Abstract
A single Atoh1 basic-helix-loop-helix transcription factor specifies multiple neuron types in the mammalian cerebellum and anterior hindbrain. The zebrafish genome encodes three paralagous atoh1 genes whose functions in cerebellum and anterior hindbrain development we explore here. With use of a transgenic reporter, we report that zebrafish atoh1c-expressing cells are organized in two distinct domains that are separated both by space and developmental time. An early isthmic expression domain gives rise to an extracerebellar population in rhombomere 1 and an upper rhombic lip domain gives rise to granule cell progenitors that migrate to populate all four granule cell territories of the fish cerebellum. Using genetic mutants we find that of the three zebrafish atoh1 paralogs, atoh1c and atoh1a are required for the full complement of granule neurons. Surprisingly, the two genes are expressed in non-overlapping granule cell progenitor populations, indicating that fish use duplicate atoh1 genes to generate granule cell diversity that is not detected in mammals. Finally, live imaging of granule cell migration in wildtype and atoh1c mutant embryos reveals that while atoh1c is not required for granule cell specification per se, it is required for granule cells to delaminate and migrate away from the rhombic lip.
Keywords: Granule cell, Atoh1, Zebrafish, Cerebellum
INTRODUCTION
The cerebellum is well known for its importance in motor coordination necessary for the generation of smooth and skillful movements (Leto et al., 2015). The vertebrate cerebellum has a deeply conserved neuronal circuitry composed of excitatory glutamatergic granule cells (GCs) and inhibitory GABAergic Purkinje cells (PCs) organized into a three-layered structure consisting of a deep GC layer, an overlying PC layer, and a superficial molecular layer where GC axons bifurcate to form parallel fibers that synapse with PC dendrites (Altman and Bayer, 1997; Butts et al., 2014; Hashimoto and Hibi, 2012; Leto et al., 2015)
In mammals, GC progenitors arise in the dorsal-most anterior hindbrain, a region called the upper rhombic lip (URL), under the control of the bHLH transcription factor Atoh1 (Ben-Arie et al., 1997). While GCs are the most numerous atoh1 derivatives, genetic lineage tracing in the mouse has demonstrated that atoh1-expressing progenitors give rise sequentially to diverse excitatory neuron types within the anterior hindbrain and cerebellum from 9.5 to 19 days post conception. Early Atoh1 derivatives (E10-5-12.5) contribute to a number of cerebellar nuclei in the tegmentum while later derivatives (E13.5 onward) generate GCs (Akazawa et al., 1995; Ben-Arie et al., 1997; Ben-Arie et al., 2000; Bermingham et al., 2001; Englund et al., 2006; Gray, 2008; Green et al., 2014; Machold and Fishell, 2005; Rose et al., 2009; Wang et al., 2005). Further diversity within the atoh1 GC lineage has been suggested based on the finding that the first atoh1-derived GCs (E12.5) contribute to the anterior cerebellar lobes while later atoh1-derived GCs (E15.5 and E16.5) contribute to progressively more posterior lobes (Machold and Fishell, 2005). However no molecular or functional criteria have confirmed these distinctions (Consalez and Hawkes, 2012).
The origins and circuitry of cerebellar neurons is conserved in zebrafish with some differences (Kani et al., 2010). Zebrafish GCs migrate from the URL to populate the Corpus Cerebelli (CCe), a structure homologous to the mammalian cerebellar vermis, which is thought to control body positioning, and into an anterior progenitor zone, the Valvula (Va). GCs in fish also migrate laterally from the URL into the eminentia granularis (EG), which supplies parallel fibers to a cerebellar-like structure in the hindbrain, the Medial Octavolateralis Nucleus (MON), and to the caudal Lobus Caudalis (LCa), homologous to the mammalian nodulus, to control motor coordination in response to vestibular information (See schematic in Fig. 1) (Kani et al., 2010; Kaslin et al., 2009; Matsui et al., 2014; Volkmann et al., 2010; Volkmann et al., 2008).
Figure 1. atoh1 gene expression in the developing zebrafish cerebellum.
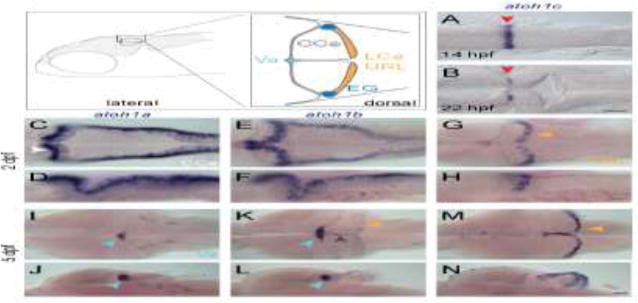
Schematic depicts regions of the zebrafish cerebellum in lateral (left panel) and dorsal (right panel) views at 5 dpf. Throughout the manuscript, white arrowheads indicate the CCe, light blue arrowheads indicate the Va, orange arrowhead indicate the URL (at early stages) or the LCa (at later stages) and dark blue arrowheads indicate the EG. Red arrowheads indicate the position of the MHB. A–N: RNA in situ hybridization in wild-type embryos with atoh1a (left column), atoh1b (middle column), and atoh1c (right column) at 14 hpf (A) 22 hpf (B), 2 dpf (C–H), and 5 dpf (I–N). Dorsal (A,B,C,E,G,I,K,M) or lateral (D,F,H,J,L,N) views are shown with anterior to the left. Gray arrowhead indicates midline of CCe. CCe, corpus cerebelli; EG, eminentia granularis; LCa, lobus caudalis cerebelli; URL, upper rhombic lip; Va, valvula cerebelli; Scale bars: 50 uM.
Zebrafish have three atoh1 genes: atoh1a, 1b, and 1c, which are expressed in overlapping but distinct progenitor domains within the rhombic lip. Initial studies have described the expression of the three zebrafish atoh1 genes, and lineage tracing of atoh1a-expressing progenitors showed that they give rise to diverse neuronal cell types including tegmental neurons, cerebellar output neurons and GCs in the CCe and Va (Chaplin et al., 2010; Kani et al., 2010). The absence of atoh1a-derived GCs in the EG and LCa suggested that other atoh1 genes may be responsible for GC diversity in fish, however this has not been further explored.
We sought to discover the role of zebrafish atoh1 genes in the generation of neuronal diversity in the cerebellum, and to take advantage of the live imaging possible in zebrafish to study how atoh1 genes control cerebellar progenitor migration and differentiation at high spatial and temporal resolution. Using single and compound mutants we describe a predominant role for atoh1c and a lesser role for atoh1a in the specification of cerebellar GCs in zebrafish. Using long-lived atoh1a and atoh1c reporters to follow their derivatives through larval development, we find that atoh1a and atoh1c specify non-overlapping GC and tegmental populations, indicating that fish use multiple atoh1 genes rather than using a single atoh1 gene over an extended developmental time to generate excitatory neuron diversity in the cerebellum, and revealing an unexpected diversity within the GC lineage. With the use of live imaging, we discovered that atoh1c expression at the rhombic lip is required not for cell cycle exit or initial GC differentiation but for GC progenitors to delaminate from the URL epithelium and initiate migration, a critical early event in neurogenesis.
RESULTS
Zebrafish atoh1 genes are expressed sequentially at the URL
All three atoh1 homologues are expressed in the URL beginning from 1 day post fertilization (dpf) and persist beyond 5 dpf (Fig. 1) (Chaplin et al., 2010; Kani et al., 2010). In addition, we identified an early transient atoh1c expression domain at the mid-hindbrain boundary (MHB), anterior to the presumptive URL. This expression domain is detected starting at 14 hpf (hours post fertilization) until 30 hpf (Fig. 1A,B) at which point it is extinguished and expression in the URL begins at 2 dpf and persists until our analysis end point, 5 dpf (Fig. 1G,H,M,N). Both atoh1a and atoh1b are expressed in the URL beginning at 24 hpf (not shown), but by 5 dpf their expression is largely restricted to the Va, a progenitor zone in the anterior-most region of the cerebellum (Fig. 1C–F, I–L) (Kani et al., 2010; Kaslin et al., 2009). Although atoh1b is primarily expressed in the Va region at 5 dpf, weak expression in also detected in the upper rhombic lip and at the midline of CCe (Fig. 1K,L, gray and orange arrowheads) where atoh1c is strongly expressed (Fig. 1M,N).
