Abstract
BRAF is the most frequently mutated gene in melanoma. Constitutive activation of mutant BRAFV600E leads to aberrant Ras-independent MAPK signaling and cell transformation. Inhibition of mutant BRAF is a current frontline therapy for such cases, with improved survival compared with chemotherapy. Unfortunately, reactivation of MAPK signaling by several mechanisms has been shown to cause drug resistance and disease recurrence. In this work, we describe the co-occurrence of an in-frame deletion within an amplified BRAFV600E locus and a missense point mutation of the transcriptional repressor BCORL1 in vemurafenib-resistant A375 melanoma cells. Functional data confirmed that truncated p47BRAFV600E and mutant BCORL1Q1076H both contribute to resistance. Interestingly, either endogenous BCORL1 silencing or ectopic BCORL1Q1076H expression mimicked the effects of a CRISPR/Cas9-edited BCORL1Q1076H locus, suggesting a complex mixture of loss- and gain-of-function effects caused by the mutation. Transcriptomic data confirmed this hypothesis. Finally, we show that the pan-RAF inhibitor sorafenib is not affected by expression of BRAF deletion variant and effectively synergizes with vemurafenib to block resistant cells, suggesting a possible intervention for this class of mutants.
Introduction
Molecularly targeted drugs have emerged as the most promising approach to cancer therapy in the last decade. Several selective treatments are currently approved for specific subsets of cancer patients carrying well-defined driver lesions [1], [2], [3], [4]. The serine/threonine kinase BRAF was the first gene to be found mutated in cancer by high-throughput sequencing technologies [5]. The Raf proteins family (ARAF, BRAF, CRAF) provides key effectors along the RAS/MAPK cellular pathway, whose activation controls cell division, survival, and differentiation and is altered in a large fraction of tumors. In particular, mutations at codon 600 of BRAF are present in about 40% to 60% of melanoma patients (mostly V600E), making BRAF a strong candidate for targeted therapy [6]. Indeed, BRAF silencing or pharmacological inhibition leads to melanoma cell death and tumor regression in preclinical models and in patients [7], [8]. Selective BRAF inhibitors, vemurafenib or dabrafenib, induce 50% to 70% responses and prolong progression-free survival compared to chemotherapy [9], [10]; both drugs are now approved for first-line treatment of BRAF-mutated melanoma. Unfortunately, responses are short-lived due to the acquisition of drug resistance. In contrast to other tumors in which resistance mainly arises through mutations of the drug target, resistance to vemurafenib seems to be very heterogeneous, as several alternative ways to reactivate the MAPK pathway have been described, including aberrant BRAFV600E splicing, NRAS or MEK1/2 mutations, activation of RTKs, NF1 loss, activation of bypass kinases, or alternative survival pathways such as PI3K/AKT [6], [11], [12], [13]. Moreover, adaptive resistance has been observed when cells activate a reversible, drug-induced stress response that allows MAPK reactivation [14]. Clearly, understanding the mechanisms of resistance is a key step in devising new therapeutic strategies.
Here we describe the co-occurrence of an amplified, in-frame internally deleted BRAF locus and a BCORL1 point mutation (Q1076H) in vemurafenib-resistant cells. BCORL1 is a transcriptional co-repressor, homologous to BCOR, whose function is poorly understood. It is known to interact with histone deacetylases, CtBP, and PCGF1 [15], [16]. Specifically, BCORL1 was shown to repress E-cadherin expression via interaction with CtBP. BCORL1 has been found mutated in hematologic disorders and implicated in hepatocellular carcinoma gene fusion events and tumor progression [17], [18], [19], [20], [21], [22]. In order to functionally validate our findings, both genetic alterations were introduced in parental, vemurafenib-sensitive A375 cells to reproduce the resistant phenotype. The truncated p47BRAFV600E protein appeared to be the dominant driver of resistance; however, manipulation of BCORL1 function conferred a small but consistent shift in sensitivity to parental cells, suggesting that BCORL1 mutation may cooperate in the induction of resistance.
Methods
Cell Lines
The A375 malignant melanoma cell line carrying a homozygous V600E mutation in BRAF was purchased from the American Type Culture Collection, where cells are routinely genotyped to verify their identity. The cells were maintained in RPMI cell culture media supplemented with 100 U/ml penicillin, 100 mg/ml gentamicin, and 2 mM glutamine. The GFP-BCoR-L1 plasmid was a kind gift of Dr. K. K. Khanna (Queensland Institute of Medical Research, Australia). The Q1076H mutation was introduced by site-directed mutagenesis as described [23] using the following sense: 5′-TGGCCTCCCAGTGGCTCCCCATAGGGGCCAAGCTGAAGGTTC-3′ and the corresponding antisense oligo. Truncated p47BRAFV600E sequence was amplified from A375-R1 cells using the primers listed in Supplementary Table 1 and cloned in phCMV2 vector in frame with the N-terminal HA tag. The fragment encoding for HA-tagged p47BRAFV600E was then subcloned into the pCDH-EF1-Puro vector. The cells were transfected using FuGENE 6 Transfection Reagent (Promega) according to the manufacturer's instructions. Cells transfected with GFP-BCoR-L1 wild-type and Q1076H mutated plasmids were selected with G418 (1 mg/ml), and the GFP-positive population was sorted by FACScan. The cells transfected with pCDH-HA-p47BRAFV600E were selected by puromycin (1.25 μg/ml) and subcloned by limited dilution to isolate several clones, which were then assayed for levels of p47BRAFV600E expression. A clone expressing intermediate BRAF levels (clone 03E9) was used for the generation of double transfectants (BRAF/BCORL1).
Antibodies
The following antibodies were used for Western blotting experiments: anti-BRAF (H-145) from Santa Cruz Biotechnology, anti-HA (clone HA.11) from Covance, and anti-actin from Sigma-Aldrich. All the other antibodies were from Cell Signaling Technology: phospho-MEK1/2 (S217-S221), total MEK1/2, phospho-ERK1/2(T202-Y204), total ERK1/2, phospho-c-Raf(S338; clone 56A6), and total c-Raf (clone C-12).
