Abstract
The transcript levels of heavy-chain zein genes (zH1 and zH2) and the occurrence of the zH polypeptides in different opaque-2 (o2) lines were investigated by RNA-blot analyses and by sodium dodecylsulfate-polyacrylamide gel electrophoresis or two-dimensional gel electrophoresis protein fractionations. Four mutant alleles o2R, o2T, o2It, and o2-676 introgressed into different genetic backgrounds (GBs) were considered. The mono-dimensional gel electrophoresis zein pattern can be either conserved or different among the various GBs carrying the same o2 allele. Likewise, in the identical GB carrying different o2 alleles, the zein pattern can be either conserved or differentially affected by the different mutant allele. Zein protein analysis of reciprocal crosses between lines with different o2 alleles or the same o2 showed in some case a more than additive zH pattern in respect to the o2 parent lines. Electrophoretic mobility shift assay approaches, with O2-binding oligonucleotide and endosperm extracts from the above o2 lines, failed to reveal o2-specific retarded band in any of the o2 extracts. The results suggest that the promoter of some zH1 and zH2 contains motif(s) that can respond to factors other than O2.
Since the discovery of the first mutation at the Opaque-2 locus of maize (Zea mays; Emerson et al., 1935) several other o2 alleles have been recovered by genetic (Schmidt et al., 1987; Motto et al., 1988; Aukerman and Schmidt, 1993) or chemical mutagenesis (Aukerman et al., 1991) and by screening analysis of genetic stocks within the collections of different laboratories (Bernard et al., 1994). Apart from those recently induced by insertion elements (Schmidt et al., 1987; Motto et al., 1988; Aukerman and Schmidt, 1993), the remaining o2 alleles fall into two categories, the first producing mutated o2 polypeptides such as the o2T, o2It, and o2-676 alleles, and the second defective in transcription, known as o2 null-transcript allele and named o2R (Schmidt et al., 1987; Bernard et al., 1994; Lazzari et al., 1994, 1995; Hoschek et al., 1996). In the screening analysis, o2 alleles were often recovered in different genetic backgrounds (GBs) and found in different collections (Schmidt et al., 1987; Bernard et al., 1994). In a similar manner, on the basis of their differences in restriction fragment-length polymorphism (RFLP) and amino acid sequence, at least three different wild-type alleles were recovered in the maize lines investigated so far (Schmidt et al., 1987; Varagona et al., 1991; Bernard et al., 1994).
In lines homozygous for any of the o2s the seed opacity is correlated with a reduced zein polypeptide content in respect to the O2 lines (Schmidt, 1993). Moreover, in crosses among different o2 alleles, none of them has been shown to complement each other in recovering the translucent and vitreous phenotype that is conferred to the seed by the O2 alleles. The zein multigene family of maize is a set of genes whose expression is tissue-specific and developmentally and spatially regulated in the endosperm by means of a still undefined set of regulatory loci (Dolfini et al., 1992; Schmidt, 1993) with the exception of the well-characterized Opaque-2 locus (Schmidt, 1993). This locus contains a single copy of the O2 gene that encodes a bZIP transcriptional activator responsible for the transcription of one of the zein gene subsets (Dolfini et al., 1992; Schmidt, 1993). This subset, identified by the M1 family of sequences, actually codes for mature zein polypeptides of 246 to 241 amino acids and explains the size variability within the heavy zein subclass that is subdivided into zH1 and zH2, respectively (Viotti et al., 1985). In vitro experiments showed that Opaque-2 binds as homodimer to the motif 5′-TCCACGTAGA-3′ (O2 box) that occurs once in the promoter of the heavy chain zein genes about −300 bp from the ATG start codon (Schmidt et al., 1990) and data from in vivo experiments support this binding specificity (Unger et al., 1993). However, evidences for promiscuity in DNA-motif binding for this type of bZIP factor (Izawa et al., 1993) have been reported (Yunes et al., 1994, 1998). Furthermore, in vitro experiments demonstrate that O2 can form heterodimers with another O2-like bZIP polypeptide that specifically binds to the O2 box as homodimer or heterodimer with O2 (Pysh et al., 1993). However, the in vivo occurrence of this or other heterodimers has never been demonstrated. So far most of the o2 alleles have been shown to severely reduce the expression of the zH1 and zH2 subset of genes that code for most of the zH1 and zH2 subclasses of polypeptides (Viotti et al., 1985). However, exceptions to this observation have been reported (Soave et al., 1981b; Viotti et al., 1982; Higgins, 1984; Dolfini et al., 1992; Schmidt, 1993) and include partial effects on the other zein subclasses (zL1, zL2, and zL3).
In this paper we demonstrate that in different inbreds having the same o2 allele, the endosperm may either be almost null for the zH1 and zH2 or contain one or both the two zH classes in variable relative amounts. In most cases this correlates with the levels of the heavy type zein transcripts suggesting that some zH genes may be regulated by factors other than O2. Moreover, in some crosses between GBs containing different o2 alleles or the same o2 allele, patterns of zein expression are obtained that are distinct from expectations based on the patterns observed among the parent lines. This corroborates the above suggestion and further indicates that in some of the o2 lines, other regulatory protein(s) may overcome the loss of the O2 zein-transactivation function.
RESULTS
Wild-Type and o2 Mutant Lines Have Specific Zein Polypeptide Patterns
Alcoholic extracts from endosperms of mature or immature maize seeds contain two major protein fractions: the sulfur-rich glutelins known as β-, γ-, and δ-zeins, and the zein polypeptides sensu stricto, known as α-zein (Shewry and Tatham, 1990; Shewry et al., 1995). The latter, by size (SDS-PAGE) or charge fractionation (isoelectric focusing [IEF]) shows several bands that are resolved into several individual polypeptides by combining the two electrophoretic techniques. The patterns are line-specific and typical examples were already reported (Viotti et al., 1985; Lund et al., 1995).
