Abstract
Inflammasomes are multiprotein structures that activate caspase-1, support secretion of pro-inflammatory cytokines, IL-1β and IL-18, and also induce inflammatory programmed cell death, termed pyoptosis. Inflammasomes are activated in response to the detection of endogenous and microbially derived danger signals and are mediated by several classes of inflammasome-forming sensors. These include several nucleotide-binding proteins of the NOD-like receptor (NLR) family, including NLRP1, NLRP3 and NLRC4, as well as the proteins Absent in Melanoma 2 (AIM2) and Pyrin. Mutations in genes encoding some of these sensors have been found to be associated with gain-of-function monogenetic inflammatory disorders in humans. Genetic, biochemical and structural studies have begun to demonstrate how these proteins sense danger signals and to shed light on the step-by-step processes that are necessary for the assembly of inflammasomes, in both physiologic responses to pathogens and potentially in autoinflammatory conditions. Recent biochemical studies of pro-caspase-1 and an adapter protein known as ASC suggest that inflammasomes act to initiate self-generating effector filaments responsible for activating caspase-1 and initiating downstream signaling. These studies have suggested a model of molecular events from sensor activation to inflammasome formation that may describe processes that are universal to inflammasome formation.
Keywords: caspase-1, IL-1β, inflammasome, NOD-like receptor
Introduction
Inflammasomes are multiprotein complexes that induce inflammation by activating the cysteine proteinase, caspase-1, which is responsible for the maturation of the pro-cytokine IL-1β into its secreted active form. Inflammasome-mediated caspase-1 activation is also responsible for maturation of IL-1-related pro-cytokines, particularly IL-18, and induction of a pro-inflammatory type of programmed cell death known as pyroptosis.
The activation and oligomerization of inflammasome-forming innate immune sensors is the critical molecular step in forming active inflammasomes. After this key step, many studies suggest that additional inflammasome components with death fold-containing domains, particularly the pyrin domain of the adapter molecule ASC or the caspase activation and recruitment domain (CARD) of pro-caspase-1, will propagate the initiating signal. Death fold-containing domains are found in many proteins that are involved with apoptosis or inflammation-related processes. Once nucleated, these death domain-containing structures are able to self-assemble into molecular fibrils, forming massive complexes that support activation of many pro-caspase-1 molecules (1, 2). This self-assembly occurs with purified proteins in a cell-free system and appears to utilize a prion-like mechanism and assembly. The resulting fibrillar structures can be isolated in vitro and from cells in which inflammasome activation has been initiated (1, 2).
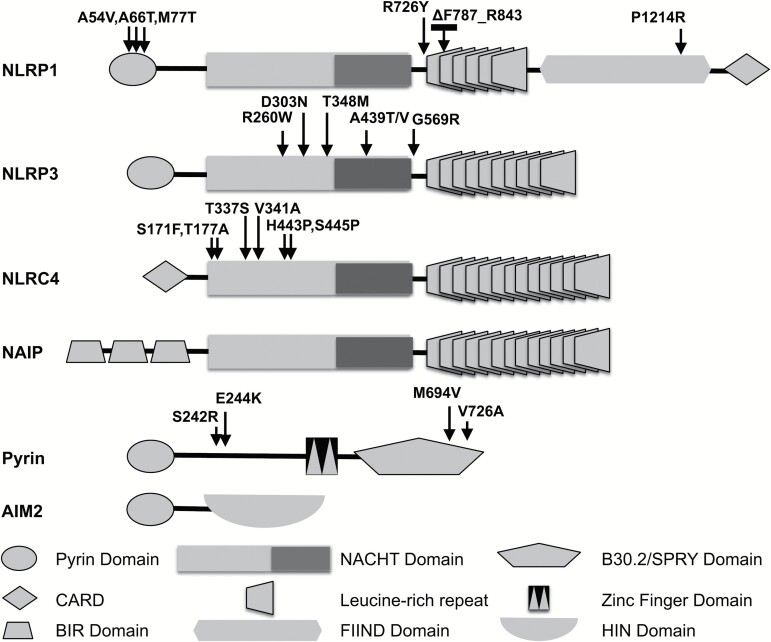
Many inflammasome-forming innate immune sensors fall into the family of proteins known as nucleotide-binding and leucine-rich repeat (LRR)-containing proteins, also known as NOD-like receptors (NLRs, formerly known as NALPs). While there are roughly 20 mammalian NLR proteins, three (NLRP1, NLRP3 and NLRC4) have been well-characterized in their role in initiating inflammasome assembly. Other proteins containing pyrin domains, a subset of Death-fold domains, are also known to facilitate inflammasome formation, including Absent in Melanoma 2 (AIM2) and Pyrin, the founding member of pyrin domain-containing proteins. Mutations in all three of these NLR proteins as well as Pyrin have been linked to autoinflammatory diseases (Fig. 1).
Fig. 1.
Mutations in inflammasome-forming sensor proteins are associated with autoinflammatory syndromes. This schematic demonstrates the domain structure of the indicated inflammasome-forming sensor proteins including the NLR proteins: NLRP1, NLRP3, NLRC4 and NAIP as well as Pyrin and AIM2. Arrows indicate approximate location of known mutations that are associated with autoimmune diseases, for example in NLRP1 (linked with NLRP1-associated autoinflammation with arthritis and dyskeratosis, multiple self-healing palmoplantar carcinoma and familial keratosis lichenoides chronica), in NLRP3 (associated with Muckle–Wells syndrome, familial cold urticaria and neonatal-onset multisystem inflammatory disease), in NLRC4 (associated with macrophage-activating syndrome) and in Pyrin (associated with familial Mediterranean fever and Pyrin-associated autoinflammation with neutrophilic dermatosis).
NLRP3 mutations are strongly associated with a disease spectrum of inherited autoinflammatory disorders collectively referred to as cryopyrin-associated periodic fever syndrome that include familial cold autoinflammatory syndrome (FCAS), Muckle–Wells syndrome and neonatal-onset multisystem inflammatory disease. Since the initial description of three mutations in NLRP3 associated with these diseases, over a hundred disease causing mutations have been identified in patients with both hereditary and spontaneously arising inflammatory syndromes (3–6). Six different missense mutations in NLRC4 associated with autoinflammatory diseases. Mutations in Thr337 (T337S), His443 (H443P), Val341 (V341A), Ser171 (S171F), Thr177 (T177A) and Ser445 (S445P) are associated with a spectrum of disease manifestations that include macrophage activation syndrome (MAS), FCAS-like disease and some have features of neonatal-onset enterocolitis, and periodic systemic inflammatory episodes (7–12). Recently, a mutation in NLRP1 was found associated with a syndrome that was called NLRP1-associated autoinflammation with arthritis and dyskeratosis (NAIAD) and two skin disorders: multiple self-healing palmoplantar carcinoma and familial keratosis lichenoides chronica (13, 14). Pyrin is the protein product of the MEFV gene, which was first identified as the site of mutations associated with the autoinflammatory condition hereditary Mediterranean fever syndrome (15, 16). Subsequently, a mutation in MEFV has been found associated with another inflammatory disorder, pyrin-associated autoinflammation with neutrophilic dermatosis (17). A list of mutations that are linked to autoinflammatory disorders is found at http://fmf.igh.cnrs.fr/ISSAID/infevers/.
All of these autoinflammatory diseases are associated with high levels of IL-1β and/or IL-18, linking their pathogenesis to constitutive activation or gain-of-function in these inflammasome-forming proteins. Interestingly, mutations in NLRC4 are associated with disproportionate increases in IL-18 over IL-1β, when compared to diseases associated with mutations in NLRP3 or NLRP1. Although NLRP3, NLRP1 and NLRC4 all share the common structural feature of a central nucleotide-binding domain (NBD), the physiologic stimuli that activate each are different. NLRP3 is activated by a wide variety of stimuli at the cellular level, including pore-forming toxins, activation of purogenic-gated potassium channels and other membrane-destabilizing stimuli. NLRP3 is also activated by microscopic crystals, including monosodium urate, calcium pyrophosphate, asbestos, silica and alum. On the other hand, NLRP1 has been shown to be activated by a limited set of stimuli, including muramyl dipeptide, a small molecule derived from bacterial cell walls, and by proteolysis of the amino-terminal domain by anthrax lethal toxin and possibly other bacteria-derived proteolytic toxins or endogenous host proteinases (18–21). Despite its identification long before the NLR family of sensors, Pyrin has only recently been identified as a sensor for disruption of the host signaling system that involves the rho GTPases, which are involved in phagocytosis of pathogens and are targeted by many pathogen-derived toxins. By detecting disruption of the host signaling pathway, Pyrin can initiate host inflammatory signaling in response to a variety of pathogens without depending on recognition of a specific pathogen-associated molecule (22).