An early isthmic domain of atoh1c expression gives rise to tegmental neurons
We were intrigued by the early MHB expression domain of atoh1c in light of recent work that described a novel early Atoh1 domain at the MHB in chick and mouse that gives rise to an early population of tegmental neurons (Green et al., 2014). We considered whether atoh1c expression at the MHB represents an evolutionarily conserved progenitor population that gives rise to tegmental neurons. In order to visualize the atoh1c-derived cell populations in vivo, we generated an atoh1c transgenic reporter, TgBAC(atoh1c:gal4FF)fh430 by BAC recombineering (Fig. 2A–L) (Bussmann and Schulte-Merker, 2011). When crossed to Tg(UAS:kaede)s1999t (Scott et al., 2007), our TgBAC(atoh1c:gal4FF)fh430 driver (hereafter referred to as Tg(atoh1c∷kaede) for simplicity) recapitulates all aspects of endogenous atoh1c expression at the MHB (Fig. 2A) and subsequently in GCs (Fig. 2G). Taking advantage of the long-lived nature of the photoconvertible Kaede fluorescent protein (Ando et al., 2002; Caron et al., 2008) we were able to follow the fate of the MHB atoh1c+ progenitor pool after atoh1c mRNA expression at the MHB was extinguished. Starting at 20 hpf, the MHB Tg(atoh1c∷kaede)+ population migrates ventrocaudally to rhombomere 1 (r1) ventral to the presumptive cerebellum (the tegmentum; Fig. 2C) and gives rise to ventral bilateral comma-shaped nuclei consisting of about 20 neurons by 2 dpf (Fig. 2E,H,K – gray arrowheads). Since the onset of atoh1c expression at the URL at 3 dpf complicates the subsequent lineage analysis of MHB-derived atoh1c neurons (see below for further discussion of atoh1c URL derivatives), we distinguished the MHB-derived atoh1c lineage by photoconverting the Kaede+ MHB domain at 22 hpf, before the onset of URL expression, and found that the majority of neurons present in ventral r1 are Kaedered indicating that they originated from the atoh1c+ MHB progenitor domain before this stage (Fig. 2M–P).
Figure 2. atoh1c-expressing progenitors give rise to to ventral r1 and cerebellar granule neurons.
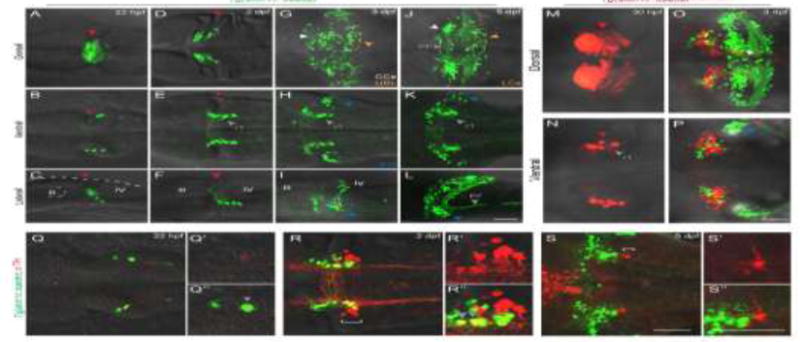
A–L: Tg(atoh1c∷kaede) transgene expression in fixed embryos stained with anti-Kaede antibody in dorsal (A,D,G,J), ventral (B,E,H,K) and lateral (C,F,I,L) views at 22 hpf (A–C), 2 dpf (D–F), 2 dpf (G–I) and 5 dpf (J–L). Arrowheads follow the color code described in Fig. 1 legend or are labeled as follows: PF: parallel fibers, r1: MHB-derived neurons in ventral r1. M–P: live imaging after photoconversion of Kaede (green to red) of MHB atoh1c+ cells at 22 hpf confirms that this Tg(atoh1c∷kaede)+ progenitor domain gives rise to ventral r1 neurons. Dorsal (M) and ventral focal planes (N–P). R–T: Tg(atoh1c∷kaede)+ cells (green) transiently express TH (red; gray arrowheads) from 22 hpf to 2 dpf and lie adjacent to the LC (indicated by white bracket). All images oriented with anterior to the left at time points as indicated. Scale bars: 50 μM.
The migratory path from the MHB of these atoh1c+ neurons and their final position in r1 are highly reminiscent of the Locus Coeruleus (LC) (Chiu and Prober, 2013; Guo et al., 1999)). The LC is a noradrenergic neuronal population present in all vertebrates that controls arousal. At 2 and 5 dpf, the Tg(atoh1c∷kaede)+ neurons lie immediately anterior to the LC, marked by tyrosine hydroxylase (TH) expression, and have overlapping contralateral and longitudinal projections with them (Fig. 2R,S) (Guo et al., 1999). Furthermore, the Tg(atoh1c∷kaede)+ r1 neurons express the biosynthetic enzyme tyrosine hydroxylase (TH) during their migration (Fig. 2Q,R, gray arrowheads), However, the atoh1c+ r1 neurons are not the LC themselves, as they eventually turn off TH while the neurons of the LC maintain it (Fig. 2S). Given the similarities between these two populations, we hypothesize that the Tg(atoh1c∷kaede)+ r1 neurons may be functionally connected to the arousal circuit.
The MHB is an important signaling center required for early development of both the midbrain and hindbrain (Rhinn and Brand, 2001). The position of the MHB lies at the interface of Otx2 and Gbx2 and is reciprocally maintained by Wnt1 expression anterior to the boundary and Fgf8 expression posterior to the boundary. Double RNA in situ hybridization with otx2, gbx2, fgf8a and wnt1 demonstrated that atoh1c is expressed in a few cells immediately posterior to the boundary (Fig. 3A–D). Not surprisingly, MHB atoh1c expression is lost under conditions where FGF or Wnt signaling are blocked early in development by heat-inducible expression of dominant negative (dn) FGFR1 or dnTCFΔC, respectively (Fig. 3E–G) (Lee et al., 2005; Martin and Kimelman, 2012). The MHB domain of Atoh1 described in the chick similarly requires Fgf8 (Green et al., 2014). To investigate the nature of this requirement, made chimeras in which cells from Tg(hs:dnFGFR1);Tg(atoh1c∷Kaede) or Tg(hs:dnTCFΔC); Tg(atoh1c∷Kaede) embryos contributed sparsely to the MHB region of a non-transgenic host embryo. After heat-shock, dn-expressing cells in these chimeras are cell-autonomously blocked for reception of FGF or Wnt signals, but the MHB morphogenetic program occurs normally in the surrounding wild-type cells. Whereas non-dn-expressing cells at the MHB expressed KaedeRed (n=10/10 chimeric embryos; Fig. 3H), cells expressing dnFGFR1 or TCFΔC contributed to the MHB but never expressed Kaede (n=0/32 chimeric embryos; Fig. 3I,J). Thus, atoh1c expression is tightly restricted to progenitors at the MHB because of a cell-autonomous requirement for both Wnt and FGF signaling to initiate atoh1 expression; in other words, atoh1c is a “coincidence detector” for Wnt and Fgf signals.
Figure 3. Specification of the atoh1c MHB domain.
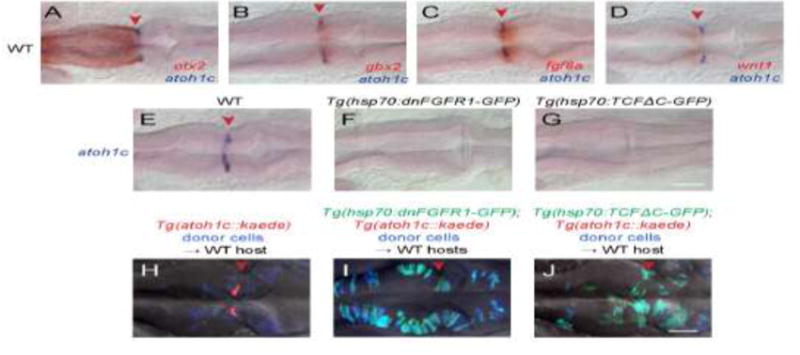
A–D: Double RNA in situs for atoh1c (blue) and otx2 (A), gbx2 (B), fgf8 (C), and wnt1 (D) (red) show that the atoh1c expression in a few cells immediately posterior to the MHB. E–G: atoh1c expression is absent when Fgf (F) or Wnt (G) signaling is blocked with heat-inducible dominant-negative transgenes activated at 10 hpf. H–J: atoh1c expression requires cell-autonomous Fgf and Wnt signaling. Live imaging of chimeras with donor-derived cells (blue) expressing dn-FGFR1 (I, green) or dn-TCFΔC (J, green) that are unable to express Tg(atoh1c∷kaede) (red in the control chimera in H) even if they lie at the MHB (red arrowheads). All embryos are at 22 hpf and are shown in dorsal views with anterior to left. Scale bars: 50 μM.