These fractionation analyses were applied to total zein extracts of the genotypes listed in Table I. In most wild-type lines, α-zeins can be resolved by SDS-PAGE (Fig. 1) into at least five discrete bands: two of heavy type (H1 and H2) and three of light type (L1, L2, and L3). Inbreds W22 and Mo17, in which the two heavy classes are poorly resolved, represent exceptions. These five size classes show in some cases a different relative abundance among wild types that, however, remains constant for a given genotype during the various developmental stages of endosperm growth (see NYR+ in Fig. 1B; data for other genotypes not shown). For convenience, the various GBs with the different O2 or o2 alleles were abbreviated as reported in Table I.
Table I.
List of the wild-type and o2 mutant alleles present in the various genetic backgrounds
| Genetic Background | Wild-Type and o2 Allelesa | Abbreviationsb | Presence of o2 Transcriptc | o2 Cellular Locationd | Stock/Referencee |
|---|---|---|---|---|---|
| W64A | O2w1 | W+ | 1fg | ||
| o2T | Wo2T | Yes | Cytoplasm | 1,3fgh | |
| A69Y | O2w3 | Y+ | 1fg | ||
| o2It | Yo2It | Yes | Nuclear | 1fgi | |
| o2T | Yo2T | Yes | Cytoplasm | 6fgh | |
| o2-676 | Y676 | Yes | Nuclear | 6jk | |
| o2R | Yo2R | No | 6gh | ||
| W22 | O2w2 | X+ | 1,4fgl | ||
| o2It | Xo2It | Yes | Nuclear | 1fgi | |
| o2-676 | X676 | Yes | Nuclear | 4jk | |
| o2R | Xo2R | No | 4gh | ||
| Bianchi | o2It | Bo2 | Yes | Nuclear | 2g |
| NYR | O2w3 | N+ | |||
| o2It | No2It | Yes | Nuclear | 2g 2fg | |
| 3316 | O2w3 | 33+ | 2g | ||
| o2It | 33o2It | Yes | Nuclear | 2gi | |
| Oh43 | O2w1 | Oh+ | 4,5g | ||
| o2-676 | Oh676 | Yes | Nuclear | 4jk | |
| o2R | Oho2R | No | 4,5gh | ||
| B37 | O2w1 | 37+ | 3g | ||
| o2R | 37o2R | No | 3 | ||
| B14 | O2w3 | 14+ | 3g | ||
| o2R | 14o2R | No | 3 | ||
| Rossman | o2R | Ro2R | No | 2fg | |
| R802 | ndm | 802+ | No | 4j | |
| o2R | 802o2R | 4j | |||
| Mo17 | O2w3 | Mo+ | 3g | ||
| o2R | Moo2 | No | 3g |
The wild-type and mutant alleles occurring in the various GBs are reported.
For convenience each line was abbreviated as reported.
These columns report, respectively, the occurrence of o2 transcripts and the relative o2 cellular location deduced from the references reported in the last column and from unpublished data (B. Lazzari).
Report of the nos. of the laboratories and centers from which the various lines have been obtained as listed in the “Materials and Methods” of Bernard et al. (1994). No. 6 identifies the Istituto Biosintesi Vegetali in which the introgression into the A69Y of the o2T, from W64Ao2T, and the o2-676 and o2R alleles from the Oh43 GB was obtained by six backcrosses.
nd, Not determined.
Figure 1.
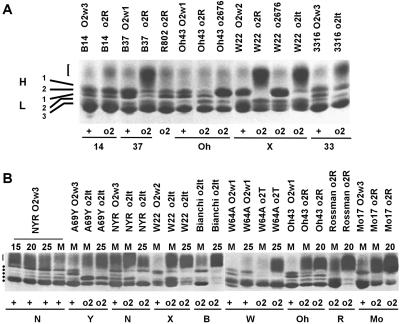
SDS-PAGE analysis of zein polypeptides from wild-type and o2 inbreds. Approximately 15 μg of zein protein extracts was loaded in each lane of A and B. Genotypes are reported at the top (see also Table I). For a proper comparison, wild-type (+) and opaque-2 (o2) in a given GB, abbreviated as in Table I, are also indicated at the bottom of each panel. A, Electrophoretic mobility of zein polypeptides extracted from mature seeds. The relative mobility of the H1 and H2 heavy chains and the L1, L2, and L3 light chains are indicated at the left. Bracket indicates the mobility of the sulfur-rich γ-zein polypeptides. The R802 O2w1 line was not inserted in this analysis; it shows, however, a pattern identical to the one of the Oh43 O2w1. B, Electrophoretic mobility of zeins extracted from seeds at maturity (M) or at different DAP as indicated at the top of each lane.
In a first global comparison among the O2 and o2 lines, the following can be deduced. The pattern of each o2 genotype is different in respect to the one of the corresponding GB with an O2 allele and all together show a wide spectrum of combinations while considering the relative abundance of the H1 and H2 bands. In some cases, as in Xo2R and Ro2R, the H1 and the H2 are almost undetectable; in Wo2T, Moo2R, and Yo2It they are severely reduced; in No2It, Xo2It, and 33o2It the H2 is absent, whereas the H1 is only partially affected in the first two lines and almost unaffected in the third.
As in O2 lines, in the o2 genotypes the pattern that occurs at maturity is almost identical to the one present in immature seeds at 20 or 25 days after pollination (DAP; Fig. 1B). In any case the zein pattern observed for each line is constant in seeds obtained from field- or greenhouse-grown plants harvested during different years (data not shown). This suggests that each pattern is GB-specific and is not affected by normal environmental growth conditions.
In the case of Bo2It, the mutant line from which the o2-Italian allele has been recovered and first described in the sixties (Nelson, 1967), a proper comparison is not possible because its O2 isogenic line is not available in any collection. The two GBs, W22 and Oh43, carrying the o2-676 allele (Aukerman et al., 1991) show a pattern almost devoid of the H2 band.