The activation of the NLRC4 inflammasome differs from NLRP3 and NLRP1 because the core oligomer of the NLRC4 inflammasome is composed of two NLR family members, NLRC4 and NAIP (NLR family of apoptosis inhibitory protein). The NLRC4 inflammasome detects the presence of flagellin and two components of the type III secretion system (T3SS), the rod and needle protein (23), which act as ligands for NAIP proteins. The specificity of the NLRC4 inflammasome is dictated by its NAIP, which directly interacts with a bacterial ligand in a receptor–ligand fashion. Four different mouse NAIPs (NAIP1, NAIP2, NAIP5 and NAIP6) and one human NAIP have been characterized. Two of these NAIPs, NAIP5 and NAIP6, interact with bacterial flagellin and stimulate the NLRC4 inflammasome (23). NAIP2 interacts with the T3SS rod protein, PrgJ. The fourth mouse NAIP, NAIP1, interacts with the T3SS needle protein (23, 24). In immortalized monocyte-derived cells, human NAIP was found to be required for T3SS needle protein-induced NLRC4 inflammasome activation (24). However, recently human primary monocyte-derived macrophages (MDMs) were found to express an alternative NAIP transcript that encodes an extended NAIP protein. The NLRC4 inflammasome is responsive to flagellin in these MDMs and expression of the extended NAIP transcript in immortalized monocyte-derived cells allowed those cells to generate IL-1β in response to cytosolic flagellin delivery (25). Once NAIP binds its specific ligand, it can bind to NLRC4, causing oligomerization of NLRC4 protomers. Like the NLRC4 inflammasome, the AIM2 inflammasome also assembles in response to pathogen-derived molecules, DNA in the host cell cytosol for AIM2. However, unlike NLRC4, which requires NAIP for its recognition of bacterial peptide ligands, the AIM2 inflammasome consists of AIM2 homooligomers.
NLR proteins have a tripartite organization, with a C-terminal LRR domain and a central NACHT (NAIP, CIITA, HET-E, TEP1) domain (Fig. 1). NLR proteins have some diversity in the N-terminal domain: NLRP1 and NLRP3 have pyrin domains, NLRC4 has a CARD, and NAIPs have a BIR (baculovirus inhibitor of apoptosis protein repeat) domain.
The LRR domain of NLRs has clearly been shown to play an auto-inhibitory role in NLR activation as expression of several NLR proteins lacking their cognate LRR has been found to lead to constitutive activation of the protein (26, 27). Because these constitutively active LRR-less NLR proteins lose ligand-sensitive activation and because LRR domains of the toll-like receptor (TLR) proteins are known to act as ligand-binding domains, the hypothesis that the LRR also acts as a ligand-binding domain for NLRs has largely been accepted as dogma. However, direct ligand-binding assays in NOD2 (another CARD-containing NLR) and specificity studies in NAIPs suggest that the NACHT domain contributes part or all of the ligand binding in at least some NLR proteins (28, 29). NLRP1 has an additional CARD on the C-terminus that is not found in other NLR proteins. The NACHT domain of NLR proteins is composed of the NBD, hinged domain 1 (HD1), the winged helix domain (WHD) and hinged domain 2 (HD2).
The NACHT domains of NLRC4, NLRP3 and NLRP1 have all been shown to bind and hydrolyze adenosine triphosphate (ATP), and many published studies suggest that ATP binding or hydrolysis plays an important role in NLR-inflammasome activation (30–33). Over-expression of mutant NLRC4 and NLRP3 that have loss of ATP binding fails to trigger IL-1β secretion (30, 31). The ability of NLRP3 to stimulate in vitro ASC oligomerization was enhanced by the addition of ATP to the reaction (1). NLRP1 inflammasome assembly reconstituted in vitro required ATP, though other nucleotide triphosphates could also support NLRP1 inflammasome formation (34). Interestingly, a recent study investigating the role of proteolysis in NLRP1 inflammasome activation showed that over-expression of a mutant NLRP1 predicted to have poor ATP binding still supported IL-1β secretion (19). Though ATP binding seems to be critical for each NLR to form an inflammasome, whether the steps of nucleotide binding and hydrolysis play the same role in activation of each different NLR has not been determined.
After assembly of the sensor protein-containing inflammasomes is initiated, nucleation of a death domain-containing protein that triggers self-propagating assembly proceeds. For pyrin domain-containing NLRs (e.g. NLRP1 and NLRP3), the N-terminal pyrin domain interacts with the pyrin domain of the adapter molecule ASC (which consists of a pyrin domain and a CARD). As ASC pyrin domains are assembled into a fibril, the CARDs can interact with the CARD of pro-caspase-1, nucleating a second polymerization event that generates branching fibrils of pro-caspase-1 that ultimately lead to activation of the pro-enzyme into active caspase-1 that is able to liberate itself from the assembling fibrils through autoproteolysis (1). For the NLRC4 inflammasome, the CARD of NLRC4 in the assembling inflammasome can initiate pro-caspase-1 oligomerization through CARD–CARD interactions resulting in direct caspase-1 activation (1). Activation of caspase-1 leads to proteolytic processing of inflammatory cytokines, pro-IL-1β and pro-IL-18, as well as pyroptosis, which is defined as caspase-1-dependent programmed cell death (35).
Extensive research has been conducted on the activation and assembly of the NLRC4 inflammasome, which has been recently aided by structural studies of the inactive protein as well as the assembled inflammasome (36, 37). In this review, we discuss the various stages of the inflammasome assembly process for inflammasomes, starting with initial activation that is strongly regulated by post-translational modification, ligand-binding, or both. We will also review the downstream assembly events including activation of NLRC4 monomers, oligomerization of NLRC4 and finally the interaction of NLRC4 with pro-caspase-1. Although this review focuses on the physiologic activation of the NLRC4 inflammasomes by bacterial ligands, there is likely substantial overlap between the mechanisms of autoinflammatory mutation induced NLRC4 inflammasome assembly and the physiologic activation of this complex. It seems likely that many of the molecular events of NLRC4 inflammasome assembly are conserved between other NLR-containing inflammasomes, although the mechanisms by which these events are achieved may vary between proteins.
Regulation of inflammasome activation by post-translational modifications
The regulation of the activation of inflammasomes is also affected by the post-translational modifications of key inflammasome components. Phosphorylation/dephosphorylation and proteolysis processes have well-documented roles in inflammasome activation. Other processes like ubiquitination/deubiquitination, nitrosylation and ribosylation are also reported to impact inflammasome activation, but are less well defined. For some sensor proteins, like NLRC4 and NLRP3, post-translational modification seems to be one step of many ultimately required for inflammasome assembly. For others, particularly Pyrin and NLRP1, post-translational modifications appear to be the key signaling mechanism leading to inflammasome activation.
Phosphorylation of inflammasome-activating sensors
Phosphorylation is a key regulator of Pyrin inflammasome activation. Pyrin is a microtubule-binding protein with five distinct domains (38). Two of these domains, the pyrin domain and the B30.2 domain, directly interact with ASC and caspase-1, respectively, to promote inflammasome assembly. The assembly of the Pyrin inflammasome is regulated by binding of phosphorylated pyrin with protein 14-3-3, which prevents association of the protein with ASC and caspase-1 (17, 39, 40). Dephosphorylation of S205 and S241 in murine Pyrin (S208 and S242 in human Pyrin) causes the dissociation of 14-3-3, permitting ASC binding, inflammasome assembly, and caspase-1 activation (39). One mutation in the human Pyrin protein, S242R, causes a disruption of 14-3-3 binding to Pyrin and is associated with pyrin-associated autoinflammation with neutrophilic dermatosis (17). Another mutation in human Pyrin that was also recently identified in patients with pyrin-associated autoinflammation with neutrophilic dermatosis, E244K, has also been shown to disrupt the 14-3-3 binding motif associated with phospho-S242 (41). In the absence of activating mutations, pyrin inflammasome assembly can be induced by cytotoxin TcdB from Clostridium difficile. This toxin glycosylates RhoA GTPase (22), which is responsible for activation of two serine-threonine kinases, PKN1 and PKN2. The basal activation of PKN1 and PKN2 is sufficient to maintain phosphorylation of pyrin, promoting 14-3-3 binding to the protein (42). The binding of 14-3-3 and PKN1/PKN2 to pyrin is weaker for mutant pyrin (FMF) as compared to wild type (42). Although dephosphorylation and 14-3-3 disassociation is a critical signal in Pyrin inflammasome activation, Pyrin inflammasome assembly also requires association with microtubules and can be blocked by inhibition of microtubule polymerization with colchicine (43). These data suggest that, like other inflammasome-forming sensors, Pyrin inflammasome assembly is also controlled by multiple regulatory processes within cells.