In Vitro BRAF Activity Assay
BRAF was immunoprecipitated from total lysates of A375 parental and resistant cells using anti-BRAF polyclonal antibody and incubated with 1 μg of recombinant human inactive MEK1 (Millipore), 2 mM ATP, and different vemurafenib concentrations in appropriate reaction buffer (25 mM Hepes pH 7.0, 5 mM MnCl2, and 5 mM MgCl2, 1 mM Na3VO4) for 20 minutes at 30°C. The reaction was stopped by addition of Laemmli buffer and heating at 95°C for 10 minutes. The samples were then run on SDS-PAGE, transferred to nitrocellulose membrane, and probed with anti–phospho-MEK1/2 antibody. The membrane was stripped and reprobed with anti-BRAF antibody.
Cell Proliferation Assay
The cells were seeded in 96-well plate and incubated for 72 hours with semi-log dilutions of inhibitors. Cell growth and viability were assayed by using the MTS CellTiter 96 AQueous Assay (Promega) according to instructions. Background signal was subtracted, and all values were normalized to DMSO-treated controls.
Next-Generation Sequencing
Whole-exome sequencing was performed on Illumina platform and analyzed as described earlier [24]. Variants were considered if present in >35% of reads from A375-R1 and in <10% of parental A375 sample reads within regions of at least 20× coverage. Synonymous substitutions, noncoding changes, and known SNPs were filtered out. The BCORL1 somatic mutation identified by WES was then validated by Sanger sequencing at GATC Biotech (see Supplementary Table 1). For RNA-sequencing experiments, total RNA was isolated from A375 cells stably expressing wild-type or mutant BCORL1, or empty vector. Three independent samples were collected per cell line. Library preparation was performed at Galseq srl (Italy) starting from 1 μg of total RNA using a TruSeq Stranded mRNA Library Prep Kit (Illumina). RNA quality was assessed by using a Tape Station instrument (Agilent). To avoid overrepresentation of 3′ ends, only high-quality RNA with an RNA Integrity Number ≥ 8 was used. The libraries were sequenced in paired-end mode (2×150bp) at a depth of 10 million read pairs per sample. Fastq files were aligned to the human genome (GRCh38/hg39) by using STAR, a splice junction mapper for RNA-Seq data, together with the corresponding splice junctions Ensembl GTF annotation. Differential gene expression and statistical analysis were performed with DESeq2 [25] and SPSS (IBM), respectively. Functional enrichment was performed with DAVID [26]. Gene expression heatmap and unsupervised hierarchical clustering were performed with GENE-E (Broad Institute).
Silencing and CRISPR/Cas9
Gene-specific siRNA pools (siGENOME SMARTpool; BCORL1, #M-019215-01; BRAF, #M-003460-03; RAF1, #M-003601-02) and control nontargeting siRNA (#D-001210-01) were purchased from Dharmacon and used for transient transfections following manufacturer's recommendations. For the generation of stably silenced cell lines, a BCORL1-specific mission shRNA plasmid clone targeting the 3′-UTR was purchased from Sigma-Aldrich (#TRCN0000033474). Transfected cells were selected with puromycin (1.25 μg/ml) and subcloned by limiting dilution to isolate individual clones. For CRISPR/Cas9 system, the plasmid pGS-CMV-hCas9 (GeneScript) was employed, together with different guide RNAs: for disruption of BCORL1 gene, a plasmid encoding a gRNA targeting the beginning of the coding sequence (5′-TTGTGCACGCCGCTGTAGAG-3′; antisense orientation; Supplementary Figure 9) under the U6 promoter was obtained from Horizon Discovery. For specific point gene editing, a gRNA corresponding to the region adjacent to the Q1076H mutation was purchased from GeneScript (5′-AACTTCAGCTTGGCCCCTC; antisense orientation) and used in combination with a 181-mer donor DNA oligonucleotide containing the desired mutation (Supplementary Figure 9).
Inhibitors
Vemurafenib was kindly provided by Plexxikon. Trametinib, dabrafenib, GDC-0879, sorafenib, and Cdk4 Inhibitor II (NSC 625987) were purchased from Selleck Chemicals. All compounds were dissolved in DMSO, aliquoted, and stored at −20°C until used.
Drug Uptake/Efflux
Quantitative real-time PCR of SLC22A1, ABCB1, and ABCC1 genes was performed using the primers and cycling conditions described by Redaelli et al. [27]. To quantify the intracellular concentration of vemurafenib, parental A375 and resistant A375-R1 cells were treated in triplicate for 2 hours with 1 μM vemurafenib and then washed. Samples were collected before (time 0) and 30 and 120 minutes after washout. Quantification was then performed by high-performance liquid chromatography–tandem mass spectrometry method as described [28], with partial modifications.
Quantitative PCR
Real-time quantitative PCR was performed on Agilent Mx3005P QPCR System using the primers listed in Supplementary Table 1 and Brilliant III Ultra-Fast SYBR QPCR Master Mix (Agilent), following the recommended protocol. The GAPDH housekeeping gene was used as an internal reference. All experiments were performed in triplicate.
Results
To investigate the mechanisms leading to acquired resistance to BRAF inhibition, we cultured A375 melanoma cells (BRAFV600E/−) in the presence of vemurafenib, starting from 1 μM, a concentration that fully suppresses MAPK pathway activation and cell growth in A375 cells [7]. Drug concentration was increased each time the cell population, after passing through a bottleneck, restored a normal growth rate, as described previously [23]. After various dose increases, a vemurafenib-resistant cell line was selected (A375-R1), displaying an IC50 value over 100-fold that of the parental line (Figure 1A). A375-R1 cells maintained a highly active MAPK pathway in the presence of vemurafenib (Figure 1B).
