ABSTRACT
Corynebacterium diphtheriae, a Gram-positive, aerobic bacterium, is the causative agent of diphtheria and cutaneous infections. While mechanisms required for heme iron acquisition are well known in C. diphtheriae, systems involved in the acquisition of other metals such as zinc and manganese remain poorly characterized. In this study, we identified a genetic region that encodes an ABC-type transporter (iutBCD) and that is flanked by two genes (iutA and iutE) encoding putative substrate binding proteins of the cluster 9 family, a related group of transporters associated primarily with the import of Mn and Zn. We showed that IutA and IutE are both membrane proteins with comparable Mn and Zn binding abilities. We demonstrated that the iutABCD genes are cotranscribed and repressed in response to iron by the iron-responsive repressor DtxR. Transcription of iutE was positively regulated in response to iron availability in a DtxR-dependent manner and was repressed in response to Zn by the Zn-dependent repressor Zur. Electrophoretic mobility shift assays showed that DtxR does not bind to the iutE upstream region, which indicates that DtxR regulation of iutE is indirect and that other regulatory factors controlled by DtxR are likely responsible for the iron-responsive regulation. Analysis of the iutE promoter region identified a 50-bp sequence at the 3′ end of the iutD gene that is required for the DtxR-dependent and iron-responsive activation of the iutE gene. These findings indicate that transcription of iutE is controlled by a complex mechanism that involves multiple regulatory factors whose activity is impacted by both Zn and Fe.
IMPORTANCE Vaccination against diphtheria prevents toxin-related symptoms but does not inhibit bacterial colonization of the human host by the bacterium. Thus, Corynebacterium diphtheriae remains an important human pathogen that poses a significant health risk to unvaccinated individuals. The ability to acquire iron, zinc, and manganese is critical to the pathogenesis of many disease-causing organisms. Here, we describe a gene cluster in C. diphtheriae that encodes a metal importer that is homologous to broadly distributed metal transport systems, some with important roles in virulence in other bacterial pathogens. Two metal binding components of the gene cluster encode surface exposed proteins, and studies of such proteins may guide the development of second-generation vaccines for C. diphtheriae.
KEYWORDS: ABC transporters, Corynebacterium, diphtheria, iron activation, iron regulation, iron transport, zinc regulation
INTRODUCTION
The etiological agent of diphtheria, Corynebacterium diphtheriae, causes severe upper respiratory and cutaneous infections in humans. Diphtheria toxin (DT) accounts for much of the morbidity and mortality observed in respiratory infections (1). While the diphtheria toxoid, an inactive form of DT, is an effective vaccine antigen for the prevention of disease caused by DT-producing C. diphtheriae, colonization and infection by nontoxinogenic strains are still observed in highly vaccinated populations (2, 3). The gene for DT synthesis, tox, is carried on the βtox+ bacteriophage and can be acquired by nontoxigenic strains of C. diphtheriae through infection and lysogeny by phage carrying the toxin gene. Although tox is carried on a phage genome, its transcription is regulated by the chromosomally encoded Fe-responsive repressor DtxR (4–6). Since it is well established that the Fe-regulated DT is produced during infection by C. diphtheriae, it seems likely that the bacteria colonize an Fe-restricted niche within the host. Mechanisms for Fe restriction within the human host include the host proteins transferrin and lactoferrin, which bind free Fe with high affinity in the extracellular environment (7). Much of the intracellular Fe is bound by hemin, which is associated with proteins such as hemoglobin. The ability to acquire Fe is essential for many bacterial pathogens to cause disease, and the competition for Fe between the host and invading bacterial pathogens has resulted in the evolution of numerous mechanisms that bacteria utilize to scavenge this vital element (8). This sequestration of Fe from invading pathogens by the host was termed nutritional immunity (7). Our perspective of nutritional immunity has since broadened from Fe-withholding strategies to include mechanisms that restrict the availability of other metals, such as Zn and Mn (8, 9). Although Fe acquisition in several bacterial pathogens has been thoroughly studied, limited information is available as to how bacterial pathogens transport Mn and Zn.
Manganese, like Fe, is a critical metal involved in cellular processes (10, 11), including oxidative stress tolerance (12, 13), DNA replication (14), and central metabolism (15). In bacteria, the import of Mn involves two distinct classes of transport systems, the MntH family and the ABC-type transporters. MntH family proteins are proton-driven metal ion transporters with homology to eukaryotic NRAMP proteins (10, 16–18). The ABC transporters involved in Mn uptake include metal binding receptor proteins of the cluster 9 group (19). In the ABC systems, Mn is first bound by a substrate binding protein (SBP) and then passed through the membrane by an ATP-driven permease. The SBPs are localized to the periplasmic space in Gram-negative bacteria and anchored to the membrane in Gram-positive organisms. In the Gram-positive bacterium Staphylococcus aureus, the MntH and MntABC transport systems acquire sufficient Mn for host infection despite sequestration by calprotectin (20), a soluble host protein with high binding affinity for Zn and Mn (21, 22). Bacteria regulate expression of Mn uptake systems through Mn-responsive regulatory proteins such as MntR, a member of the DtxR family of metal-dependent repressors (23).
Zinc also serves indispensable roles for bacteria (11, 24), and acquisition of Zn is important in the virulence of numerous pathogens (8). The availability of Zn to invading bacterial pathogens is limited within the host, with calprotectin sequestering free Zn (21). Zn at high levels can be toxic to bacteria, and macrophages use Zn intoxication as a mechanism for pathogen elimination (8, 25). Much like for Fe and Mn, pathogens must balance Zn sufficiency and toxicity. Although Gram-positive bacteria import Zn through ABC transporters of the cluster 9 group, certain members of the group are specific to Zn, while others are more promiscuous and can bind and transport multiple metals (26, 27). Furthermore, Zn-specific SBPs typically possess a loop region that is rich in acidic and His residues (His-rich region) and that is not found in the Mn or Fe transporters (19, 27). In Streptococcus pneumoniae, two separate SBPs, AdcA and AdcAII, function in Zn acquisition (26), and both SBPs use a shared ABC transporter to import Zn. Much like for Fe and Mn, regulation of Zn import is controlled by Zn-dependent transcriptional regulators such as Zur (28, 29) and the analogous protein AdcR (30, 31). High levels of intracellular Zn activate Zur, which in turn represses transcription of genes involved in Zn uptake. Collectively, studies on Mn and Zn acquisition and the genetic regulation of these systems illustrate that these metals, like Fe, are crucial to the survival and pathogenesis of many bacteria (8).
In C. diphtheriae, the Mn- and Zn-responsive regulators MntR (32) and Zur (33) have been identified, but the systems involved in acquisition of these essential nutrients are poorly defined. C. diphtheriae MntA (32) and a TroA homolog, Dip0438, (33), are SBPs of the cluster 9 family, which have been identified from screens for Mn- and Zn-regulated genes. While both share significant homology to metal import systems, mutations in mntA had no effect on growth in Mn-depleted medium (32), suggesting that other transporters allow for sufficient import of Mn in the mntA mutant. In this study, we identified a C. diphtheriae gene cluster, dip0169 to dip0173 (iutABCDE), which is predicted to encode two cluster 9 family SBPs, IutA (Dip0169) and IutE (Dip0173). Both proteins were shown to be membrane localized and were able to bind Mn2+ and Zn2+. Despite the proximity and organization of these genes on the chromosome, differential regulation of these two SBPs was observed in response to Fe and Zn but not Mn. Transcription of iutA was repressed by Fe alone, and analysis of the iutE promoter indicated that transcription was activated by Fe and repressed by Zn. The Fe activation of iutE transcription was dependent on DtxR and did not require DtxR binding to the iutE upstream region. These findings suggest that transcription of iutE is regulated by a novel and complex mechanism that involves multiple metal-dependent regulatory factors.
RESULTS
Identification of the iut gene cluster as a potential cation transport system.
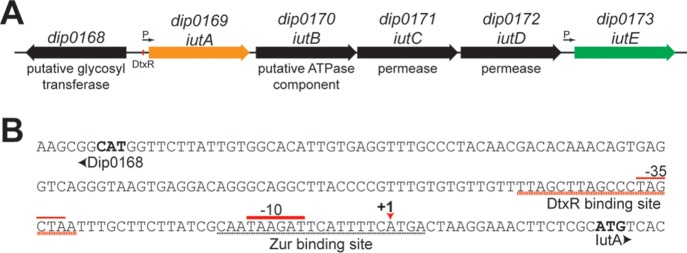
In a search for putative Corynebacterium diphtheriae Zn and Mn uptake mechanisms, we focused on proteins with homology to Treponema pallidum TroA (34), one of the first characterized SBPs of the cluster 9 family. In C. diphtheriae strain NCTC13129, a BLAST search (35) identified four proteins with significant amino acid similarity to TroA, i.e., Dip0169 (88% coverage and 35% identity), Dip0173 (88% coverage and 33% identity), MntA (dip0615) (89% coverage and 30% identity), and Dip0438 (88% coverage and 26% identity). Dip0169 (designated IutA) and Dip0173 (designated IutE) were of interest due to the chromosomal proximity of the genes and their presence in an unusual genetic cluster; iutA and iutE flank a putative ABC transporter locus (designated iutBCD) (Fig. 1A). IutA and IutE exhibit high amino acid homology (58% identity and 70% similarity), and both proteins possess an N-terminal lipoprotein signal peptidase II recognition sequence and the associated cysteine residue required for membrane anchoring (see Fig. S1 in the supplemental material, red). IutA and IutE also possess three highly conserved histidine residues that are known to coordinate metal binding in other cluster 9 proteins (29, 36) (Fig. S1, yellow). Despite high homology between IutA and IutE, IutE alone possesses a His-rich region (Fig. S1, bold). These charged regions are common among Zn transporters in the cluster 9 family, while those without this sequence are less specific and are associated with the transport of Mn and other divalent cations (19, 27, 29, 36).
FIG 1.
iut gene cluster organization. (A) Organization of the iut gene cluster in C. diphtheriae. The red box denotes the predicted DtxR binding motif within the iutA promoter region. Arrows indicate the promoters identified here. (B) dip0168-iutA intergenic region with notable elements, including the DtxR binding site, putative Zur binding site, transcription initiation site (+1), and promoter elements (−10 and −35). The +1 site was identified through 5′ RACE and DNA sequencing. The start codons for Dip0168 and IutA are in bold.