In a more detailed analysis we compared seven GBs in which the o2R allele was introgressed. In the first four lines, 14o2R, 37o2R, 802o2R, and Oho2R, both heavy classes are present and only partially reduced in respect to the light classes when compared with the corresponding GB with an O2 allele; whereas, in the other three lines, Xo2R, Ro2R, and Moo2R, a severe reduction or absence is observed.
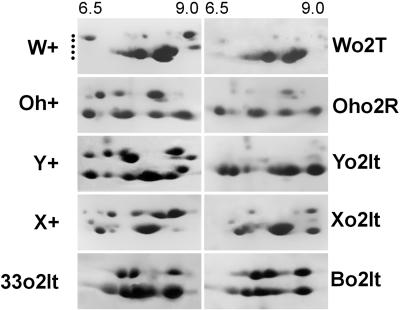
Two-dimensional analysis of a selected set of o2 lines reveals that each spot contributing to the relative abundance of the H1 or H2 class is either absent, partially expressed, or almost unaffected when compared with the corresponding wild-type genotype (Fig. 2). This corroborates the above observations and strongly suggests that some zein genes of heavy type are expressed both in the absence of the O2 protein (e.g. Oho2R) or in the presence of defective o2 polypeptides, as in the o2T, o2It,or o2-676 lines (see also Fig. 5).
Figure 2.
Two-dimensional (2D) gel analysis of zein protein extracts. Approximately 100 μg of zein protein extracts was loaded for each genotype. In the first dimension (IEF), the equilibrium pH gradient from left to right was between 6.5 and 9.0 as reported at the top and this applies for all the panels. Genotypes, abbreviated as reported in Table I, are indicated at the left or at the right of each panel. Dots on the left identify the five major size classes of zeins.
Figure 5.
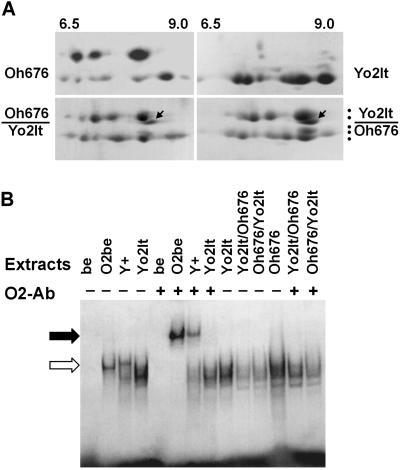
A, 2D analysis of zein extracts from the Yo2It and Oh676 lines and from their reciprocal crosses as indicated at the side of each panel. The pH gradient (from 6.5–9.0) is indicated at the top and applies for all the panels. Dots indicate the five major size classes. Arrow indicates the zein spot expressed in the reciprocals. B, EMSAs of double strand oligonucleotide containing the O2-binding site. Protein extracts from endosperms and bacterial protein extracts from E. coli, expressing (O2be) or nonexpressing (be) the O2 polypeptide, were used in this analysis. Wild-type and o2 maize lines or o2 hybrids are indicated at the top of each lane. Addition (+) of O2-antiserum (O2-Ab) is indicated. Open arrow indicates the migration of the oligonucleotide bands shifted by O2 protein in O2be and Y+. Closed arrow indicates the super-shift determined by the addition of O2-antiserum to the mixture.
Using mono-dimensional gel electrophoresis (1D) and 2D analyses we further considered the lines in which the o2It allele Bianchi-o2It (Bo2It) was introgressed into several inbreds: No2It, Xo2It, 33o2It, and Yo2It (Figs. 1, A and B and 2). All these lines, apart from Yo2It, show in 1D the H1 band and absence of the H2 band. The 2D analyses (Fig. 2) show that in Xo2It the H1 band consists of two main spots that are the same observed for No2It (data not shown) and of three main spots in both the Bo2It and 33o2It. The two Xo2It spots have, however, different pIs from the three spots of the Bo2It and 33o2It. The Yo2It has faint H1 and H2 bands; each constituted by two or three very faint spots as revealed by 2D. This again puts forward the hypothesis that combinations of specific zHs and specific-GB factor(s) may in some way give rise to specific zH accumulation in o2 lines.
Heavy Chain Zein Genes in o2 Lines
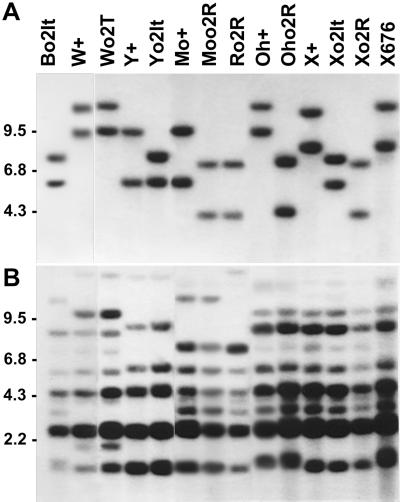
The genotypes described above and the others considered in the following sections were characterized both for proper introgression of the various o2 alleles and for the RFLP patterns specific for those zein genes that specify H1 and H2 polypeptides. DNAs from the genotypes of interest were digested with EcoRI, a restriction enzyme diagnostic for both the O2 locus and the H1 and H2 zein loci (Dolfini et al., 1992; Bernard et al., 1994; Lund et al., 1995). The Southern filter (Fig. 3) was first hybridized to O2 cDNA and then to M1 cDNA, a zein probe representative of both heavy zein size classes (Viotti et al., 1985) of zein-SF4 sequences (Rubenstein and Geraghty 1986; Lund et al., 1995). The O2 or o2 alleles have their own specific pattern (Fig. 3A) that identifies each of them and confirms the correct introgression for a given o2 allele in each GB. The line Bo2It, in which the o2It allele was first recovered (Bernard et al., 1994), is also reported for proper comparison.
Figure 3.
Southern analysis of genomic DNA from different maize lines digested with EcoRI and probed with O2 cDNA (A) and with the heavy chain zein cDNA M1 (B). Maize lines, abbreviated as in Table I, are reported on the top and apply for both panels. Molecular mass markers in kb are on the left. Approximately 10 μg of DNA was digested to completion as described by Bianchi and Viotti (1988) and applied in each lane.