Figure 1.
Characterization of vemurafenib-resistant cells. (A) Dose-response curves of A375 and A375-R1 cells in the presence of increasing concentrations of vemurafenib. The proliferation rate of vehicle-treated controls is set as 100%. (B) Western blot analysis of A375 and A375-R1 cell lysates after 4-hour treatment with the indicated concentrations of vemurafenib. The membranes were probed with antibodies recognizing phosphorylated (pMEK) and total MEK1/2 proteins. (C) Quantitative real-time PCR showing similar BRAF transcription in parental and resistant cells. GAPDH was used for normalization. (D) In vitro kinase activity of immunoprecipitated BRAF from A375 and A375-R1 cells on a GST-MEK1 substrate. Left: The reaction was run in the absence or presence of ATP. Right: ATP was added in all samples in the presence of the indicated concentrations of vemurafenib. Reactions were stopped by Laemmli buffer, run on SDS-PAGE, and probed with anti-pMEK antibody. Anti-BRAF blot is shown for loading check.
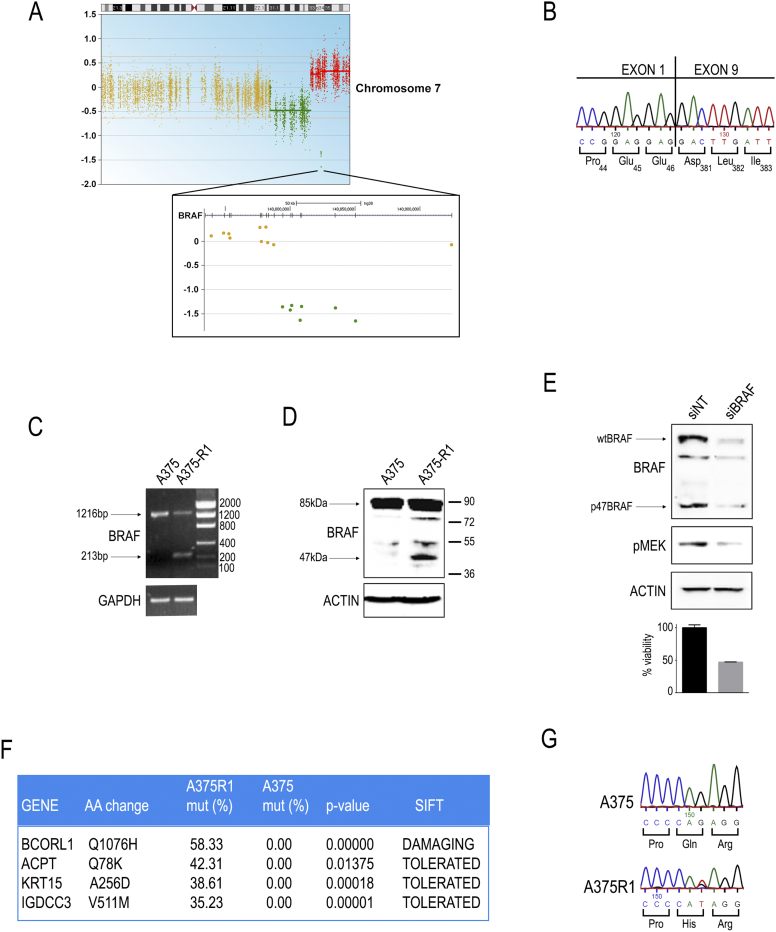
Initial characterization of A375-R1 cells showed that BRAF expression was not changed (Figure 1C). Similarly, quantitative expression analysis of selected factors that may impact on MAPK activation showed only very modest increases in COT, MOS, and LCK expression (Supplementary Figure S1). We further excluded mutations in Ras family proteins, as well as the acquisition of secondary BRAF point mutations, while confirming the persistence of BRAFV600E (data not shown). Drug uptake and efflux were not altered (Supplementary Figure S2). Interestingly, immunoprecipitated BRAF was refractory to inhibition in in vitro kinase assays, suggesting intrinsic drug resistance (Figure 1D). A375-R1 cells maintained their drug-resistant phenotype even after 1 month off drug, suggesting a stable, irreversible mechanism (Supplementary Figure S3). Whole-exome sequencing analysis revealed the amplification of an internally in-frame deleted BRAFV600E gene (Figure 2A). The loss of BRAF exons 2 to 8, with in-frame joining of exon 1 to exon 9 (Figure 2, B-C), generated a shorter protein lacking residues 47 to 380, including the whole Ras-binding domain and the cysteine-rich region, of a theoretical molecular weight of approximately 47 kDa (p47BRAFV600E; Figure 2D, Supplementary Figure S4). In addition to the truncated p47BRAFV600E, we could still detect the full-length 85-kDa BRAFV600E kinase in A375-R1 cells (Figure 2, C-D). Moreover, chromosome 7 appeared to have undergone complex gain/loss events as determined by copy number analysis (Figure 2A). Interestingly, in A375-R1 cells, RAF1 was constitutively phosphorylated on Ser338; however, RAF1 knockdown did not affect MAPK activationor sensitivity to vemurafenib (Supplementary Figure S5). In contrast, siRNA-mediated silencing of BRAF suppressed MEK phosphorylation and cell viability (Figure 2E). Furthermore, heterozygous point mutations in four additional loci were found in A375-R1 cells at >35% frequency, used as a cutoff for the identification of prevalent alterations (Figure 2F). Three of four identified point mutations were predicted to have no impact on protein function. In addition, we could not detect expression of these three genes in A375 or A375-R1 cells by quantitative PCR or RNA-seq (data not shown). In contrast, the BCORL1 gene was expressed at a good level (RPKM ≈200). A375-R1 cells were found to carry a mutation in BCORL1 that caused the substitution of a histidine for a glutamine (Q1076H; Figure 2, F-G), within a linear motif predicted to be recognized by SH3 domains (LPVAPQR, aa 1071-1077). Glutamine 1076 is very conserved across species (Supplementary Figure S6), and the Q1076H substitution was indicated as possibly damaging by SIFT and PolyPhen-2 algorithms. According to OncoScore [29], BCORL1 is a cancer-causing gene, with a score of 62.2 (values >21 are considered oncogenic).