To address whether this system is required for metal transport in C. diphtheriae, a deletion of the operon (iutABCDE) was introduced into wild-type C. diphtheriae strain 1737. The Δiut mutant displayed no differences in growth compared to the wild-type parent strain when grown in metal-depleted mPGT medium. This finding suggests that other systems may also be functioning in the transport of Mn or Zn, and we reasoned that likely candidates for this metal uptake activity are the putative metal transporters encoded by mnt (mntABCD) and tro (dip0438 to dip0441). Nonpolar deletions of these two uptake systems were constructed in C. diphtheriae, and these mutations were both moved into the Δiut strain to construct the Δiut Δmnt Δtro triple deletion mutant. The triple deletion mutant also showed no growth deficiency relative to the wild-type strain when grown in either rich or metal-limited mPGT medium (M. P. Schmitt, unpublished results), suggesting the presence of additional mechanisms for Zn and/or Mn import.
IutA and IutE are membrane proteins with Mn2+ and Zn2+ binding properties.
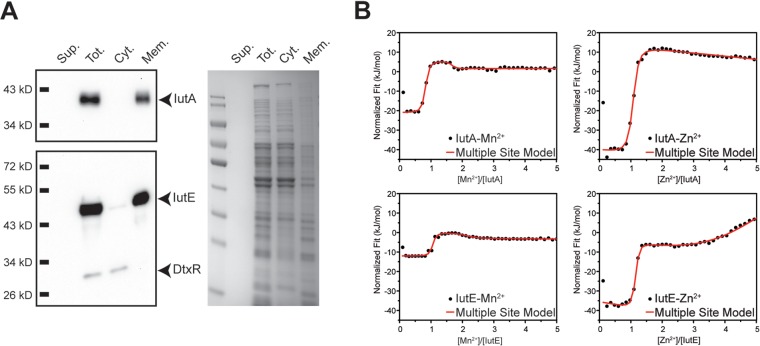
In Gram-positive bacteria, the SBPs of metal transport systems are associated primarily with metal uptake, but they may also function as adhesins and have been examined as components in certain bacterial vaccines (31, 37, 38). To determine the cellular localization of IutA and IutE, we used ultracentrifugation to separate the cytosolic and membrane fractions (Fig. 2A). Fractions, including the total cell lysate and filtered culture supernatant, were separated by SDS-PAGE and transferred for Western blotting. Using protein-specific antisera, IutA and IutE were detected predominantly in the total cell lysate (membrane and cytosol fractions) and membrane fractions. As expected, the cytosolic control, DtxR, was found in the total lysate and cytosolic fraction.
FIG 2.
Subcellular fractionation and ITC for IutA and IutE. (A) Wild-type C. diphtheriae proteins were separated following lysis and ultracentrifugation. Filtered culture supernatant (Sup.), total lysate (Tot.), and the cytoplasmic (Cyt.) and membrane (Mem.) fractions were probed using antisera against IutA, IutE, and DtxR or stained with Coomassie blue. (B) Representative fitted ITC binding profiles for IutA and IutE with Mn2+ and Zn2+. For Mn2+ binding to IutA and IutE (left upper and lower panels, respectively), two binding sites are evident for each protein titration profile as two inflection points at ∼1- and ∼2-fold excess of Mn2+ with 1:1 protein/ligand stoichiometry. However, for Zn2+ binding to IutA and IutE, only a single high-affinity site is observed for each protein at stoichiometric ligand/protein amounts (right upper and lower panels, respectively). At >2-fold ligand excess, weak metal binding is observed for both proteins as evidenced by a negative slope and a positive slope for IutA and IutE, respectively. Thermodynamic parameters obtained after fittings are shown in Table 1.
While a growth phenotype for the C. diphtheriae Δiut mutant was not observed, we sought to identify a possible function for IutA and IutE by examining the metal binding ability of these proteins. Isothermal titration calorimetry (ITC) on purified recombinant IutA and IutE was performed to discern metal binding properties in the presence of either Mn2+ or Zn2+ (Fig. 2B; Table 1). We identified the presence of two binding sites for Mn2+ in IutA and IutE, with the higher-affinity site showing comparable binding to Mn2+ between the two proteins (Kd1IutA = 1.1 ± 0.5 nM and Kd1IutE = 2 ± 1 nM). The binding affinity of the second Mn2+ binding site was also similar between IutA and IutE, but the Kd was at least 200-fold lower than that for the high-affinity site (Kd2IutA = 0.2 ± 0.1 μM and Kd2IutE = 0.7 ± 0.3 μM). Compared to the affinity for Mn2+, binding to Zn2+ was roughly 300- and 35-fold weaker for IutA (Kd = 350 ± 40 nM) and IutE (Kd = 70 ± 30 nM), respectively. When the concentration of Zn2+ exceeded the protein concentration by 2-fold, a second weak binding event was observed for both proteins. However, as opposed to the case for the high-affinity metal binding site, ITC data fitting for the second, low-affinity site yielded inconsistent stoichiometry. Taken together, these results suggest that the second Zn binding site is likely nonspecific binding to IutA and IutE.
TABLE 1.
Isothermal titration calorimetry of IutA and IutE with Mn2+ and Zn2+
| Parameter | Valuea for: |
|||
|---|---|---|---|---|
| Mn2+ |
Zn2+ |
|||
| IutA | IutE | IutA | IutE | |
| Kd1 (nM) | 1.1 ± 0.5 | 2 ± 1 | 350 ± 40 | 70 ± 30 |
| ΔH1 (kJ/mol) | −17.7 ± 0.5 | −18 ± 1 | −55 ± 1 | −34 ± 5 |
| ΔS1 (J/mol · K) | 112 | 106 | −62 | 24.5 |
| Stoichiometry | 0.98 ± 0.01 | 1.01 ± 0.01 | 1.023 ± 0.005 | 1.01 ± 0.01 |
| ΔG1 (kJ/mol) | −51 | −50 | −37 | −41 |
| Kd2 (μM) | 0.2 ± 0.1 | 0.7 ± 0.3 | 6 ± 3 | |
| ΔH2 (kJ/mol) | 3.1 ± 0.5 | 6 ± 4 | −58 ± 25 | |
| ΔS2 (J/mol · K) | 137 | 136 | −94 | |
| Stoichiometry | 1.06 ± 0.08 | 1.1 ± 0.2 | 5.2 ± 0.2 | |
| ΔG2 (kJ/mol) | −38 | −35 | −30 | |
The errors reported for Kd, ΔH, and stoichiometry were obtained from ITC data-fitting software, whereas the errors for ΔG and ΔS were derived from Kd and ΔH error values.
Zinc and iron regulate IutA and IutE expression through Zur and DtxR.
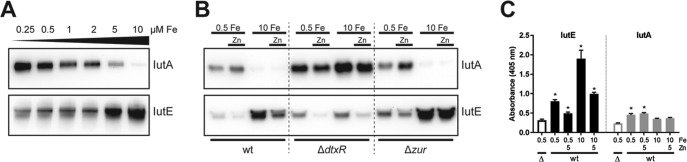
The iutA promoter region contains a putative DtxR binding motif with a strong consensus to the known DtxR binding site (39) (Fig. 1B), suggesting that transcription of iutA is repressed by Fe via DtxR. To address whether Fe, and consequently DtxR, impacts gene expression of the iutABCDE cluster, wild-type C. diphtheriae was grown in metal-limited medium (mPGT) with various levels of Fe supplementation (0.25 to 10 μM FeCl3). Consistent with DtxR-mediated regulation, IutA protein levels were reduced as the available Fe increased; surprisingly, however, IutE levels increased with Fe supplementation (Fig. 3A). Given the differences in protein abundance and the similarity of IutA and IutE to proteins involved in Mn and Zn transport, we assessed the roles of other metals in regulating the expression of these two proteins.
FIG 3.
Expression of IutA and IutE is controlled by DtxR and Zur. (A) Protein levels of IutA and IutE were assessed following growth in mPGT with iron supplementation (final concentrations are indicated) by Western blotting. (B) Protein levels of IutA and IutE were assessed in the wild-type C. diphtheriae and isogenic ΔdtxR and Δzur mutants. Iron (μM) and Zn were supplemented where indicated; Zn was added to a final concentration of 5 μM. (C) ELISA on whole C. diphtheriae 1737 cells (wild type [wt] or Δiut mutant [Δ]) adsorbed to plastic microtiter plates following growth in mPGT with metal supplementation as indicated (μM). Antiserum against IutE or IutA was used for detection. Statistical significance was determined by two-way analysis of variance (ANOVA) and multiple-comparison test. *, P < 0.05 when comparing the indicated sample against the respective Δiut sample (n = 3).
We hypothesized that the expression of iutA and iutE may also be controlled by the Zn-activated Zur (33) and Mn-activated MntR (32). The presence of high levels of Zn resulted in the repression of IutE expression but did not significantly alter IutA abundance (Fig. 3B). Mn had no discernible effect on either IutA or IutE levels (not shown). Reduced protein expression in response to Fe and Zn is consistent with DtxR (5, 6, 32, 40)- and Zur (33)-dependent activity in C. diphtheriae. We addressed the roles of these regulators by assessing protein levels in isogenic ΔdtxR and Δzur mutant backgrounds (Fig. 3B). In the ΔdtxR strain, IutA expression was increased relative to wild-type levels, while the expression profile of IutA in the Δzur strain was similar to that in the wild-type strain. IutE expression was higher in the Δzur mutant regardless of the Zn levels, suggesting that transcription of iutE is repressed by Zur in the presence of Zn. However, in the ΔdtxR mutant, IutE expression was reduced under high-Fe conditions, and stimulation of expression by Fe was not observed, suggesting that DtxR is required for the increase in expression under high-Fe conditions.
Although our data indicate that both IutA and IutE are localized to the membrane (Fig. 2A), it was not known whether these proteins are exposed at the cell surface. To determine whether IutA and IutE are surface exposed, we performed whole-cell enzyme-linked immunosorbent assays (ELISAs) using the wild-type C. diphtheriae or the Δiut mutant that was grown in high- and low-Fe media and in the presence and absence of Zn. IutE was detected above background levels (Δiut) under all conditions examined, and the signal intensity for IutE was consistent with our protein expression results, where IutE levels were increased in high Fe relative to low Fe and reduced in the presence of Zn (Fig. 3C). IutA was detected on the cell surface following growth in low-Fe medium but was not significantly detected on cells grown in Fe-replete medium, which is also consistent with the protein expression results (Fig. 3B). It is unclear why IutA detection is comparatively weaker at the cell surface than the levels observed for IutE; the epitopes recognized by our polyclonal antisera may not be available for antibody binding, or IutA itself may not be as well exposed as IutE at the cell surface.
iutABCD are coregulated by iron, while iutE is transcribed independently.