The only GB that shows the same pattern between the wild type and the mutant o2 is the W64A in which the o2T originated as spontaneous mutation of the O2w1 (Emerson et al., 1935; Mertz et al., 1965; Dolfini et al., 1992; Bernard et al., 1994; Table I). The restriction patterns of M1 homologous sequences are in most cases specific for each GB and appear identical within the O2 and o2 lines with the same GB, thus suggesting the absence of gross deletion or rearrangement for M1 sequences in the lines under comparison (Fig. 3B). Moreover, the results of Figure 3B show that the M1 pattern of the two GBs A69Y (Y) and W22 (X), in which the o2It has been introgressed from the Bo2It, corresponds to the one specific of each GB rather than to that of the o2It progenitor.
Heavy Chain Zein Transcripts Occur in o2 Lines
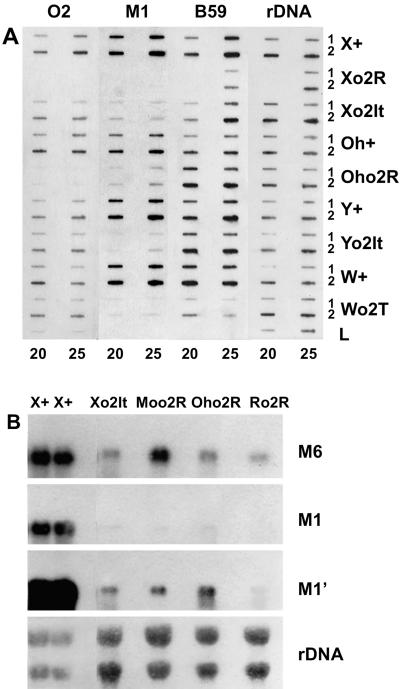
It has been shown that in some GB (Viotti et al., 1982, 1985; Messing, 1987) few genes of the SF1, SF2, or SF3 subset, which usually code for 213 or 219 amino acid zL chains because of proper intragenic duplication within the central coding region, bring their coding capacity up to 240 or 245 amino acids. If properly transcribed, these genes would generate heavy zeins that are, however, not regulated by O2. This may impair the data and interpretations on heavy chain expression that occurs in those o2 lines showing H1 and H2 polypeptides accumulation. With respect to this, some of the lines with a differential expression of H polypeptides were investigated for the presence of transcripts homologous to M1. For each line the M1 transcript level was also compared with the ones of the light type corresponding to the prototype M6 and B59 zein sequences belonging to SF1 and SF3, respectively (Viotti et al., 1985; Messing, 1987; Aukerman et al., 1991). RNA analysis by slot-blot hybridization (Fig. 4A) of a set of wild-types and corresponding o2 genotypes confirms the absence of o2 transcript in Oho2R and Xo2R and, as expected on the basis of the o2 allele type, its occurrence in Xo2It, Yo2It, and Wo2T. Hybridization with M1 shows in o2 lines in respect to the corresponding wild-type: (a) Absence of signal in the Xo2R that is almost devoid of H polypeptides; (b) a faint signal in Wo2T and Yo2It that are significantly affected in H polypeptide accumulation; (c) a slightly higher signal detection in Xo2It, which shows a partial H1 accumulation; and (d) finally, a significant signal in Oho2R in which both H1 and H2 are accumulated. These results are corroborated by the data obtained in the northern analysis of Figure 4B in which the M1 transcripts, almost undetectable in a short exposure, are revealed in a longer exposure. Also in this case, the level of M1 transcript parallels the amount of H polypeptides present at maturity in the o2 line considered. In fact Ro2R, which is almost devoid of H polypeptides, shows a very faint signal only in the longer exposure. On the other hand the other three o2 lines (Xo2It, Moo2R, and Oho2R) that do partially or significantly accumulate them show a higher signal. It should be noted, however, that the amount of heavy chain polypeptides determined at a given stage of development or at maturity represents an accumulation. On the contrary the transcript datum represents a steady-state level, which should not necessarily reflect the final abundance of H polypeptides, as cytoplasmic control mechanisms for the translational efficiency of zH mRNA have been put forward (Spena et al., 1985; Kodrzycki et al., 1989).
Figure 4.
A, Slot-blot analysis of total RNAs of maize genotypes as reported on the right. RNA was isolated from endosperms at 20 and 25 DAP, as indicated at the bottom. For each genotype and DAP, 1 and 2 μg loads were analyzed as indicated on the right. The total RNA at 20 DAP from the Xo2R line was not available in our stock of frozen seeds. Lane L contains total RNA from maize leaf in 1- and 2-μg horizontal loads from left to right, respectively. Probes are reported at the top of each strip. The filter was subjected to a series of sequential hybridization-stripping cycles using the probes indicated from left to right. B, Northern analysis of total RNA from endosperms of maize lines as indicated at the top. The wild-type RNA from endosperms at 20 and 25 DAP from left to right was 5 μg for each load and the o2 RNAs (20 DAP) were 10 μg for each load. Probes are indicated at the right of each panel. M1′ shows a longer exposure than M1. The filter was subjected to a series of sequential hybridizations to the four probes (from top to bottom) and stripping. Exposure time, at −80°C with intensifying screens, was 6 d for O2 and 5 h for zein probes. rDNA blots were exposed for 1 h at room temperature.