Figure 2.
BRAF and BCORL1 mutations in resistant cells. (A) Whole-exome sequencing copy number variation analysis revealed focal imbalances of the distal region of chromosome 7 in A375-R1 cells compared to parental cells. BRAF locus (zoomed area) lies within the amplified region but shows loss of exons 2 to 8. (B) Sanger sequencing of the deleted BRAF allele showing in-frame junction of exon 1 with exon 9 in A375-R1 cells. (C) PCR amplification of the region across the break point reveals the presence of two BRAF transcripts in A375-R1 cells: full-length wild-type (1216-bp band) and truncated (213-bp band). (D) Anti-BRAF Western blotting shows the presence of a smaller band of approximately 47kDa in A375-R1 cells. Actin is shown as a loading control. (E) siRNA-mediated silencing of BRAF in A375-R1 cells leads to downregulation of both full-length and truncated BRAF proteins, suppression of MEK1/2 phosphorylation, and decrease in cell viability. (F) Whole-exome sequencing comparative analysis of A375-R1 versus A375 cells revealed acquired mutations in the four indicated genes at frequency >35%. SIFT prediction of mutation impact on protein function is shown. (G) Sanger validation of BCORL1 heterozygous genomic mutation in A375-R1 cells.
In order to verify the biological relevance of these findings, p47BRAFV600E and BCORL1Q1076H were introduced in parental A375 cells (Figure 3 and Supplementary Figure S7) transiently or stably. Both mutants were able to induce a small shift in sensitivity to vemurafenib (increased IC50 and increased MEK/ERK phosphorylation) when expressed alone, and this effect was further enhanced by simultaneous co-expression (Figure 3, A-C, transient expression; Figure 3, D-G, stable clones). However, p47BRAFV600E expression levels were found to correlate with resistance (Figure 3H and Supplementary Figure S8): clones with high p47BRAFV600E expression were highly resistant, and their sensitivity was not further altered by expression of BCORL1Q1076H. Interestingly, acute silencing of BCORL1 in A375-R1 cells slightly reduced BRAF protein levels and, consequently, MEK activation (Figure 3I) but did not cause any significant change in drug sensitivity. On the other hand, overexpression of BCORL1Q1076H, but not BCORL1WT, increased BRAF in parental cells (Figure 3I). This effect was not observed at transcript level, suggesting a posttranslational type of control (data not shown). Altogether, these data suggest that expression of BCORL1Q1076H can only confer partial resistance to vemurafenib in A375 parental cells, while p47BRAFV600E may explain most of the resistant phenotype in A375-R1 cells.
Figure 3.
Functional validation of mutations. (A-C) Transient co-transfection of p47BRAFV600E and BCORL1Q1076H conferred partial resistance to vemurafenib-mediated inhibition of A375 cell growth (A) and MAPK signaling (C). Forty-eight hours after transfection, the cells were challenged for additional 48 (A) or 4 (C) hours with vemurafenib and harvested. Cell proliferation shown in A was detected by thymidine incorporation. (B) Histogram plot from two proliferation experiments (mean ± SEM). MAPK pathway activity shown in C was detected by Western blotting as MEK and ERK phosphorylation. (D-G) Stably transfected A375 clones expressing HA-tagged p47BRAFV600E (D, Western blot, clone E9) and wild-type or mutated BCORL1 (E, qPCR), singularly or combined, were isolated and tested in proliferation assays for vemurafenib sensitivity. Data from at least four independent experiments are reported as mean ± SEM IC50 values (F); the red bar represents A375-R1 cells, for comparison; ev, empty vector. (G) Representative Western blot showing MEK1/2 phosphorylation (pMEK) in transfected cells treated with the indicated vemurafenib doses for 4 hours. Actin is shown as a control. (H) Correlation between p47BRAFV600E expression (x-axis, log scale) determined by p47-specific qPCR and vemurafenib IC50 (y-axis, log scale). Clone E9, used for all experiments, is indicated. (I) Upper panel: siRNA-mediated silencing of BCORL1 (siBCORL1) and BRAF (siBRAF) in A375-R1 cells; a nontargeting scrambled siRNA (siNT) was used as a control. Lysates were probed with the indicated antibodies. Lower panel: Parental A375 cells were transiently transfected with empty vector, wild-type (WT), or mutated (Q1076H) BCORL1 and checked for BRAF expression. Actin is shown for loading control.
Since neither ectopic BCORL1Q1076H overexpression in parental cells nor its silencing in A375-R1 cells caused a major phenotype and considering that most of BCORL1 mutations reported in literature [17], [18], [22] are loss-of-function type (nonsense/frameshift), we hypothesized that BCORL1 loss, rather than gain, might induce drug resistance in A375 cells. To test this hypothesis, BCORL1 was knocked out by RNA interference (Figure 4, A-B andD-E) or by CRISPR/Cas9 (Figure 4, C andF and Supplementary Figure S9A) in the A375 parental cell line and in A375 cells stably expressing p47BRAFV600E before testing for vemurafenib sensitivity. In all cases, we noted a small increase of IC50, indicating a mild induction of drug resistance (Figure 4H). These data suggested that loss of BCORL1 function may, at least in part, contribute to resistance to BRAF inhibition. In line with the initial observation, BCORL1-silenced or knockout cells showed reduced BRAF protein expression (Supplementary Figure S10A). The same effect was seen in stably silenced A375-R1 cells, with no effect on cell growth. In order to assess the effects of the exact mutation found in A375-R1 resistant cells, CRISPR/Cas9 system was employed to introduce a BCORL1Q1076H substitution at the endogenous locus in parental cells (Supplementary Figure S9B). We obtained clones with heterozygous Q1076H mutation and a frameshift on the other allele (BCORL1Q1076H/fs). Two such clones were tested and proved moderately resistant to vemurafenib, while a third clone carrying a frameshift and a wild-type allele (BCORL1WT/fs) showed exactly the same sensitivity as the parental cell line (Figure 4, G-H). The edited clones showed a reduction of total BCORL1 transcript compared to parental cells (Supplementary Figure 10B). However, the direct comparison between mutant and wild-type clones rules out any gene dosage effect on drug sensitivity. These results confirmed that mutant BCORL1 affects sensitivity to BRAF inhibition.