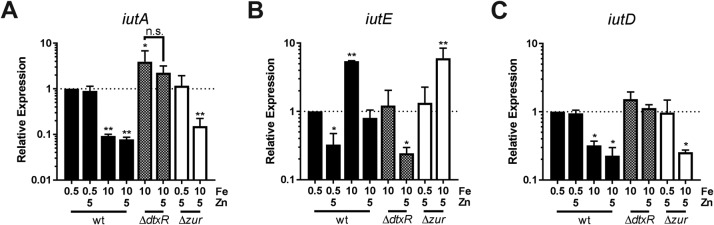
To confirm transcriptional regulation of iutA and iutE, we used quantitative PCR (qPCR) to assess relative transcript levels. In wild-type C. diphtheriae, transcription of iutA was reduced under Fe-replete conditions relative to low-Fe conditions (Fig. 4A), while transcript levels with iutA were derepressed in the ΔdtxR mutant with high Fe, supporting Fe- and DtxR-mediated repression of iutA expression. Transcript levels of iutA were not affected by Zn supplementation in the wild type or altered in the Δzur mutant, suggesting no role for Zn or Zur in regulation of iutA. Conversely, for iutE, transcript levels were reduced following growth with Zn supplementation and increased following growth in high Fe compared to low Fe (Fig. 4B). iutE transcript levels in the ΔdtxR mutant were reduced compared to levels detected in the wild type under similar growth conditions, suggesting that the Fe activation of iutE transcription is dependent upon DtxR. In the Δzur mutant, transcript levels were higher following growth under Fe-replete conditions. Taken together, these results indicate that the Fe/DtxR-dependent and Zn/Zur-dependent mechanisms act independently to regulate transcription of iutE.
FIG 4.
Relative expression of iutA, iutE, and iutD. Relative transcript levels for iutA (A), iutE (B), and iutD (C) were determined in the C. diphtheriae 1737 wild-type (wt) strain and isogenic ΔdtxR and Δzur mutants. Relative expression was determined using the ΔΔCq method comparing against the respective wild-type transcript levels following low-iron growth (wt 0.5 Fe set to 1); statistical significance was determined by two-way ANOVA and multiple-comparison test on ΔCq values. *, P < 0.05; **, P < 0.01 (when comparing the indicated sample against the wt 0.5 Fe sample; n = 3). n.s., not significant. Final concentrations of metals (micromolar) are indicated.
Operons for ABC transporters typically contain a single promoter that controls the transcription of all the genes in the operon. Given the unusual nature of the iutABCDE gene cluster and evidence for distinct promoters upstream of iutA and iutE, we sought to determine whether the iutABCD region was expressed as a single transcriptional unit by assessing transcript levels of the iutD gene, the terminal gene in the iutABCD genetic region. Relative quantification of iutD mRNA showed a profile that was almost identical to that observed for iutA under all conditions tested, suggesting that iutABCD are cotranscribed and that iutE is transcribed from a separate promoter (Fig. 4A and C).
Zur binds at both iutA and iutE promoter regions.
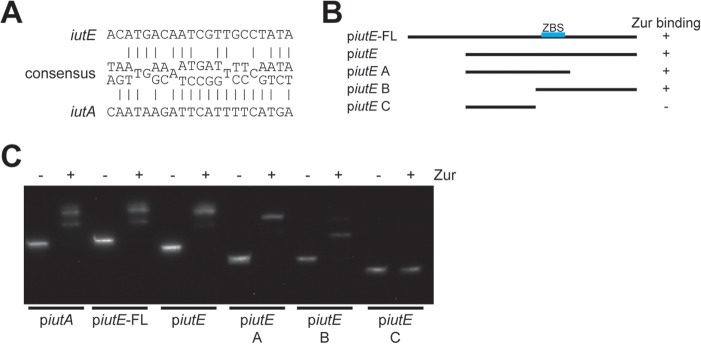
A study assessing the Zur regulon for Corynebacterium glutamicum described a consensus Zur binding motif (41) (Fig. 5A). We used this consensus sequence to identify a putative Zur binding site upstream of iutE, as well as a possible Zur binding site upstream of iutA, which was unexpected, since there was no previous evidence for Zn and Zur regulation for iutA. To test Zur binding to both promoter regions and validate the location of the iutE Zur binding site, we performed electrophoretic mobility shift assays (EMSAs) using the C. diphtheriae Zur protein with various DNA fragments for the iutE promoter (Fig. 5B) and a single fragment for iutA. The addition of Zur altered the migration of DNA fragments predicted to contain the Zur binding sites for both iutA and iutE (Fig. 5C), indicating that Zur binds to both regions; dissection of the iutE promoter region confirms the predicted location of the Zur binding site. While Zur binds both promoter regions in this in vitro approach, it is not clear whether the binding of Zur at the iutA promoter region is biologically relevant, as our expression data suggest that Zn and Zur do not regulate iutA (Fig. 3B and 4A). Although the predicted iutA Zur binding site is not expected to affect the divergently transcribed gene dip0168 (Fig. 1B), we nevertheless assessed expression of dip0168 and found no significant change in transcription in response to Zn (not shown). The role for the iutA Zur binding site may not have any biological relevance, or our conditions may not be sufficient to discern an effect.
FIG 5.
Verification of the Zur binding site in IutE and IutA. (A) Putative iutE and iutA Zur binding sites compared to the Actinobacteria Zur consensus site determined previously (41); many positions within the consensus are variable, and the most prominent bases are shown. (B) Relative positions of different promoter fragments for iutE and summary of the results shown in the EMSA with recombinant Zur (C). (C) EMSA with recombinant Zur incubated with the iutA promoter region and with different portions of the iutE promoter region. Recombinant Zur was added where indicated (+).
Identification of promoter elements involved in iutE transcription.
While DtxR repression of gene expression has been described (5, 6, 40, 42), its role in activation of gene expression has been observed only for the hrtAB locus in C. diphtheriae, and the mechanism for this activation was not determined (43). In related organisms, several mechanisms have been described. In Mycobacterium, the DtxR ortholog IdeR activates ferritin expression by binding the ferritin promoter and antagonizing activity of a separate DNA binding protein, Lsr2 (44). Alternatively, DtxR may control a separate regulator such as an Fe-responsive small RNA (sRNA) (45) or a DNA binding protein, such as the DtxR-regulated RipA protein in C. glutamicum (46). We assessed each of these potential mechanisms for activation of iutE expression.
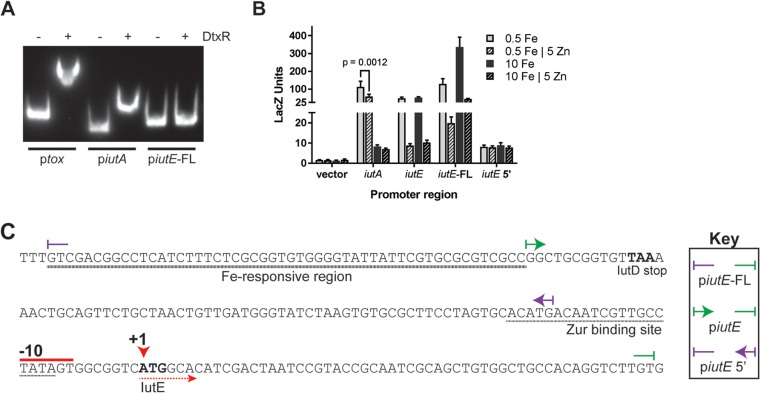
Given the prediction of a DtxR binding site upstream of iutA and DtxR-dependent activation on iutE, we hypothesized that DtxR could bind to the regions upstream of iutA and iutE. We assessed the effect of DtxR on the mobility of DNA fragments harboring the iutA and iutE promoter regions by EMSA alongside the tox promoter region, which serves as a positive control for DtxR binding (Fig. 6A). While DtxR binds the iutA promoter, the presence of DtxR did not alter the mobility of the DNA fragment harboring the iutE promoter and upstream region, suggesting that DtxR does not bind upstream of the iutE coding region. The binding of DtxR at the iutA promoter is consistent with DtxR regulation of iutA transcription (5, 6, 40). The inability of DtxR to bind the iutE promoter and upstream region suggests that the effect of DtxR on iutE transcription is indirect and that DtxR may control the expression of a second factor that directly impacts iutE transcription.
FIG 6.
IutE iron activation requires a DNA region upstream of the promoter. (A) EMSA with purified DtxR and promoter regions of tox, iutA, and iutE; DtxR was added where indicated (+). (B) Beta-galactosidase units observed from promoter fusion constructs following growth in mPGT with metal supplementation as indicated. The means and standard deviations from three independent experiments are shown. (C) DNA sequence of the iutE promoter region with notable features marked. The boundaries of the promoter fusions and DNA fragments used for EMSA are indicated (key). +1, start site of transcription; −10, putative promoter element.
To assess the location of promoter elements necessary for iutA and iutE expression, we generated promoter-lacZ fusions. For iutA, a single construct was designed that demonstrated strong Fe repression but also weak Zn repression in the wild-type strain. The location of the predicted Zur binding site overlaps the putative −10 and the experimentally determined +1 site (Fig. 1B), suggesting a potential role for Zur in regulating iutA expression. However, when we assessed the protein abundance and relative RNA levels under in vivo conditions (Fig. 3B and 4A), zinc did not repress iutA expression. The reason for the inconsistency in the results among assays to assess regulation of iutA is not known. It is possible that a novel regulatory factor, potentially titrated away in the promoter fusion plasmid studies, prevented the interaction of Zur with the iutA Zur binding site.