In addition to regulation by Wnt and Fgf signaling pathways, we have found that the early atoh1c expression domain is subject to Notch-mediated lateral inhibition. This well-studied process starts when a proneural gene non-autonomously limits its own expression by transcriptional activation of Delta, which in turn stimulates Notch in adjacent cells to inhibit proneural gene expression there. This process is amplified through a field a cells until a single progenitor expresses high levels of the proneural gene and acquires the “primary” cell fate – in the nervous system this is the neuronal fate (Artavanis-Tsakonas et al., 1999). When we pharmacologically inhibit Notch signaling with γ-secretase inhibitor DAPT (Geling et al., 2002) we find that the atoh1c expression domain is expanded to approximately double the number of expressing cells compared to vehicle-treated controls at 22 hpf (Suppl. Fig. 1). The expansion of the atoh1c expression domain in DAPT-treated embryos results in the generation of ectopic tegmental neurons which are visualized using Tg(atoh1c∷kaede) transgenic line. Atoh1c expression is still restricted to the MHB because of its requirement for MHB-derived Wnt and Fgf signaling. Our results indicate a proneural function for atoh1c at the MHB and its regulation by classical lateral inhibition.
Atoh1c is required for the specification of multiple granule cell populations in the zebrafish cerebellum
By 3 dpf, the majority of the Tg(atoh1c∷kaede)+ cells in the cerebellum have migrated to regions populated by GCs: the CCe, EG and LCa (Fig. 2G–L, Movie 1). Their axons appear starting at 3.5 dpf and resemble GC parallel fibers (Hibi and Shimizu, 2012; Takeuchi et al., 2015). In addition to the parallel fibers across the surface of the cerebellum (Fig. 2J, gray arrowhead), GCs of the EG and LCa also project parallel fibers out of the cerebellum to a cerebellar-like structure in the dorsal hindbrain, the MON (Fig. 2L, gray arrowhead) (Bae et al., 2009; Takeuchi et al., 2015). Given the location and projections of the Tg(atoh1c∷kaede)+ population, we conclude that they are GCs of the CCe, EG, and LCa. atoh1c-derived GCs are not detected in the Va, where many atoh1a-derived cells are located (Kani et al., 2010). Interestingly, we detected no overlap in Tg(atoh1c∷kaede) and Tg(atoh1a:dtomato)-expressing cells in double transgenic fish (Fig. 4) as well as by double RNA in situ hybridization (not shown). We conclude that the Tg(atoh1a:EGFP)+ GCs within the CCe described by Kani et al. (Kani et al., 2010) represent a population of GCs distinct from the majority Tg(atoh1c∷kaede)+ population. It remains to be seen whether these atoh1a+ and atoh1c+ GC populations do indeed play functionally distinct roles within the cerebellum.
Figure 4. atoh1a- and atoh1c-derived neurons specify distinct progenitor pools.
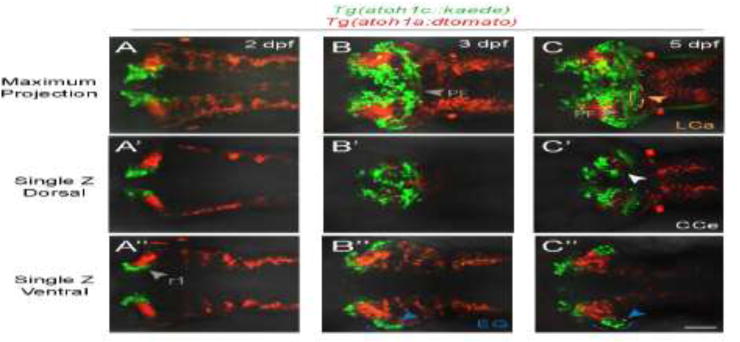
Live imaging of wild-type embryos with Tg(atoh1c∷kaede) (green) and Tg(atoh1a:dtomato) (red) transgenes indicate atoh1a and atoh1c-derived neurons are distinct cerebellar populations. Maximum projections of a z-stack (A–C) and single z-slices at dorsal (A′–C′) or ventral (A″–C″) focal planes with anterior to the left. A: 2 dpf, B: 3 dpf, C: 5 dpf. Scale bar: 50 μM.
To directly assess the function of Atoh1c, we used transcription activator-like effector nucleases (TALENs) to generate a 122 bp deletion that results in a premature stop codon before the Atoh1c DNA binding domain (atoh1cfh367). Expression of cerebellin12 (cbln12), a marker of mature GCs (Takeuchi et al., 2016), is strongly reduced in the CCe, EG and LCa in atoh1c mutants (Fig. 5A,B). Expression of the vesicular glutamate transporter 1 (vglut1) mRNA and protein, in glutamatergic neurons in the cerebellum, is similarly reduced (Fig. 5C,D and data not shown). The majority of the Tg(atoh1c∷kaede)+ cells accumulate at the URL in atoh1cfh367 mutants, where they strongly express atoh1c mRNA (Fig. 5I,J), retain a neural progenitor-like morphology, and elaborate comparatively few parallel fibers (Fig. 5E–H, Movie 2). Thus atoh1c contributes to the specification of GCs located in the CCe, LCa, and EG.
Figure 5. atoh1c is required for cerebellar granule cell fate.
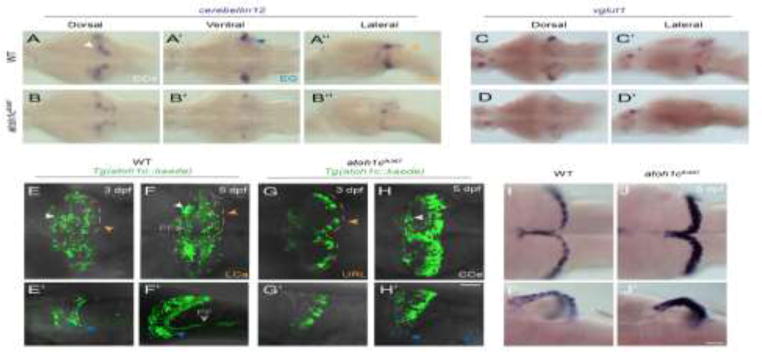
A–D: Reduction in the number of mature GCs in atoh1c mutant as indicated by decreased staining for cerebellin12 (B) and vglut1 (D), at 5 dpf. E–H: The majority of mutant cells expressing the Tg(atoh1c∷kaede) remain as an non-migrated population in the URL at 3 dpf (G, G′ compare to wild-type in E, E′) and 5 dpf (H, H′ compare to wild-type in F, F′). I–J: Upregulation of atoh1c mRNA in URL at 5 dpf in atoh1c mutant embryos (J, J′ compare to I, I′). Dorsal (A,B,C,D,E,F,G,H,I,J), ventral (A′,B′) and lateral (A″,B″,C′,D′,E′,F′,G′,H′,I′,J′) views are shown with anterior to the left. Scale bars: 50 μM.
If atoh1c is required for the development of GCs, then we predict one or more aspects of GC maturation are defective in the mutant. Using markers of post-mitotic neurons (HuC/D; (Kim et al., 1996)), committed GC precursors (NeuroD1; (Kani et al., 2010)) and cell proliferation (EdU incorporation) we determined the differentiation state of the atoh1cfh367 cells (Fig. 6). It is important to note that due to epigenetic silencing of the UAS element (Akitake et al., 2011), not all of the atoh1c-derived GCs are detectable in a given fish using the atoh1c∷kaede transgene. In 5 dpf wild-type larvae, the majority of Tg(atoh1c∷kaede)+ cells have migrated away from their birthplace at the URL although a small population of HuC/D+ (Fig. 6A), NeuroD1+ (Fig. 6C) GCs remain near the URL in the LCa. In contrast, in the atoh1c mutant, the majority of the Tg(atoh1c∷kaede)+ cells remain at the URL but do not express HuC/D or elaborate axons (Fig. 6B), but they do express NeuroD1 (Fig. 6D) suggesting that they are committed but undifferentiated GC precursors. To determine whether these are proliferating progenitors, we exposed 3 dpf larvae to 5-ethynyl-2′-deoxyuridine (EdU) continually for two days before fixing at 5 dpf (Fig 6E,F). Any cells that were proliferating during that period will incorporate EdU. In the atoh1c mutant, the massively expanded Tg(atoh1c∷kaede)+ population in the URL is post-mitotic during this period (Fig. 6F). This post-mitotic but undifferentiated progenitor state with strongly increased atoh1c mRNA expression in atoh1c mutants (Fig. 5I–J) is consistent with a classical proneural function for Atoh1c in the URL, whereby Atoh1c promotes GC differentiation cell-autonomously and inhibits it in neighboring cells through upregulation of Notch ligand(s) (Artavanis-Tsakonas et al., 1999). A classical proneural function for Atoh1 in the URL as has previously been proposed (Gazit et al., 2004; Machold et al., 2007; Millimaki et al., 2007).