Several studies suggest that phosphorylation/dephosphorylation plays a role in the activation of NLR-based inflammasomes as well. Qu et al. (44) showed that PKCδ phosphorylates mouse NLRC4 on serine residue 533. Murine macrophages expressing the NLRC4 mutant, S533A, were unable to activate caspase-1 in response to the presence of proper NLRC4 inflammasome stimuli. It was further shown that the phosphorylation of NLRC4 is independent of NAIP proteins or NLRC4 inflammasome-activating ligands (45). More recently, the leucine-rich repeat kinase 2 (LRRK2), a protein involved with various inflammatory diseases, directly interacts with NLRC4 and phosphorylates serine 533, leading to the activation of the NLRC4 inflammasome (46). Unfortunately, the molecular role phosphorylation of serine 533 plays in NLRC4 inflammasome assembly has not been further elucidated by the detailed structural studies of NLRC4 that have been published to date.
Many different kinases have been found to be required for optimal NLRP3 inflammasome activation. Several studies examining NLRP3 inflammasome activation by fungi, plasmodium parasites and Mycobacterium tuberculosis show that blockade of signaling by tyrosine kinases Syk and Lyn reduces NLRP3-inflammasome-dependent IL-1β secretion through impact on inflammasome activation (47–49). Pharmacologic or genetic reduction of expression studies of other kinases, including TAK1, DAPK and ERK1, have also suggested that in some circumstances, these kinases contribute to NLRP3 activation (50–52). A role for the double-stranded RNA-dependent protein kinase, PKR, in activation of NLRP3 remains controversial in the literature, with studies suggesting PKR can activate, inhibit or play no role at all in activation of NLRP3 (53–56). Despite the implication of so many kinases in NLRP3 activation, there are very limited studies demonstrating a role of NLRP3 phosphorylation in NLRP3 inflammasome assembly. A 2017 study by Stutz et al. demonstrated that the pyrin domain of NLRP3 is phosphorylated at baseline and that phosphatase 2A (PP2A)-mediated dephosphorylation was required for NLRP3–ASC association and subsequent inflammasome assembly (57). Phosphomimetic mutations with basic residues in place of phosphorylated serines prevented association between NLRP3 and ASC pyrin domains and structural predictions suggested that a patch of negative surface charge localized at the site of the phorphorylated serines should disrupt the inter-domain contacts needed for pyrin domain oligomerization (57–59). At this point, the identity of the kinase or kinases required for maintaining NLRP3 pyrin domain phosphorylation in the basal state are unknown.
Using a variety of unbiased genetic screens, several research groups found that NIMA-related kinase 7 (NEK7) is required for optimal NLRP3 inflammasome activation. NEK7 and NLRP3 directly interact with one another, and the deletion of NEK7 leads to the absence of caspase-1 activation and IL-1 β secretion in both peritoneal and bone marrow-derived macrophages (BMDMs) treated with NLRP3 inflammasome-activating compounds (58, 59). Abrogation of NEK7 expression had no effect on NLRC4 or AIM2 inflammasome activation (58, 59). The kinase inactive NEK7 mutant (K64M) still leads to secretion of IL-1β and can immunoprecipitate NLRP3 in a similar fashion to wild-type NEK7, the kinase activity of NEK7 has been shown to be dispensable for NLRP3 inflammasome activation (59). Thus, despite these multiple correlated reports of a protein kinase being a critical component in NLRP3 signaling, evidence for a role of phosphorylation of NLRP3 in modulating the proteins activity remains limited.
Proteolysis in inflammasome activation
The activation of the NLRP1 inflammasome can occur through proteolytic cleavage. The anthrax lethal toxin from Bacillus anthracis can directly cleave NLRP1b and activate the NLRP1 inflammasome, leading to the secretion of IL-1β and pyroptosis (20, 21, 60). Lethal factor toxin cleavage of NLRP1 occurs in the pyrin domain of NLRP1, which is also the site of mutations in NLRP1 that are associated with autoinflammatory disorders (14). These data suggest that unlike most NLR proteins in which the N-terminal CARD or pyrin domain serves to bind to downstream effector molecules, the C-terminal CARD domain of NLRP1 may actually serve the effector binding function for NLRP1. Chavarria-Smith et al. also recently demonstrated that artificial introduction of a TEV protease cleavage site in the pyrin domain of NLRP1 created a TEV protease activated inflammasome (19). These findings support the concept posed by Chavarria-Smith et al. that the pyrin domain of NLRP1 may act as a regulator of NLRP1 activity and serve as a protease sensor to detect pathogen-derived toxins that have evolved to destroy pyrin domain-containing innate immune sensors. It remains to be determined if other inflammasome-forming sensors are substantially regulated by proteolysis.
Interaction and activation of inflammasome-forming sensors by ligands
There are two well-documented examples of inflammasome-forming sensors being directly activated by ligand binding events: activation of the AIM2 inflammasome and the NLRC4 inflammasome. The AIM2 is a receptor that senses cytosolic, double-stranded DNA (dsDNA) and leads to the activation of caspase-1 (61–64). AIM2 interacts with dsDNA through the electrostatic charges from amino acids in the HIN domain of AIM2 and the sugar phosphate backbone of dsDNA (65), thus there is no substantial impact of DNA sequence on AIM2 activation. Autoinhibition of AIM2 occurs from the interaction between the HIN and pyrin domains, and this autoinhibition is released when the HIN domain binds dsDNA (65). In addition to requiring direct DNA binding for activation, AIM2 inflammasome assembly is regulated in cells by a second HIN domain-containing protein, p202 (66, 67). The HIN domain of p202 has a higher DNA binding affinity than the HIN domain of AIM2, and the interaction of p202 with AIM2 can lead to the inhibition of the AIM2 inflammasome (67).
Early research into the NLRC4 inflammasome suggested this complex was important for sensing bacterial flagellin. Several published reports demonstrated that genetic inactivation of NLRC4 reduced or eliminated IL-1β release induced by flagellin and flagellin-producing bacteria (68–71). Several early studies also demonstrated that NAIP5 was required for flagellin-induced NLRC4 inflammasome activation (72–74). Flagellin proteins from a number of bacterial species were shown, using co-immunoprecipitation and yeast two-hybrid assays, to interact with NAIP5. These flagellin proteins failed to interact with NLRC4 in similar assays. The ability of each flagellin to stimulate IL-1β secretion from BMDMs correlated with binding to NAIPs in these assays (23).
Studies into the primary structure of flagellin required for activating the NLRC4 inflammasome showed that a change in three amino acids (L470A, L472A and L473A) in the C-terminus eliminated flagellin-induced pyroptosis (75) and interaction with NAIP5 (23). The same study used NAIP5-deficient mice to show a direct role for NAIP5/NLRC4 in response to the C-terminus of flagellin. Interestingly, the full-length flagellin activated the NLRC4 inflammasome in the absence of NAIP5 (75). Although the N-terminus of flagellin alone does not cause pyroptosis in mouse BMDMs, the authors determined that the N-terminus of flagellin in the context of full-length flagellin, or when co-expressed with the C-terminus, caused activation of the NLRC4 inflammasome in a NAIP5-independent fashion (76). Interestingly, either the N-terminal 65 amino acids or C-terminal 65 amino acids of flagellin were found to activate NAIP5-dependent NLRC4 inflammasome formation when NAIP5, NLRC4 and pro-caspase-1 were co-expressed in HEK293 cells, which do not express a native NLRC4 inflammasome. It remains unclear why full-length flagellin was found to stimulate NLRC4 inflammasome formation in a NAIP5-independent fashion in BMDMs, but the authors question whether the N-terminus of flagellin can alter or enhance the NLRC4-dependent response to the C-terminus of flagellin so that NAIP5 is no longer necessary, pointing to an alternate mechanism to activate the NLRC4 inflammasome.