For iutE, three separate constructs spanning different regions of the promoter were designed (Fig. 6C). In initial studies of the iutE promoter, we designed a fusion construct (piutE) that contained sequences encompassing the intergenic region upstream of iutE. Expression from the piutE construct in the wild-type strain was repressed by Zn, consistent with the presence of the Zur binding site, but was not affected by high Fe levels (Fig. 6B and C). Considering the qPCR results, which showed both Zn and Fe regulation, we designed an iutE promoter fusion construct that extended into the coding region of iutD (piutE-FL); this same construct was used for our EMSAs (Fig. 5B and C and 6A). The piutE-FL promoter fusion construct exhibited overall enhanced promoter activity compared to the piutE construct and showed increased LacZ activity under Fe-replete conditions relative to levels observed after growth in low-Fe medium (∼2.6-fold increase) (Fig. 6B). Zn-dependent repression was also observed for the piutE-FL construct, consistent with the location of the Zur binding site (Fig. 6B and C). Very low, constitutive expression was detected for the piutE 5′ construct, which contains the region upstream of the Zur binding site (Fig. 6B and C). Together, these findings suggest that transcription at the iutE promoter is regulated by both Fe and Zn, and the region required for the Fe-responsive activation of transcription includes the 3′ end of the iutD coding region, which we have designated an “Fe-responsive region” (Fig. 6C).
If the Fe-responsive region is far removed from the transcription initiation site (TIS), then it is unlikely that an sRNA is responsible for Fe activation of iutE expression, since sRNAs typically act upon transcripts (45). The Fe-responsive region we identified was within the coding region of iutD, over 100 bases from the start codon of iutE (Fig. 6C). We identified the 5′ end of the iutE mRNA by 5′ rapid amplification of cDNA ends (RACE) and DNA sequencing, which indicated that the TIS of iutE coincides with the start codon (Fig. 6C). Because of this unusual finding, we confirmed the RACE result in control studies in which total cDNA was used as the template for PCR using primers at different locations relative to the putative TIS. Control primers were designed to overlap the putative TIS to confirm the RACE result, 20 bases upstream of the TIS to capture potential transcription initiation upstream of the observed +1 site, and further upstream as a negative control. In agreement with the RACE result, a PCR product was observed from total cDNA when a reverse primer was paired with a forward primer overlapping the TIS (+1 primer) but not when it was paired with a forward primer designed to anneal 20 bases upstream of the transcriptional start (upstream primer) (see Fig. S2 in the supplemental material). Together, these data indicate that the mRNA for iutE does not have a 5′ untranslated region (UTR) and is a leaderless transcript, a class of transcripts that have only recently been identified through high-throughput sequencing (47, 48). Furthermore, the lack of a 5′ UTR and the identification of an Fe-responsive DNA region upstream of transcription initiation suggest that an sRNA is likely not responsible for the Fe- and DtxR-dependent stimulation of iutE transcription.
RipA does not regulate iutE expression.
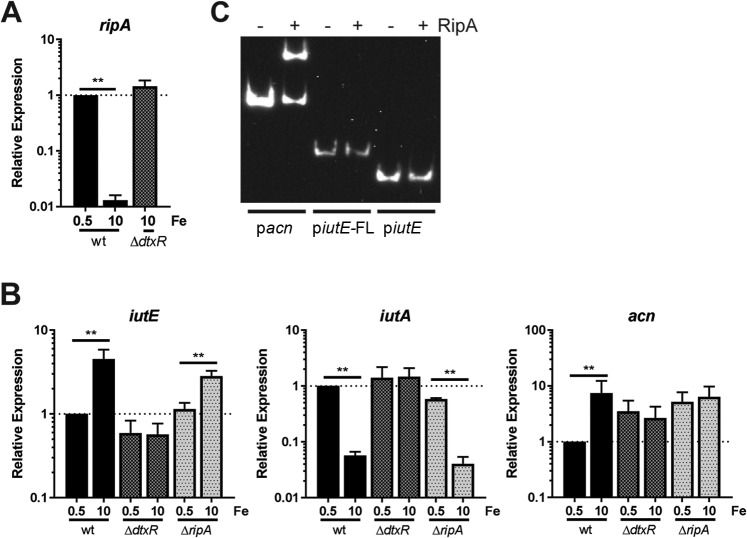
Our results indicate that DtxR is required but acts indirectly to increase iutE transcript levels in response to increased Fe availability. In C. glutamicum, RipA is an AraC-type transcriptional regulator, which is itself regulated by DtxR (46). The combined function of DtxR and RipA allows for the derepression of genes under Fe-replete conditions and could explain the regulation of iutE in C. diphtheriae. In C. diphtheriae, RipA (Dip0922) was initially identified as iron-regulated protein 3 (Irp3) (40). We confirmed the DtxR-dependent Fe regulation of ripA by qPCR and observed >70-fold repression by Fe in the wild-type strain (Fig. 7A). Transcript levels of ripA in the ΔdtxR mutant grown with high Fe were comparable to levels observed with low Fe for the wild type, consistent with DtxR-dependent regulation.
FIG 7.
Assessing expression of iutE in a ΔripA mutant. (A) The relative transcript levels for ripA were determined by comparing the C. diphtheriae 1737 wild-type (wt) and ΔdtxR mutant strains following growth in mPGT with iron supplementation as indicated. (B) The relative transcript levels for iutE, iutA, and the putative RipA-regulated acn gene were measured following growth in modified mPGT with Fe supplementation as indicated. Relative expression was determined using the ΔΔCq method; statistical significance was determined by two-way ANOVA and multiple-comparison test on ΔCq values. **, P ≤ 0.0001 (n = 3). (C) EMSA performed with recombinant RipA and the promoter regions pacn, piutE-FL, and piutE.
Next, we generated a nonpolar deletion of ripA in C. diphtheriae to assess transcript levels for iutE in parallel with the wild-type parent and isogenic ΔdtxR mutant. Levels of iutE mRNA in the wild type and the ΔdtxR mutant were consistent with results described previously showing that iutE transcription is activated by Fe in a DtxR-dependent manner (Fig. 7B). We then assessed RipA regulation of iutE; if RipA regulates iutE in a manner similar to how it regulates genes in C. glutamicum, then iutE transcript levels in the ΔripA mutant should be at constitutively high levels regardless of the Fe concentration in the growth medium. As shown in Fig. 7B, expression of iutE in the ΔripA mutant was still regulated by Fe, with significantly more transcripts detected under Fe-replete conditions than in low-Fe medium. This finding suggests that iutE is not regulated by RipA or at least is not regulated in a manner consistent with the function of RipA previously described in C. glutamicum (46). Consistent with prior observations, iutA was repressed by Fe in a DtxR-dependent manner and was unaffected by the deletion of ripA. As a control for RipA function, we assessed relative transcript levels of acn (dip1283), a gene that encodes aconitase and is known to be RipA regulated in C. glutamicum (46, 49, 50). Comparing low- and high-Fe growth, acn transcript levels were more abundant with high Fe in the wild type and did not differ in the ΔripA mutant (Fig. 7B), consistent with the observed function of RipA in C. glutamicum (46). However, in the ΔdtxR mutant, acn transcript levels were not comparable to wild-type levels, which would be expected for a RipA-regulated gene. Rather, elevated levels of acn transcript were detected under both high- and low-Fe conditions, suggesting the presence of additional regulators, which has been previously reported for acn expression in C. glutamicum (51). In C. glutamicum studies, expression of acn has been assessed in the wild-type, ΔripA (46), and ΔdtxR (50) strains, but side-by-side analyses comparing low- and high-Fe conditions were not done. As such, it is unclear if acn expression in C. glutamicum has a profile similar to our observations in C. diphtheriae. To determine whether RipA binds the iutE promoter region, we purified recombinant C. diphtheriae RipA and performed EMSAs with the acn promoter region (pacn) and two iutE promoter fragments: the longer piutE-FL promoter fragment, which showed Fe regulation, and the shorter fragment (piutE), which displayed regulation only by Zn (Fig. 6B). The pacn showed binding to RipA (Fig. 7C), while neither iutE promoter fragment was affected by the presence of recombinant RipA, indicating that RipA does not bind the iutE upstream region. Together, our results suggest that RipA does not regulate transcription of iutE and that other factors, yet to be identified, mediate regulation of the Fe-dependent activation of iutE transcription.
DISCUSSION
Under high-Fe conditions, bacteria repress expression of Fe acquisition systems while upregulating certain Fe-containing proteins (49, 50, 52) and various Fe storage proteins such as ferritins (44). These same Fe-containing proteins are downregulated under Fe-limited conditions to reserve Fe for essential functions in what is termed the “iron-sparing” response (45, 53). Fe-responsive regulators such as DtxR, found in some Gram-positive bacteria, and Fur, which is found in both Gram-negative and Gram-positive organisms, control much of the Fe regulon in numerous bacteria, including many pathogens (5, 44, 54). These regulatory proteins are activated upon Fe binding, where they interact with DNA at specific binding sites to inhibit transcription of genes involved in functions ranging from Fe metabolism to virulence (40, 42, 44, 49, 50, 55–58). While the primary activity associated with these Fe-responsive regulators is to repress gene expression, DtxR and Fur have also been shown to activate gene transcription (44–46). In this study, we provide evidence for the increased transcription of the iutE gene in response to Fe availability. Furthermore, we show that this elevated transcription is DtxR dependent and requires sequences that reside over 70 bp upstream of the iutE promoter.
The presence of increased transcript levels in response to Fe in association with either Fur or DtxR has been described in other organisms and includes at least three distinct mechanisms: (i) direct binding of DtxR upstream of a promoter region, (ii) regulation of a small RNA (sRNA), and (iii) regulation of a second regulatory protein. An example of direct activation by a DtxR-like protein occurs in Mycobacterium tuberculosis, where induction of ferritin (BfrB) expression involves the direct binding of the Fe-bound form of IdeR, a DtxR homolog, to the bfrB promoter region (44). In this study, we found no evidence for DtxR binding upstream of iutE, suggesting that transcription of iutE is not regulated by Fe in a manner analogous to that for bfrB.