Figure 6. atoh1cfh367 cerebellar cells accumulate as post-mitotic but undifferentiated granule cell progenitors.
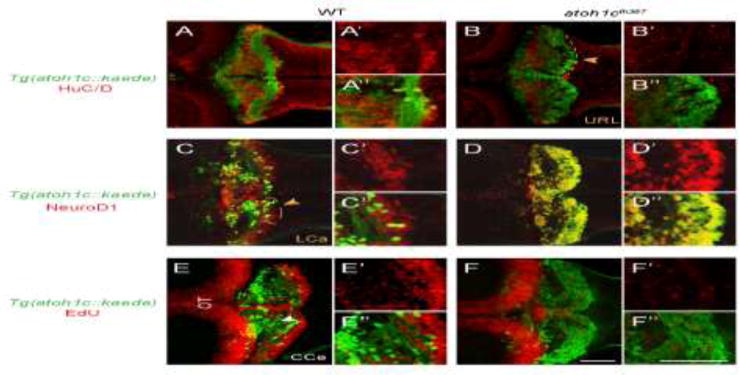
A,B: The majority of Tg(atoh1c∷kaede)+ cells (green) in the atoh1c mutant are located within the URL and do not express HuC/D (red) at 5 dpf (B) indicating that they are not post-mitotic neurons. C,D: atoh1cfh367 cells (green) in the URL are positive for NeuroD1 (red, D). E,F: atoh1cfh367 cells (green) in the URL do not incorporate EdU (red, F). Strong EdU incorporation anterior to the cerebellum in both wild-type and mutant is in the optic tectum (OT). Dorsal views with anterior to the left. Scale bars: 50 μM.
Atoh1c is required for delamination from the URL
Loss of atoh1c results in the accumulation atoh1c-expressing GC precursors in the URL that are unable to terminally differentiate. How does a differentiation-arrested committed precursor behave? In order to address this, we performed high-resolution time-lapse imaging of both wild-type and atoh1cfh367 embryos at 3 dpf, a time when we can continuously capture the birth and migration of GCs in wild-type embryos. Before migrating, wild-type atoh1c-expressing progenitors are epithelial, with their apical end feet along the ventricle-facing surface of the URL. They elaborate a basal process and very soon thereafter they release their apical contact and move away from the URL following this process (Koster and Fraser, 2001; Volkmann et al., 2010). This transition is accomplished in a period of six hours (Fig. 7A–D, Movie 3). In contrast, atoh1c mutant progenitors at the URL elaborate highly dynamic basal processes but fail to detach from the epithelium during the same period (Fig. 7E–H, Movie 4). Interestingly, “escaper” neurons in atoh1c mutants complete GC differentiation and elaborate parallel fibers (Fig. 5F, cells in CCe). Our observations suggest that Atoh1c promotes the release of the GC precursors from the URL epithelium, an essential step in neuronal differentiation (Hartenstein et al., 1992; Pacary et al., 2012).
Figure 7. Atoh1c is required for release of granule neuron progenitors from URL.
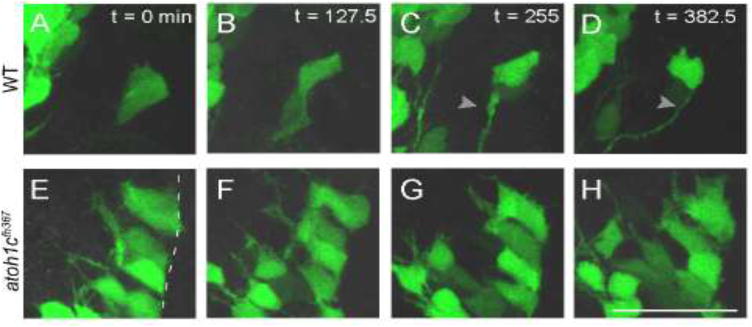
Still images from confocal timelapses taken at 3 dpf. Dotted line indicates the URL where the apical surfaces of GC progenitors attach. In wild-type, Tg(atoh1c∷kaede)+ progenitors detach from the URL over a six-hour interval (A–D) while in atoh1c mutants, Tg(atoh1c∷kaede)+ cells remain attached at the URL (E–H). Time points indicated in upper right corner in minutes. Scale bar: 25 μM.
Atoh1a is required for a subset of GCs that arise independently of atoh1c
Given that GCs were strongly reduced but not absent in atoh1c mutants (Fig. 5A–D) and that all three atoh1 paralogs are expressed in the URL (Fig. 1C–N) but that Tg(atoh1a:dtomato) and Tg(atoh1c∷kaede) mark different GC populations (Fig. 4), we were interested in understanding the functional relationships between the atoh1 paralogues. We generated mutant alleles for atoh1a and 1b: atoh1afh282 is a missense mutation within the bHLH DNA domain that causes the loss of sensory hair cells in the zebrafish lateral line (Pujol-Marti et al., 2012) and atoh1bfh473 is a 55 bp deletion that truncates the protein upstream of the bHLH domain. atoh1a and atoh1c are expressed independently of each other whereas the expression of atoh1b is regulated by atoh1c in regions of overlapping expression: the URL and midline of the CCe (not shown).
Loss of neither atoh1a or atoh1b had any detectable effect on cbln12 expression or on the migration and differentiation of Tg(atoh1c∷kaede)+ cells (Fig. 8B,C). It is likely that the remaining cbln12+ GCs in the atoh1cfh367 mutant (Fig. 8D′) are due to redundancy amongst atoh1 genes in GC differentiation. We therefore generated all possible atoh1−/− allelic combinations. We found that in atoh1afh282; atoh1cfh367 double mutant embryos there was a near-complete loss of cbln12 expression (Fig. 8E′). This was not further enhanced in the atoh1afh282; atoh1cfh367; atoh1bfh473 triple mutant (Fig. 8H′) nor did we observe additional GC defects in the atoh1bfh473;atoh1cfh367 double mutant (Fig. 8F) suggesting that atoh1b does not contribute detectably to GC development. Given that both atoh1a and atoh1c both contribute to cerebellin12 GC expression, but that Tg(atoh1a:dtomato) and Tg(atoh1c∷kaede) are expressed in non-overlapping GC populations (Fig. 4), we conclude that the two atoh1 genes specify distinct GC populations within the CCe. The function of atoh1a in GC specification is only detectable in the absence of the larger population of atoh1c-dependent GCs. This is consistent with previous lineage analysis that showed only a minority of GCs in the CCe are derived from atoh1a progenitors (Kani et al., 2010)(and Fig. 4 and 8).
Figure 8. atoh1a, atoh1b, and atoh1c have non-overlapping roles in cerebellar development.
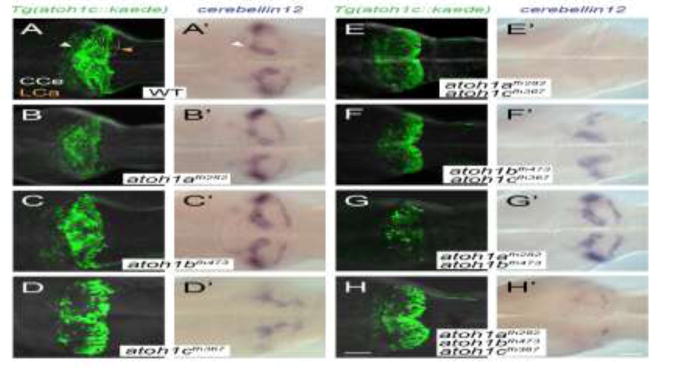
Images of 5 dpf wild-type (A), single (B–D), and compound (E–H) atoh1 mutants with the Tg(atoh1c:kaede) (left column) or cbln12 RNA in situ hybridization (right column). Dorsal views with anterior to the left. Scale bars: 50 uM.
atoh1a can rescue the atoh1cfh367 phenotype
A classic model for the maintenance of duplicated genes posits that regulatory changes render both genes essential for a subset of their pre-duplicated functions (Force et al., 1999). If this is the case for atoh1a and atoh1c, we predict that either atoh1a or atoh1c could rescue GC differentiation when expressed in the atoh1c progenitor domain in an atoh1cfh367 mutant. We sought to rescue the atoh1cfh367 phenotype by injection of a DNA construct encoding the full-length atoh1a cDNA under atoh1c regulation. We injected homozygous atoh1c mutant embryos carrying the atoh1c:gal4ff transgene at the one-cell stage with an UAS:atoh1a-p2a-mcherryCAAX construct or a positive (UAS:atoh1c-p2a-mcherryCAAX) or negative (UAS:mappleCAAX) control construct, and determined the position of mcherry or mapple-expressing cells at 4 dpf, using migration away from the URL as a proxy for GC differentiation. In these experiments, we defined “migrated” by both the location of the expressing cell as well as the presence of an axon, scoring any cells with an axon or cells located in the CCe or EG as “migrated”. We found that atoh1a (91% cells migrated; n=141 cells, 21 embryos) and atoh1c (99% cells migrated; n=105 cells, 12 embryos) were equally effective in rescuing the migration of atoh1c mutant GCs, compared to UAS:mappleCAAX alone (67% cells migrated; n=270 cells, 27 embryos) We conclude that atoh1a and atoh1c have equivalent functions in GC progenitors, at least with respect to promoting delamination from the URL, and that their distinct functions are the result of their different spatiotemporal regulation.
