Abstract
The cysteine protease separase opens the cohesin ring by cleaving its kleisin subunit and is a pivotal cell cycle factor for the transition from metaphase to anaphase. It is inhibited by forming a complex with the chaperone securing, and in vertebrates, also by the Cdk1-cyclin B1 complex. Separase is activated upon the destruction of securin or cyclin B1 by the proteasome, after ubiquitination by the anaphase-promoting complex/cyclosome (APC/C). Here we review recent structures of the active protease segment of Chaetomium thermophilum separase in complex with a substrate-mimic inhibitor and full-length Saccharomyces cerevisiae and Caenorhabditis elegans separase in complex with securin. These structures define the mechanism for substrate recognition and catalysis by separase, and show that securin has extensive contacts with separase, consistent with its chaperone function. They confirm that securin inhibits separase by binding as a pseudo substrate.
Separase is a large (140–250 kD) eukaryotic endopeptidase belonging to the CD clan of cysteine proteases, which also includes caspases and gingipain [1], reviewed in [2–5••]. It cleaves the kleisin subunit (Scc1/Rad21/Mcd1 for mitosis and Rec8 for meiosis) of the cohesin complex that entraps sister chromatids during cell division, and therefore it has essential roles in chromosome segregation [1,6–11]. While most of the cohesins located on the chromosome arms are removed through a phosphorylation-dependent “prophase pathway” [6,9], centromeric cohesins are protected by shugoshin and are subjected to separase cleavage for chromosome segregation during the transition from metaphase to anaphase [1,10,12,13]. Over-expression of separase is linked to aneuploidy and tumorigenesis, making it a potential target for anti-cancer drug discovery [14,15•].
Besides its roles in chromosome segregation, separase also has important functions in other cellular events, such as stabilizing the anaphase spindle by cleaving and localizing the kinetochore-associated protein Slk19 [16], regulating centriole disengagement in mammals by cleaving pericentrin/kendrin [17–19], DNA damage repair [20], membrane trafficking [21], telomere protection [22], and Cdk1 inhibition [23].
Consistent with its crucial cellular functions, the activity of separase is tightly regulated. Securin, a natively unfolded protein in solution [24,25], is the first reported regulator of separase and acts as both a chaperone and an inhibitor [26–33]. Securin binds to nascent separase protein co-translationally to help its proper folding and forms a stable complex with separase until the onset of anaphase. In vertebrates, the Cdk1-cyclin B1 complex is another regulator of separase activity [23,33–35]. Cdk1 phosphorylates separase and then forms a stable complex with it through interactions between cyclin B1 and a Cdc6-like sequence in the N-terminal regulatory region of separase, and this process is dependent on the isomerization of separase by Pin1 [36•].
Separase is activated by the destruction of securin [37–39] and cyclin B1 [23] via the proteasome pathway upon ubiquitination in their N-terminal region by the anaphase-promoting complex/cyclosome (APC/C) [40,41]. APC/C-mediated reduction in securin level in aged female mice is linked to premature chromosome segregation in meiosis II [42]. Besides the two key mechanisms mentioned above, other regulatory processes have also been reported. For example, auto-cleavage of separase in higher eukaryotes occurs upon activation, which affects mitosis progression but not the protease activity of separase [31,43–46]. Protein phosphatase 2A (PP2A) binds to a region of separase adjacent to the auto-cleavage sites [47], which stabilizes separase-associated securin through dephosphorylation [48]. Phosphorylation of securin in yeast enhances its interaction with separase and promotes the nuclear localization of separase [49].
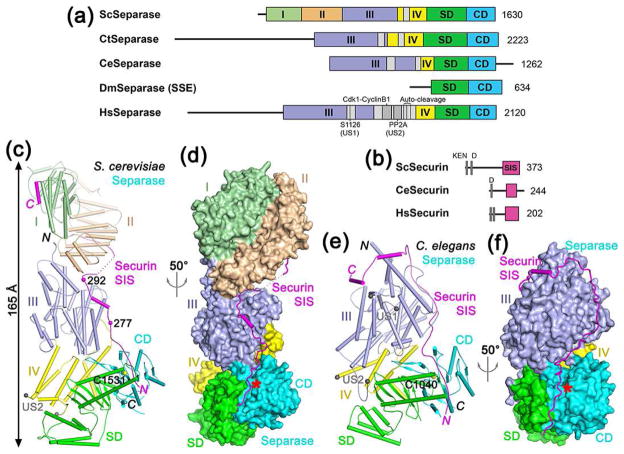
The primary structure of separase contains a C-terminal caspase-like catalytic domain (CD) of ~200 residues and an N-terminal α-helical regulatory region (Fig. 1a). An additional domain is located between the helical region and CD, and this domain has been named the substrate-binding domain (SD) [50••] or the pseudo-protease domain (PPD) [51••]. The CD is conserved among eukaryotes, with 34% sequence identity between yeast and human separase. The conservation of the SD is weaker, with 24% identity between yeast and human separase. In contrast, the α helical region is poorly conserved, both in sequence and in length, contributing to the extensive size variations among these enzymes (Fig. 1a). Some separases also contain C-terminal extensions beyond the CD, further increasing the size variation. Drosophila separase is distinct in being composed of two separate subunits [52,53] (Fig. 1a).