In many bacterial species, the transcription of regulatory sRNAs is controlled by Fur in an Fe-dependent manner (45), and while these systems have been extensively studied in Gram-negative bacteria, little is known about Fe-regulated sRNAs in Gram-positive organisms. In Corynebacterium species, sRNAs associated with gene regulation have not been identified, although a recent transcriptome sequencing (RNA-Seq) analysis in C. glutamicum identified numerous noncoding sRNAs, which may have the potential to function in the regulation of gene expression (59). Regulatory sRNAs typically control gene expression by associating with target transcripts directly through sequence complementarity, resulting in RNase-dependent degradation of the target (60). With regard to the transcription of iutE, the absence of a 5′ UTR does not exclude a role for an sRNA; however, we identified a region in iutD (the Fe-responsive region) (Fig. 6C) that is partly, if not solely, responsible for Fe-dependent activation of iutE. For iutE, the requirement of this nontranscribed region for the Fe activation of transcription of iutE does not support a role for an sRNA in the regulation of iutE but instead suggests that a DtxR-regulated DNA binding protein, which binds far upstream of the transcriptional start site, controls expression of iutE (Fig. 6C).
RipA is a DNA binding protein whose expression is known to be controlled by DtxR (40), and we investigated the possibility that RipA is involved in controlling expression of iutE in response to Fe. We confirmed that expression of RipA is downregulated by DtxR under high-Fe conditions, which suggested that RipA may be responsible for the increased transcription of iutE in Fe-replete medium. However, gene expression studies using a ripA deletion mutant and EMSA analysis with purified RipA showed that RipA does not appear to be involved in controlling the elevated expression of iutE observed in high-iron environments. Collectively, the data suggest that an additional DtxR-regulated protein is required to activate transcription of iutE under Fe-replete conditions. The Fe-dependent regulation of iutE transcription is clearly complex and involves multiple regulators and metal cofactors. Additional studies will be needed to unravel the complexities of this regulatory system and ultimately identify the regulatory factor(s) involved in the Fe- and DtxR-dependent activation of iutE.
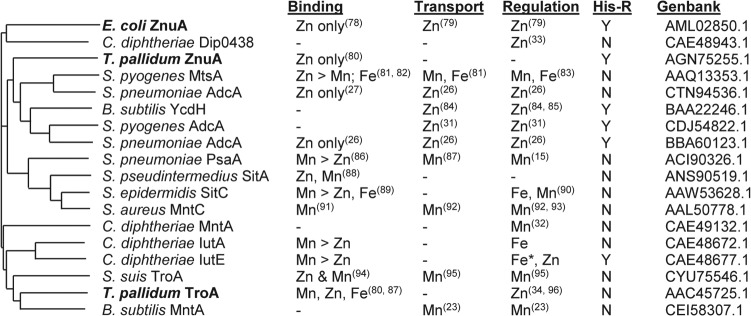
While a function for IutE was not determined, the findings from this study suggest that IutE may have a role in the transport of Zn and/or Mn. The evidence for a role in Zn and/or Mn transport for IutE includes the following: (i) the sequence of IutE shows high homology to cluster 9 family SBPs and the presence of a His-rich region (see Fig. S1 in the supplemental material), which is strongly correlated with cluster 9 SBPs that transport Zn (Fig. 8); (ii) the ITC studies showed that purified IutE binds Mn and Zn with high affinity; and (iii) transcription of iutE is repressed by Zn in a Zur-dependent manner. While these observations are consistent with a role for IutE in Zn and/or Mn transport, we were unable to show a phenotype for the iutABCDE mutant (or the ΔiutE single mutant [M. Schmitt, unpublished observations]) during growth in metal-depleted medium. We also examined the two other C. diphtheriae cluster 9 family ABC transporters (MntABCD and Dip0438 to 0441) in our search for a phenotype, but a triple mutant with all the known cluster 9 family ABC transporters deleted did not exhibit a growth defect in metal-depleted medium relative to the wild-type parent strain. This suggests that other systems are involved in the transport of Zn and Mn. C. diphtheriae strain 1737 does not encode an MntH homolog, which serve as Mn transporters in numerous other bacteria (10, 16–18, 20).
FIG 8.
Dendrogram and properties of related substrate binding proteins. Substrate binding proteins (GenBank accession numbers are indicated) were aligned using ClustalO (77) and the resultant dendrogram drawn using MSwordtree (97). Proteins in bold are derived from Gram-negative organisms. Published metal binding properties with relative binding affinities if known, experimentally established metal transport, regulation in response to the indicated metal ion(s), and the presence (Y) or absence (N) of a His-rich region (His-R) are indicated (15, 23, 26, 27, 31–34, 78–96). —, information not available. *, activation by the indicated metal.
An unusual feature of the IutABCDE system is the opposite effect that Fe exerts on the expression of IutE and IutA. Most surprising is the increased expression of IutE under Fe-replete conditions. Most proteins that exhibit elevated expression in high-Fe environments are Fe-containing enzymes (46, 52, 53), Fe storage proteins (44), or proteins associated with stress responses (45, 50). While IutE is not predicted to have a function that is similar to that of any of these protein families, it is possible that IutE is expressed under high-Fe conditions to counter the deleterious effects of increased intracellular Fe. In aerobic environments, Fe can contribute to the formation of reactive oxygen species (ROS), which cause severe damage to cellular macromolecules (61). Zn can protect sulfhydryl groups from free radicals and inhibits free radical formation by competing with redox-active metals such as Fe (62, 63). The protective function of Mn under oxidative stress conditions is also well established (64), and a recent report showed that the SloC substrate binding protein, an Mn transporter, has a critical role in the oxidative stress tolerance response in Streptococcus mutans (65). In this study, we showed that IutE binds Mn and Zn with high affinity and is a member of the cluster 9 family of proteins, which are involved in Mn and Zn transport. Based on these observations, it is possible that IutE facilitates the transport of Zn and/or Mn into the cell, where these metals serve a protective function against the toxicity caused by high levels of Fe. While IutA has sequence similarity to SBPs involved in Mn and Zn transport, the downregulation of iutABCD transcription by Fe and DtxR suggests a potential role in Fe transport. Numerous bacterial ABC-type Mn transporters are known to also acquire Fe (18), so the uptake of Fe by IutA would not be unusual. The function of the IutBCD ABC transporter is unclear, and future studies will be required to determine which metals are utilized by the IutBCD system and whether IutA, IutE, or both function with this ABC transporter to mobilize metals through the membrane.
MATERIALS AND METHODS
Strains, media, and growth conditions.
C. diphtheriae strains were routinely grown in heart infusion broth with 0.2% (vol/vol) Tween 80 (HIB-TW) or on heart infusion agar (1.5% [wt/vol] agar) at 37°C. E. coli strains were grown in Luria-Bertani (LB) medium or on LB agar (1.5% [wt/vol] agar). Strains were stored at −80°C in their respective culture media with 20% (vol/vol) glycerol. mPGT medium is a semidefined minimal medium that contains 0.5% (wt/vol) Casamino Acids that is pretreated with Chelex to remove divalent cations (66). The mPGT medium used in this study was prepared with an additional Chelex treatment (2 g/liter) for 2 h prior to filter sterilization. mPGT for the ΔripA mutant and for wild-type 1737 or the ΔdtxR mutant grown in parallel was modified with additional Casamino Acids (1.5% [wt/vol]) and no second Chelex treatment. Spectinomycin was used at 100 μg/ml for both C. diphtheriae and Escherichia coli strains. Kanamycin and ampicillin were used at 100 μg/ml and 50 μg/ml, respectively.
Cloning and plasmids.
The primers listed in in Table S1 in the supplemental material were used for the amplification from the C. diphtheriae strain 1737 genomic DNA template. Restriction sites or splice-overlap PCR was used to generate constructs for ligation into indicated vectors. Cloned inserts were confirmed by DNA sequencing (Macrogen). Relevant information for each plasmid is listed in Table 2.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Description or use | Reference or source |
|---|---|---|
| C. diphtheriae strains | ||
| 1737 | Wild type, Gravis biotype, Tox+ | 70 |
| 1737 ΔdtxR | Deletion of dtxR in 1737 | 71 |
| 1737 Δzur | Deletion of zur in 1737 | This study |
| 1737 ΔripA | Deletion of ripA in 1737 | This study |
| 1737 Δiut | Deletion of iut gene cluster in 1737 | This study |
| 1737 Δiut Δmnt | Deletion of iut and mnt in 1737 | This study |
| 1737 Δiut Δmnt Δtro | Deletion of iut, mnt, and tro in 1737 | This study |
| E. coli strains | ||
| DH5α | Cloning and protein expression | 72 |
| BL21(DE3) | Protein expression | 73 |
| S17-1 λpir | Mating strain | 74 |
| Plasmids | ||
| pGP1-2 | Encodes temp-inducible T7 RNA polymerase; Kanr | 75 |
| pMS298 | Encodes C. diphtheriae dtxR under T7 control; Ampr | 42 |
| pET30zur | C. diphtheriae zur cloned into pET30a | This study |
| pET30ripA | C. diphtheriae ripA cloned into pET30a | This study |
| pIutAHis | C. diphtheriae iutA cloned into pET30a | This study |
| pIutAStrepII | Strep-tagged iutA cloned into pET51b | This study |
| pIutEHis | C. diphtheriae iutE cloned into pET30a | This study |
| pIutEStrepII | Strep-tagged iutE cloned into pET51b | This study |
| pKΔiut | Suicide vector for deletion of iut gene cluster (dip0169–dip0173) | This study |
| pKΔripA | Suicide vector for deletion of ripA (dip0922) | This study |
| pKΔtro | Suicide vector for deletion of tro operon (dip0438–dip0441) | This study |
| pKΔmnt | Suicide vector for deletion of mnt operon (dip0615–dip0618) | This study |
| pK19mobsacBΔzur | Suicide vector for deletion of zur (dip1710) | 33 |
| pSPZ | Carries lacZ; Spcr | 76 |
| pSPZiutA | iutA-lacZ promoter fusion | This study |
| pSPZiutE | iutE-lacZ promoter fusion | This study |
| pSPZiutE(FL) | iutE(FL)-lacZ promoter fusion | This study |
| pSPZiutE5′ | iutE5′-lacZ promoter fusion | This study |
Mutation construction in C. diphtheriae.
Mutations in C. diphtheriae were created by allelic exchange using the E. coli strain S17-1 λpir for conjugation (described in reference 67). Deletion of genes was verified by PCR across the gene locus using primers external to the deletion construct. Plasmids pKΔiut, pKΔripA, pKΔtro, and pKΔmnt were used to introduce in-frame deletions of each respective gene. Plasmids pKΔtro and pKΔmnt were constructed by GenScript Biotech.
Generation of antisera against IutA and IutE.