Genetic Background Combinations Differently Affect Heavy Chain Gene Transcription
These results indicate that in certain GBs some zein genes belonging to the zH type, a group usually positively regulated in its transcription by O2, can be transcribed even in the absence of the O2 product (o2R allele) or in the presence of non-functional o2, as in the case of the o2T, o2It, or o2-676 alleles. This was further tested by analyzing the zein polypeptides accumulated in the crosses between the Yo2It and the Oh676 or X676 lines. These alleles produce o2 polypeptides that are nuclearly localized (Varagona and Raikhel, 1994; B. Lazzari, unpublished data). The zein extracts of the three parent lines and of the four reciprocals to Yo2It were analyzed by SDS-PAGE. In the 1D analysis of the reciprocals a more than additive zein pattern was revealed in respect to the parent lines (data not shown). The 2D fractionation of the extracts from the reciprocals between Yo2 and Oh676 (Fig. 5A) reveals indeed an additive spot of H2 size. This indicates that in combining specific o2 GBs with certain zein sequences coding for heavy polypeptides there is a recovered transcription of some of them. Furthermore, this puts forward the possibility that o2 defective polypeptides that are nuclearly localized, as is the case of the o2It and o2-676, can functionally complement each other or form functional in vivo heterodimers with other bZIP proteins (Pysh et al., 1993). This was tested in electrophoretic mobility shift assays (EMSA) experiments with an oligonucleotide containing the O2-binding site (Fig. 5B). Whole cell extracts from both the o2 parents and their reciprocals were compared with a whole cell protein extract from a wild-type endosperm and to a bacterial extract from an Escherichia coli strain expressing the O2 polypeptide (O2be). The O2 endosperm extract shows three retarded bands one of which is O2-specific as it is super-shifted by the addition of anti-O2 polyclonal antibodies. This specific retarded band is never observed in any of the o2 extracts, nor does a super-shifted band appear when they are challenged with the O2 polyclonal antibodies. In all the endosperm extracts two additional retarded bands are also revealed. These bands might represent two other O2 box-binding bZIP polypeptides expressed in maize endosperm (Pysh et al., 1993; Carlini et al., 1999).
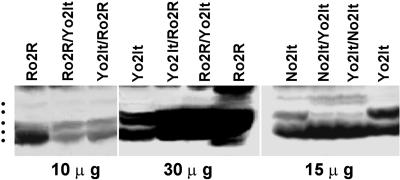
An additional evidence that specific combinations of GBs can specifically and positively affect zH transcription in o2 lines derives from the results reported in Figure 6. The 1D analysis of the zeins extracted from the reciprocals between Yo2It and No2It or Yo2It and Ro2R in fact reveal in respect to the parent lines an additional H2 band that occurs only in the crosses involving the Yo2It and No2It lines. This suggests that the A69Y and NYR GBs complement each other for factor(s) that transactivate some of the zH2 sequences occurring in one of the two or in both the parental genotypes.
Figure 6.
SDS-PAGE analysis of zein polypeptides from o2 genotypes and their reciprocal crosses. Zein protein extracts were obtained from mature seeds and loaded in the amounts indicated at the bottom. Dots on the left side identify the five major size classes of zeins.
DISCUSSION
The wild-type and o2 lines investigated in this study derive from stocks of different groups (Nelson, 1967; Bianchi and Viotti, 1988; Schmidt, 1993; Bernard et al., 1994) in which the introgression of the various o2 alleles in a given GB has been obtained with at least six backcrosses. For a given line, the 1D and 2D gel patterns of the zein extracts obtained from the original seeds and from seeds of our stocks in different harvests were shown to be constant, indicating a high reproducibility of results with this approach. Moreover, the data presented in this paper and previous publications (Bianchi and Viotti, 1988; Aukerman et al., 1991; Dolfini et al., 1992; Schmidt, 1993; Bernard et al., 1994) about the molecular analyses of each line carried out at the level of the DNA and of the O2 and o2 transcripts and polypeptides establish an unequivocal identification and characterization of each genotype.
For a given genetic background, the introgression of an o2 allele might have, to a certain extent, modified the zein gene subset identified by the M1-like sequences, since the O2 locus, located on chromosome 7, lies close to zein gene clusters (Soave et al., 1981a). This, however, does not seem to be the case since at least six back-crosses were performed, the M1 RFLP remains the same of the introgressed GB, and the M1 sequences are mostly if not all located on chromosome 4 (Viotti et al., 1982).
In the various o2 lines analyzed by RNA blots, the steady-state transcript levels of M1-like sequences show lower and different intensities in respect to the corresponding wild type with a severe reduction as in Ro2R and Xo2R or only a partial reduction as in Xo2It and Yo2It. However, in most cases the transcript levels correlate with the level of H1 and H2 polypeptides accumulated in the o2 lines considered. To be more confident with this type of correlation, amino-terminal microsequencing of two purified spots from an o2 line still accumulating heavy type zeins was performed. Apart from one amino acid difference detected in each sequence, the first 22 amino acids of each spot (data not shown) were identical to the one of the polypeptides identified by the M1 clone (Viotti et al., 1985). This strongly correlates the occurrence of heavy type transcripts to the accumulation of H polypeptides in o2 lines. At the beginning of zein polypeptide analysis most of the published data about o2 lines were obtained with the W64Ao2T, W22o2R, and A69Yo2It genotypes (Di Fonzo et al., 1977; Schmidt et al., 1987; Motto et al., 1988; Kodrzycki et al., 1989), which show an almost complete or severe reduction of H polypeptides that led to the improper generalization that all the o2 lines have to be H1 and H2 null.
As reported by others (Kodrzycki et al., 1989; Dolfini et al., 1992), the steady-state levels of the transcripts of the other zein gene subsets (Fig. 4) appear in some genotypes to be also affected by the o2 mutation. This may suggest a direct involvement of O2 in the transcription of some members of the other subsets (presently under investigation) or an indirect effect of the lowered zH polypeptide accumulation on the intracellular stability of macromolecules and of other storage proteins because of improper filling in of the protein bodies. In this respect translatability and stability of zein mRNA could impact zein polypeptides accumulation (Spena et al., 1985; Plotnikov and Bakaldina, 1996) and partly explain zein pattern variations among GBs. The studies on light- and heavy-zein transcript stabilities in the W64A GB carrying the O2w1 allele or the o2T allele indicated that the half-life of the two transcript populations is different with, however, an higher stability for the light transcripts in the mutant genotype. Data on heavy transcripts in the mutant genotype were not obtained in that study (Plotnikov and Bakaldina, 1996) due to their highly reduced level (less than 20%) in respect to the normal line. However, based on these observations it is reasonable to think that the heavy transcripts detected in the various GBs carrying any of the o2 alleles described in this paper have similar half-life as: (a) Specific sequences in the 5′-untranslated region and 3′-untranslated region that are responsible of mRNA instability (Green, 1993) are absent in any of the zein transcripts and (b) computer searches indicated that the various heavy-zein transcripts and genes share a high homology of sequence in these regions (B. Lazzari, unpublished data).