Legionella infection was found to cause NLRC4-mediated IL-1β secretion in a flagellin/NAIP5-dependent fashion whereas Salmonella activation of NLRC4 was found to be partially NAIP5-independent, suggesting that Salmonella produced a factor with an alternate pathway to NLRC4 activation (76). The presence of a second NLRC4 activation pathway was supported by the observation that NLRC4 activation and IL-1β secretion depended entirely on NAIP5 in cells infected with Legionella in which the flagellin gene from Legionella was replaced with the Salmonella flagellin gene. Miao et al. (77) noted that the T3SS rod protein, PrgJ, found in Salmonella but not Legionella, had some sequence homology to flagellin. They went on to demonstrate that PrgJ stimulated NLRC4-dependent caspase-1 activation and IL-1β secretion.
To determine the region of rod protein necessary for NLRC4 activation, the amino acid sequences of PrgJ homologs were compared and strong identity was seen in the C-terminus of the proteins. Deletion of the last seven amino acids (amino acids 95–101) prevented IL-1β secretion in macrophages primed with LPS and stimulated with PrgJ, and substituting alanine for valine at position 95 also led to the abrogation of IL-1β secretion (77).
The discovery that PrgJ activated NLRC4 led to the hypothesis that PrgJ might act on NLRC4 through a separate NAIP protein. Kofoed et al. (78) used short-hairpin RNA (shRNA) knockdowns of NAIP2 in mouse macrophages to determine the role NAIP2 plays in PrgJ-induced NLRC4 inflammasome activation. The authors showed Listeria exogenously expressing Salmonella PrgJ or Salmonella lacking flagellin could induce pyroptosis and activation of caspase-1 in BMDMs from both wild-type and Naip5–/– mice. Knockdown of NAIP2 expression by shRNA blocked pyroptosis and caspase-1 activation in these cells. Zhao et al. (23) demonstrated that caspase-1 activation in response to PrgJ or BsaK, a T3SS rod protein from Burkholderia thailandensis, was diminished by knockdown of NAIP2 in macrophages. They also found that BsaK interacted with NAIP2, but not NAIP1, NAIP5, NAIP6 or NLRC4 in a yeast two-hybrid system (23). These studies demonstrated a direct role for NAIP2 in recognizing T3SS rod protein and stimulating the formation of the NLRC4 inflammasome.
A more detailed study was performed to determine which regions of NAIP govern its ligand specificity. NAIP chimeras were created by combining part of NAIP5/6 with complementing regions from NAIP2, and using the chimeric NAIP, along with NLRC4 and specific ligand, for co-expression in HEK293T cells for the inflammasome reconstitution assays (29). Results showed that the LRR domain was not involved in the ligand specificity of each NAIP, whereas the NBD, HD1, WHD and HD2 regions were involved in NAIP specificity, leading to the hypothesis that binding of bacterial ligand to HD2 of NAIPs may cause steric inhibition or assumption of a conformation that blocks association between the NACHT domain and the inhibitory LRR domain. Two recent publications have clarified the interactions between L. pneumophila flagellin (FlaA), NAIP5 and NLRC4. Tenthorey et al. (79) used cryo-electron microscopy (cryo-EM) to determine the contact points between flagellin and NAIP5. They determined that six different regions of NAIP5 directly interact with FlaA. Helices in the HD1 domain, HD2 domain, BIR domain, a region N-terminally from the BIR domain and two regions of the LRR domain are involved in the binding of FlaA. Point mutations in these regions of NAIP5 reduced its binding to FlaA. The authors also mutated regions of FlaA and determined that amino acids L470, L472 and L473 were crucial to binding to NAIP5, similar to a previous report (75), along with residues 31–33 and 458–460. The interactions between PrgJ and NAIP2 were also examined, and mutational analysis showed that the C-terminal portion of PrgJ was crucial for recognition by NAIP2, along with residues 32–34 and 65–88. The authors concluded that multiple point mutations in both FlaA and PrgJ were necessary for the evasion of NAIP5 or NAIP2, but these multiple mutations caused disrupted function for FlaA and PrgJ. A second study also used cryo-EM to investigate the interactions between S. typhimurium flagellin (FliC), NAIP5 and NLRC4 (80). The authors determined that two parallel helices of flagellin interact with several regions of NAIP5, including the BIR and LRR domains, similar to the Tenthorey study (79). The binding of FliC to NAIP5 acts to stabilize the active conformation of NAIP5, and this interaction is necessary for NLRC4 activation. The initial recognition of FliC by NAIP5 seems to occur in a binding pocket that is formed by the BIR1 and HD1 domains (80). The need for multisurface recognition between NAIP and these virulence factors limits the ability for pathogens to evade recognition by the NLRC4 inflammasome.
Much of the data surrounding NAIP protein bacterial ligand specificity arise from studies of mouse proteins derived from multiple NAIP genes harbored by this species. Humans only have a single NAIP gene, raising questions surrounding the ligand specificity and role in host defense that the human NLRC4 inflammasome plays. Early studies found human monocyte-derived cells induced IL-1β secretion in response to T3SS needle protein, but not flagellin or rod protein (23, 24, 81). Kortmann et al. (25) demonstrated that two commonly used human monocytic cell lines express a spliced NAIP transcript lacking nucleotides 3990–4160. This group further demonstrated that primary monocytes express full-length NAIP protein and are responsive to Salmonella flagellin. The complementation of U-937 cells with the full-length hNAIP transcript rendered the cells responsive to challenge from S. typhimurium and S. typhi (25). Two recent studies also support the capacity of hNAIP to recognize inner rod proteins and flagellin. Treatment with inner rod and needle proteins (PscI and PscF) from Pseudomonas aeruginosa induced secretion of IL-1β from both THP1 cells driven towards macrophage differentiation and from fresh PBMCs in response to the bacteria. Reduction in NLRC4 or hNAIP using siRNA reduced this IL-1β secretion (82). A second study revealed that primary human macrophages can activate the hNAIP/NLRC4 inflammasome response to PrgJ, and several other inner rod proteins from different bacteria can stimulate the hNAIP/NLRC4 inflammasome. This study further demonstrated that the full-length hNAIP transcript was capable of conferring NLRC4 inflammasome activation in response to a flagellin, inner rod and needle proteins (83). Thus, it appears that unlike murine NAIPs, numerous structurally related bacterial ligands can be sensed by a single human NAIP, encoded by a single NAIP gene.
NLRC4 inflammasome structure provides a model for NLR-inflammasome assembly mechanisms
An important tool used to study the interactions between the different members of the NLRC4 inflammasome is the reconstituted inflammasome assay. Co-expression of a NAIP, NLRC4, specific bacterial ligand, pro-caspase-1 and pro-IL-1β (or a subset of the five proteins) in HEK293 cells is commonly used to generate NLRC4 inflammasome complexes in cells. Multiple studies using co-expression of a NAIP and NLRC4 and subsequent co-precipitation experiments have shown an interaction between the NAIP and NLRC4 (23, 29, 78, 84). These interactions are detected by this assay methodology only in the presence of a NAIP-specific ligand. Self-association of NAIPs did not occur, either in the presence or absence of ligand (29, 84).
Initial studies to determine the stoichiometry of NAIP, ligand and NLRC4 in the NLRC4 inflammasome were conducted using immunoprecipitation of ‘reconstituted’ inflammasome components from transfected cells combined with densitometric analysis of immunoblots. These studies suggested that there was 1 NAIP molecule for each bacterial ligand and for every 2.5 NLRC4 molecules in the inflammasome complex (29).
Two subsequent studies employed nanogold-labeled NAIP and cryo-EM to study isolated recombinant inflammasome complexes demonstrated that there is only 1 NAIP molecule present in NLRC4 inflammasome complexes that molecular modeling suggested contained 10 NLRC4 protomers (36, 37). Because the initial reported studies examined the structure of the multiprotein complexes isolated by immunoprecipitation, it is possible that many of the isolated complexes were only partially formed, leading to a discounted ratio of NLRC4 to NAIP. It is also possible there is differential efficiency in membrane capture or detection of NAIP and NLRC4 in the immunoblots performed. The currently accepted models generally follow the structurally supported model of an 11-member macro-structure containing 1 NAIP and 10 NLRC4 protomers.