Plasmids pIutAHis and pIutEHis were used for the purification of recombinant, N-terminally His-tagged IutA and IutE. Plasmids were transformed into E. coli BL21(DE3), and protein was purified following the HisTALON protocol (Clontech Laboratories, Inc.). Eluted protein fractions were dialyzed against phosphate-buffered saline (PBS) and used to immunize guinea pigs for antiserum production (Cocalico Biologicals, Inc.).
Subcellular fractionation.
Overnight HIB cultures of wild-type C. diphtheriae were passaged into mPGT supplemented with 1 μM FeCl3 and grown for 6 h. Cultures were then diluted to an optical density at 600 nm (OD600) of approximately 0.03 into mPGT medium supplemented with 1 μM FeCl3 and incubated at 37°C overnight. Bacterial cells were harvested by centrifugation, while the supernatant (supernatant fraction) was collected and centrifuged at 21,000 × g to remove bacteria. The cell pellet was lysed in 10 mM NaH2PO4–5 mM MgSO4 (pH 7) using 0.1-mm glass beads (Fisher Scientific). The crude lysate was centrifuged at 21,000 × g for 10 min at 4°C. The supernatant (clarified lysate) was collected and subjected to centrifugation at 135,000 × g for 90 min at 4°C. The supernatant (cytoplasmic fraction) was collected, and the pellet (membrane fraction) was resuspended in 1× Laemmli buffer. Fractions were separated by SDS-PAGE.
SDS-PAGE and Western blotting.
All protein samples were mixed with Laemmli buffer to a final concentration of 1× and boiled. The sample loading volume was normalized to OD600. Separation of proteins was done by 12% SDS-PAGE. Proteins were detected by Coomassie blue staining or Western blotting. Guinea pig IutA and IutE antisera were used at a dilution of 1:5,000. Goat anti-guinea pig horseradish peroxidase-labeled antibody was used at a dilution of 1:50,000.
Purification and ITC of Strep-tagged IutA and IutE.
Recombinant IutA and IutE proteins were designed to contain a streptavidin (Strep) binding region at their N termini. Plasmids for the induction and purification of Strep-IutA and Strep-IutE were constructed by GenScript Biotech. Regions encoding IutA and IutE that lacked the signal sequences were cloned into pET51b. Plasmids were transformed into BL21(DE3) for protein production. Protein purification was carried out using a gravity flow Strep-Tactin Sepharose column (IBA Lifesciences) following manufacturer guidelines after mechanical lysis with silica beads. Purified Strep-IutA and Strep-IutE were dialyzed against 20 mM HEPES (pH 7.3) containing 20 mM EDTA for 24 h at 4°C to remove metal ions that may be present during protein purification. Subsequently, each protein was dialyzed against 20 mM HEPES buffer (pH 7.3) for 24 h at 4°C and concentrated with 4-ml Amicon tubes (Darmstadt, Germany) to a final concentration of either 30 μM (IutE) or 50 μM (IutA). Ligand solutions were prepared in the buffer resulting from the last dialysis step. Samples were degassed and equilibrated at 25°C for 1 h prior to the titration experiments.
Isothermal titration calorimetry (ITC) experiments were performed at 25°C on a nano ITC instrument (TA Instruments, New Castle, DE). For IutA, 1 mM MnCl2 or ZnSO4 was used as the titrant; for IutE, 0.5 mM each ligand was used. For the respective ligand solutions, 49 μl was injected into the 300-μl instrument sample cell in 1-μl or 1.5-μl injection aliquots with a 4-min delay between injections. The raw ITC data were analyzed with NanoAnalyze V3.6 software (TA instruments, New Castle, DE) by fitting calorimetric data to either single or multiple binding site models with a linear ITC response correction for the buffer.
ELISA.
Wild-type C. diphtheriae 1737 or an isogenic Δiut mutant derivative was grown in mPGT with metal supplementation as indicated; the Δiut mutant was grown in mPGT with 0.5 μM FeCl3 only. Following growth, equivalent numbers of whole cells (as determined by OD600 measurement and normalization) in PBS were adsorbed onto plastic microtiter plates for 18 h. Wells were washed using PBS with 0.05% Tween 20 (PBST) to remove nonadherent cells. Wells were blocked with 5% (wt/vol) blocking buffer in PBST (PBST-BB) for 1 h at 37°C. Wells were washed with PBST before diluted primary antibody (IutA, 1:100; IutE, 1:1,000) in PBST-BB was added; PBST-BB alone was added to wells for treatments with secondary antibody only. The primary antibody was washed away with PBST and incubated with an alkaline phosphatase-linked secondary antibody (1:1,000 in PBST-BB). Plates were washed using PBST, and the substrate p-nitrophenyl phosphate (pNPP) (Sigma) was used for detection at 405 nm following incubation at room temperature. All antibody incubations were performed for 1 h at 37°C.
RNA isolation and qPCR.
Bacteria were grown to mid-logarithmic growth phase in 5 ml of mPGT with metal supplementation, and then 4.5 ml of culture was added to 500 μl 95% (vol/vol) ethanol with 5% (vol/vol) phenol, mixed briefly by vortexing, and centrifuged to pellet bacterial cells. Cell pellets were resuspended in 250 μl of PBS with 9.5% (vol/vol) ethanol, 0.5% (vol/vol) phenol, and 13.4 mM β-mercaptoethanol and subsequently lysed in Lysing Matrix B (MP Biomedicals) for 40 s at 4°C. TRIzol LS reagent (750 μl; Invitrogen) was added to the cell lysate and mixed by brief vortexing. The TRIzol-lysate mixture was centrifuged at 21,000 × g for 5 min at 4°C to remove cell debris. Approximately 700 μl of the TRIzol-lysate mixture was processed using the Zymo DirectZOL purification kit. Eluted RNA was subjected to Ambion Turbo DNase I treatment (Invitrogen) and either stored at −80°C or used immediately for cDNA synthesis using the ProtoScript II first-strand cDNA synthesis kit (New England BioLabs, Inc.). qPCR was performed by adding diluted cDNA (1:200 in double-distilled water [ddH2O]) with the primers to the genes of interest (see Table S2 in the supplemental material) designed using Primer3 (68) and Luna Universal qPCR master mix (New England BioLabs, Inc.). Data were collected with a Roche LightCycler 96 and analyzed using the ΔΔCq method. gyrB was used for normalization across samples.
Beta-galactosidase assays.
C. diphtheriae with the indicated plasmids was passaged into mPGT following overnight growth in HIB-TW. mPGT cultures were then diluted into fresh mPGT with metal supplementation as indicated. Cells were pelleted and treated with 10 mg/ml lysozyme in PBS at 37°C for 30 m. Following treatment, beta-galactosidase activity assays were performed as described by Miller (69).
5′ RACE.
Following the manufacturer's guidelines for the 5′ RACE System for Rapid Amplification of cDNA Ends, version 2.0 (Invitrogen), total RNA isolated from C. diphtheriae grown in mPGT with either 0.5 μM or 10 μM FeCl3 was subjected to reverse transcription using an iutA- or iutE-specific primer (see Table S3 in the supplemental material, GSP1), respectively. Poly(C) tails were added to each reverse-transcribed product and used to prime PCR with a second gene-specific primer. PCR products were purified using the GeneClean kit (MP Biomedicals) and submitted for sequencing (Macrogen). Validation of the iutE transcriptional start site was performed through PCR of cDNA using primers indicated in Table S3.
Purification of DtxR.
DtxR was purified as described in reference 42 with modifications. Briefly, a culture of E. coli DH5α carrying plasmids pGP1-2 and pMS298 was grown to mid-logarithmic growth phase in LB (with ampicillin and kanamycin) and induced by heat shock at 42°C for 30 m followed by incubation at 37°C for 3 h. Cells were pelleted by centrifugation and lysed by sonication in DtxR lysis buffer (42 mM NaH2PO4, 58 mM Na2HPO4, 50 mM NaCl, 5 mM MgCl2). Cell debris was removed by centrifugation, and the clarified cell lysate was incubated with Ni-nitrilotriacetic acid (Ni-NTA) (Qiagen) at 4°C with rotation for 3 h. The Ni-NTA resin was washed repeatedly with lysis buffer, and DtxR was eluted using elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). Elution fractions were dialyzed against PBS overnight and subsequently dialyzed against PBS with 15% glycerol. Purified proteins were aliquoted and stored at −80°C. A His tag on DtxR was not required, since DtxR's innate Fe binding properties allow the protein to bind to the Ni-NTA resin.
Purification of His-Zur.
A single colony of BL21(DE3) carrying pET30zur was used to inoculate an overnight culture of Overnight Express TB medium (Millipore). Bacterial cells were harvested by centrifugation, lysed in HisTALON xTractor buffer (Clontech Laboratories, Inc.), and processed following the HisTALON gravity column purification protocol. Elution fractions with His-Zur were dialyzed against PBS overnight and subsequently dialyzed against PBS with 15% glycerol. Purified proteins were aliquoted and stored at −80°C.
Purification of His-RipA.
An overnight culture of BL21(DE3) carrying pET30ripA was diluted into fresh LB with kanamycin. At mid-logarithmic phase, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.2 mM. After incubation at 37°C for 5 h, cells were pelleted by centrifugation and cell pellets frozen at −20°C. Pellets were thawed and resuspended in DtxR lysis buffer. Cells were lysed by sonication and the lysate clarified by centrifugation. Clarified lysate was incubated with Ni-NTA (Qiagen). The suspension was transferred to a gravity flow column and the resin washed repeatedly with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0). RipA was eluted using the DtxR elution buffer. Elution fractions were dialyzed against PBS overnight and subsequently dialyzed against PBS with 15% (vol/vol) glycerol. Elution fractions were combined and concentrated using Amicon Ultra 0.5-ml centrifugal filters (Millipore).
EMSA.
For all electrophoretic mobility shift assays (EMSAs) used here, biotinylated DNA probes were generated through PCR amplification with biotinylated primers (Integrated DNA Technologies). Primers used for the EMSAs are described in Table S4 in the supplemental material. Biotinylated DNA was detected using the LightShift chemiluminescent EMSA kit (Thermo Scientific).