Discussion
Here we have defined the roles of atoh1 genes in zebrafish cerebellar development with the use of live imaging of transgenic reporters combined with mutant analysis. We find that of the three atoh1 genes in the zebrafish genome, atoh1c plays the most prominent role in the development of cerebellar GCs, both in the cerebellar corpus (CCe) required for postural control and in the caudolateral lobes (EG and LCa) involved in vestibular function. We show that atoh1c is required not for GC specification per se, but for GC progenitors to lose their epithelial character, migrate away from the upper rhombic lip, and terminally differentiate. Finally, we show that atoh1c functions as an Wnt and FGF “coincidence detector” at the mid-hindbrain boundary to specify an early population of neuronal progenitors that give rise to a ventral r1 neurons that transiently take on a noradrenergic fate.
More atoh1 genes for more granule cell diversity
Lineage tracing experiments in mouse have shown that atoh1-expressing progenitors give rise to a range of excitatory neuron types in the tegmentum and cerebellum (Machold and Fishell, 2005; Rose et al., 2009; Wang et al., 2005). The main determinant of which neuron is generated is developmental time: tegmental neurons are generated first, then deep cerebellar projection neurons, then GCs (Fig. 9); if changing signals in the URL environment are responsible for these changes in fate, they have yet to be identified. Our work, together with earlier lineage tracing experiments (Kani et al., 2010) show that zebrafish atoh1 genes generate similar neuronal populations and in addition generate a larger diversity of GC types (Fig. 9). An early population of atoh1c-expressing progenitors at the MHB generate tegmental neurons; Kani et al. showed that atoh1a-expressing progenitors generate cerebellar projection neurons (eurydendroid cells) that are equivalent of the deep cerebellar neurons of mammals, and finally both atoh1a and atoh1c-expressing progenitors at the URL generate a range of GC types.
Figure 9. atoh1-derivatives in zebrafish and mouse.
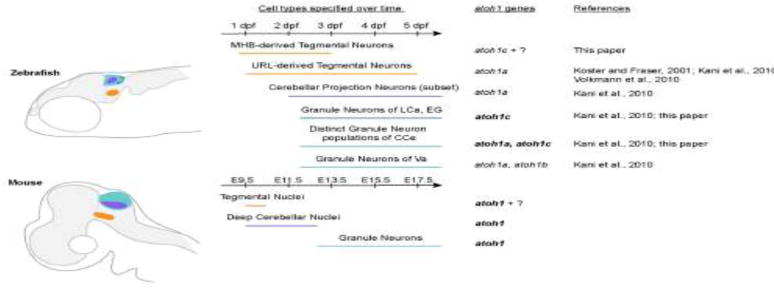
Schematic outlines atoh1-derivatives generated over time in the anterior hindbrain of zebrafish (top) and mouse (bottom). Bold text indicates gene has been shown to be required for development of the neuronal population. Homologous neuronal populations are indicated with the colored regions and lines. Dpf: days post fertilization; E: embryonic day; CCe, corpus cerebelli; EG, eminentia granularis; LCa, lobus caudalis cerebelli; Va, valvula cerebelli.
We consider this diversity of GC types first. Based on atoh1 expression patterns and lineage tracing of atoh1a+ cells in zebrafish which showed the presence of atoh1a-derived GCs in the Va and CCe but not the EG or LCa, Kani et al. predicted that atoh1c and/or atoh1b would be required for the GCs in these structures. Our work confirms this prediction: zebrafish atoh1c is required for the differentiation of EG and LCa GCs. Thus, additional atoh1 genes that arose during teleost evolution may have allowed for the formation of cerebellar structures such as the EG and the LCa that are not observed in mammals.
Unexpectedly, our work reveals an additional degree of GC diversity within the CCe, a homologous structure to the mammalian cerebellar vermis. We find that atoh1c is required for a population of GCs in the CCe that does not overlap with the atoh1a population, so that atoh1c and atoh1a together are required for the full complement of GCs in the CCe. Cerebellar GCs have been described as one singular population in vertebrates. The possibility of further diversity within the atoh1 GC lineage as suggested based on the finding that the earliest atoh1-derived GCs contribute to the anterior cerebellar lobes while later atoh1-derived GCs contribute to progressively more posterior lobes (Machold and Fishell, 2005). However no molecular or functional criteria have confirmed these distinctions (Consalez and Hawkes, 2012). Our data reveals distinct atoh1a and atoh1c populations within the CCe (Fig. 9). Both populations express the pan-GC marker cerebellin12, as do the EG and the LCa, and our overexpression experiment suggests that atoh1a can substitute for atoh1c in the atoh1c-derived population. While it is possible that the two populations function identically in the cerebellar circuit, in a recent RNA-Seq screen for GC-specific genes, Takeuchi et al. (Takeuchi et al., 2016) identified several GC markers that have distinct patterns of expression within the CCe, suggesting heterogeneity in that population. It remains to be determined whether these domains correspond to the atoh1a-derived and atoh1c-derived GCs. The use of long-lived transgenic lines generated in this study and in Kani et al. 2010 will allow for the careful dissection of the morphology and projection patterns of these two GC populations, which will yield further information about whether the two populations are functionally distinct within the cerebellar circuit.
atoh1 genes and the neuroepithelial progenitor state
We have observed that in the absence of atoh1c, progenitors fail to migrate away from the URL and differentiate into GCs. Instead, Tg(atoh1c∷kaede) cells accumulate at the URL in a post-mitotic, partially differentiated (NeuroD1-positive, HuC/D-negative) state. A similar accumulation of GC progenitors at the URL was detected in mouse atoh1 mutants (Ben-Arie et al., 1997; Ben-Arie et al., 2000). Our finding that non-migrated atoh1c mutant progenitors at the URL express NeuroD1 is interesting in light of the fact that in mice, GCs do not turn on NeuroD1 until after they complete their transit-amplifying divisions in the external GC layer (Miyata et al., 1999). The expression of NeuroD1 in undifferentiated GC progenitors at the URL in atoh1c mutants may reflect the finding that most GCs in fish differentiate directly after leaving the URL, without going through a transit-amplifying phase (Butts et al., 2014; Chaplin et al., 2010).
The accumulation of atoh1c+ cells is typical of classical Notch-dependent lateral inhibition, whereby proneural genes limit their own expression by transcriptional activation of Delta, which in turn stimulates Notch in adjacent cells to inhibit proneural gene expression (Artavanis-Tsakonas et al., 1999). We have shown that the early expression domain of atoh1c is subject to lateral inhibition indicated by an increase of the atoh1c-expressing population and resulting in the generation of ectopic tegmental neurons when Notch-signaling is pharmacologically inhibited (Suppl. Fig. 1). We observe an increase of atoh1c-expressing cells at the URL by RNA in situ hybridization in atoh1c−/− mutant embryos (Fig. 5I–J) which supports the idea that atoh1c-expressing progenitors at the URL are also subject to Notch-mediated lateral inhibition. The dramatic accumulation of Tg(atoh1c∷kaede) cells at the URL in the atoh1c−/− mutant embryos are likely due to two factors: the lack of migration of the atoh1c+ progenitors, which we believe is the largest contributor to the mutant phenotype, as well as the failure of Notch signaling and loss of lateral inhibition which would result in a larger population of atoh1c-expressing unable to proceed with maturation.