Figure 4.
Stable knockdown of BCORL1 in A375 cells (A-C) and A375-p47BRAFV600E [clone E9] (D-F). (A, D) Efficiency of shRNA-mediated BCORL1 silencing as shown by quantitative PCR using GAPDH as a reference gene. (B, E) Dose-response curves obtained in the presence of increasing concentrations of vemurafenib, with cells expressing a nontargeting (shNT, blue curves) or a BCORL1-specific (shBCORL1, red curves) shRNA. (C, F) CRISPR/Cas9 system was used to disrupt BCORL1 gene; vemurafenib dose-response curves are shown comparing knockout (KO) with parental (WT) cells. (G) CRISPR/Cas9-mediated gene editing was used to introduce the Q1076H substitution in the endogenous BCORL1 locus; vemurafenib dose-response curves are shown comparing two mutated clones (C8 and D7) with a wild-type clone (WT) and with parental A375 cells (Par). All curves are representative of at least three experiments. For all panels, extra-sum-of-squares F test was run to compare the two curves; Pvalues are indicated at the lower-left corner, where P < .05 indicates that the curves are significantly different. (H) Summary of IC50 data (mean ± SEM) obtained from all experiments. For each comparison, control cells IC50 (blue bars, shNT or WT) is set to 1; test cells IC50 value (red bars, shBCORL1 or KO) is normalized over its control. Student's t test was used to compare IC50s. *P < .05; **P < .01.
Since we had only indirect evidence toward the consequences of the Q1076H mutation on BCORL1 function, which is described as a transcriptional repressor, we investigated how the transcriptional landscape changed in A375 cells overexpressing the wild-type or the mutant form of BCORL1 (Figure 5). Overall, 213 and 132 genes were significantly down- and upregulated, respectively, by forced overexpression of wild-type BCORL1 compared to parental cells (Figure 5, A-D). Only about one fourth of these changes were conserved in the mutant-expressing cells, suggesting a loss of function. Analysis of replicate consistency by Pearson correlation [30] again suggested a loss-of-function impact on gene function, although not significant due to low replicates number (not shown). However, a relevant number of genes were deregulated (40 down, 62 up) by the mutant only, indicating that a change of function is also associated with the mutation. When the signatures were analyzed by gene ontology (Figure 5, E-F), the top terms where similar for both datasets: nervous system development in the upregulated gene list and kidney development for downregulated genes. However, two relevant exceptions were found: the mutant suppressed negative regulation of MAPK activity, whereas wild-type BCORL1 induced melanocyte differentiation genes (Figure 5, E-F and Supplementary Figure 11A).
Figure 5.
Transcriptomic analysis of A375 stably overexpressing wild-type (WT) or mutated (MUT) BCORL1 compared to parental (Par) A375. Venn diagrams (A, C) show the number of differentially expressed genes (DEGs) versus Par. Heatmaps (B, D) show hierarchical clustering of genes significantly dysregulated by WT versus Par (B, down; D, up). Note that regulation obtained by WT samples is partially lost in MUT samples. (E-F) Functional annotation of significant DEGs in WT (E) and MUT (F) samples; red, upregulated genes; green, downregulated genes; a dotted line indicates P = .05.
Next, we asked whether it is possible to circumvent resistance to vemurafenib by using alternative inhibitors or drug combinations. First, we explored cross-resistance of A375-R1 cells to other RAF inhibitors [31], [32], [33]. These cells were resistant to dabrafenib (Supplementary Figure S12A) but sensitive to GDC-0879(Figure 6A) and sorafenib (Figure 6B). Interestingly, sorafenib synergized with vemurafenib in proliferation assays and in blocking MEK1/2 activation (Figure 6, C-E), suggesting that the combination of two RAF inhibitors with a different mode of action can result in enhanced efficacy against BRAF deletion mutants. Next, we investigated the sensitivity of A375-R1 cells to a MEK1/2 inhibitor to test if intervention downstream of p47BRAFV600E would be able to block the oncogenic pathway and cell growth. Trametinib showed picomolar activity in both parental and drug-resistant cells, with superimposable dose-response curves (Figure 6F), suggesting that targeting the pathway downstream of the mutation may overcome resistance. Finally, since A375 cells carry a deletion of CDKN2A locus, encoding for the CDK inhibitor protein p16Ink4A, we tested a pharmacological blockade of CDK4/Cyclin D1 using the Cdk4 Inhibitor II (NSC 625987). Parental and vemurafenib-resistant cells displayed similar sensitivity to this inhibitor, with A375-R1 cells being slightly more sensitive, suggesting another way to bypass resistance (Supplementary Figure S12B). However, the CKD4 inhibitor did not show any cooperative effect with vemurafenib (data not shown).
Figure 6.
(A) Western blot shows inhibition of pMEK by GDC-0879, but not by vemurafenib (vem), in A375-R1 cells. (B) Dose-response curves of sorafenib showing equal sensitivity of A375 (blue) and A375-R1 (red) cells. (C) A375 and A375-R1 cells were treated for 4 hours with the indicated drugs and analyzed by Western blot. In A375-R1 cells, MAPK signaling is sensitive to sorafenib (1 μM) and fully suppressed by combined treatment. (D) In vitro kinase assay of immunoprecipitated BRAF (see Figure 1D) in the presence of vemurafenib, sorafenib, or both. (E) Dose-response curves of vemurafenib alone (red) or in the presence of sorafenib 0.3 μM (blue) or 1 μM (green) in A375-R1 cells. (F) Dose-response curves of trametinib showing equal sensitivity of A375 (blue) and A375-R1 (red) cells.