Figure 1.
Structures of separase-securin complexes. (a). Domain organization of separase. The helical region of yeast separase is divided into four domains (I–IV) and given different colors, which is followed by SD (substrate-binding domain) and CD (catalytic domain). The domain boundaries for the N-terminal region of C. thermophilum and human separase are not known and therefore are not indicated. The unstructured segments (US) in the helical region are shown in gray. For Drosophila separase, only the subunit containing the SD-CD (known as SSE) is shown. Sc: S. cerevisiae (yeast), Ct: Chaetomium thermophilum, Ce: C. elegans, Dm: D. melanogaster, Hs: Homo sapiens. (b). Domain organization of securin. The separase interaction segment (SIS) is shown in magenta. The N-terminal KEN and D-boxes are indicated. (c). Schematic drawing of the structure of the yeast separase-securin complex. The domains of separase are colored as in Fig. 1a, and the securin SIS is in magenta. The catalytic Cys1531 is shown as a sphere model. Two of the phosphorylation sites in securin are indicated with spheres. The ends of the unstructured segment (US2) are indicated by the two gray spheres. (d). Structure of the yeast separase-securin complex, with separase shown as a molecular surface, viewed after 50° rotation around the vertical axis from panel c. The active site of separase is indicated with the red star. (e). Schematic drawing of the structure of the C. elegans separase-securin complex. The catalytic Cys1040 is shown as a sphere model. (f). Structure of the C. elegans separase-securin complex, with separase shown as a molecular surface. All structure figures were produced with PyMOL (www.pymol.org).
Securin has a KEN-box and a D-box in its N-terminal region which are crucial for ubiquitination by APC/C, while its C-terminal region mediates the binding and inhibition of separase (Fig. 1b). This region has been named the separase interaction segment (SIS) [50••] or the separase-binding motif (SBM) [54••].
While separase was first characterized nearly two decades ago, detailed understanding of this central player for chromosome segregation was hampered by the lack of atomic structural information. Only low-resolution electron microscopy (EM) maps of human and C. elegans separase-securin complex were available [55,56]. Remarkably, three atomic structures were reported since 2016, including the crystal structures of the active protease segment of the separase from the thermophilic fungus C. thermophilum at up to 1.85 Å resolution [51••], the crystal structures of the yeast S. cerevisiae separase-securin complex at up to 2.6 Å resolution [50••], and the cryo-EM structure of the C. elegans separase-securin complex at 3.8 Å resolution [54••]. These structures represent significant breakthroughs for the field, and the observations from them are reviewed here.
Overall structures of the separase-securin complex
The overall structure of the yeast separase-securin complex assumes a highly-elongated shape, with overall dimensions of 65 × 70 × 165 Å (Fig. 1c, 1d). The shape of this complex is generally similar to that observed for the human separase-securin complex at low resolution [54••,55], suggesting that the human complex may have a similar organization. The overall structure of the C. elegans separase-securin complex is less elongated, about 110 Å for the longest dimension (Fig. 1e, 1f), because C. elegans separase (1262 residues) is much smaller than yeast separase (1630 residues). In fact, C. elegans separase also contains a C-terminal extension, and the CD actually terminates at residue 1140 (Fig. 1a). In both structures, securin assumes a mostly unfolded conformation and runs in an anti-parallel direction along the entire length of separase, from its active site to the N-terminal region at the opposite end of the structure.