Solving the crystal structure of NLRC4 provided an initial glimpse into the protein in its inactive form (85). The crystallized protein lacked the N-terminal CARD and a short region deleted to increase solubility (Δ622–644), but still retained ATPase activity. NLRC4 was bound to ADP, even though ADP was not supplemented in the preparation, suggesting that ADP exchange from the inactive protein is likely relatively slow.
Analysis of the crystal structure of NLRC4 suggests the front edge of the ADP-binding site is formed by the WHD of NLRC4, including His443. His443 is a key residue implicated in a gain-of-function mutation that leads to familial cold autoinflammatory like syndrome (9). His443 forms hydrogen bonds with the β-phosphate group of ADP, which could stabilize the closed or inactive conformation of NLRC4. A similar key histidine residue, His438, is also found in apoptotic protease activating factor 1 (Apaf-1) (86). Lys175 and Ser176 make up the P loop of a Walker A motif (GXXXGKT/S) in the NLRC4 NBD that hydrogen bonds with ADP as predicted for nucleotide-binding proteins that contain this structural motif. Further highlighting the potential role for this nucleotide-binding site in inflammasome activation, two mutations associated with inflammatory disease (S171F and T177A) are within or adjacent to this Walker A motif (8, 10). Two other hydrogen bonds form between the N1 and N6 of adenine and Thr135 of NLRC4 (85). Strangely, though expressed in an insect cell line, the recombinant NLRC4 used for these structural studies was phosphorylated at S533. In the crystal structure, this phosphorylated residue appears to stabilize a HD2-LRR that would serve to maintain the closed, inactive structure of NLRC4. It is possible that S533 phosphorylation has effects on NLRC4 interaction with additional cellular factors that are critical to in vivo inflammasome assembly and that are missed by the static view of the inactive protein afforded by x-ray crystallography. This finding highlights that despite the tremendous value in vitro structural studies provide, they still have substantial limitations in explaining in vivo dynamic processes.
Transfection of NLRC4 with mutation of His443 to leucine increased IL-1β processing in the absence of NAIP5 or Legionella flagellin. Using size-exclusion chromatography, this mutant protein was also found to run in a large molecular weight complex similar in size to the complex wild-type NLRC4 ran in when it was expressed with FliC and NAIP5. Thus the H443P autoinflammatory disease-associated mutant of NLRC4 may be capable of reconstituting inflammasome formation without requirement for the bacterial ligand sensor, NAIP (85). The deletion of the LRR domains also led to an increase in IL-1β processing, and removal of LRRs and HD2 produced an even greater increase when compared with removal of LRRs. The mutation of Tyr617, a residue in the region between the NBD and LRRs whose packing in the structure appears to potentially stabilize the closed structure of the ADP-bound NLRC4, to alanine also led to constitutive NLRC4 inflammasome activation. The crystal structure of ADP-bound NLRC4 suggests that HD2 comes in contact with α8 of the NBD and this interaction keeps NLRC4 in its inactive form (85).
Electron micrographic analysis of negatively stained NLRP1 inflammasomes that were assembled in vitro demonstrated that the NLRP1 inflammasome appeared to have a bladed ring-like structure that is reminiscent of the structure of the caspase-3-activating structure known as the apoptosome (34, 87). The primary protein unit of the apoptosome is Apaf-1, a protein that shares the central NACHT domain primary structure found in NLR proteins. Initial EM studies of NAIP5–NLRC4 inflammasome complexes isolated from transfected cells revealed ring-like structures with 11- or 12-fold symmetry (84).
These initial studies lacked enough structural detail to provide substantial insight into the mechanisms of their assembly. Two later studies using cryo-EM and fitting with the known domain structures generated from crystallographic studies of inactive NLRC4 to deduce the structure of assembled PrgJ–NAIP2–NLRC4 inflammasome complexes have offered insight into the molecular events involved in inflammasome assembly (36, 37). A ligand-bound NAIP interacts with an inactive NLRC4 molecule as the initial step of NLRC4 inflammasome assembly (Fig. 2). In preparations of PrgJ–NAIP2–NLRC4 inflammasomes, fractions from size-exclusion chromatography correlating with complexes smaller than the full inflammasome demonstrated partially assembled structures with the terminal NLR density in the open or activated state. The deduced structure of inflammasome-associated, active NLRC4 determined by cryo-EM suggests that NLRC4 undergoes structural remodeling during activation, with the rotation of the WHD–HD2–LRR regions around the hinge region of the molecule (region between HD1 and WHD) (37). All six known gain-of-function mutations in NLRC4 are found within areas of these subdomains that must undergo extensive changes in their structure between the inactive and activated conformations of the protein (7, 9, 11).
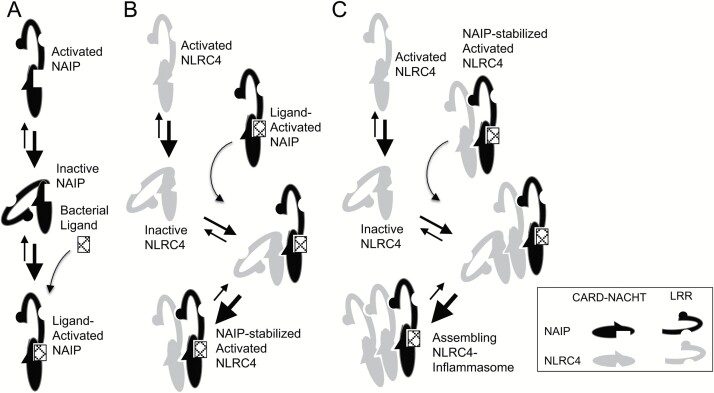
Fig. 2.
A model for assembly of the NLRC4 inflammasome includes sequential association with inactive NLR monomers followed by stabilization of the activated conformation of the added monomer. (A) NAIP favors its inactive conformation at baseline. In the presence of appropriate bacterial ligand, NAIP is stabilized in its open, activated conformation. (B) Ligand-mediated activation of NAIP uncovers an NLRC4-interacting surface on the NACHT domain, which binds the inactive NLRC4 protomer. Like NAIP, unliganded NLRC4 favors its inactive conformation but, once bound to NAIP, the open NLRC4 conformation is stabilized through intermolecular LRR interactions. (C) Activation of NLRC4 reveals a previously buried homo-oligomerization surface, which can interact with both active and inactive NLRC4 conformations. Once associated with an active NLRC4 monomer, the newly added NLRC4 is also stabilized in its active conformation, which can now associate with the next inactive NLRC4 monomer. Step C is repeated until inflammasome assembly is complete.
The surface of inactive NLRC4 that comes in contact with NAIP is the ‘receptor’ or ‘B’ surface (made up of structural elements from the NBD and HD1, including key amino acids I123/I124/D125). This surface is basic in charge and largely accessible in both the active and inactive conformations. The surface of the ligand-bound active NAIP or inflammasome-associated NLRC4 that interacts with the neighboring NLRC4 is termed the ‘catalytic’ surface or ‘A’ surface (including key amino acids R288/Q433/L435) and is largely acidic (37). The LRR of the inactive molecule projects over the region of the NLR that comprises the ‘catalytic’ surface of the activated molecule and also partially occupies the area of the adjacent NLRC4 molecule in the inflammasome structure (37). Disruption of the interaction between and bound ADP by mutation of H433 could destabilize the closed conformation of the protein. The opened structure of the mutant would have an exposed catalytic surface that could interact with other NLRC4 molecules and promote inflammasome assembly in a manner analogous to ligand-bound NAIP, suggesting a mechanism for the constitutive activity of this autoinflammatory disease-associated mutation.
Oligomerization of the wheel-like structure occurs unidirectionally, with the ‘catalytic’ surface of the activated molecule interacting with the ‘receptor’ surface of an inactive molecule. The newly added NLRC4 molecule would then be stabilized in its active conformation through additional contacts between LRR domains. The stabilized open conformation of the newly added NLRC4 would then reveal the protomer’s previously hidden ‘catalytic’ surface that would be capable of interacting with the next inactive NLRC4 molecule to be added to the forming inflammasome.