Discussion
Acquisition of pharmacological resistance to targeted therapies is the great challenge of current oncology. Specific inhibition of mutant BRAF has shown tremendous efficacy in BRAF-mutated cancer, particularly in malignant melanoma. However, drug resistance has so far limited its clinical success. We describe here the co-occurrence of two genetic lesions in BRAF inhibitor-resistant cells. Indeed, melanoma cells appear to possess several ways to evade drug-induced cell death. Increased RAF1 expression has been suggested as one such mechanism [34]. We noted RAF1 hyperactivation in our vemurafenib-resistant cells; however, this did not seem to contribute to drug resistance. Likely, it is a by-product of aberrant dimerization properties of the truncated p47BRAFV600E protein through allosteric transactivation of the second protomer in BRAF-RAF1 heterodimers [35], [36]. A modest increase of COT (MAP3K8), previously associated to reactivation of MAPKs under vemurafenib treatment [37], as well as c-Mos [38] and Lck [39] expression was observed. However, these minor changes are unlikely to provide a significant contribution to drug resistance. Johannessen et al. found that cells expressing less than five-fold COT overexpression were still sensitive to inhibition [37]. Other possible mediators of resistance, such RAS or SRC [40], [41], were also ruled out.
Poulikakos et al. discovered an alternatively spliced BRAF transcript in vemurafenib-resistant cells and patients [42]. This aberrant BRAFV600E variant shows enhanced dimerization properties that confer inhibitor resistance through negative allostery. The authors excluded genomic deletion and ascribed this variant to aberrant splicing. Another report described amplification of the entire V600E-mutated BRAF locus as a mechanism of acquired drug resistance [43]. In this work, we identified a genomic rearrangement of the BRAF locus, resulting in an internally deleted mutant BRAF gene resembling splice variants, within an amplified region of chromosome 7. While this manuscript was under review, a clinical case was reported to carry a similar genomic deletion, providing clinical relevance to our finding [44]. Interestingly, since we could still detect the full-length gene in a cell line known to be BRAF hemizygous [45], we hypothesize that the amplification occurred before the deletion or, alternatively, that two populations are present in the culture. The deleted BRAF gene leads to the expression of a truncated p47BRAFV600E protein, which was sufficient to confer resistance to vemurafenib and dabrafenib. In contrast, GDC-0879 and sorafenib were able to inhibit the MAPK pathway in cells expressing p47BRAFV600E. In this regard, recent structural data highlight the importance of inhibitor binding mode within the active site: compounds that stabilize the αC helix in the inactive conformation, such as vemurafenib or dabrafenib, cannot bind to the second BRAF molecule within a dimer, while inhibitors that do not cause such movement, like GDC-0879, will bind and inhibit both protomers [46]. Analysis of BRAFV600E structure in complex with sorafenib (PDB: 3OG7) revealed that this inhibitor belongs to the latter class, explaining our synergistic data (Supplementary Figure S13).
BCORL1 has been implicated in cancer progression both as a tumor suppressor [17], [18] and as an oncogene [20], indicating a complex behavior in different settings. Approximately 8% of acute myeloid leukemia patients were found to carry BCORL1 mutations, mostly (87%) frameshift or nonsense [18]. On the other hand, in hepatocellular carcinoma, high BCORL1 levels are associated with advanced tumor stage and multiple tumor nodes [20]. In the cBioPortal database [47], BCORL1 mutations are spread throughout the entire coding sequence, suggesting either passenger or inactivating mutations. Notably, 30% are truncating. In particular, in the provisional TCGA melanoma dataset, BCORL1 missense or deletion mutations were found in 18/367 (4.90%) patients. Half of these cases also harbor BRAF mutations, which likely reflect the general incidence of BRAF mutations in melanoma. As a comparison, mutation frequencies of three housekeeping noncancer genes (ACTA1, GAPDH, GUSB) were 0.7% to 1.5%. Survival curves do not differ between BCORL1-mutated and nonmutated cases. The Broad Institute study [48] reported the same frequency of BCORL1 mutations (6/121 cases, 4.96%). In this case, 5/6 patients carried a BRAF mutation as well. Overall, our data point to a complex combination of loss-of-function, gain-of-function, and possibly other secondary effects, where most wild-type activity is lost, as suggested by transcriptome data and silencing/knockout experiments, but new functions are likely acquired, explaining why both overexpressing the mutant and silencing the wild-type allele have a similar small but significant effect on drug sensitivity. Gene ontology analysis revealed mostly similar signatures of wild-type versus parental and mutant versus parental comparisons, with the interesting exception of a significant loss of MAPK negative regulation by mutant BCORL1. In particular, IL1B gene, which is known to suppress ERK activity [49], was strongly downregulated by mutant but not by wild-type BCORL1, pointing to a gain-of-function type of effect. Whether IL1B repression is a cause of drug resistance remains to be elucidated. On the other hand, when looking at top regulated genes in wild-type transcriptome, whose regulation is lost by the mutant (loss-of-function type), no obvious direct relation to MAPK pathway could be observed, except GFRA1, a neurotrophic factor co-receptor able to activate MAPKs, which was kept at low levels by wild-type BCORL1 (Supplementary Figure 11B).