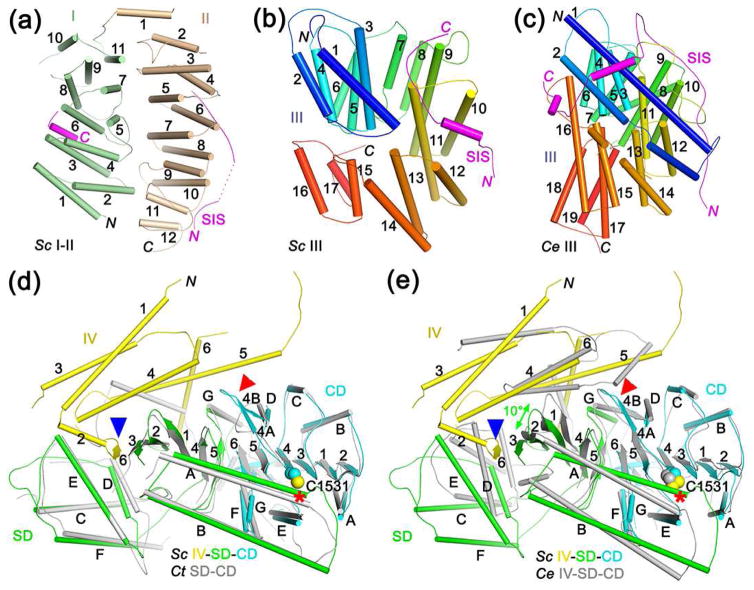
The structure reveals that the N-terminal helical region of yeast separase can be divided into four domains, I–IV, with domains I and II located far away from the active site in CD (Fig. 1c, 1d). These two domains are positioned side by side, such that the loops connecting the helical hairpins at one end of each domain are facing each other (Fig. 2a). C. elegans separase lacks these two domains, explaining its smaller size (Fig. 1a). The arrangements of the remaining domains in C. elegans separase are generally similar to those in yeast separase, although there are also detailed differences.
Figure 2.
Structures of separase domains and their interaction with securin. (a). Schematic drawing of domains I and II of yeast separase. The segment of SIS that interacts with these domains are also shown (magenta). (b). Schematic drawing of domain III of yeast separase, colored from blue at the N-terminus to red at the C-terminus. (c). Schematic drawing of domain III of C. elegans separase. (d). Overlay of the structures of yeast IV-SD-CD (in color) and the SD-CD of C. thermophilum separase (gray). The catalytic Cys residue is indicated with the red asterisk. Blue arrowhead points to the additional β-strand provided by domain IV to the SD in yeast separase. Red arrowhead points to conformational differences in the loop L4 region. (e). Overlay of the structures of yeast IV-SD-CD (in color) and the IV-SD-CD of C. elegans separase (gray). The superposition is based on the CD only, and a 10° difference is observed for the forientation of the SD β sheet (green arrow).
Domain III of yeast separase consists of helical hairpins arranged in a right-handed superhelix (Fig. 2b). While the architecture of this domain in C. elegans separase is generally similar, the locations of the helices (Fig. 2c) as well as the orientation of the domain relative to the rest of the structure (Fig. 1e) are different.
The SD has intimate contacts with the CD and is crucial for substrate binding by separase (Fig. 2d, see below), hence its name [50••]. The CD and SD have also been named the active protease domain (APD) and pseudo-protease domain (PPD), respectively [51••], although the backbone fold of SD has no similarity to caspases or other proteases. The structure of SD contains a five-stranded β-sheet with an RNase H-like fold, with inserts of a hairpin of two long helices (αA and αB) that contribute to substrate binding as well as a four-helical bundle. One face of the β-sheet in SD is covered by domain IV, which also provides an additional strand to the β-sheet in SD and contacts the CD in yeast separase (Fig. 2d), suggesting that domains IV-SD-CD together might form a stable module that mediates the protease activity of separase. Such an active module has also been named the separase protease domain (SPD) [51••].
The SD-CD structure of yeast separase is highly similar to that of C. thermophilum separase (Fig. 2d), with rms distance of 1.5 Å for their equivalent Cα atoms (39% sequence identity), indicating good conservation between these fungal enzymes and also that there are no significant conformational changes in these domains caused by the helical region. In comparison, there are substantial structural differences between the IV-SD-CD of yeast and C. elegans separase (Fig. 2e). With the CD of the two structures superposed, a 10° difference in the orientation of the SD β-sheet is observed between them, and there are also structural differences in the helical hairpin and especially the four-helical bundle in the SD. Moreover, C. elegans separase domain IV does not provide an extra β-strand to the SD and is located in a different position compared to that in yeast separase, although the domain still covers the β-sheet in SD and contacts the CD. These observed structural changes may reflect actual differences between fungal and animal separases.
The helical region in human separase contains two unstructured segments (US1 and US2), which harbor post-translational modification and protein binding sites that are important for the function of separase (Fig. 1a). US1 contains two phosphorylation sites, Ser1126 and Ser1153, that lead to inhibition of separase activity [34] and cause a tendency of inactivation by aggregation or precipitation [33], which can be counteracted by the Cdk1-cyclin B1 complex with the help of isomerization by Pin1 of the Ser1126 site [36•]. US2 contains the binding sites for Cdk1-cyclin B1 [23,35,57] and PP2A [47,48], and also the auto-cleavage sites [31,43] (Fig. 1a).
These two segments also exist in many of the other separases, although US1 may be absent in yeast separase (Fig. 1a). US1 has ~60 residues and corresponds to a loop in domain III, while US2 has ~250 residues in human separase but ~50 residues in others and is near the connection between domain IV and SD (Fig. 1e). Both segments are away from the active site of separase and the bound securin, although they might be able to reach the active site [54••]. The exact mechanism for their action will require further studies.