Our high-resolution live imaging of these cells in atoh1c mutants shows that they exhibit a migratory behavior, extending dynamic basal processes similar to wild-type GC progenitors do. However, while wild-type GC progenitors are able to release their apical contacts and migrate away from the URL, atoh1c mutant cells cannot release these contacts and remain trapped at the URL. Recent studies have shown that eliminating N-cadherin-containing apical junctions is a critical step in neuronal differentiation (Matsuda et al., 2016; Pacary et al., 2012; Rousso et al., 2012). Interestingly, atoh1c mutant progenitors that escape the URL appear to complete differentiation and generate parallel fibers, suggesting that a key function of atoh1c may be to promote apical detachment of neural progenitors, possibly by directly or indirectly repressing N-cadherin expression or activity.
atoh1c-expressing cells at the MHB boundary give rise to tegmental neurons
We have identified a novel population of atoh1c-derived neurons that lie in close association with the Locus Coeruleus, an evolutionarily ancient noradrenergic population involved in states of arousal and sleep-wake cycles. Like the LC neurons, the atoh1c neurons are specified immediately posterior to the MHB in cells that require the co-incident reception of Wnt and FGF signals. They migrate ventrally and caudally into ventral r1, express tyrosine hydroxylase, and elaborate rostral, caudal, and cross-midline projections. However, unlike the LC, they do not express subsequent enzymes required for norepinephrine synthesis such as dopamine beta-hydroxylase (DBH, data not shown) and they turn off TH expression soon after arriving in ventral r1.
Lineage tracing in mouse has revealed an array of atoh1-derivatives including a number of tegmental pontine and deep cerebellar nuclei that appear before GC precursors emerge from the URL (Fig. 9) (Machold and Fishell, 2005; Rose et al., 2009; Wang et al., 2005). These include TH-positive cholinergic neurons in the lateral parabrachial and pedunculopontine tegmental nuclei, which lie adjacent to the Locus Coeruleus, and are vital for arousal and attention and have been implicated in the generation of REM sleep (Rose et al., 2009). In a separate study, early-born atoh1-derived neurons in the peri-LC region were shown to regulate sleep states in mice (Hayashi et al., 2015). We hypothesize that the MHB population of LC-associated atoh1c neurons may play an ancient role in regulating states of arousal, possibly analogous to the lateral parabrachial or pedunculopontine tegmental nuclei. However, because these atoh1c+ r1 neurons persist in all atoh1 mutant combinations we have generated, we are as yet unable to assess their functions in mutant fish. Interestingly, Machold and Fishell also noted that some atoh1-derived tegmental neurons are present in atoh1 mouse mutants, suggesting the presence of compensating mechanisms for the specification of some Atoh1-derived neuronal populations in mouse.
Concluding remarks
We have shown that atoh1 genes contribute to the development of neuronal diversity in the zebrafish cerebellum, however we have not addressed the functions of atoh1 neurons in zebrafish behavior. Registering atoh1-derived populations with recent zebrafish neuroanatomical atlases will help to predict their functions (Marquart et al., 2015; Randlett et al., 2015), as will functional studies using genetically-encoded neuronal activity reporters under different behavioral paradigms (Muto and Kawakami, 2016). Surprisingly, atoh1c mutants are viable and do not exhibit obvious behavioral abnormalities as adults, however we anticipate that behaviors such as the vestibulo-ocular reflex, in which the eye rotates to compensate for changes in body position (Bianco et al., 2012), may be affected. Likewise, the hypothesized role of the atoh1c-expressing r1 neurons in arousal can be measured based on their activity in sleep paradigms (Chiu et al., 2016).
MATERIAL AND METHODS
Zebrafish Lines and Maintenance
Zebrafish (Danio rerio) were staged and maintained according to standard procedures as previously described (Kimmel et al., 1995). Experiments using zebrafish followed the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee standards and guidelines (IACUC#1392). All transgenic lines were maintained in the *AB background. Transgenic lines used in this study include Tg(UAS:kaede)s1999 (Davison et al., 2007), gift of the Baier lab, Tg(hsp70l:dnfgf1r-EGFP)pd1 (Lee et al., 2005), Tg(hsp70:TCFΔC-GFP)w74 (Martin and Kimelman, 2012), gifts of the Kimelman lab, Tg(atoh1a:EGFP)nns7 (Kani et al., 2010) and Tg(atoh1a:dTomato)nns8 (Wada et al., 2010), gifts of the Hibi lab.
Fgf and Wnt signaling inhibition
Fgf and Wnt signaling was blocked by incubating 10 hpf embryos containing the heat-inducible Tg(hsp70l:dnfgfr-EGFP)pd1 or Tg(hsp70:TCFΔC-GFP)w74 for 15 minutes at 38°C or 40°C, respectively (Lee et al., 2005; Martin and Kimelman, 2012).
Notch signaling inhibition
Notch signaling was blocked by incubating 4 hpf embryos in a final concentration of 50 μM DAPT (Sigma D5942) diluted in embryo media for the amount of time as indicated in the supplemental figure 1 legend. Vehicle control embryos were incubated in embryo media containing a final concentration of 0.5% DMSO for the same amount of time as DAPT-treated embryos.
Generation of mutant alleles
atoh1afh282 was generated by TILLING (Draper et al., 2004) and has been previous described (Pujol-Marti et al., 2012)…atoh1bfh473 was generated with use of CRISPR/Cas9 as outlined in (Shah et al., 2015). The CRISPR gRNA sequences for atoh1b were: 5′-GGCTGACCCGGAGTGACCCG and 5′-GGTGCGTGCGTAATTCTCCA. atoh1cfh367 was generated with the use of TALENs as outlined in (Sanjana et al., 2012). The TALEN target sequences for atoh1c were as follows: 5′-GGAGAGAGACTGACAGATC and 5′-CCAGCCCATTGGGGCTTT. In each experiment in this paper, embryos were phenotyped blind and then later genotyped by PCR using the following protocols: atoh1afh282: forward primer 5′-ATGGATGGAATGAGCACGGA and reverse primer 5′-GTCGTTGTCAAAGGCTGGGA followed by digestion with AvaI (New England Biolabs) generates a 195 bp + 180 bp WT allele and a 195 bp + 258 bp mutant allele; atoh1bfh473: forward primer 5′-TGGACACTTTCGGGAGGAGT and reverse primer 5-CTTCAGAGGCAGCTTGAGGG generates a 180 bp WT allele and a 125 bp mutant allele; atoh1cfh367: forward primer 5′-GACTCCCTGTGGTCATTATCAA and reverse primer 5′-AGCTCACTCAGGGTGCTGAT generates a 510 bp WT allele and a 388 bp mutant allele.
Plasmid construction and injection
TgBAC(atoh1c:gal4ff)fh430 was generated according to the protocol outlined in (Bussmann and Schulte-Merker, 2011) and using BAC recombineering plasmids provided by the Shulte-Merker lab. A BAC containing atoh1c (#38G3) was obtained from the BACPAC Resources Center at Children’s Hospital Oakland Research Institute. This BAC was recombineered to insert gal4ff 114bp downstream of the atoh1c ATG and to add tol2 arms flanking the genomic DNA insert. Tg(UAS:kaede)s1999 embryos were injected at the one-cell stage with 180 pg of the modified BAC along with 90 pg of tol2 transposase mRNA. Expressing embryos were raised to adulthood for the establishment of stable transgenic lines.
For the rescue experiments, the following constructs were generated using gateway cloning: UAS:atoh1a-p2a-mcherryCAAX, UAS:atoh1c-p2a-mcherryCAAX), UAS:mAppleCAAX). TgBAC(atoh1c:gal4ff)fh430 embryos were injected at the one-cell stage with 120 pg of DNA along with 25 pg of tol2 transposase mRNA.
RNA in situ hybridization
For all in situ hybridizations, embryos were fixed in 4% paraformaldehyde with 1× PBS (phosphate-buffered saline) and 4% sucrose at 4°C overnight. RNA in situ hybridization was performed as described (Thisse et al., 1993). Embryos were mounted in glycerol on coverslips and transmitted light images were taken on a Zeiss Axioplan2 microscope.
Live imaging
Embryos were anesthetized with 0.4% ethyl 3-aminobenzoate methanesulfonate (ms-222) (Sigma) and immobilized in 1.2% low-melting point agarose (Gibco). All embryos were imaged on a Zeiss LSM700 inverted confocal microscope.
EdU Labeling
Starting at 3 dpf, embryos were incubated in final concentration of 0.5 mM F-ara-EdU (Sigma T511293) diluted in fish water. Embryos were anesthetized and fixed at 5 dpf. For EdU visualization, fixed whole embryos were permeablized in PBSTr (phosphate-buffered saline + 0.5% Triton X-100) for 30 mins at RT and then incubated in a solution containing 10uM Cy5-azide (Lumiprobe A2020), 2 mM copper(II) sulfate (Sigma 45167), and 20 mM sodium ascorbate (Sigma A7631) for 1 hour at RT. After 3 PBS washes, samples were processed for immunofluorescence as described below.