It is unclear whether BCORL1 mutation preceded or followed BRAF deletion in our A375-R1 cell population. All A375-R1 subclones isolated carried both alterations. Considering the strong phenotype induced by p47BRAFV600E, it is possible to speculate that BCORL1Q1076H was selected during an early phase of stepwise drug increase, while p47BRAFV600E may have subsequently taken over to provide strong advantage at high vemurafenib doses. Further studies may help clarify the relative contribution of the two alterations in drug resistance: for example, one could revert BCORL1 to wild-type in A375-R1 cells to check how sensitivity to vemurafenib changes. However, we tried to manipulate BCORL1 expression in A375-R1 cell line by silencing or overexpressing the gene, but this did not affect significantly cell viability, nor its sensitivity to the drug. We believe this is due to the fact that truncated p47BRAFV600E has taken the lead in driving resistance in this cell line. Indeed, altering BCORL1 expression in transfectant clone E9 (expressing intermediate p47BRAFV600E levels) has a slight impact on resistance (Figures 3 and 4), while doing so in clone H1 (high p47BRAFV600E) does not (not shown). Another independent vemurafenib-resistant cell line was selected in our laboratory which did not carry the BCORL1 mutation nor a BRAF deletion (data not shown), indicating that it is not a general route to resistance in this cell population.
We noticed that manipulation of BCORL1 levels had an effect on BRAF protein abundance. We do not know what is the significance of such phenomenon, as this did not translate into a different sensitivity to BRAF inhibition in A375-R1 cells. We might hypothesize that partial loss of BCORL1 function (by missense mutation) was selected rather than total gene loss because of this effect.
In conclusion, we described here a novel mechanism by which melanoma cells can acquire aberrant, drug-resistant BRAF variants, and the selection of a compound mutational status involving BRAF and the transcriptional factor BCORL1 in vemurafenib-resistant cells, adding to the known complexity of MAPK reactivation mechanisms in this disease.
The following are the supplementary data related to this article.
List of Primers Used in This Study. Cloning Sites Are Underlined.
Supplementary figures
Footnotes
This work was supported by the Italian Association for Cancer Research (AIRC; grant no. IG-14249 to C.G.P.; grant no. IG-17727 to R.P.) and by the EU Commission (Horizon 2020 Marie Sklodowska-Curie Innovative Training Network (ITN-ETN) Grant, Award No. 675712).
References
- 1.Gambacorti-Passerini C, Piazza R. How I treat newly diagnosed chronic myeloid leukemia in 2015. Am J Hematol. 2015;90(2):156–161. doi: 10.1002/ajh.23887. [DOI] [PubMed] [Google Scholar]
- 2.Gambacorti Passerini C, Farina F, Stasia A, Redaelli S, Ceccon M, Mologni L, Messa C, Guerra L, Giudici G, Sala E. Crizotinib in advanced, chemoresistant anaplastic lymphoma kinase-positive lymphoma patients. J Natl Cancer Inst. 2014;106(2) doi: 10.1093/jnci/djt378. [DOI] [PubMed] [Google Scholar]
- 3.Krajewska J, Olczyk T, Jarzab B. Cabozantinib for the treatment of progressive metastatic medullary thyroid cancer. Expert Rev Clin Pharmacol. 2016;9(1):69–79. doi: 10.1586/17512433.2016.1102052. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14(7):455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sala E, Mologni L, Truffa S, Gaetano C, Bollag GE, Gambacorti-Passerini C. BRAF silencing by short hairpin RNA or chemical blockade by PLX4032 leads to different responses in melanoma and thyroid carcinoma cells. Mol Cancer Res. 2008;6(5):751–759. doi: 10.1158/1541-7786.MCR-07-2001. [DOI] [PubMed] [Google Scholar]
- 8.Hoeflich KP, Gray DC, Eby MT, Tien JY, Wong L, Bower J, Gogineni A, Zha J, Cole MJ, Stern HM. Oncogenic BRAF is required for tumor growth and maintenance in melanoma models. Cancer Res. 2006;66(2):999–1006. doi: 10.1158/0008-5472.CAN-05-2720. [DOI] [PubMed] [Google Scholar]
- 9.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ascierto PA, Minor D, Ribas A, Lebbe C, O'Hagan A, Arya N, Guckert M, Schadendorf D, Kefford RF, Grob JJ. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J Clin Oncol. 2013;31(26):3205–3211. doi: 10.1200/JCO.2013.49.8691. [DOI] [PubMed] [Google Scholar]
- 11.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19(11):1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 12.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4(1):94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4(1):80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagan JK, Arnold J, Hanchard KJ, Kumar R, Bruno T, Jones MJ, Richard DJ, Forrest A, Spurdle A, Verdin E. A novel corepressor, BCoR-L1, represses transcription through an interaction with CtBP. J Biol Chem. 2007;282(20):15248–15257. doi: 10.1074/jbc.M700246200. [DOI] [PubMed] [Google Scholar]
- 16.Junco SE, Wang R, Gaipa JC, Taylor AB, Schirf V, Gearhart MD, Bardwell VJ, Demeler B, Hart PJ, Kim CA. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure. 2013;21(4):665–671. doi: 10.1016/j.str.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damm F, Chesnais V, Nagata Y, Yoshida K, Scourzic L, Okuno Y, Itzykson R, Sanada M, Shiraishi Y, Gelsi-Boyer V. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013;122(18):3169–3177. doi: 10.1182/blood-2012-11-469619. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Collins R, Jiao Y, Ouillette P, Bixby D, Erba H, Vogelstein B, Kinzler KW, Papadopoulos N, Malek SN. Somatic mutations in the transcriptional corepressor gene BCORL1 in adult acute myelogenous leukemia. Blood. 2011;118(22):5914–5917. doi: 10.1182/blood-2011-05-356204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Totoki Y, Tatsuno K, Yamamoto S, Arai Y, Hosoda F, Ishikawa S, Tsutsumi S, Sonoda K, Totsuka H, Shirakihara T. High-resolution characterization of a hepatocellular carcinoma genome. Nat Genet. 2011;43(5):464–469. doi: 10.1038/ng.804. [DOI] [PubMed] [Google Scholar]
