the single-copy chromosomal reference gene, dxs (d-1-deoxyxylulose 5-phosphate synthase), using a method similar to that reported by Lee et. al. (47). However, the reverse primer for dxs was moved downstream 1 nucleotide, based on mismatches detected for other E. coli dxs sequences in GenBank. Native Taq DNA polymerase isolated from Thermus aquaticus (Invitrogen/Thermo Fisher Scientific) was utilized in all reaction mixtures due to the possible contamination of recombinant Taq preparations with blaTEM-1 DNA. Absolute quantification analysis was performed using LightCycler software v3.5.3 with the automated method. An internal calibrator of 1.5 × 105 copies of pBR322/μl spiked into 1.5 × 105 CFU/µl of DH5α was included in each run to control for PCR efficiency and was used to normalize the target and reference results from each sample. To determine the concentrations of blaTEM-1 or dxs in each sample, normalized crossing point (Cp) values were compared to a standard curve generated by performing qPCR on known quantities of each target. The dxs standard curve was generated by performing qPCR on DH5α lysates made from samples with a range of established CFU per microliter (4.7 × 103 CFU/µl up to 6 × 105 CFU/µl). The blaTEM-1 standard curve was generated by qPCR on samples containing a range of pBR322 DNA concentrations (1.3 × 103 plasmids/µl up to 1.0 × 108 plasmids/µl) spiked into DH5α. The number of double-stranded copies per microliter of blaTEM-1 was then divided by the number of double-stranded copies of dxs per microliter to determine the relative number of copies of blaTEM-1 per cell.
Bacterial genome sequencing and data interpretation.
E. coli 907355 genomic DNA was isolated from cultures containing 0, 4/4 µg/ml, or 8/8 µg/ml TZP by using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer’s instructions. A whole-genome shotgun library was then constructed from each sample by using a TruSeq Nano DNA High-Throughput Library Prep kit (Illumina) according to Illumina’s standardized protocol. Dual indexed paired-end libraries with an average insert size of 800 bp were made by shearing 200 ng genomic DNA with a Q800R sonicator system (QSonica) in a total volume of 50 µl 1× Tris-EDTA buffer. The sheared DNA was cleaned and size selected using sample purification beads provided in the library preparation kit. Universal adapters for paired-end sequencing and indexing were then added according to an optimized protocol from Illumina. The indexed libraries were pooled and sequenced on a HiSeq 2500 system (Illumina) with a target of 100× coverage per genome.
PacBio sequencing libraries were prepared using an SMRTbell Template Prep kit 1.0 (Pacific Biosciences) according to the manufacturer’s standard protocol. Genomic DNA was extracted using an extraction kit (Qiagen) and 1 µg of DNA was used as the template to prepare SMRTbell libraries. DNA was sheared using a Covaris g-TUBEs apparatus at 4,000 rpm for 3 to 4 min and cleaned using 0.75× AMPure XP beads (Beckman Coulter, Inc.) to size select the desired range of fragments and remove all DNA fragments of <200 bp. SMRTbell-adapted libraries were sequenced on one SMRT cell, using P6C4v2 chemistry. Output files were processed and assembled into CCS reads by using PacBio RSII SMRT portal software (v2.3.0) default settings with minimum passes at three and minimum predicted accuracy of 0.9.
MinION sequencing libraries were prepared from unsheared genomic DNA using the SQK-MAP005 2D library preparation kit (Oxford Nanopore Technologies) and the EXP-LWI001 low-input expansion kit (Oxford Nanopore Technologies) when less than 1 µg of DNA was available. To ensure minimal shearing during the library preparation process, the general procedure provided by Oxford Nanopore Technologies was adapted to use of wide-bore pipette tips, as well as a HulaMixer (Thermo Fisher Scientific) in place of vortexing. Libraries were sequenced using R7.3 chemistry, and poretools was used to extract fastq files from the native HDF5 format (48).