Mechanism of substrate recognition by separase
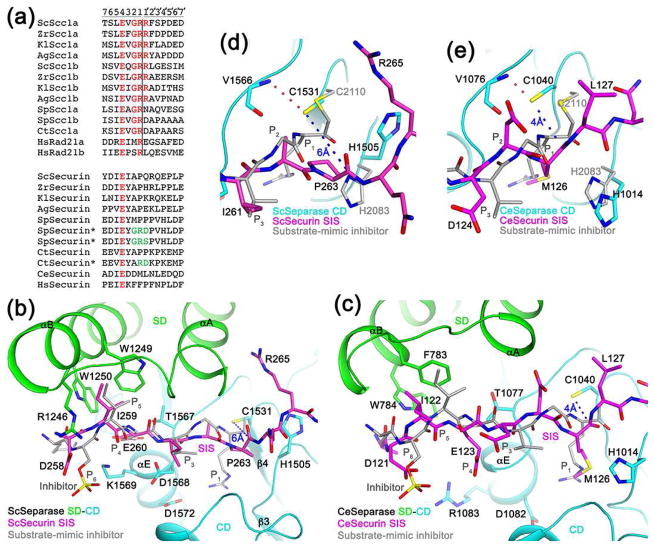
Separase substrates share the motif (D/E/S)x(D/E)xxR, with the cleavage site located after the Arg residue (Fig. 3a) [7,10,58]. The structure of C. thermophilum SD-CD in complex with a substrate-mimic inhibitor has defined the molecular basis for substrate recognition [51••]. The P1 Arg is ion-paired with an Asp residue, while the P4 Glu interacts with the main-chain amides at the N-terminus of helix αE of CD. The P5 residue contacts two aromatic side chains in helix αB from the helical hairpin insert of SD (Fig. 3b). Phosphorylation of the P6 Ser enhances the cleavage by separase [59], and the phosphate group interacts with a collection of Arg and Lys residues. The observed binding mode is supported by extensive structure-based mutagenesis studies [51••]. Mutations of S. pombe and C. thermophilum securin to introduce residues found in the substrate, especially an Arg residue at P1 (Fig. 3a), allowed its cleavage by separase [32,51••], indicating that securin functions as a pseudo substrate and inhibits separase by binding in the active site. Human separase cannot cleave yeast Scc1 and vice versa [31], suggesting that there is species specificity in the recognition.
Figure 3.
Molecular mechanism for the inhibition of separase by securin. (a). Alignment of the cleavage sites in separase substrates. The two cleavage sites in each protein are named a and b. The equivalent residues in securin are also shown. The asterisks indicate securin mutants (mutations in green) that become substrates of separase. The P and P′ residues are labeled at the top, and the cleavage site is indicated with the vertical line. (b). Binding mode of residues 258-265 of yeast securin SIS (magenta) in the active site of yeast separase. Side chains of residues in the interface are shown as stick models and labeled. The bound position of a substrate-mimic inhibitor to C. thermophilum separase SD-CD is shown in gray. (c). Binding mode of residues 121-127 of C. elegans securin SIS (magenta) in the active site of C. elegans separase. (d). Closeup of the active site region of yeast separase. The catalytic Cys1531 side chain is hydrogen-bonded to the main-chain amide of Val1566 (dashed line in red) and 6 Å from the carbonyl carbon of Pro263 (dashed line in blue). (e). Close-up of the active site region of C. elegans separase.
Mechanism for the inhibition of separase by securin
Consistent with biochemical studies, the structures show that securin inhibits separase as a pseudo substrate, with the N-terminal region of the SIS located in the active site of separase (Fig. 1c–1f). Securins share the P4 Glu residue with the substrate (Fig. 3a). The position of this segment of securin in the yeast separase-securin complex is similar to that of the substrate-mimic, and the recognition of the P4 and P5 residues are similar as well (Fig. 3b). In the C. elegans separase-securin complex, the position of securin has recognizable differences compared to that of the substrate-mimic (Fig. 3c). In addition, the P4 Glu side chain is pointed away from helix αE, and the positions of P5 and the helical hairpin it contacts are also different (Fig. 3c).
Most importantly, the P1 Arg of the substrate is replaced by Pro263 in yeast securin, and a large conformational difference is observed for the binding mode of this residue. In fact, the distance between the side chain of the nucleophilic Cys1531 and the carbonyl carbon of Pro263 is 6 Å (Fig. 3d), defining one reason why securin is not cleaved even though it binds in the active site. In addition, the carbonyl oxygen of Pro263 is pointed toward the Cys1531 thiolate rather than the oxyanion hole, and therefore this carbonyl group is not in the correct conformation for nucleophilic attack by Cys1531, whose thiolate is stabilized by a hydrogen-bond with the main-chain amide of Val1566. Moreover, the side chain of the second member His1505 of the catalytic machinery assumes a different rotamer to avoid steric clash with Pro263 and is pointed nearly in the opposite direction compared to His2083 in the C. thermophilum substrate-mimic complex (Fig. 3d), and this conformation is unlikely to support catalysis either.