To test the importance of amino acids predicted to interact between NAIPs and NLRC4, the authors mutated several residues (R586, R590 in NAIP5; R631, R634 in NAIP2; E122, D123, D125 in NLRC4) and showed that these amino acids are involved in the NAIP–NLRC4 interaction, greatly reducing the amount of NAIP co-precipitated with NLRC4 (36). To evaluate the role of the basic amino acid residues on the NLRC4 ‘catalytic’ surface in promoting oligomerization, two residues were replaced with acidic residues to generate the R285D NLRC4 and double-mutant R288D/H289D NLRC4. Less of the mutant proteins co-immunoprecipitated with NAIP2 when they were over-expressed in HEK293 cells, suggesting that the stoichiometry of NLRC4:NAIP was reduced by these mutations (36). Hu et al. also investigated the role of R288 in promoting NLRC4 oligomerization. When NLRC4 with the mutation of R288 to alanine was co-expressed with PrgJ and NAIP2, the resulting proteins formed PrgJ–NAIP2–NLRC4 complexes rather than the PrgJ–NAIP2–NLRC410 inflammasome complexes that formed when wild-type NLRC4 was expressed. These data suggest that acidic amino acids found in the ‘catalytic’ surface of both active NAIP and NLRC4 are not exposed in the inactive crystallized conformation of the proteins and play a key role promoting assembly of the inflammasome complex through their interaction with the ‘receptor’ surface of inactive NLRC4 molecules that are to be added to the assembling complex.
Although these detailed cryo-EM studies provide the best characterized structural information about the assembled NLRC4 inflammasome, other studies have provided additional data with both consistencies and inconsistencies with these models. In the earlier referenced studies by Halff et al. of in vitro NLRC4 inflammasome assembly and ligand recognition, high concentrations of purified NLRC4 were found to assemble into elongated fibrils suggesting that NLRC4 could potentially assemble into a helical structure rather than a closed disc (84). A later study using cryo-electron tomography (cryo-ET) to examine isolated FliC-D0L–NAIP5–NLRC4 inflammasomes (FliC-D0L is a flagellin fragment) demonstrated the presence of rod-like structures that fit the model of extending helices of NLRC4 rather than closed discs reported for the PrgJ–NAIP2–NLRC4 inflammasome (88).
It is unclear from the current published data whether there are actual differences in the structure of the inflammasome wheel versus helix as a result of the NAIP–ligand combination that initiates assembly. It is also possible that these structural differences stem from the expression systems, methods and conditions used to isolate actual complexes. Regardless, the detailed structural data from these studies support a model where a single-liganded NAIP activates NLRC4, which in turn can induce activation of the next unit to assemble in the oligomer. This model allows a single bacterial ligand detection event to trigger signaling-complex assembly. This inflammasome assembly model differs from the Apaf-1 apoptosome assembly model where each Apaf-1 molecule is independently activated by cytochrome C ligand and the liganded Apaf-1 assumes a conformation required for apoptosome assembly, a process that requires a cytochrome C: Apaf-1 stoichiometry of 1:1 (89).
Of the known NLRs that form inflammasome complexes in humans, only NLRC4 directly interacts with caspase-1 using its CARD, thus eliminating the requirement for ASC. In order to confirm that CARD from NLRC4 activates caspase-1, a fluorescence polarization assay was used, similar to one used to examine the polymerization of ASC (1). The CARD from NLRC4 (NLRC4CARD) was tagged with green fluorescent protein, along with full-length NLRC4 (NLRC4FL). NLRC4CARD was able to cause caspase-1CARD polymerization, whereas NLRC4FL did not show significant caspase-1CARD polymerization. Polymerization of caspase-1CARD also occurred with the PrgJ–NAIP2–NLRC4 inflammasome (36). The authors conclude that ASC-independent NAIP/NLRC4 inflammasomes use a similar mechanism for the activation of caspase-1 that is used for ASC-dependent inflammasomes. The formation of the NLRC4 inflammasome may use its oligomerization of CARD to directly nucleate caspase-1 filaments and cause activation of caspase-1 (90).
Interestingly, EM studies of NLRC4-containing inflammasomes carried out with full-length NLRC4 that includes its N-terminal CARD domain have revealed stacked disc structures that resemble the double-disc structure observed in studies of Drosophila apaf-1-related killer (DARK)-containing apoptosomes. It is likely these double discs are formed by homotypic interactions between the protruding CARD domains from the structural proteins. Though there are definitive studies, it is likely these homotypic interactions would not occur in the setting of the NLR (or DARK) CARD domain interacting with downstream effector molecules like pro-caspase-1 through CARD–CARD interactions (91).
It is generally believed that, like the CARD domain of NLRC4, the pyrin domain of NLRP3 would occupy the center of an NRLP3 inflammasome and serve to nucleate ASC via pyrin–pyrin domain interactions. The resulting ASC fibril that extends from the NLRP3 inflammasome would then initiate pro-caspase-1 nucleation via CARD–CARD interactions that result in formation of active caspase-1. NLRP1 appears capable of activating pro-caspase-1 directly through interaction with the C-terminal CARD domain or through N-terminal pyrin domain interactions with ASC as an adapter for pro-caspase-1 interaction in in vitro inflammasome assembly assays (34).
However, these in vitro inflammasome assembly studies preceded the recent discovery of C-terminal autoproteolytic processing that removes the C-terminal CARD domain and exogenous NLRP1-activating signals from proteolytic bacterial toxins that remove the N-terminal pyrin domain. In light of these more recent findings, it is unclear in these models how the NLRP1 inflammasome might interact with pro-caspase-1 after in vivo assembly (18, 19). It is possible that the proteolyzed activated NLRP1 might act as an initiator, catalyzing assembly of non-proteolyzed NLRP1 protomers that have intact domains for initiating pro-caspase activation. It is also possible the proteolyzed, activated NLRP1 could form a hetero-oligomer with other NLR proteins or adapter molecules that interact with pro-caspase-1. Tremendous progress has been made on understanding NLR and effector molecule oligomerization in recent years, but open questions remain regarding how the self-assembling effector complexes are kept in inactive states within cells and how these inhibitory signals are relieved by assembled inflammasomes.
Conclusions
A model for the activation and assembly of the NAIP/NLRC4 inflammasome (Fig. 2) is now supported by substantial structural data (36, 37). First, an inactive NAIP molecule recognizes a specific ligand (flagellin, rod protein or needle protein). This leads to stabilization of the open, activated form of NAIP (79, 80). Second, the ligand-bound, activated NAIP interacts with an inactive NLRC4, which in the absence of the NAIP binding is stabilized in an inactive conformation by interactions between the NBD and WHD. Binding of ligand-bound NAIP by inactive NLRC4 stabilizes the active conformation of NLRC4 with the rotated LRR domain interacting with the LRR of the open NAIP. The newly activated NLRC4 uses its catalytic surface to associate with inactive NLRC4 and stabilize the activate conformation of the newly added NLRC4 molecule. This process is repeated until the inflammasome disk is complete. Third, oligomerization of the CARDs from the various NLRC4 subunits leads to the recruitment of pro-caspase-1 and causes dimerization, autoproteolysis and activation of caspase-1.
It is unclear whether other NLR inflammasomes use the same mechanism by which the last activated NLR protein creates a binding surface that stabilizes the active conformation of the next NLR protein to be added to the assembling complex or if they require ligand binding to activate each NLR that will be added to the inflammasome complex. It is likely that all NLR inflammasomes will share a common-bladed ring structure, perhaps with some variation in the number of monomeric NLR required to complete the ring. Finally, the role of ADP–ATP exchange and ATP hydrolysis has yet to be firmly established and this may also vary between different NLR proteins.
Given that either ASC or Caspase-1 are capable of self-assembly into caspase-1-activating structures once the initial nucleation of their pyrin or CARD domain is achieved, it seems likely that inflammasome formation is not dynamic and, once formed, the NLR-inflammasome becomes dispensable to signal propagation. This calls into question whether there is a deactivation process and inflammasome disassembly, or whether, once initiated, the entire signaling cascade is irreversible. Many of these open questions will likely be answered in the near future as the researchers in the field continue to apply a combination of genetic, biochemical and structural methodologies to the study of inflammasome biology.
Funding
J.A.D. and W.G.F. were supported by the National Institutes of Health through U19 AI109965 and R01 AI088255. J.A.D. was an awardee of the Burroughs Wellcome Fund Career Award for Medical Sciences.