A hybrid de novo assembly of PacBio and Illumina reads was generated using SPAdes 3.9 with the “multiple-kmer” and “careful” options (49). All assembled contigs were annotated using the automated annotation software Prokka v1.11 (50). A BLAST search against NCBI GenBank identified the closest homologous E. coli completed genome, and then a subsequent BLAST against this completed genome was performed to manually refine annotation of the blaTEM-1-containing element (51).
Determination of gene copy number by analysis of WGS data.
Copy numbers of individual genes were determined by calculating ratios of the coverage of the gene of interest to the mean coverage of seven E. coli MLST genes (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). These MLST genes are involved in housekeeping functions and are known to be present in single copy within the genome. The copy number analysis was performed separately for the Illumina and PacBio sequencing data. To estimate the copy number of the p1ESCUM-like plasmid, the copy number of the traI, traJ, and traK genes was averaged and compared to the mean MLST gene copy number. Read mapping and coverage estimation were performed by using BWA v0.77 and SAMtools v0.1.19 (52, 53).
Efflux inhibitor assay.
An assay using PAβN (Sigma-Aldrich) was performed as previously described, except TZP was used (54). A ≥4-fold decrease in TZP MIC in the presence of PAβN was considered a significant decrease.
Outer membrane protein detection.
Samples for outer membrane protein detection were prepared by diluting a 2.0 McFarland suspension 1:200 in 5 ml CAHMB. Cultures were grown to mid-log phase (~2.5 to 3.0 McFarland), and equal volumes of cells based on McFarland readings (~1 ml) were pelleted in a microcentrifuge. The pellets were then suspended in 100 µl 2× Laemmli sample buffer and frozen at −20°C.
Outer membrane proteins were detected according to the Western immunoblotting procedure described above, except using different antibodies. Primary antibodies, either anti-OmpF or anti-OmpC polyclonal IgG (Biorbyt), were used at a 1∶1,000 dilution. Secondary goat anti-rabbit IgG–horseradish peroxidase conjugate antibodies (Bio-Rad) were used at a 1∶3,000 dilution.
Accession number(s).
All sequence and assembly data are accessible through NCBI BioProject PRJNA431448.
ACKNOWLEDGMENTS
We thank Jennifer Hix for laboratory assistance, as well as Sylvain Orenga, Mark Adams, and Richard Goering for helpful discussions.
Funding for this research was provided by BioMérieux and the Jackson Laboratory.
Footnotes
Citation Schechter LM, Creely DP, Garner CD, Shortridge D, Nguyen H, Chen L, Hanson BM, Sodergren E, Weinstock GM, Dunne WM Jr., van Belkum A, Leopold SR. 2018. Extensive gene amplification as a mechanism for piperacillin-tazobactam resistance in Escherichia coli. mBio 9:e00583-18. https://doi.org/10.1128/mBio.00583-18.