In C. elegans securin the P1 Arg is replaced by Met126, and its side chain is inserted into the S1 pocket, occupying roughly the same position as the Arg side chain (Fig. 3e). The Asp1082 residue that would normally recognize the Arg side chain assumes a different rotamer (Fig. 3c). Interestingly, the main chain of Met126 assumes a conformation that appears to be ready for cleavage. The thiolate of Cys1040 is positioned directly above the Re face of the carbonyl group, with a distance of 4 Å to its carbon atom, and the second member His1014 assumes the catalytically competent rotamer (Fig. 3e). On the other hand, the separation between Cys1040 and His1014 is ~2.5 Å longer in the securin complex as compared to the substrate-mimic complex (Fig. 3e), likely due to the presence of Met at the P1 position in securin. This may be the reason why C. elegans securin is not cleaved in the active site of separase [54••].
Securin also has extensive contacts with the helical region of separase
In both the yeast and the C. elegans separase-securin complexes, securin has extensive contacts with the helical region of separase, outside of its active site. In fact, securin makes contact with every domain of separase in both structures (Fig. 1c–1f), and 4,600 Å2 of the surface area of yeast securin is buried in the interface with separase. The contacts involve ionic, hydrogen-bonding and van der Waals interactions.
In both structures, a helix at the C-terminal end of the securin SIS interacts with the most N-terminal domain of separase (domain I in yeast separase and domain III in C. elegans separase) (Fig. 2a, 2c). This is consistent with biochemical data showing that the C-terminus of securin binds to the N-terminal region in both yeast and human separase [30,55]; deletion of the first 155 residues of yeast separase disrupts complex formation with securin [30] (although deleting these residues could also disrupt the folding of separase); as well as that securin binds separase co-translationally as early as when the N-terminal region is translated [33].
The extensive contacts between separase and securin is consistent with the chaperone function of securin in promoting and stabilizing the folding of separase. Leaving out any region of the yeast securin SIS impairs the production of soluble separase [33,50••], demonstrating the importance of the entire SIS for the chaperone function of securin. Especially, a securin SIS lacking the region in the separase active site also failed to produce soluble separase, indicating that securin binding in the active site of separase is also important for its proper folding.
However, the exact mode of interaction between separase and securin is rather different between the yeast and the C. elegans complexes. For example, the locations of securin on the surface of domain III is dramatically different in the two complexes (Fig. 1c, 1e). This is likely a reflection of the weak sequence conservations of securin as well as the helical region of separase.
Comparison to caspase and other cysteine proteases
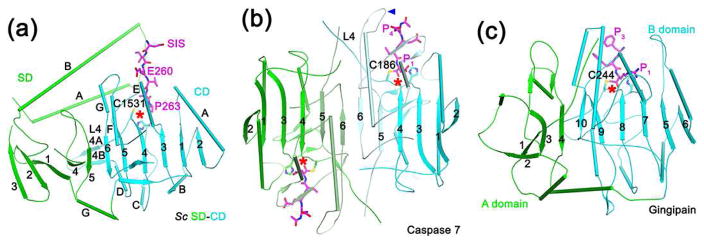
The structure of separase CD (Fig. 4a) is similar to that of caspase [60] (Fig. 4b), gingipain [61] (Fig. 4c) and other cysteine proteases. They share conserved catalytic machinery and oxyanion hole, and the substrate binding modes are similar as well. At the same time, there are also substantial differences among them.
Figure 4.
Structural comparisons with caspase and gingipain. (a). Schematic drawing of SD-CD of yeast separase (green and cyan, respectively), together with the portion of securin SIS (magenta) that is in the CD active site (indicated with the red star). The four-helical bundle in SD is omitted for clarity. (b). Schematic drawing of caspase 7 in a covalent complex with a substrate-mimic inhibitor (magenta). One molecule is colored in light cyan and cyan for its two fragments, while the other in light green and green. The blue arrowhead indicates the loop preceding strand β6 that is in the active site. The orientation of the first molecule is the same as that of separase CD. (c). Schematic drawing of gingipain in a covalent complex with a substrate-mimic inhibitor (magenta). The A and B domains are colored in green and cyan, respectively.