Nucleotide sequence comparison of the zH published sequences within the promoter region containing the O2-binding site reveals microheterogeneity with base substitution and/or deletion. DNA fragments with such substitution and/or deletion when tested in O2-binding assays lead to non-functional in vitro recognition by O2 (Schmidt et al., 1992). This, however, does not necessarily reflect the in vivo situation as other unidentified GB-specific factors, or the bZIP factors (Pysh et al., 1993; Carlini et al., 1999), or still a combination of all of these may substitute for the loss of O2 activity by recognizing and transactivating zH genes with canonical (O2ts) or non canonical (o2ts) O2 target sequence.
On the other hand, data on (a) the occurrence of the two other retarded bands in EMSA (Schmidt et al., 1992; Pysh et al., 1993; Fig. 5B), (b) the detection of few other polypeptides (Ciceri et al., 1997; P. Ciceri and A. Viotti, unpublished data) in southwestern experiments capable to bind the canonical O2-target sequence (O2ts) in O2-null lines, (c) the capacity of O2 to complement gcn4-mutant yeast strain (Mauri et al., 1993; GCN4 binding site, 5′-ATGACTCAT-3′), and (d) the capacity of O2 to cooperatively recognize and bind in in vivo/in vitro experiments non-canonical O2 target sequence (Yunes et al., 1994, 1998) in addition to the in vitro binding to the ACGT core of G- or C-box type motif (Izawa et al., 1993) have been reported. All together, these data represent clear evidence of promiscuity in O2-binding specificity and suggest the presence in maize endosperm of other factors recognizing the canonical or non-canonical O2 target sequence occurring in the zH gene promoter.
Supporting evidence of such a scenario derives from our in vivo data: (a) the four o2R lines in the GBs Oh43, B14, B37, and R802 (absence of any O2 product) transcribe most of the zH genes and significantly accumulate H1 and H2 zeins, and (b) the results of the crosses reported in Figures 5 and 6 indicate that some genes coding for heavy-type zeins are expressed in o2 hybrids only in specific GB combinations; among these there is the Oh43 GB. To make this happen, proper combinations must occur. That we cannot detect any specific or additional complex with O2ts in Oh43 GB (Fig. 5) suggests that the interactions among these proteins might be transient or unstable under the conditions for the EMSA.
The occurrence of factor(s) that can substitute O2 activity in o2 maize endosperm has been reported, even though they are activated just by particular growth conditions (Müller et al., 1997). Still, this factor(s) might be constitutively expressed in certain GBs, whereas in others, it may have to be activated as reported. It is interesting that in this respect, the o2 line investigated in Müller et al. (1997) is the A69Yo2It.
A schematic representation of these findings and interpretations is reported in Table II. The maize genotypes are subdivided into two main groups with different capacity in expressing zH genes in the presence of O2 or o2 allele. This takes into account both the occurrence of zH genes with wild-type (O2ts) or mutated (o2ts) O2 target sequence and the possibility that some GB with any o2 allele, but having O2-vicarious factor gene(s), can per se transactivate most of the zH genes (group-I, O2VF lines) or complement the absence/loss of function of group-II (o2vf lines) when properly crossed.
Table II.
Grouping of maize genotypes according to their zein pattern and capacity to transactivate zH genes with canonical (O2ts-zH) or non-canonical (o2ts-zH) O2 target sequence
| Zein Gene Type | Group I, O2VFs
|
Group II, o2vfs
|
||
|---|---|---|---|---|
| Oh43, R802, B37a, 3316, B14, NYR
|
W22, Mo17, A69Yb, W64Ac, Rossman
|
|||
| Allele O2 | Allele o2R or o2It | Allele O2 | Allele o2R, o2T, or o2It | |
| O2ts-zH | +, Hs | (+, Hs) | +, Hs | ((+, Hs)) |
| o2ts-zH | +, Hs | +, Hs | +, Hs | − |
Grouping is based on presence/activity or absence/inactivity of regulatory gene(s) coding for O2 vicarious factor(s), respectively, O2VF or o2vf. Data are derived from the results reported in Figures 1, 5, and 6 of present work and from unpublished data (M. Lauria and A. Viotti). + and Hs, Normal zH transcription and heavy zein polypeptide accumulation, respectively; however, only partially or severely attenuated when occurring in single or double parentheses, respectively. −, Absence or severe reduction in transcription.
This GB with the o2It allele shows a zein pattern identical to the B37o2R line (data not shown).
This GB with an o2T allele shows a zein pattern identical to the A69Yo2It line (data not shown).
This GB with the o2It allele shows a zein pattern identical to the W64Ao2T line (data not shown).
An alternative explanation that could apply for the o2It or o2-676 genotypes is that these factor(s) in combination with the o2It or o2–676 that are nuclearly localized might partially complement for their defective functions in transactivating some of the zH genes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Maize (Zea mays) plants were grown in the field or in the greenhouse during 1993, 1995, and 1998. Seeds were harvested at maturity or at 15, 20, and 25 DAP around noon as described (Bernard et al., 1994). A list of genetic stocks is reported in Table I.
Nucleic Acid Isolation and Blot Analyses
DNA and RNA were extracted and purified as described (Dolfini et al., 1992; Bernard et al., 1994). DNA Southern analysis and RNA analyses by northern and slot blots were carried out as reported (Bernard et al., 1994). Probes were labeled with the Rediprime system (Amersham, Buckinghamshire, UK). The O2 cDNA, the zein cDNA (M1, M6, and B59), and rDNA probes were those already described (Viotti et al., 1985; Dolfini et al., 1992). Washing conditions of the nucleic acid filters were performed at high stringency (Viotti et al., 1982, 1985), which discriminates homologies lower than 95%.