REFERENCES
- 1.Drawz SM, Bonomo RA. 2010. Three decades of β-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li XZ, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonomo RA. 2017. β-lactamases: a focus on current challenges. Cold Spring Harb Perspect Med 7:a025239. doi: 10.1101/cshperspect.a025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K, Jacoby GA, Medeiros AA. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 39:1211–1233. doi: 10.1128/AAC.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambler RP. 1980. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci 289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 7.Datta N, Kontomichalou P. 1965. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208:239–241. doi: 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- 8.Heffron F, Sublett R, Hedges RW, Jacob A, Falkow S. 1975. Origin of the TEM beta-lactamase gene found on plasmids. J Bacteriol 122:250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidet P, Basmaci R, Guglielmini J, Doit C, Jost C, Birgy A, Bonacorsi S. 2016. Genome analysis of Kingella kingae strain KWG1 reveals how a β-lactamase gene inserted in the chromosome of this species. Antimicrob Agents Chemother 60:703–708. doi: 10.1128/AAC.02192-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lescat M, Calteau A, Hoede C, Barbe V, Touchon M, Rocha E, Tenaillon O, Médigue C, Johnson JR, Denamur E. 2009. A module located at a chromosomal integration hot spot is responsible for the multidrug resistance of a reference strain from Escherichia coli clonal group A. Antimicrob Agents Chemother 53:2283–2288. doi: 10.1128/AAC.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu PJ, Shannon K, Phillips I. 1994. Effect of hyperproduction of TEM-1 β-lactamase on in vitro susceptibility of Escherichia coli to beta-lactam antibiotics. Antimicrob Agents Chemother 38:494–498. doi: 10.1128/AAC.38.3.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu PJ, Shannon K, Phillips I. 1995. Mechanisms of hyperproduction of TEM-1 β-lactamase by clinical isolates of Escherichia coli. J Antimicrob Chemother 36:927–939. doi: 10.1093/jac/36.6.927. [DOI] [PubMed] [Google Scholar]
- 13.Shannon K, Williams H, King A, Phillips I. 1990. Hyperproduction of TEM-1 β-lactamase in clinical isolates of Escherichia coli serotype O15. FEMS Microbiol Lett 55:319–323. [DOI] [PubMed] [Google Scholar]
- 14.Stapleton P, Wu PJ, King A, Shannon K, French G, Phillips I. 1995. Incidence and mechanisms of resistance to the combination of amoxicillin and clavulanic acid in Escherichia coli. Antimicrob Agents Chemother 39:2478–2483. doi: 10.1128/AAC.39.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seetulsingh PS, Hall LM, Livermore DM. 1991. Activity of clavulanate combinations against TEM-1 β-lactamase-producing Escherichia coli isolates obtained in 1982 and 1989. J Antimicrob Chemother 27:749–759. doi: 10.1093/jac/27.6.749. [DOI] [PubMed] [Google Scholar]
- 16.Martinez JL, Cercenado E, Rodriguez-Creixems M, Vincente-Perez MF, Delgado-Iribarren A, Baquero F. 1987. Resistance to β-lactam/clavulanate. Lancet ii:1473. [DOI] [PubMed] [Google Scholar]
- 17.Waltner-Toews RI, Paterson DL, Qureshi ZA, Sidjabat HE, Adams-Haduch JM, Shutt KA, Jones M, Tian GB, Pasculle AW, Doi Y. 2011. Clinical characteristics of bloodstream infections due to ampicillin-sulbactam-resistant, non-extended-spectrum β-lactamase-producing Escherichia coli and the role of TEM-1 hyperproduction. Antimicrob Agents Chemother 55:495–501. doi: 10.1128/AAC.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Wang Y, Ni Y. 2013. Hyperproduction of TEM-1 β-lactamase mediates the resistance of Escherichia coli to piperacillin-tazobactam and cefoperazone. Chin J Infect Chemother 13:167–172. [Google Scholar]