Caspase functions as a dimer of intra-chain cleaved hetero-dimers (Fig. 4b). The dimer contains a central β-sheet, with the strands roughly in the same plane. Similarly, a large, mostly planar β-sheet exists in gingipain, formed by its A and B domains (Fig. 4c). Separase also contains a large β-sheet for SD-CD, with the SD occupying the same position as the second molecule in caspase and the A domain in gingipain. However, the β-strands in SD are nearly perpendicular to those in CD, as well as their equivalents in caspase and gingipain. In addition, the second molecule of caspase and the A domain of gingipain do not contribute directly to catalysis in the active site, while the SD is crucial for substrate binding in separase.
Four surface loops are important for constituting the active site of caspase [62]. Among these, the loop preceding strand β6 in caspase (Fig. 4b) is provided by the helical insert of SD in separase (Fig. 4a). Loop L4 in caspase [63], also known as loop L2 [62], contains the catalytic Cys and the intra-chain cleavage site, and is important for the activation of caspase. The equivalent loop in separase (loop L4) does not need to undergo proteolysis for activation and is not in contact with the P side of the substrate, although mutations in this region can affect catalysis [51••]. This loop assumes a different conformation in the structure of separase-securin complexes, with a small two-stranded β-sheet (Fig. 2d–2e, 4a). It is placed between the CD and the helical region, especially domain III, and therefore the helical region may regulate the conformation of this loop.
Summary and Perspectives
The three recent structures have provided the first molecular insights into separase and securin. They have illuminated the molecular mechanism for substrate recognition and catalysis by separase and defined the domain organization and overall architecture of yeast and C. elegans separase. They have also revealed that securin inhibits separase by binding as a pseudo substrate in the active site, confirming earlier biochemical data. Moreover, securin contacts all the domains in separase, consistent with its chaperone function. The human separase-securin complex has a similar overall shape as the yeast separase-securin complex, indicating that it may have a similar architecture as well.
The structures confirm the expectation that separase is related to caspase, gingipain and other CD clan cysteine proteases, and also reveal significant differences to them. While the active site in caspase and gingipain is composed of residues from a single domain, the active site in separase requires contributions from two distinct domains. Separase also contains an extended N-terminal helical region, making it much larger than other cysteine proteases. The exact functions of this region are still not fully understood, and it has no direct contribution to the active site in the current structures. This helical region may regulate the catalytic activity of separase (for example controlling the conformation of the L4 loop in CD), and it may also interact with the Scc1 substrate to facilitate the cleavage [58]. The helical region may also support other functions of separase, for example by mediating the activation of human separase by DNA of 100 bp or larger [64]. The presence of many post-translational modification and protein-binding sites in the unstructured segments is also consistent with a regulatory role for the helical region. It will be interesting to elucidate the mechanism of how prolyl isomerization, phosphorylation and auto-cleavage affect the structure and activity of separase.
The structures of separase show extensive contacts (1,000 Å2 surface area burial) among its domains, suggesting that these structures may be stable on their own, in the absence of securin. However, it remains to be seen whether the active separase, after the destruction of securin, assumes the same conformation as that observed in the securin complex.
The current structures have defined the molecular mechanism for the inhibition of separase by securin. Further studies will be needed to reveal the mechanism of separase inhibition by Cdk1-cyclin B1 in vertebrates. In addition, an atomic structure of the human separase-securin complex would also be of great interest, as it may form the foundation for the design and development of inhibitors that could be efficacious for treating human diseases.
Highlights.
Atomic structures of separase and its complex with securin have become available.
Securin inhibits separase by binding as a pseudo substrate to its active site.
Securin has extensive interactions with separase, consistent with its chaperone function.
The catalytic domain of separase resembles cysteine proteases, but has unique features.