Immunofluorescence
For whole-mount immunostaining, embryos were fixed in 4% paraformaldehyde with 1× PBS (phosphate-buffered saline) and 4% sucrose at 4°C overnight. The fixed embryos were washed with PBSTr (PBS + 0.5% Triton X-100), dissected and incubated in acetone at −20 °C for 7 min. Dissected brains were washed once with PBSTr and twice with PDT (PBS, 1% BSA, 1% DMSO, 0.5% Triton X-100), and incubated in 5% goat serum in PDT at RT for 2 h. The samples were incubated with the primary antibody solution at 4 °C overnight. After four washes with PBST, the tissues were incubated with secondary antibodies (1/250 dilution, Alexa Fluor 488 and/or Alexa Fluor 594 goat anti-mouse and/or goat anti-rabbit IgG (H + L), (Molecular Probes, Invitrogen). Following staining, tissue was cleared step-wise in a glycerol series and mounted for confocal imaging. The following antibodies were used: chicken anti-GFP (1:500, Abcam, ab13970); rabbit anti-Kaede (1:500, MBL Co. Ltd., PM012M); mouse anti-HuC/D (1:500, Invitrogen, A-21271); mouse anti-TH (1:500, Millipore, MAB318); mouse anti-NeuroD1(1:500, gift from the Hibi Lab).
Chimeric Analysis
atoh1cfh367, dnFgfr, or TCFΔC embryos or wild-type controls to be used as donors in transplantation experiments were injected at the 1-cell stage with either Cascade Blue-dextran or Rhodamine-dextran (10,000 mw, Invitrogen). Cells were transplanted from these donor embryos into wild-type or atoh1cfh367 mutant host embryos at the early gastrula stage as described (Kemp et al., 2009), targeting donor cells to the dorsal CNS at the mid-hindbrain level of host recipients. Donor embryos were raised to 3 dpf to determine which transgene(s) they carried, and/or were genotyped for the presence of the atoh1cfh367 allele. Hosts of donors of the relevant genotype(s) were imaged as described above (“Live Imaging”).
Supplementary Material
Maximum projection time lapse movie of wild-type embryo expressing the atoh1c∷kaede transgene. Movie starts at 3.5 dpf. Time stamp indicated in hours. White dash line indicates location of the URL. Scale bar: 50 uM.
Maximum projection time lapse movie of atoh1cfh367 embryo expressing the atoh1c∷kaede transgene. Movie starts at 3.5 dpf. Time stamp indicated in hours. White dash line indicates location of the URL. Scale bar: 50 uM.
Maximum projection time lapse movie of wild-type Tg(atoh1c∷kaede) embryo at 3 dpf. Time stamp indicated in hours. White dash line indicates location of the URL. Scale bar: 50 uM.
Maximum projection time lapse movie of atoh1cfh367; Tg(atoh1c∷kaede) embryo at 3 dpf. Time stamp indicated in hours. White dash line indicates the location of the URL. Scale bar: 50 uM.
Pharmacological inhibition of Notch signaling with γ-secretase inhibitor DAPT blocks restriction of atoh1c expression domain (B) resulting in an overproduction of Tg(atoh1c∷kaede)+ neurons (D) compared to stage-matched vehicle treated control embryos (A, C). 22 hpf embryos (A,B) treated for 18 hours or 30 hpf embryos (C,D) treated for 26 hours in orientations as indicated. Red arrowheads indicate location of MHB. Scale bars: 50 uM.
Summary statement.
atoh1 genes specify distinct populations of tegmental and granule neurons in the zebrafish hindbrain and promote their delamination from the neuroepithelium, a process critical for neuronal maturation.
Highlights.
Three atoh1 genes are expressed in the cerebellum
atoh1c is the principal gene required for granule cell specification
atoh1c also marks an early mid-hindbrain-derived tegmental neuron population
atoh1c is required for the delamination of granule cell precursors from the upper rhombic lip
atoh1a and atoh1c specify distinct granule cell populations in the cerebellar corpus
Acknowledgments
We thank David Kimelman, Rachel Wong, David Raible, Herwig Baier, and Stephan Schulte-Merker for transgenic lines, constructs and reagents. David Prober and Owen Randlett provided valuable help regarding the identity of atoh1c r1 neurons. We would like to thank the members of the Moens lab for helpful discussions and comments on this manuscript, Jason Stonick for technical help, and Rachel Garcia for excellent zebrafish care. We also thank Luyuan Pan, Arish Shah, and Daniel Berman for their help with the isolation of the atoh1cfh367 mutant.
FUNDING:
This work was supported by National Institutes of Health R01 NS082567 to C.B.M. and National Institutes of Health Training Grant T32 HD007183 to C.U.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS:
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS:
C.B.M. and C.U.K. designed and performed experiments, analyzed data, and wrote the manuscript. C.Y.S. performed experiments for Figure 3. M.H. provided key reagents prior to publication as well as intellectual input into the interpretation of the data.
References
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. The Journal of biological chemistry. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- Akitake CM, Macurak M, Halpern ME, Goll MG. Transgenerational analysis of transcriptional silencing in zebrafish. Developmental biology. 2011;352:191–201. doi: 10.1016/j.ydbio.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the cerebellar system: in relation to its evolution, structure, and functions. CRC Press; Boca Raton: 1997. [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bae YK, Kani S, Shimizu T, Tanabe K, Nojima H, Kimura Y, Higashijima S, Hibi M. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Developmental biology. 2009;330:406–426. doi: 10.1016/j.ydbio.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, Matzuk M, Bellen HJ, Zoghbi HY. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Bianco IH, Ma LH, Schoppik D, Robson DN, Orger MB, Beck JC, Li JM, Schier AF, Engert F, Baker R. The tangential nucleus controls a gravito-inertial vestibulo-ocular reflex. Curr Biol. 2012;22:1285–1295. doi: 10.1016/j.cub.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann J, Schulte-Merker S. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development. 2011;138:4327–4332. doi: 10.1242/dev.068080. [DOI] [PubMed] [Google Scholar]
- Butts T, Green MJ, Wingate RJ. Development of the cerebellum: simple steps to make a ‘little brain’. Development. 2014;141:4031–4041. doi: 10.1242/dev.106559. [DOI] [PubMed] [Google Scholar]
- Caron SJ, Prober D, Choy M, Schier AF. In vivo birthdating by BAPTISM reveals that trigeminal sensory neuron diversity depends on early neurogenesis. Development. 2008;135:3259–3269. doi: 10.1242/dev.023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin N, Tendeng C, Wingate RJ. Absence of an external germinal layer in zebrafish and shark reveals a distinct, anamniote ground plan of cerebellum development. J Neurosci. 2010;30:3048–3057. doi: 10.1523/JNEUROSCI.6201-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CN, Prober DA. Regulation of zebrafish sleep and arousal states: current and prospective approaches. Front Neural Circuits. 2013;7:58. doi: 10.3389/fncir.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CN, Rihel J, Lee DA, Singh C, Mosser EA, Chen S, Sapin V, Pham U, Engle J, Niles BJ, Montz CJ, Chakravarthy S, Zimmerman S, Salehi-Ashtiani K, Vidal M, Schier AF, Prober DA. A Zebrafish Genetic Screen Identifies Neuromedin U as a Regulator of Sleep/Wake States. Neuron. 2016;89:842–856. doi: 10.1016/j.neuron.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consalez GG, Hawkes R. The compartmental restriction of cerebellar interneurons. Front Neural Circuits. 2012;6:123. doi: 10.3389/fncir.2012.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Developmental biology. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, McCallum CM, Stout JL, Slade AJ, Moens CB. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- Englund C, Kowalczyk T, Daza RA, Dagan A, Lau C, Rose MF, Hevner RF. Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J Neurosci. 2006;26:9184–9195. doi: 10.1523/JNEUROSCI.1610-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit R, Krizhanovsky V, Ben-Arie N. Math1 controls cerebellar granule cell differentiation by regulating multiple components of the Notch signaling pathway. Development. 2004;131:903–913. doi: 10.1242/dev.00982. [DOI] [PubMed] [Google Scholar]
- Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO reports. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PA. Transcription factors and the genetic organization of brain stem respiratory neurons. J Appl Physiol (1985) 2008;104:1513–1521. doi: 10.1152/japplphysiol.01383.2007. [DOI] [PubMed] [Google Scholar]