For Zur binding assays, purified Zur was incubated at room temperature with biotinylated DNA in 1× binding buffer (Thermo Scientific) (10 mM Tris, 50 mM KCl, 1 mM dithiothreitol [DTT], pH 7.5) supplemented with 0.1 M KCl, 0.25 mM ZnCl2, 0.05 μg/μl poly(dI · dC), 0.05% NP-40, 0.1 μg/μl bovine serum albumin (BSA), and 2.5% (vol/vol) glycerol. A reaction in which Zur was omitted was run in parallel. Loading buffer was added directly to samples following incubation, and the entire reaction product was separated by gel electrophoresis (6% acrylamide with 45 mM Tris-borate [0.5× TB]) and transferred onto nylon in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA).
For DtxR binding assays, purified DtxR was incubated at room temperature with biotinylated DNA in DtxR binding buffer [20 mM Na2HPO4, 50 mM NaCl, 2 mM DTT, 5 mM MgCl2, 0.2 μg/μl BSA, 0.05 μg/μl poly(dI · dC), and 0.5 mM FeSO4 in 10% glycerol at pH 7.0]. A reaction in which DtxR was omitted was run in parallel. Samples were separated by gel electrophoresis (5% acrylamide with 50 mM Na2HPO4 and 1 mM DTT, pH 7.0) and transferred onto a nylon membrane in 0.5× TBE.
For RipA binding assays, gel shifts were performed as described in reference 46 with modifications. Purified RipA was incubated at room temperature with biotinylated DNA in 20 mM Tris-HCl (pH 7.4), 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, 5 mM MgCl2, 2.5 mM CaCl2, 0.2 μg/μl BSA, 0.05 μg/μl sonicated salmon sperm DNA, 0.5% NP-40, and 5% glycerol for 15 min. A reaction in which RipA was omitted was run in parallel. Reaction products were separated by gel electrophoresis (6% acrylamide with 0.5× TBE) and transferred onto nylon in 0.5× TBE.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the intramural research program at the Center for Biologics Evaluation and Research, Food and Drug Administration.
We thank Scott Stibitz and Paul Carlson for helpful comments on the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00051-18.
REFERENCES
- 1.Pappenheimer AM., Jr 1977. Diphtheria toxin. Annu Rev Biochem 46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 2.May ML, McDougall RJ, Robson JM. 2014. Corynebacterium diphtheriae and the returned tropical traveler. J Travel Med 21:39–44. doi: 10.1111/jtm.12074. [DOI] [PubMed] [Google Scholar]
- 3.Farfour E, Badell E, Zasada A, Hotzel H, Tomaso H, Guillot S, Guiso N. 2012. Characterization and comparison of invasive Corynebacterium diphtheriae isolates from France and Poland. J Clin Microbiol 50:173–175. doi: 10.1128/JCM.05811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fourel G, Phalipon A, Kaczorek M. 1989. Evidence for direct regulation of diphtheria toxin gene transcription by an Fe2+-dependent DNA-binding repressor, DtoxR, in Corynebacterium diphtheriae. Infect Immun 57:3221–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt MP, Twiddy EM, Holmes RK. 1992. Purification and characterization of the diphtheria toxin repressor. Proc Natl Acad Sci U S A 89:7576–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd J, Oza MN, Murphy JR. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci U S A 87:5968–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg ED. 1975. Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA 231:39–41. [DOI] [PubMed] [Google Scholar]
- 8.Palmer LD, Skaar EP. 2016. Transition metals and virulence in bacteria. Annu Rev Genet 50:67–91. doi: 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zackular JP, Chazin WJ, Skaar EP. 2015. Nutritional immunity: S100 proteins at the host-pathogen interface. J Biol Chem 290:18991–18998. doi: 10.1074/jbc.R115.645085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juttukonda LJ, Skaar EP. 2015. Manganese homeostasis and utilization in pathogenic bacteria. Mol Microbiol 97:216–228. doi: 10.1111/mmi.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nies DH, Grass G. 1 October 2009, posting date Transition metal homeostasis. EcoSal Plus 2009. doi: 10.1128/ecosalplus.5.4.4.3. [DOI] [PubMed] [Google Scholar]
- 12.Hopkin KA, Papazian MA, Steinman HM. 1992. Functional differences between manganese and iron superoxide dismutases in Escherichia coli K-12. J Biol Chem 267:24253–24258. [PubMed] [Google Scholar]
- 13.Jakubovics NS, Smith AW, Jenkinson HF. 2002. Oxidative stress tolerance is manganese (Mn2+) regulated in Streptococcus gordonii. Microbiology 148:3255–3263. doi: 10.1099/00221287-148-10-3255. [DOI] [PubMed] [Google Scholar]
- 14.Auling G, Thaler M, Diekmann H. 1980. Parameters of unbalanced growth and reversible inhibition of deoxyribnucleic acid synthesis in Brevibacterium ammoniagenes ATCC 6872 induced by depletion of Mn2+. Inhibitor studies on the reversibility of deoxyribonucleic acid synthesis. Arch Microbiol 127:105–114. [DOI] [PubMed] [Google Scholar]
- 15.Ogunniyi AD, Mahdi LK, Jennings MP, McEwan AG, McDevitt CA, Van der Hoek MB, Bagley CJ, Hoffmann P, Gould KA, Paton JC. 2010. Central role of manganese in regulation of stress responses, physiology, and metabolism in Streptococcus pneumoniae. J Bacteriol 192:4489–4497. doi: 10.1128/JB.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. 1995. NRAMP defines a family of membrane proteins. Proc Natl Acad Sci U S A 92:10089–10093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makui H, Roig E, Cole ST, Helmann JD, Gros P, Cellier MF. 2000. Identification of the Escherichia coli K-12 NRAMP orthologue (MntH) as a selective divalent metal ion transporter. Mol Microbiol 35:1065–1078. doi: 10.1046/j.1365-2958.2000.01774.x. [DOI] [PubMed] [Google Scholar]
- 18.Papp-Wallace KM, Maguire ME. 2006. Manganese transport and the role of manganese in virulence. Annu Rev Microbiol 60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- 19.Claverys JP. 2001. A new family of high-affinity ABC manganese and zinc permeases. Res Microbiol 152:231–243. doi: 10.1016/S0923-2508(01)01195-0. [DOI] [PubMed] [Google Scholar]
- 20.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP. 2013. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun 81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 22.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ. 2013. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A 110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Que Q, Helmann JD. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 24.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charriere GM, Waddell SJ, Foti M, Lugo-Villarino G, Gao Q, Maridonneau-Parini I, Butcher PD, Castagnoli PR, Gicquel B, de Chastellier C, Neyrolles O. 2011. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10:248–259. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plumptre CD, Eijkelkamp BA, Morey JR, Behr F, Counago RM, Ogunniyi AD, Kobe B, O'Mara ML, Paton JC, McDevitt CA. 2014. AdcA and AdcAII employ distinct zinc acquisition mechanisms and contribute additively to zinc homeostasis in Streptococcus pneumoniae. Mol Microbiol 91:834–851. doi: 10.1111/mmi.12504. [DOI] [PubMed] [Google Scholar]
- 27.Loisel E, Jacquamet L, Serre L, Bauvois C, Ferrer JL, Vernet T, Di Guilmi AM, Durmort C. 2008. AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: biochemical and structural Analysis. J Mol Biol 381:594–606. doi: 10.1016/j.jmb.2008.05.068. [DOI] [PubMed] [Google Scholar]
- 28.Choi SH, Lee KL, Shin JH, Cho YB, Cha SS, Roe JH. 2017. Zinc-dependent regulation of zinc import and export genes by Zur. Nat Commun 8:15812. doi: 10.1038/ncomms15812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hantke K. 2005. Bacterial zinc uptake and regulators. Curr Opin Microbiol 8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Reyes-Caballero H, Guerra AJ, Jacobsen FE, Kazmierczak KM, Cowart D, Koppolu UM, Scott RA, Winkler ME, Giedroc DP. 2010. The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J Mol Biol 403:197–216. doi: 10.1016/j.jmb.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makthal N, Nguyen K, Do H, Gavagan M, Chandrangsu P, Helmann JD, Olsen RJ, Kumaraswami M. 2017. A critical role of zinc importer AdcABC in group A Streptococcus-host interactions during infection and its implications for vaccine development. EBioMedicine 21:131–141. doi: 10.1016/j.ebiom.2017.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt MP. 2002. Analysis of a DtxR-like metalloregulatory protein, MntR, from Corynebacterium diphtheriae that controls expression of an ABC metal transporter by an Mn2+-dependent mechanism. J Bacteriol 184:6882–6892. doi: 10.1128/JB.184.24.6882-6892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith KF, Bibb LA, Schmitt MP, Oram DM. 2009. Regulation and activity of a zinc uptake regulator, Zur, in Corynebacterium diphtheriae. J Bacteriol 191:1595–1603. doi: 10.1128/JB.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardham JM, Stamm LV, Porcella SF, Frye JG, Barnes NY, Howell JK, Mueller SL, Radolf JD, Weinstock GM, Norris SJ. 1997. Identification and transcriptional analysis of a Treponema pallidum operon encoding a putative ABC transport system, an iron-activated repressor protein homolog, and a glycolytic pathway enzyme homolog. Gene 197:47–64. doi: 10.1016/S0378-1119(97)00234-5. [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee S, Wei B, Bhattacharyya-Pakrasi M, Pakrasi HB, Smith TJ. 2003. Structural determinants of metal specificity in the zinc transport protein ZnuA from Synechocystis 6803. J Mol Biol 333:1061–1069. doi: 10.1016/j.jmb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Anderson AS, Scully IL, Timofeyeva Y, Murphy E, McNeil LK, Mininni T, Nunez L, Carriere M, Singer C, Dilts DA, Jansen KU. 2012. Staphylococcus aureus manganese transport protein C is a highly conserved cell surface protein that elicits protective immunity against S. aureus and Staphylococcus epidermidis. J Infect Dis 205:1688–1696. doi: 10.1093/infdis/jis272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kallio A, Sepponen K, Hermand P, Denoel P, Godfroid F, Melin M. 2014. Role of Pht proteins in attachment of Streptococcus pneumoniae to respiratory epithelial cells. Infect Immun 82:1683–1691. doi: 10.1128/IAI.00699-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen CE, Schmitt MP. 2009. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J Bacteriol 191:2638–2648. doi: 10.1128/JB.01784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JH, Wang T, Ault K, Liu J, Schmitt MP, Holmes RK. 1997. Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect Immun 65:4273–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schroder J, Jochmann N, Rodionov DA, Tauch A. 2010. The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genomics 11:12. doi: 10.1186/1471-2164-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt MP, Holmes RK. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect Immun 59:1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bibb LA, Schmitt MP. 2010. The ABC transporter HrtAB confers resistance to hemin toxicity and is regulated in a hemin-dependent manner by the ChrAS two-component system in Corynebacterium diphtheriae. J Bacteriol 192:4606–4617. doi: 10.1128/JB.00525-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurthkoti K, Tare P, Paitchowdhury R, Gowthami VN, Garcia MJ, Colangeli R, Chatterji D, Nagaraja V, Rodriguez GM. 2015. The mycobacterial iron-dependent regulator IdeR induces ferritin (bfrB) by alleviating Lsr2 repression. Mol Microbiol 98:864–877. doi: 10.1111/mmi.13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oglesby-Sherrouse AG, Murphy ER. 2013. Iron-responsive bacterial small RNAs: variations on a theme. Metallomics 5:276–286. doi: 10.1039/c3mt20224k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wennerhold J, Krug A, Bott M. 2005. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J Biol Chem 280:40500–40508. doi: 10.1074/jbc.M508693200. [DOI] [PubMed] [Google Scholar]