Acknowledgments
This research is supported by grant R35GM118093 from the NIH to LT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 3.Uhlmann F. Separase regulation during mitosis. Biochem Soc Trans. 2003;70:243–251. doi: 10.1042/bss0700243. [DOI] [PubMed] [Google Scholar]
- 4.Yanagida M. Cell cycle mechanisms of sister chromatid separation; role of Cut1/separin and Cut2/securin. Genes to Cells. 2000;5:1–8. doi: 10.1046/j.1365-2443.2000.00306.x. [DOI] [PubMed] [Google Scholar]
- 5••.Kamenz J, Hauf S. Time to split up: dynamics of chromosome separation. Trends Cell Biol. 2017;27:42–54. doi: 10.1016/j.tcb.2016.07.008. A recent review on chromosome segregation and separase function. [DOI] [PubMed] [Google Scholar]
- 6.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 8.Buonomo SBC, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I dependes on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- 9.Waizenegger IC, Hauf S, Meinke A, Peters J-M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 10.Hauf S, Waizenegger IC, Peters J-M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 11.Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell. 2010;18:397–409. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 14.Zhang N, Scorsone K, Ge G, Kaffes CC, Dobrolecki LE, Mukherjee M, Lewis MT, Berg S, Stephan CC, Pati D. Identification and characterization of separase inhibitors (Sepins) for cancer therapy. J Biomol Screen. 2014;19:878–889. doi: 10.1177/1087057114520972. [DOI] [PubMed] [Google Scholar]
- 15•.Zhang N, Pati D. Biology and insights into the role of cohesin protease separase in human malignancies. Biol Rev. 2017 doi: 10.1111/brv.12321. A recent review on the involvement of separase in tumor development. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan M, Lehane C, Uhlmann F. Orchestrating anaphase and mitotic exit: Separase cleavage and localization of Slk19. Nat Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 18.Lee K, Rhee K. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle. 2012;11:2476–2485. doi: 10.4161/cc.20878. [DOI] [PubMed] [Google Scholar]
- 19.Matsuo K, Ohsumi K, Iwabuchi M, Kawamata T, Ono Y, Takahashi M. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr Biol. 2012;22:915–921. doi: 10.1016/j.cub.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 20.Nagao K, Adachi Y, Yanagida M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature. 2004;430:1044–1048. doi: 10.1038/nature02803. [DOI] [PubMed] [Google Scholar]
- 21.Richie CT, Bembenek JN, Chestnut B, Furuta T, Schumacher JM, Wallenfang M, Golden A. Protein phosphatase 5 is a negative regulator of separase function during cortical granule exocytosis in C. elegans. J Cell Sci. 2011;124:2903–2913. doi: 10.1242/jcs.073379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cipressa F, Morciano P, Bosso G, Mannini L, Galati A, Daniela Raffa G, Cacchione S, Musio A, Cenci G. A role for separase in telomere protection. Nat Commun. 2016;7:10405. doi: 10.1038/ncomms10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorr IH, Boos D, Stemmann O. Mutual inhibition of separase and Cdk1 by two-step complex formation. Mol Cell. 2005;19:135–141. doi: 10.1016/j.molcel.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Puig N, Veprintsev DB, Fersht AR. Human full-length securin is a natively unfolded protein. Prot Sci. 2005;14:1410–1418. doi: 10.1110/ps.051368005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Csizmok V, Felli IC, Tompa P, Banci L, Bertini I. Structural and dynamic characterization of intrinsically disordered human securin by NMR spectroscopy. J Amer Chem Soc. 2008;130:16873–16879. doi: 10.1021/ja805510b. [DOI] [PubMed] [Google Scholar]
- 26.Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- 27.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 28.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 29.Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters J-M, Kinzler KW, Vogelstein B, Lengauer C. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 30.Hornig NCD, Knowles PP, McDonald NQ, Uhlmann F. The dual mechanism of separase regulation by securin. Curr Biol. 2002;12:973–982. doi: 10.1016/s0960-9822(02)00847-3. [DOI] [PubMed] [Google Scholar]
- 31.Waizenegger IC, Gimenez-Abian JF, Wernic D, Peters J-M. Regulation of human separase by securin binding and autocleavage. Curr Biol. 2002;12:1368–1378. doi: 10.1016/s0960-9822(02)01073-4. [DOI] [PubMed] [Google Scholar]
- 32.Nagao K, Yanagida M. Securin can have a separase cleavage site by substitution mutations in the domain required for stabilization and inhibition of separase. Genes to Cells. 2006;11:247–260. doi: 10.1111/j.1365-2443.2006.00941.x. [DOI] [PubMed] [Google Scholar]
- 33.Hellmuth S, Pohlmann C, Brown A, Bottger F, Sprinzl M, Stemmann O. Positive and negative regulation of vertebrate separase by Cdk1-cyclin B1 may explain why securin is dispensable. J Biol Chem. 2015;290:8002–8010. doi: 10.1074/jbc.M114.615310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–726. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- 35.Boos D, Kuffer C, Lenobel R, Korner R, Stemmann O. Phosphorylation-dependent binding of cyclin B1 to a Cdc6-like domain of human separase. J Biol Chem. 2008;283:816–823. doi: 10.1074/jbc.M706748200. [DOI] [PubMed] [Google Scholar]
- 36•.Hellmuth S, Rata S, Brown A, Heidmann S, Novak B, Stemmann O. Human chromosome segregation involves multi-layered regulation of separase by the peptidyl-prolyl-isomerase Pin1. Mol Cell. 2015;58:495–506. doi: 10.1016/j.molcel.2015.03.025. A role for Pin1 and prolyl cis-trans isomerization in the regulation of separase by Cdk1-cyclin B1 and securin. [DOI] [PubMed] [Google Scholar]