- 19.Sanders CC, Iaconis JP, Bodey GP, Samonis G. 1988. Resistance to ticarcillin-potassium clavulanate among clinical isolates of the family Enterobacteriaceae: role of PSE-1 β-lactamase and high levels of TEM-1 and SHV-1 and problems with false susceptibility in disk diffusion tests. Antimicrob Agents Chemother 32:1365–1369. doi: 10.1128/AAC.32.9.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pérez-Moreno MO, Pérez-Moreno M, Carulla M, Rubio C, Jardí AM, Zaragoza J. 2004. Mechanisms of reduced susceptibility to amoxycillin-clavulanic acid in Escherichia coli strains from the health region of Tortosa (Catalonia, Spain). Clin Microbiol Infect 10:234–241. doi: 10.1111/j.1198-743X.2004.00766.x. [DOI] [PubMed] [Google Scholar]
- 21.Lartigue MF, Leflon-Guibout V, Poirel L, Nordmann P, Nicolas-Chanoine M-H. 2002. promoters P3, Pa/Pb, P4, and P5 upstream from blaTEM genes and their relationship to β-lactam resistance. Antimicrob Agents Chemother 46:4035–4037. doi: 10.1128/AAC.46.12.4035-4037.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez JL, Vicente MF, Delgado-Iribarren A, Perez-Diaz JC, Baquero F. 1989. Small plasmids are involved in amoxicillin-clavulanate resistance in Escherichia coli. Antimicrob Agents Chemother 33:595. doi: 10.1128/AAC.33.4.595-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.San Millan A, Escudero JA, Gifford DR, Mazel D, MacLean RC. 2016. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat Ecol Evol 1:10. doi: 10.1038/s41559-016-0010. [DOI] [PubMed] [Google Scholar]
- 24.Creely D, Zambardi G, van Belkum A, Dunne WM Jr, Peyret M, Gayral JP, Shortridge D, Shubert C. 2013. International dissemination of Escherichia coli strains with discrepant behaviour in phenotypic antimicrobial susceptibility tests. Eur J Clin Microbiol Infect Dis 32:997–1002. doi: 10.1007/s10096-013-1837-5. [DOI] [PubMed] [Google Scholar]
- 25.Shubert C, Slaughter J, Creely D, van Belkum A, Gayral JP, Dunne WM, Zambardi G, Shortridge D. 2014. Population analysis of Escherichia coli isolates with discordant resistance levels by piperacillin-tazobactam broth microdilution and agar dilution testing. Antimicrob Agents Chemother 58:1779–1781. doi: 10.1128/AAC.02181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI 2017. Performance standards for antimicrobial susceptibility testing, 27th ed. Supplement M100. CLSI, Wayne, PA. [Google Scholar]
- 27.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Firth N, Ippen-Ihler K, Skurray R. 1996. Structure and function of the F factor and mechanism of conjugation. In Neidhardt F, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 29.Reguera JA, Baquero F, Pérez-Díaz JC, Martínez JL. 1991. Factors determining resistance to β-lactam combined with β-lactamase inhibitors in Escherichia coli. J Antimicrob Chemother 27:569–575. doi: 10.1093/jac/27.5.569. [DOI] [PubMed] [Google Scholar]
- 30.Nelson EC, Segal H, Elisha BG. 2003. Outer membrane protein alterations and blaTEM-1 variants: their role in beta-lactam resistance in Klebsiella pneumoniae. J Antimicrob Chemother 52:899–903. doi: 10.1093/jac/dkg486. [DOI] [PubMed] [Google Scholar]
- 31.Beceiro A, Maharjan S, Gaulton T, Doumith M, Soares NC, Dhanji H, Warner M, Doyle M, Hickey M, Downie G, Bou G, Livermore DM, Woodford N. 2011. False extended-spectrum β-lactamase phenotype in clinical isolates of Escherichia coli associated with increased expression of OXA-1 or TEM-1 penicillinases and loss of porins. J Antimicrob Chemother 66:2006–2010. doi: 10.1093/jac/dkr265. [DOI] [PubMed] [Google Scholar]
- 32.Sun S, Berg OG, Roth JR, Andersson DI. 2009. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182:1183–1195. doi: 10.1534/genetics.109.103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renau TE, Léger R, Flamme EM, Sangalang J, She MW, Yen R, Gannon CL, Griffith D, Chamberland S, Lomovskaya O, Hecker SJ, Lee VJ, Ohta T, Nakayama K. 1999. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J Med Chem 42:4928–4931. doi: 10.1021/jm9904598. [DOI] [PubMed] [Google Scholar]
- 35.Sandegren L, Andersson DI. 2009. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat Rev Microbiol 7:578–588. doi: 10.1038/nrmicro2174. [DOI] [PubMed] [Google Scholar]