- 47.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray TA. 2015. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet 11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albersmeier A, Pfeifer-Sancar K, Ruckert C, Kalinowski J. 2017. Genome-wide determination of transcription start sites reveals new insights into promoter structures in the actinomycete Corynebacterium glutamicum. J Biotechnol 257:99–109. doi: 10.1016/j.jbiotec.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 49.Brune I, Werner H, Huser AT, Kalinowski J, Puhler A, Tauch A. 2006. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics 7:21. doi: 10.1186/1471-2164-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wennerhold J, Bott M. 2006. The DtxR regulon of Corynebacterium glutamicum. J Bacteriol 188:2907–2918. doi: 10.1128/JB.188.8.2907-2918.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emer D, Krug A, Eikmanns BJ, Bott M. 2009. Complex expression control of the Corynebacterium glutamicum aconitase gene: identification of RamA as a third transcriptional regulator besides AcnR and RipA. J Biotechnol 140:92–98. doi: 10.1016/j.jbiotec.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 52.Bott M, Niebisch A. 2003. The respiratory chain of Corynebacterium glutamicum. J Biotechnol 104:129–153. doi: 10.1016/S0168-1656(03)00144-5. [DOI] [PubMed] [Google Scholar]
- 53.Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song K-B, Smaldone GT, Helmann JD. 2008. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A 105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torres VJ, Attia AS, Mason WJ, Hood MI, Corbin BD, Beasley FC, Anderson KL, Stauff DL, McDonald WH, Zimmerman LJ, Friedman DB, Heinrichs DE, Dunman PM, Skaar EP. 2010. Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun 78:1618–1628. doi: 10.1128/IAI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunkle CA, Schmitt MP. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J Bacteriol 185:6826–6840. doi: 10.1128/JB.185.23.6826-6840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitt MP, Holmes RK. 1994. Cloning, sequence, and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. J Bacteriol 176:1141–1149. doi: 10.1128/jb.176.4.1141-1149.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol 188:3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kunkle CA, Schmitt MP. 2005. Analysis of a DtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J Bacteriol 187:422–433. doi: 10.1128/JB.187.2.422-433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mentz A, Neshat A, Pfeifer-Sancar K, Puhler A, Ruckert C, Kalinowski J. 2013. Comprehensive discovery and characterization of small RNAs in Corynebacterium glutamicum ATCC 13032. BMC Genomics 14:714. doi: 10.1186/1471-2164-14-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8. [PubMed] [Google Scholar]
- 62.Gaballa A, Helmann JD. 2002. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol Microbiol 45:997–1005. doi: 10.1046/j.1365-2958.2002.03068.x. [DOI] [PubMed] [Google Scholar]
- 63.Bray TM, Bettger WJ. 1990. The physiological role of zinc as an antioxidant. Free Radic Biol Med 8:281–291. doi: 10.1016/0891-5849(90)90076-U. [DOI] [PubMed] [Google Scholar]
- 64.Eijkelkamp BA, McDevitt CA, Kitten T. 2015. Manganese uptake and streptococcal virulence. Biometals 28:491–508. doi: 10.1007/s10534-015-9826-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crepps SC, Fields EE, Galan D, Corbett JP, Von Hasseln ER, Spatafora GA. 2016. The SloR metalloregulator is involved in the Streptococcus mutans oxidative stress response. Mol Oral Microbiol 31:526–539. doi: 10.1111/omi.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tai SP, Krafft AE, Nootheti P, Holmes RK. 1990. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb Pathog 9:267–273. doi: 10.1016/0882-4010(90)90015-I. [DOI] [PubMed] [Google Scholar]
- 67.Ton-That H, Schneewind O. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol 50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 68.Koressaar T, Remm M. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 69.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 70.Popovic T, Kombarova SY, Reeves MW, Nakao H, Mazurova IK, Wharton M, Wachsmuth IK, Wenger JD. 1996. Molecular epidemiology of diphtheria in Russia, 1985-1994. J Infect Dis 174:1064–1072. doi: 10.1093/infdis/174.5.1064. [DOI] [PubMed] [Google Scholar]
- 71.Lyman LR, Peng ED, Schmitt MP. 8 January 2018. The Corynebacterium diphtheriae iron-regulated surface protein HbpA is involved in the utilization of the hemoglobin-haptoglobin complex as an iron source. J Bacteriol doi: 10.1128/jb.00676-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 73.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 74.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 75.Tabor S, Richardson CC. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A 82:1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oram DM, Jacobson AD, Holmes RK. 2006. Transcription of the contiguous sigB, dtxR, and galE genes in Corynebacterium diphtheriae: evidence for multiple transcripts and regulation by environmental factors. J Bacteriol 188:2959–2973. doi: 10.1128/JB.188.8.2959-2973.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yatsunyk LA, Easton JA, Kim LR, Sugarbaker SA, Bennett B, Breece RM, Vorontsov II, Tierney DL, Crowder MW, Rosenzweig AC. 2008. Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. J Biol Inorg Chem 13:271–288. doi: 10.1007/s00775-007-0320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol 28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 80.Desrosiers DC, Sun YC, Zaidi AA, Eggers CH, Cox DL, Radolf JD. 2007. The general transition metal (Tro) and Zn2+ (Znu) transporters in Treponema pallidum: analysis of metal specificities and expression profiles. Mol Microbiol 65:137–152. doi: 10.1111/j.1365-2958.2007.05771.x. [DOI] [PubMed] [Google Scholar]
- 81.Janulczyk R, Pallon J, Bjorck L. 1999. Identification and characterization of a Streptococcus pyogenes ABC transporter with multiple specificity for metal cations. Mol Microbiol 34:596–606. doi: 10.1046/j.1365-2958.1999.01626.x. [DOI] [PubMed] [Google Scholar]
- 82.Janulczyk R, Ricci S, Bjorck L. 2003. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect Immun 71:2656–2664. doi: 10.1128/IAI.71.5.2656-2664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hanks TS, Liu M, McClure MJ, Fukumura M, Duffy A, Lei B. 2006. Differential regulation of iron- and manganese-specific MtsABC and heme-specific HtsABC transporters by the metalloregulator MtsR of group A Streptococcus. Infect Immun 74:5132–5139. doi: 10.1128/IAI.00176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gaballa A, Wang T, Ye RW, Helmann JD. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J Bacteriol 184:6508–6514. doi: 10.1128/JB.184.23.6508-6514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gaballa A, Helmann JD. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol 180:5815–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. 2011. A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog 7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dintilhac A, Alloing G, Granadel C, Claverys JP. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol 25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 88.Abate F, Malito E, Cozzi R, Lo Surdo P, Maione D, Bottomley Matthew J. 2014. Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius. Biosci Rep 34:e00154. doi: 10.1042/BSR20140088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ridley K. 2004. Functional characterisation of ABC-type metal ion uptake systems in the staphylococci. Ph.D. thesis University of Nottingham, Nottingham, United Kingdom. [Google Scholar]
- 90.Cockayne A, Hill PJ, Powell NB, Bishop K, Sims C, Williams P. 1998. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect Immun 66:3767–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gribenko A, Mosyak L, Ghosh S, Parris K, Svenson K, Moran J, Chu L, Li S, Liu T, Woods VL Jr, Jansen KU, Green BA, Anderson AS, Matsuka YV. 2013. Three-dimensional structure and biophysical characterization of Staphylococcus aureus cell surface antigen-manganese transporter MntC. J Mol Biol 425:3429–3445. doi: 10.1016/j.jmb.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 92.Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Peacock S, Foster SJ. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol 44:1269–1286. doi: 10.1046/j.1365-2958.2002.02944.x. [DOI] [PubMed] [Google Scholar]
- 93.Handke LD, Hawkins JC, Miller AA, Jansen KU, Anderson AS. 2013. Regulation of Staphylococcus aureus MntC expression and its role in response to oxidative stress. PLoS One 8:e77874. doi: 10.1371/journal.pone.0077874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng B, Zhang Q, Gao J, Han H, Li M, Zhang J, Qi J, Yan J, Gao GF. 2011. Insight into the interaction of metal ions with TroA from Streptococcus suis. PLoS One 6:e19510. doi: 10.1371/journal.pone.0019510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wichgers Schreur PJ, Rebel JM, Smits MA, van Putten JP, Smith HE. 2011. TroA of Streptococcus suis is required for manganese acquisition and full virulence. J Bacteriol 193:5073–5080. doi: 10.1128/JB.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hazlett KR, Rusnak F, Kehres DG, Bearden SW, La Vake CJ, La Vake ME, Maguire ME, Perry RD, Radolf JD. 2003. The Treponema pallidum tro operon encodes a multiple metal transporter, a zinc-dependent transcriptional repressor, and a semi-autonomously expressed phosphoglycerate mutase. J Biol Chem 278:20687–20694. doi: 10.1074/jbc.M300781200. [DOI] [PubMed] [Google Scholar]
- 97.Savva G, Conn J, Dicks J. 2004. Drawing phylogenetic trees in LATEX and Microsoft Word. Bioinformatics 20:2322–2323. doi: 10.1093/bioinformatics/bth228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.