Zein Extraction and Analysis
Zeins were extracted and analyzed by fractionation on SDS-PAGE, IEF, and 2D gels as reported in Viotti et al. (1985) and Lund et al. (1995). Gels were stained with Coomassie Brilliant Blue R250.
EMSAs
The double strand DNA probe was prepared as described (Ciceri et al., 1997) by annealing the oligonucleotides 5′-GGCATTCCACGTAGATAA-3′ and 5′-GGTTATCTACGTGGAATG-3′ containing the O2 core binding site (underlined). Fill-in labeling with [α32P-dCTP] was performed by Klenow. Escherichia coli extracts expressing or nonexpressing O2 polypeptide were prepared as reported in Ciceri et al. (1997). Protein extracts from endosperm at 20 DAP were obtained by homogenizing three to four endosperms in 0.2 mL of 10 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.9, 50 mm KCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and “1× protease inhibitor cocktail” (Boehringer Mannheim, Basel). The homogenate was centrifuged in the cold room at 15,000g for 10 min. Glycerol was added to the supernatant to a final concentration of 10% (w/v) and aliquots were immediately frozen and stored at −80°C until use. Aliquots were used once and then discarded. Frozen aliquots retain their DNA-binding activity for at least 4 months. Five to ten microliters of endosperm protein extracts was mixed to 0.5 ng of labeled probe in a final volume of 20 μL containing 10 mm HEPES, pH 7.9, 50 mm KCl, 10 mm dithiothreitol, 40 μg mL−1 sonicated salmon sperm DNA, 2 mg mL−1 bovine serum albumin, and 10% (w/v) glycerol. The mixture was incubated at 25°C for 30 min. When indicated, at the end of the incubation time, 0.1 μL of rabbit polyclonal anti-O2 serum was added to the mixture and the incubation was extended for 30 more min. Mixtures were analyzed on a 5% (w/v) polyacrylamide gel according to Schmidt et al. (1992).
ACKNOWLEDGMENTS
We are grateful to Dr. Emanuele Quattrini for technical help in growing maize plants in the green house and in the field.
Footnotes
This work was partially supported by a grant from Piano Nazionale Biotecnologie Vegetali, Ministero per le Politiche Agricole (to A.V.).
LITERATURE CITED
- Aukerman MJ, Schmidt RJ. A 168-bp derivative of Suppressor-mutator/Enhancer is responsible for the maize o2-23 mutation. Plant Mol Biol. 1993;21:355–362. doi: 10.1007/BF00019950. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Schmidt RJ, Burr B, Burr FA. An arginine to lysine substitution in the bZIP domain of an opaque-2 mutant in maize abolishes specific DNA binding. Genes Dev. 1991;5:310–320. doi: 10.1101/gad.5.2.310. [DOI] [PubMed] [Google Scholar]
- Bernard L, Ciceri P, Viotti A. Molecular analysis of wild-type and mutant alleles at the Opaque-2 regulatory locus reveals different mutations and types of O2 products. Plant Mol Biol. 1994;24:949–959. doi: 10.1007/BF00014448. [DOI] [PubMed] [Google Scholar]
- Bianchi M, Viotti A. DNA methylation and tissue-specific transcription of the storage protein genes of maize. Plant Mol Biol. 1988;11:203–214. doi: 10.1007/BF00015672. [DOI] [PubMed] [Google Scholar]
- Carlini LE, Ketudat M, Parsons RL, Prabhakar S, Schmidt RJ, Guiltiman MJ. The maize EmBP-1 orthologue differentially regulates Opaque2-dependent gene expression in yeast and cultured maize endosperm cells. Plant Mol Biol. 1999;41:339–349. doi: 10.1023/a:1006338727053. [DOI] [PubMed] [Google Scholar]
- Ciceri P, Gianazza E, Lazzari B, Lippoli G, Genga A, Hoschek G, Schmidt RJ, Viotti A. Phosphorylation of Opaque-2 changes diurnally and impacts its DNA binding activity. Plant Cell. 1997;9:97–108. doi: 10.1105/tpc.9.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fonzo N, Fornasari E, Salamini F, Soave C. SDS-protein subunits in normal, opaque-2 and floury-2 maize endosperms. Maydica. 1977;XXII:77–88. [Google Scholar]
- Dolfini S, Landoni M, Tonelli C, Bernard L, Viotti A. Spatial regulation in the expression of structural and regulatory storage-protein genes in Zea mays endosperm. Dev Genet. 1992;13:264–276. [Google Scholar]
- Emerson RA, Beadle GW, Fraser AC (1935) A summary of linkage groups in maize. Cornell Univ Agric Exp Sta Mem 180
- Green PJ. Control of mRNA stability in higher plants. Plant Physiol. 1993;102:1065–1070. doi: 10.1104/pp.102.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins TJV. Synthesis and regulation of major proteins in seeds. Annu Rev Plant Physiol. 1984;35:191–221. [Google Scholar]
- Hoschek G, Kelly AR, Schmidt RJ. Characterization of the molecular defect in a null allele of the opaque-2 locus of maize. Plant Mol Biol. 1996;32:1159–1161. doi: 10.1007/BF00041400. [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH. Plant bZIP protein DNA binding specificity. J Mol Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- Kodrzycki R, Boston R, Larkins B. The opaque-2 mutation of maize differentially reduces zein gene transcription. Plant Cell. 1989;1:105–114. doi: 10.1105/tpc.1.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzari B, Ciceri P, Carzaniga R, Genga A, Faoro F, Viotti A. Molecular characterization of o2T: a mutant allele at the opaque-2 locus. Maize Genet Coop Newslett. 1995;69:102. [Google Scholar]
- Lazzari B, Ciceri P, Cellini F, Viotti A. Sequence analysis of an opaque-2 mutant of Zea mays. Maize Genet Coop Newslett. 1994;68:60–61. [Google Scholar]
- Lund G, Ciceri P, Viotti A. Maternal-specific demethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L. Plant J. 1995;8:571–581. doi: 10.1046/j.1365-313x.1995.8040571.x. [DOI] [PubMed] [Google Scholar]