- 20.Yin G, Liu Z, Wang Y, Dou C, Li C, Yang W, Yao Y, Liu Q, Tu K. BCORL1 is an independent prognostic marker and contributes to cell migration and invasion in human hepatocellular carcinoma. BMC Cancer. 2016;16:103. doi: 10.1186/s12885-016-2154-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Duployez N, Marceau-Renaut A, Boissel N, Petit A, Bucci M, Geffroy S, Lapillonne H, Renneville A, Ragu C, Figeac M. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood. 2016;127(20):2451–2459. doi: 10.1182/blood-2015-12-688705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Yamaguchi S, Burstein MD, Terashima K, Chang K, Ng HK, Nakamura H, He Z, Doddapaneni H, Lewis L. Novel somatic and germline mutations in intracranial germ cell tumours. Nature. 2014;511(7508):241–245. doi: 10.1038/nature13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceccon M, Mologni L, Bisson W, Scapozza L, Gambacorti-Passerini C. Crizotinib-resistant NPM-ALK mutants confer differential sensitivity to unrelated Alk inhibitors. Mol Cancer Res. 2013;11(2):122–132. doi: 10.1158/1541-7786.MCR-12-0569. [DOI] [PubMed] [Google Scholar]
- 24.Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, Antolini L, Mologni L, Donadoni C, Papaemmanuil E. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45(1):18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Redaelli S, Perini P, Ceccon M, Piazza R, Rigolio R, Mauri M, Boschelli F, Giannoudis A, Gambacorti-Passerini C. In vitro and in vivo identification of ABCB1 as an efflux transporter of bosutinib. J Hematol Oncol. 2015;8:81. doi: 10.1186/s13045-015-0179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sala F, Marangon E, Bagnati R, Livi V, Cereda R, D'Incalci M, Zucchetti M. Development and validation of a high-performance liquid chromatography-tandem mass spectrometry method for the determination of the novel proteasome inhibitor CEP-18770 in human plasma and its application in a clinical pharmacokinetic study. J Mass Spectrom. 2010;45(11):1299–1305. doi: 10.1002/jms.1842. [DOI] [PubMed] [Google Scholar]
- 29.Piazza R, Ramazzotti D, Spinelli R, Pirola A, De Sano L, Ferrari P, Magistroni V, Cordani N, Sharma N, Gambacorti-Passerini C. OncoScore: a novel, Internet-based tool to assess the oncogenic potential of genes. Sci Rep. 2017;7:46290. doi: 10.1038/srep46290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger AH, Brooks AN, Wu X, Shrestha Y, Chouinard C, Piccioni F, Bagul M, Kamburov A, Imielinski M, Hogstrom L. High-throughput Phenotyping of Lung Cancer Somatic Mutations. Cancer Cell. 2016;30(2):214–228. doi: 10.1016/j.ccell.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falchook GS, Long GV, Kurzrock R, Kim KB, Arkenau TH, Brown MP, Hamid O, Infante JR, Millward M, Pavlick AC. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379(9829):1893–1901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64(19):7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 33.Hoeflich KP, Herter S, Tien J, Wong L, Berry L, Chan J, O'Brien C, Modrusan Z, Seshagiri S, Lackner M. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer Res. 2009;69(7):3042–3051. doi: 10.1158/0008-5472.CAN-08-3563. [DOI] [PubMed] [Google Scholar]
- 34.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, Dias-Santagata D, Stubbs H, Lee DY, Singh A. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68(12):4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468(7326):968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okazaki K, Sagata N. MAP kinase activation is essential for oncogenic transformation of NIH3T3 cells by Mos. Oncogene. 1995;10(6):1149–1157. [PubMed] [Google Scholar]
- 39.Li M, Ong SS, Rajwa B, Thieu VT, Geahlen RL, Harrison ML. The SH3 domain of Lck modulates T-cell receptor-dependent activation of extracellular signal-regulated kinase through activation of Raf-1. Mol Cell Biol. 2008;28(2):630–641. doi: 10.1128/MCB.00150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vergani E, Vallacchi V, Frigerio S, Deho P, Mondellini P, Perego P, Cassinelli G, Lanzi C, Testi MA, Rivoltini L. Identification of MET and SRC activation in melanoma cell lines showing primary resistance to PLX4032. Neoplasia. 2011;13(12):1132–1142. doi: 10.1593/neo.111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480(7377):387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, Ng C, Chodon T, Scolyer RA, Dahlman KB. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson DB, Childress MA, Chalmers ZR, Frampton GM, Ali SM, Rubinstein SM, Fabrizio D, Ross JS, Balasubramanian S, Miller VA. BRAF internal deletions and resistance to BRAF/MEK inhibitor therapy. Pigment Cell Melanoma Res. 2017 doi: 10.1111/pcmr.12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamoto I, Pirker C, Bilban M, Berger W, Losert D, Marosi C, Haas OA, Wolff K, Pehamberger H. Seven novel and stable translocations associated with oncogenic gene expression in malignant melanoma. Neoplasia. 2005;7(4):303–311. doi: 10.1593/neo.04514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karoulia Z, Wu Y, Ahmed TA, Xin Q, Bollard J, Krepler C, Wu X, Zhang C, Bollag G, Herlyn M. An Integrated Model of RAF Inhibitor Action Predicts Inhibitor Activity against Oncogenic BRAF Signaling. Cancer Cell. 2016;30(3):501–503. doi: 10.1016/j.ccell.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 47.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269) doi: 10.1126/scisignal.2004088. [pl1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lagathu C, Yvan-Charvet L, Bastard JP, Maachi M, Quignard-Boulange A, Capeau J, Caron M. Long-term treatment with interleukin-1beta induces insulin resistance in murine and human adipocytes. Diabetologia. 2006;49(9):2162–2173. doi: 10.1007/s00125-006-0335-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of Primers Used in This Study. Cloning Sites Are Underlined.
Supplementary figures