Conflicts of interest statement: J.A.D. is a paid Scientific Consultant for Jecure Therapeutics, Inc. W.G.F. has declared no conflicts of interest.
References
- 1. Lu, A.,, Magupalli, V. G.,, Ruan, J., et al. 2014. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156:1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cai, X.,, Chen, J.,, Xu, H., et al. 2014. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156:1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feldmann, J.,, Prieur, A. M.,, Quartier, P., et al. 2002. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am. J. Hum. Genet. 71:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dode, C., Le Du, N., Cuisset, L.et al. . 2002. New mutations of CIAS1 that are responsible for Muckle-Wells syndrome and familial cold urticaria: a novel mutation underlies both syndromes. Am. J. Hum. Genet. 70:1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoffman, H. M., Mueller, J. L., Broide, D. H., Wanderer, A. A. and Kolodner, R. D. 2001. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saito, M., Nishikomori, R., Kambe, N.et al. . 2008. Disease-associated CIAS1 mutations induce monocyte death, revealing low-level mosaicism in mutation-negative cryopyrin-associated periodic syndrome patients. Blood 111:2132. [DOI] [PubMed] [Google Scholar]
- 7. Canna, S. W., de Jesus, A. A., Gouni, S.et al. . 2014. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46:1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kawasaki, Y., Oda, H., Ito, J.et al. . 2017. Identification of a high-frequency somatic NLRC4 mutation as a cause of autoinflammation by pluripotent cell-based phenotype dissection. Arthritis Rheumatol. 69:447. [DOI] [PubMed] [Google Scholar]
- 9. Kitamura, A., Sasaki, Y., Abe, T., Kano, H. and Yasutomo, K. 2014. An inherited mutation in NLRC4 causes autoinflammation in human and mice. J. Exp. Med. 211:2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang, J.,, Alfano, D. N.,, Squires, J. E., et al. 2017. Novel NLRC4 mutation causes a syndrome of perinatal autoinflammation with hemophagocytic lymphohistiocytosis, hepatosplenomegaly, fetal thrombotic vasculopathy, and congenital anemia and ascites. Pediatr. Dev. Pathol. 20:498. [DOI] [PubMed] [Google Scholar]
- 11. Romberg, N., Al Moussawi, K., Nelson-Williams, C.et al. . 2014. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat. Genet. 46:1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Volker-Touw, C. M.,, de Koning, H. D.,, Giltay, J. C., et al. 2017. Erythematous nodes, urticarial rash and arthralgias in a large pedigree with NLRC4-related autoinflammatory disease, expansion of the phenotype. Br. J. Dermatol. 176:244. [DOI] [PubMed] [Google Scholar]
- 13. Grandemange, S., Sanchez, E., Louis-Plence, P.et al. . 2016. A new autoinflammatory and autoimmune syndrome associated with NLRP1 mutations: NAIAD (NLRP1-associated autoinflammation with arthritis and dyskeratosis). Ann Rheum Dis. 76: 1191. [DOI] [PubMed] [Google Scholar]
- 14. Zhong, F. L., Mamai, O., Sborgi, L.et al. . 2016. Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167:187. [DOI] [PubMed] [Google Scholar]
- 15.International, F. M. F. C. 1997. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell 90:797. [DOI] [PubMed] [Google Scholar]
- 16. French, F. M. F. C. 1997. A candidate gene for familial Mediterranean fever. Nat. Genet. 17:25. [DOI] [PubMed] [Google Scholar]
- 17. Masters, S. L., Lagou, V., Jeru, I.et al. . 2016. Familial autoinflammation with neutrophilic dermatosis reveals a regulatory mechanism of pyrin activation. Sci. Transl. Med. 8:332ra45. [DOI] [PubMed] [Google Scholar]
- 18. Finger, J. N.,, Lich, J. D.,, Dare, L. C., et al. 2012. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem. 287:25030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chavarría-Smith, J., Mitchell, P. S., Ho, A. M., Daugherty, M. D. and Vance, R. E. 2016. Functional and evolutionary analyses identify proteolysis as a general mechanism for NLRP1 inflammasome activation. PLoS Pathog. 12:e1006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chavarría-Smith, J. and Vance, R. E. 2013. Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog. 9:e1003452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levinsohn, J. L.,, Newman, Z. L.,, Hellmich, K. A., et al. 2012. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog. 8:e1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu, H.,, Yang, J.,, Gao, W., et al. 2014. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513:237. [DOI] [PubMed] [Google Scholar]
- 23. Zhao, Y.,, Yang, J.,, Shi, J., et al. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477:596. [DOI] [PubMed] [Google Scholar]
- 24. Yang, J., Zhao, Y., Shi, J. and Shao, F. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl Acad. Sci. USA 110:14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kortmann, J., Brubaker, S. W. and Monack, D. M. 2015. Cutting edge: inflammasome activation in primary human macrophages is dependent on flagellin. J. Immunol. 195:815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dowds, T. A., Masumoto, J., Zhu, L., Inohara, N. and Núñez, G. 2004. Cryopyrin-induced interleukin 1beta secretion in monocytic cells: enhanced activity of disease-associated mutants and requirement for ASC. J. Biol. Chem. 279:21924. [DOI] [PubMed] [Google Scholar]
- 27. Poyet, J. L., Srinivasula, S. M., Tnani, M., Razmara, M., Fernandes-Alnemri, T. and Alnemri, E. S. 2001. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276:28309. [DOI] [PubMed] [Google Scholar]
- 28. Mo, J., Boyle, J. P., Howard, C. B., Monie, T. P., Davis, B. K. and Duncan, J. A. 2012. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J. Biol. Chem. 287:23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tenthorey, J. L., Kofoed, E. M., Daugherty, M. D., Malik, H. S. and Vance, R. E. 2014. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell 54:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu, C., Wang, A., Wang, L., Dorsch, M., Ocain, T. D. and Xu, Y. 2005. Nucleotide binding to CARD12 and its role in CARD12-mediated caspase-1 activation. Biochem. Biophys. Res. Commun. 331:1114. [DOI] [PubMed] [Google Scholar]
- 31. Duncan, J. A.,, Bergstralh, D. T.,, Wang, Y., et al. 2007. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl Acad. Sci. USA 104:8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martino, L., Holland, L., Christodoulou, E., Kunzelmann, S., Esposito, D. and Rittinger, K. 2016. The biophysical characterisation and SAXS analysis of human NLRP1 uncover a new level of complexity of NLR proteins. PLoS One 11:e0164662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harris, P. A., Duraiswami, C., Fisher, D. T.et al. . 2015. High throughput screening identifies ATP-competitive inhibitors of the NLRP1 inflammasome. Bioorg. Med. Chem. Lett. 25:2739. [DOI] [PubMed] [Google Scholar]
- 34. Faustin, B.,, Lartigue, L.,, Bruey, J. M., et al. 2007. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell 25:713. [DOI] [PubMed] [Google Scholar]
- 35. Miao, E. A., Rajan, J. V. and Aderem, A. 2011. Caspase-1-induced pyroptotic cell death. Immunol. Rev. 243:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang, L.,, Chen, S.,, Ruan, J., et al. 2015. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu, Z.,, Zhou, Q.,, Zhang, C., et al. 2015. Structural and biochemical basis for induced self-propagation of NLRC4. Science 350:399. [DOI] [PubMed] [Google Scholar]
- 38. Chae, J. J., Aksentijevich, I. and Kastner, D. L. 2009. Advances in the understanding of familial Mediterranean fever and possibilities for targeted therapy. Br. J. Haematol. 146:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao, W., Yang, J., Liu, W., Wang, Y. and Shao, F. 2016. Site-specific phosphorylation and microtubule dynamics control Pyrin inflammasome activation. Proc. Natl Acad. Sci. USA 113:E4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jéru, I.,, Papin, S.,, L’hoste, S., et al. 2005. Interaction of pyrin with 14.3.3 in an isoform-specific and phosphorylation-dependent manner regulates its translocation to the nucleus. Arthritis Rheum. 52:1848. [DOI] [PubMed] [Google Scholar]
- 41. Moghaddas, F., Llamas, R., De Nardo, D.et al. . 2017. A novel pyrin-associated autoinflammation with neutrophilic dermatosis mutation further defines 14-3-3 binding of pyrin and distinction to familial Mediterranean fever. Ann. Rheum. Dis. 76:2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park, Y. H., Wood, G., Kastner, D. L. and Chae, J. J. 2016. Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat. Immunol. 17:914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van Gorp, H., Saavedra, P. H., de Vasconcelos, N. M.et al. . 2016. Familial Mediterranean fever mutations lift the obligatory requirement for microtubules in pyrin inflammasome activation. Proc. Natl. Acad. Sci. USA 113:14384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qu, Y., Misaghi, S., Izrael-Tomasevic, A.et al. . 2012. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490:539. [DOI] [PubMed] [Google Scholar]