- 37.Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 38.Cohen-Fix O, Peters J-M, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 39.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 40.Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 41.Peters J-M. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 42.Nabti I, Grimes R, Sarna H, Marangos P, Carroll J. Maternal age-dependent APC/C-mediated decrease in securin causes premature sister chromatid separation in meiosis II. Nat Commun. 2017;8:15346. doi: 10.1038/ncomms15346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou H, Stemmann O, Anderson JS, Mann M, Kirschner MW. Anaphase specific auto-cleavage of separase. FEBS Lett. 2002;528:246–250. doi: 10.1016/s0014-5793(02)03238-6. [DOI] [PubMed] [Google Scholar]
- 44.Herzig A, Lehner CF, Heidmann S. Proteolytic cleavage of the THR subunit during anaphase limits Drosophila separase function. Genes Dev. 2002;16:2443–2454. doi: 10.1101/gad.242202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chestukhin A, Pfeffer C, Milligan S, DeCaprio JA, Pellman D. Processing, localization, and requirement of human separase for normal anaphase progression. Proc Natl Acad Sci USA. 2003;100:4574–4579. doi: 10.1073/pnas.0730733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papi M, Berdougo E, Randall CL, Ganguly S, Jallepalli PV. Multiple roles for separase auto-cleavage during the G2/M transition. Nat Cell Biol. 2005;7:1029–1035. doi: 10.1038/ncb1303. [DOI] [PubMed] [Google Scholar]
- 47.Holland AJ, Bottger F, Stemmann O, Taylor SS. Protein phosphatase 2A and separase form a complex regulated by separase autocleavage. J Biol Chem. 2007;282:24623–24632. doi: 10.1074/jbc.M702545200. [DOI] [PubMed] [Google Scholar]
- 48.Hellmuth S, Bottger F, Pan C, Mann M, Stemmann O. PP2A delays APC/C-dependent degradation of separase-associated but not free securin. EMBO J. 2014;33:1134–1147. doi: 10.1002/embj.201488098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarwal R, Cohen-Fix O. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 2002;16:1371–1382. doi: 10.1101/gad.971402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Luo S, Tong L. Molecular mechanism for the regulation of yeast separase by securin. Nature. 2017;542:255–259. doi: 10.1038/nature21061. Crystal structures of the yeast separase-securin complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Lin Z, Luo X, Yu H. Structural basis of cohesin cleavage by separase. Nature. 2016;532:131–134. doi: 10.1038/nature17402. Crystal structures of the active protease segment of C. thermophilum separase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jager H, Herzig A, Lehner CF, Heidmann S. Drosophila separase is required for sister chromatid separation and binds to PIM and THR. Genes Dev. 2001;15:2572–2584. doi: 10.1101/gad.207301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jager H, Herzig B, Herzig A, Sticht H, Lehner CF, Heidmann S. Structure predictions and interaction studies indicate homology of separase N-terminal regulatory domains and Drosophila THR. Cell Cycle. 2004;3:182–188. [PubMed] [Google Scholar]
- 54••.Borland A, Martin TG, Zhang Z, Yang J, Bai X-C, Chang L, Scheres SHW, Barford D. Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution. Nat Struct Mol Biol. 2017;24:414–418. doi: 10.1038/nsmb.3386. EM structure of the C. elegans separase-securin complex and low-resolution EM structure of human separase-securin complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viadiu H, Stemmann O, Kirschner MW, Walz T. Domain structure of separase and its binding to securin as determined by EM. Nat Struct Mol Biol. 2005;12:552–553. doi: 10.1038/nsmb935. [DOI] [PubMed] [Google Scholar]
- 56.Bachmann G, Richards MW, Winter A, Beuron F, Morris E, Bayliss R. A closed conformation of the Caenorhabditis elegans separase-securin complex. Open Biol. 2016;6:160032. doi: 10.1098/rsob.160032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holland AJ, Taylor SS. Cyclin-B1-mediated inhibition of excess separase is required for timely chromosome disjunction. J Cell Sci. 2006;119:3325–3336. doi: 10.1242/jcs.03083. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan M, Hornig NCD, Porstmann T, Uhlmann F. Studies on substrate recognition by the budding yeast separase. J Biol Chem. 2004;279:1191–1196. doi: 10.1074/jbc.M309761200. [DOI] [PubMed] [Google Scholar]
- 59.Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 60.Wei Y, Fox T, Chambers SP, Sintchak J, Coll JT, Golec JMC, Swenson L, Wilson KP, Charifson PS. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol. 2000;7:423–432. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 61.Eichinger A, Beisel HG, Jacob U, Huber R, Medrano FJ, Banbula A, Potempa J, Travis J, Bode W. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 1999;18:5453–5462. doi: 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi Y. Caspase activation: revisiting the induced proximity model. Cell. 2004;117:855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 63.McLuskey K, Mottram JC. Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem J. 2015;466:219–232. doi: 10.1042/BJ20141324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun Y, Kucej M, Fan HY, Yu H, Sun QY, Zou H. Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner. Cell. 2009;137:123–132. doi: 10.1016/j.cell.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]