- Mauri I, Maddaloni M, Lohmer S, Motto M, Salamini F, Thompson R, Martegani E. Functional expression of the transcriptional activator Opaque-2 of Zea mays in transformed yeast. Mol Gen Genet. 1993;241:319–326. doi: 10.1007/BF00284684. [DOI] [PubMed] [Google Scholar]
- Mertz ET, Bates LS, Nelson OE. Second mutant gene affecting the amino acid pattern of maize endosperm proteins. Science. 1965;145:1469–1470. doi: 10.1126/science.150.3702.1469. [DOI] [PubMed] [Google Scholar]
- Messing J. Rigby PWJ, ed, Genetic Engineering. Vol. 6. London: Harcourt Brace Jovanovich Publishers; 1987. The genes encoding seed storage proteins in higher plants; pp. 1–46. [Google Scholar]
- Motto M, Maddaloni M, Ponziani G, Brembilla M, Marotta R, Fonzo ND, Soave C, Thompson RD, Salamini F. Molecular cloning of the o2–m5 allele of Zea mays using transposon marking. Mol Gen Genet. 1988;212:488–494. [Google Scholar]
- Müller M, Dues G, Balconi C, Salamini F, Thompson RD. Nitrogen and hormonal responsiveness of the 22-kDa a-zein and b-32 genes in maize endosperm is displayed in the absence of the transcriptional regulator Opaque-2. Plant J. 1997;12:281–291. doi: 10.1046/j.1365-313x.1997.12020281.x. [DOI] [PubMed] [Google Scholar]
- Nelson OE. Un gene mutante che influenza la sintesi delle proteine nell'endosperma di mais. Maydica. 1967;XII:81–96. [Google Scholar]
- Plotnikov VK, Bakaldina NB. Differential stability of zein mRNA in developing corn kernel. Plant Mol Biol. 1996;31:507–515. doi: 10.1007/BF00042224. [DOI] [PubMed] [Google Scholar]
- Pysh LD, Aukerman MJ, Schmidt RJ. OHP1: a maize basic domain/leucine zipper protein that interacts with Opaque2. Plant Cell. 1993;5:227–236. doi: 10.1105/tpc.5.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein I, Geraghty DE. Genetic organization of zeins. In: Pomeranz Y, editor. Advances in Cereal Science and Technology. VIII. American Association of Cereal Chemists Press 1986. , St. Paul, pp 297–315. [Google Scholar]
- Schmidt RJ. Opaque-2 and zein gene expression. In: Verma DPS, editor. Control of Plant Gene Expression. Boca Raton, FL: CRC Press; 1993. pp. 337–355. [Google Scholar]
- Schmidt RJ, Burr FA, Aukerman MJ, Burr B. Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc Natl Acad Sci USA. 1990;87:46–50. doi: 10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Burr FA, Burr B. Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science. 1987;238:960–963. doi: 10.1126/science.2823388. [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G. Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell. 1992;4:689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Napier JA, Tatham AS. Seed storage proteins: structures and biosynthesis. Plant Cell. 1995;7:945–956. doi: 10.1105/tpc.7.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Tatham AS. The prolamin storage proteins of cereal seeds: structure and evolution. Biochem J. 1990;267:1–12. doi: 10.1042/bj2670001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soave C, Reggiani R, Fonzo ND, Salamini F. Clustering of genes for 20-kd zein subunits in the short arm of maize chromosome 7. Genetics. 1981a;97:363–377. doi: 10.1093/genetics/97.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soave C, Tardani I, Fonzo ND, Salamini F. Regulation of zein level in maize endosperm by a protein under control of the opaque-2 and opaque-6 loci. Cell. 1981b;27:403–410. doi: 10.1016/0092-8674(81)90423-2. [DOI] [PubMed] [Google Scholar]
- Spena A, Krause E, Dobberstein B. Translation efficiency of zein messenger-RNA is reduced by hybrid formation between the 5′-untranslated and 3′-untranslated region. EMBO J. 1985;4:2153–2158. doi: 10.1002/j.1460-2075.1985.tb03909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger E, Parsons RL, Schmidt RJ, Bowen B, Roth BA. Dominant negative mutants of Opaque2 suppress transactivation of a 22-kD zein promoter by Opaque2 in maize endosperm cells. Plant Cell. 1993;5:831–841. doi: 10.1105/tpc.5.8.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona MJ, Raikhel NV. The basic domain in the bZIP regulatory protein Opaque2 serves two independent functions: DNA binding and nuclear localization. Plant J. 1994;5:207–214. doi: 10.1046/j.1365-313x.1994.05020207.x. [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV. Monocot regulatory protein Opaque-2 is localized in the nucleus of maize endosperm and transformed tobacco plants. Plant Cell. 1991;3:105–113. doi: 10.1105/tpc.3.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti A, Abildsten D, Pogna N, Sala E, Pirrotta V. Multiplicity and diversity of cloned zein cDNA sequences and their chromosomal localization. EMBO J. 1982;1:53–58. doi: 10.1002/j.1460-2075.1982.tb01123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti A, Cairo G, Vitale A, Sala E. Each zein gene class can produce polypeptides of different sizes. EMBO J. 1985;4:1103–1110. doi: 10.1002/j.1460-2075.1985.tb03746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunes JA, Neto GC, Silva MD, Leite A, Ottoboni L, Arruda P. The transcriptional activator Opaque2 recognizes two different target sequences in the 22-kD-like a-prolamin genes. Plant Cell. 1994;6:237–249. doi: 10.1105/tpc.6.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunes JA, Vettore AL, Silva MD, Leite A, Arruda P. Cooperative DNA binding and sequence discrimination by the Opaque-2 bZIP factor. Plant Cell. 1998;10:1941–1955. doi: 10.1105/tpc.10.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]