- 45. Matusiak, M., Van Opdenbosch, N., Vande Walle, L., Sirard, J. C., Kanneganti, T. D. and Lamkanfi, M. 2015. Flagellin-induced NLRC4 phosphorylation primes the inflammasome for activation by NAIP5. Proc. Natl Acad. Sci. USA 112:1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu, W.,, Liu, X.,, Li, Y., et al. 2017. LRRK2 promotes the activation of NLRC4 inflammasome during Salmonella typhimurium infection. J. Exp. Med. 214:3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gross, O., Poeck, H., Bscheider, M., Dostert, C.et al. . 2009. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature 459:433. [DOI] [PubMed] [Google Scholar]
- 48. Saïd-Sadier, N., Padilla, E., Langsley, G. and Ojcius, D. M. 2010. Aspergillus fumigatus stimulates the NLRP3 inflammasome through a pathway requiring ROS production and the Syk tyrosine kinase. PLoS One 5:e10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shio, M. T.,, Tiemi Shio, M.,, Eisenbarth, S. C., et al. 2009. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog. 5:e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chuang, Y. T.,, Lin, Y. C.,, Lin, K. H., et al. 2011. Tumor suppressor death-associated protein kinase is required for full IL-1β production. Blood 117:960. [DOI] [PubMed] [Google Scholar]
- 51. Ghonime, M. G.,, Shamaa, O. R.,, Das, S., et al. 2014. Inflammasome priming by lipopolysaccharide is dependent upon ERK signaling and proteasome function. J. Immunol. 192:3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gong, Y. N.,, Wang, X.,, Wang, J., et al. 2010. Chemical probing reveals insights into the signaling mechanism of inflammasome activation. Cell Res. 20:1289. [DOI] [PubMed] [Google Scholar]
- 53. He, Y., Franchi, L. and Núñez, G. 2013. The protein kinase PKR is critical for LPS-induced iNOS production but dispensable for inflammasome activation in macrophages. Eur. J. Immunol. 43:1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hett, E. C.,, Slater, L. H.,, Mark, K. G., et al. 2013. Chemical genetics reveals a kinase-independent role for protein kinase R in pyroptosis. Nat. Chem. Biol. 9:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lu, B., Nakamura, T., Inouye, K.et al. . 2012. Novel role of PKR in inflammasome activation and HMGB1 release. Nature 488:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yim, H. C.,, Wang, D.,, Yu, L., et al. 2016. The kinase activity of PKR represses inflammasome activity. Cell Res. 26:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stutz, A.,, Kolbe, C. C.,, Stahl, R., et al. 2017. NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J. Exp. Med. 214:1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. He, Y., Zeng, M. Y., Yang, D., Motro, B. and Núñez, G. 2016. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shi, H., Wang, Y., Li, X.et al. . 2016. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hellmich, K. A.,, Levinsohn, J. L.,, Fattah, R., et al. 2012. Anthrax lethal factor cleaves mouse nlrp1b in both toxin-sensitive and toxin-resistant macrophages. PLoS One 7:e49741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bürckstümmer, T.,, Baumann, C.,, Blüml, S., et al. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10:266. [DOI] [PubMed] [Google Scholar]
- 62. Fernandes-Alnemri, T., Yu, J. W., Datta, P., Wu, J. and Alnemri, E. S. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hornung, V.,, Ablasser, A.,, Charrel-Dennis, M., et al. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Roberts, T. L., Idris, A., Dunn, J. A.et al. . 2009. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323:1057. [DOI] [PubMed] [Google Scholar]
- 65. Jin, T., Perry, A., Jiang, J.et al. . 2012. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ru, H.,, Ni, X.,, Zhao, L., et al. 2013. Structural basis for termination of AIM2-mediated signaling by p202. Cell Res. 23:855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yin, Q., Sester, D. P., Tian, Y.et al. . 2013. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell. Rep. 4:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Franchi, L.,, Amer, A.,, Body-Malapel, M., et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in Salmonella-infected macrophages. Nat. Immunol. 7:576. [DOI] [PubMed] [Google Scholar]
- 69. Miao, E. A.,, Alpuche-Aranda, C. M.,, Dors, M., et al. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569. [DOI] [PubMed] [Google Scholar]
- 70. Ren, T., Zamboni, D. S., Roy, C. R., Dietrich, W. F. and Vance, R. E. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Miao, E. A., Ernst, R. K., Dors, M., Mao, D. P. and Aderem, A. 2008. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc. Natl Acad. Sci. USA 105:2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Molofsky, A. B.,, Byrne, B. G.,, Whitfield, N. N., et al. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203:1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zamboni, D. S.,, Kobayashi, K. S.,, Kohlsdorf, T., et al. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7:318. [DOI] [PubMed] [Google Scholar]
- 74. Lamkanfi, M.,, Amer, A.,, Kanneganti, T. D., et al. 2007. The Nod-like receptor family member Naip5/Birc1e restricts Legionella pneumophila growth independently of caspase-1 activation. J. Immunol. 178:8022. [DOI] [PubMed] [Google Scholar]
- 75. Lightfield, K. L.,, Persson, J.,, Brubaker, S. W., et al. 2008. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9:1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lightfield, K. L.,, Persson, J.,, Trinidad, N. J., et al. 2011. Differential requirements for NAIP5 in activation of the NLRC4 inflammasome. Infect. Immun. 79:1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Miao, E. A.,, Mao, D. P.,, Yudkovsky, N., et al. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 107:3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kofoed, E. M. and Vance, R. E. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477:592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tenthorey, J. L.,, Haloupek, N.,, López-Blanco, J. R., et al. 2017. The structural basis of flagellin detection by NAIP5: a strategy to limit pathogen immune evasion. Science 358:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang, X., Yang, F., Wang, W.et al. . 2017. Structural basis for specific flagellin recognition by the NLR protein NAIP5. Cell Res. 2018;28:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rayamajhi, M., Zak, D. E., Chavarria-Smith, J., Vance, R. E. and Miao, E. A. 2013. Cutting edge: mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191:3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Grandjean, T.,, Boucher, A.,, Thepaut, M., et al. 2017. The human NAIP-NLRC4-inflammasome senses the Pseudomonas aeruginosa T3SS inner-rod protein. Int. Immunol. 29:377. [DOI] [PubMed] [Google Scholar]
- 83. Reyes Ruiz, V. M.,, Ramirez, J.,, Naseer, N., et al. 2017. Broad detection of bacterial type III secretion system and flagellin proteins by the human NAIP/NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 114:13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Halff, E. F., Diebolder, C. A., Versteeg, M., Schouten, A., Brondijk, T. H. and Huizinga, E. G. 2012. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J. Biol. Chem. 287:38460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hu, Z., Yan, C., Liu, P.et al. . 2013. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341:172. [DOI] [PubMed] [Google Scholar]
- 86. Riedl, S. J., Li, W., Chao, Y., Schwarzenbacher, R. and Shi, Y. 2005. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434:926. [DOI] [PubMed] [Google Scholar]
- 87. Yu, X.,, Acehan, D.,, Ménétret, J. F., et al. 2005. A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure 13:1725. [DOI] [PubMed] [Google Scholar]
- 88. Diebolder, C. A., Halff, E. F., Koster, A. J., Huizinga, E. G. and Koning, R. I. 2015. Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: implications for NLR activation. Structure 23:2349. [DOI] [PubMed] [Google Scholar]
- 89. Yuan, S., Topf, M., Reubold, T. F., Eschenburg, S. and Akey, C. W. 2013. Changes in Apaf-1 conformation that drive apoptosome assembly. Biochemistry 52:2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lu, A. and Wu, H. 2015. Structural mechanisms of inflammasome assembly. FEBS J. 282:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yu, X., Wang, L., Acehan, D., Wang, X. and Akey, C. W. 2006. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J. Mol. Biol. 355:577. [DOI] [PubMed] [Google Scholar]