- Green MJ, Myat AM, Emmenegger BA, Wechsler-Reya RJ, Wilson LJ, Wingate RJ. Independently specified Atoh1 domains define novel developmental compartments in rhombomere 1. Development. 2014;141:389–398. doi: 10.1242/dev.099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, Mullins MC, Rosenthal A. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein soulless/Phox2a. Neuron. 1999;24:555–566. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Hartenstein AY, Rugendorff A, Tepass U, Hartenstein V. The function of the neurogenic genes during epithelial development in the Drosophila embryo. Development. 1992;116:1203–1220. doi: 10.1242/dev.116.4.1203. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Hibi M. Development and evolution of cerebellar neural circuits. Development, growth & differentiation. 2012;54:373–389. doi: 10.1111/j.1440-169X.2012.01348.x. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Kashiwagi M, Yasuda K, Ando R, Kanuka M, Sakai K, Itohara S. Cells of a common developmental origin regulate REM/non-REM sleep and wakefulness in mice. Science. 2015;350:957–961. doi: 10.1126/science.aad1023. [DOI] [PubMed] [Google Scholar]
- Hibi M, Shimizu T. Development of the cerebellum and cerebellar neural circuits. Developmental neurobiology. 2012;72:282–301. doi: 10.1002/dneu.20875. [DOI] [PubMed] [Google Scholar]
- Kani S, Bae YK, Shimizu T, Tanabe K, Satou C, Parsons MJ, Scott E, Higashijima S, Hibi M. Proneural gene-linked neurogenesis in zebrafish cerebellum. Developmental biology. 2010;343:1–17. doi: 10.1016/j.ydbio.2010.03.024. [DOI] [PubMed] [Google Scholar]
- Kaslin J, Ganz J, Geffarth M, Grandel H, Hans S, Brand M. Stem cells in the adult zebrafish cerebellum: initiation and maintenance of a novel stem cell niche. J Neurosci. 2009;29:6142–6153. doi: 10.1523/JNEUROSCI.0072-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp HA, Carmany-Rampey A, Moens C. Generating chimeric zebrafish embryos by transplantation. J Vis Exp. 2009 doi: 10.3791/1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Ueshima E, Muraoka O, Tanaka H, Yeo SY, Huh TL, Miki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett. 1996;216:109–112. doi: 10.1016/0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental dynamics: an official publication of the American Association of Anatomists. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Direct imaging of in vivo neuronal migration in the developing cerebellum. Curr Biol. 2001;11:1858–1863. doi: 10.1016/s0960-9822(01)00585-1. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Leto K, Arancillo M, Becker EB, Buffo A, Chiang C, Ding B, Dobyns WB, Dusart I, Haldipur P, Hatten ME, Hoshino M, Joyner AL, Kano M, Kilpatrick DL, Koibuchi N, Marino S, Martinez S, Millen KJ, Millner TO, Miyata T, Parmigiani E, Schilling K, Sekerkova G, Sillitoe RV, Sotelo C, Uesaka N, Wefers A, Wingate RJ, Hawkes R. Consensus Paper: Cerebellar Development. Cerebellum. 2015 doi: 10.1007/s12311-015-0724-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Machold RP, Kittell DJ, Fishell GJ. Antagonism between Notch and bone morphogenetic protein receptor signaling regulates neurogenesis in the cerebellar rhombic lip. Neural Dev. 2007;2:5. doi: 10.1186/1749-8104-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart GD, Tabor KM, Brown M, Strykowski JL, Varshney GK, LaFave MC, Mueller T, Burgess SM, Higashijima S, Burgess HA. A 3D Searchable Database of Transgenic Zebrafish Gal4 and Cre Lines for Functional Neuroanatomy Studies. Front Neural Circuits. 2015;9:78. doi: 10.3389/fncir.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL, Kimelman D. Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev Cell. 2012;22:223–232. doi: 10.1016/j.devcel.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Rand K, Palardy G, Shimizu N, Ikeda H, Dalle Nogare D, Itoh M, Chitnis AB. Epb41l5 competes with Delta as a substrate for Mib1 to coordinate specification and differentiation of neurons. Development. 2016;143:3085–3096. doi: 10.1242/dev.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Namikawa K, Babaryka A, Koster RW. Functional regionalization of the teleost cerebellum analyzed in vivo. Proc Natl Acad Sci USA. 2014;111:11846–11851. doi: 10.1073/pnas.1403105111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millimaki BB, Sweet EM, Dhason MS, Riley BB. Zebrafish atoh1 genes: classic proneural activity in the inner ear and regulation by Fgf and Notch. Development. 2007;134:295–305. doi: 10.1242/dev.02734. [DOI] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Kawakami K. Calcium Imaging of Neuronal Activity in Free-Swimming Larval Zebrafish. Methods Mol Biol. 2016;1451:333–341. doi: 10.1007/978-1-4939-3771-4_23. [DOI] [PubMed] [Google Scholar]
- Pacary E, Martynoga B, Guillemot F. Crucial first steps: the transcriptional control of neuron delamination. Neuron. 2012;74:209–211. doi: 10.1016/j.neuron.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Pujol-Marti J, Zecca A, Baudoin JP, Faucherre A, Asakawa K, Kawakami K, Lopez-Schier H. Neuronal birth order identifies a dimorphic sensorineural map. J Neurosci. 2012;32:2976–2987. doi: 10.1523/JNEUROSCI.5157-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randlett O, Wee CL, Naumann EA, Nnaemeka O, Schoppik D, Fitzgerald JE, Portugues R, Lacoste AM, Riegler C, Engert F, Schier AF. Whole-brain activity mapping onto a zebrafish brain atlas. Nat Methods. 2015;12:1039–1046. doi: 10.1038/nmeth.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M, Brand M. The midbrain–hindbrain boundary organizer. Curr Opin Neurobiol. 2001;11:34–42. doi: 10.1016/s0959-4388(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Rose MF, Ahmad KA, Thaller C, Zoghbi HY. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc Natl Acad Sci USA. 2009;106:22462–22467. doi: 10.1073/pnas.0911579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso DL, Pearson CA, Gaber ZB, Miquelajauregui A, Li S, Portera-Cailliau C, Morrisey EE, Novitch BG. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012;74:314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods. 2015;12:535–540. doi: 10.1038/nmeth.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Yamaguchi S, Sakakibara Y, Hayashi T, Matsuda K, Hara Y, Tanegashima C, Shimizu T, Kuraku S, Hibi M. Gene expression profiling of granule cells and Purkinje cells in the zebrafish cerebellum. The Journal of comparative neurology. 2016 doi: 10.1002/cne.24114. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Yamaguchi S, Yonemura S, Kakiguchi K, Sato Y, Higashiyama T, Shimizu T, Hibi M. Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish. PLoS Genet. 2015;11:e1005587. doi: 10.1371/journal.pgen.1005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Volkmann K, Chen YY, Harris MP, Wullimann MF, Koster RW. The zebrafish cerebellar upper rhombic lip generates tegmental hindbrain nuclei by long-distance migration in an evolutionary conserved manner. The Journal of comparative neurology. 2010;518:2794–2817. doi: 10.1002/cne.22364. [DOI] [PubMed] [Google Scholar]
- Volkmann K, Rieger S, Babaryka A, Koster RW. The zebrafish cerebellar rhombic lip is spatially patterned in producing granule cell populations of different functional compartments. Developmental biology. 2008;313:167–180. doi: 10.1016/j.ydbio.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Wada H, Ghysen A, Satou C, Higashijima S, Kawakami K, Hamaguchi S, Sakaizumi M. Dermal morphogenesis controls lateral line patterning during postembryonic development of teleost fish. Developmental biology. 2010;340:583–594. doi: 10.1016/j.ydbio.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum projection time lapse movie of wild-type embryo expressing the atoh1c∷kaede transgene. Movie starts at 3.5 dpf. Time stamp indicated in hours. White dash line indicates location of the URL. Scale bar: 50 uM.
Maximum projection time lapse movie of atoh1cfh367 embryo expressing the atoh1c∷kaede transgene. Movie starts at 3.5 dpf. Time stamp indicated in hours. White dash line indicates location of the URL. Scale bar: 50 uM.
Maximum projection time lapse movie of wild-type Tg(atoh1c∷kaede) embryo at 3 dpf. Time stamp indicated in hours. White dash line indicates location of the URL. Scale bar: 50 uM.
Maximum projection time lapse movie of atoh1cfh367; Tg(atoh1c∷kaede) embryo at 3 dpf. Time stamp indicated in hours. White dash line indicates the location of the URL. Scale bar: 50 uM.
Pharmacological inhibition of Notch signaling with γ-secretase inhibitor DAPT blocks restriction of atoh1c expression domain (B) resulting in an overproduction of Tg(atoh1c∷kaede)+ neurons (D) compared to stage-matched vehicle treated control embryos (A, C). 22 hpf embryos (A,B) treated for 18 hours or 30 hpf embryos (C,D) treated for 26 hours in orientations as indicated. Red arrowheads indicate location of MHB. Scale bars: 50 uM.