- 36.McGann P, Courvalin P, Snesrud E, Clifford RJ, Yoon EJ, Onmus-Leone F, Ong AC, Kwak YI, Grillot-Courvalin C, Lesho E, Waterman PE. 2014. Amplification of aminoglycoside resistance gene aphA1 in Acinetobacter baumannii results in tobramycin therapy failure. mBio 5:e00915. doi: 10.1128/mBio.00915-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallagher LA, Coughlan S, Black NS, Lalor P, Waters EM, Wee B, Watson M, Downing T, Fitzgerald JR, Fleming GTA, O’Gara JP. 2017. Tandem amplification of the staphylococcal cassette chromosome mec element can drive high-level methicillin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 61:e00869-17. doi: 10.1128/AAC.00869-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girlich D, Bonnin RA, Bogaerts P, De Laveleye M, Huang DT, Dortet L, Glaser P, Glupczynski Y, Naas T. 2017. Chromosomal amplification of the blaOXA-58 carbapenemase gene in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother 61:e01697-16. doi: 10.1128/AAC.01697-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livermore DM, Seetulsingh P. 1991. Susceptibility of Escherichia coli isolates with TEM-1 β-lactamase to combinations of BRL42715, tazobactam or clavulanate with piperacillin or amoxicillin. J Antimicrob Chemother 27:761–767. doi: 10.1093/jac/27.6.761. [DOI] [PubMed] [Google Scholar]
- 40.Monogue ML, Nicolau DP. 2016. In vitro-in vivo discordance with humanized piperacillin-tazobactam exposures against piperacillin-tazobactam-resistant/pan-β-lactam-susceptible Escherichia coli. Antimicrob Agents Chemother 60:7527–7529. doi: 10.1128/AAC.01208-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stainton SM, Monogue ML, Nicolau DP. 2017. In vitro-in vivo discordance with humanized piperacillin-tazobactam exposures against piperacillin-tazobactam-resistant/pan-β-lactam-susceptible Klebsiella pneumoniae strains. Antimicrob Agents Chemother 61:e00491-17. doi: 10.1128/AAC.00491-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shelburne SA, Kim J, Munita JM, Sahasrabhojane P, Shields RK, Press EG, Li X, Arias CA, Cantarel B, Jiang Y, Kim MS, Aitken SL, Greenberg DE. 2017. Whole genome sequencing accurately identifies resistance to extended spectrum β-lactams for major Gram-negative bacterial pathogens. Clin Infect Dis 65:738–745. doi: 10.1093/cid/cix417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.CLSI 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard- tenth edition. CLSI document M07-A10. CLSI, Wayne, PA. [Google Scholar]
- 44.Benov L, Al-Ibraheem J. 2002. Disrupting Escherichia coli: a comparison of methods. J Biochem Mol Biol 35:428–431. doi: 10.5483/BMBRep.2002.35.4.428. [DOI] [PubMed] [Google Scholar]
- 45.Steingrube VA, Wallace RJ Jr, Brown BA, Pang Y, Zeluff B, Steele LC, Zhang Y. 1991. Acquired resistance of Nocardia brasiliensis to clavulanic acid related to a change in β-lactamase following therapy with amoxicillin-clavulanic acid. Antimicrob Agents Chemother 35:524–528. doi: 10.1128/AAC.35.3.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Innis M, Gelfand D, Sninsky J, White T. 1990. PCR protocols: a guide to methods and applications. Academic Press, London, United Kingdom. [Google Scholar]
- 47.Lee C, Kim J, Shin SG, Hwang S. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J Biotechnol 123:273–280. doi: 10.1016/j.jbiotec.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 48.Loman NJ, Quinlan AR. 2014. poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kern WV, Steinke P, Schumacher A, Schuster S, von Baum H, Bohnert JA. 2006. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J Antimicrob Chemother 57:339–343. doi: 10.1093/jac/dki445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of major porins OmpF and OmpC in E. coli. Download FIG S1, DOC file, 0.1 MB (53KB, doc) .
Copyright © 2018 Schechter et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.



