Abstract
Salmonella infection is a major public health concern, and colonization in humans can be chronic and increases the risk of cancers. Wnt signaling is a key pathway for intestinal renewal and development, inflammation, and tumorigenesis. In the current study, we report a novel role of Wnt1 in infection and colon cancer using cell culture models, a Salmonella-colitis colon cancer model, and human samples. In contrast to the bacteria-induced increases in Wnt2 and Wnt11, Salmonella colonization significantly reduced the level of Wnt1 in intestinal epithelial cells in vivo and in vitro. The bacterial AvrA protein is known to activate the canonical Wnt pathway. Wnt1 expression level was downregulated by AvrA-expressing Salmonella but stabilized by AvrA-deficient Salmonella in the intestine of Salmonella-colitis mice. In a chronic Salmonella-infected cancer model, the Wnt1 protein level was decreased in the AvrA+ infected group. Thus, we further assessed the functional role of Wnt1 downregulation in the inflammatory response and colorectal cancer (CRC) progression. Moreover, downregulation of Wnt1 by the Crispr-Cas9 method promoted cancer cell invasion and migration. Interestingly, we found that Wnt1 was downregulated in human CRC tissue, and Wnt1 downregulation may be correlated with cancer progression. Our study provides insights into mechanisms by which enteric bacteria regulate Wnt1 expression and potentially contribute to infection-associated colon cancer.
Introduction
Growing interest and accumulating data on human microbiota indicate that host-microbe interplay has an important role in the development of colorectal cancer (CRC) [1], which suggests that chronic infection and inflammation contribute to tumor initiation and tumor progression [2]. Increasing evidences showed that gut microbiota can promote CRC progression through multiple processes including the deregulation of chronic inflammatory state and immune response, interacting with the host epigenetic machinery, altering stem cell dynamics, and affecting host metabolism [3], [4], [5]. However, evidence to support a direct interaction of intestinal bacteria and host genes in CRC development is still limited. Deregulation of the Wnt/β-catenin signal transduction pathway has been implicated in the pathogenesis of inflammatory bowel diseases and CRC [6], [7], [8]. Salmonella infection is a major public health concern, and colonization in humans can be chronic and increase the risk of inflammatory bowel diseases and CRC [9], [10]. Our previous studies and those by other groups have shown that Salmonella use various strategies to regulate Wnt signaling. The activation of Wnt/β-catenin by Salmonella infection is involved in cell proliferation, inflammation, apoptosis, transdifferentiation, and tumorigenesis [11], [12], [13], [14], [15], [16], [17], [18], [19].
Wnt-1 is a Wnt family member that triggers the Wnt/β-catenin signaling cascade [20]. There are a few studies concerning Wnt-1 expression in the colon that have shown overexpression or a lack of expression of Wnt-1 in tumor tissue compared to normal colonic mucosa [21], [22], [23]. These inconsistent studies suggest that Wnt-1 expression was regulated by an unknown mechanism in these analyzed cases of CRC. However, the mechanism by which enteric bacteria regulate Wnt1 and how Wnt1 modulates the host response to pathogenic bacteria remain unexplored.
This present study investigated the effects of Salmonella infection on Wnt1 repression in intestinal epithelial cells in vitro and in vivo. We found that Wnt1 protein expression was decreased after Salmonella colonization. Wnt1 is involved in protecting intestinal cells by blocking the invasion of pathogenic bacteria and suppressing inflammation. Furthermore, we found decreased Salmonella invasion in cells in which Wnt1 expression was knocked down compared to those with normal Wnt1 levels. The proinflammatory cytokines IL-8, IL-6, and granulocyte-macrophage colony-stimulating factor (GM-CSF) were significantly upregulated in response to Salmonella infection in Wnt1-knockdown cells compared to control, uninfected cells. Functionally, downregulation of Wnt1 inhibited cancer cell invasion and migration. Further analysis revealed that the downregulation of Wnt1 in cancer cells occurred via Salmonella-induced ubiquitination. Interestingly, we found the Wnt1 was downregulated in CRC patients, and Wnt1 downregulation correlated with CRC progression. Our study provides novel insights into mechanisms by which gut bacteria regulate Wnt1 expression and potentially contribute to infection-associated colon cancer.
Materials and Methods
Cell Culture
Human colonic epithelial HCT116 and Caco-2 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin, and L-glutamine at 37°C, as previously described [17].
Bacterial Strains and Growth Condition
Salmonella typhimurium strains used in this study included wild-type Salmonella SL1344 (SB300) and the nonpathogenic Salmonella mutant strain derived from PhoPc, PhoPCAvrA− [11], [15], [24] Nonagitated microaerophilic bacterial cultures were prepared by inoculating 5 ml of Luria-Bertani broth with 0.01 ml of a stationary-phase culture followed by overnight incubation (18 hours) at 37°C, as previously described [15].
Animal Experiment
The animal study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the University of Rochester University Committee on Animal Resources committee (UCAR 2007-065) when Dr. Jun Sun worked at the University of Rochester and UIC Protocol for Animal use (ACC 15231). All efforts were made to minimize suffering. The animal samples shown in Figure 5A were collected from Salmonella-infected intestine [15]. Animal experiments of Salmonella-infected colon cancer mouse model were performed by using specific pathogen-free female C57BL/6 mice (Taconic) that were 6 to 7 weeks old as previously described [19], [25]. Briefly, mice were infected with indicated Salmonella strains by oral gavage only once and then treated with azoxymethane (AOM)–dextran sodium sulfate (DSS) to develop colon cancer. AOM, 10 mg/kg body weight, intraperitoneal injection, 1% DSS in drinking water (Supplement Figure S1). At 1, 3, 6, 10, and 49 weeks after Salmonella infection, tissue samples were collected. The tumors and paired adjacent “normal” mucosa from colon shown in Figures 5, B and C and 3S were collected from mice 49 weeks postinfection.
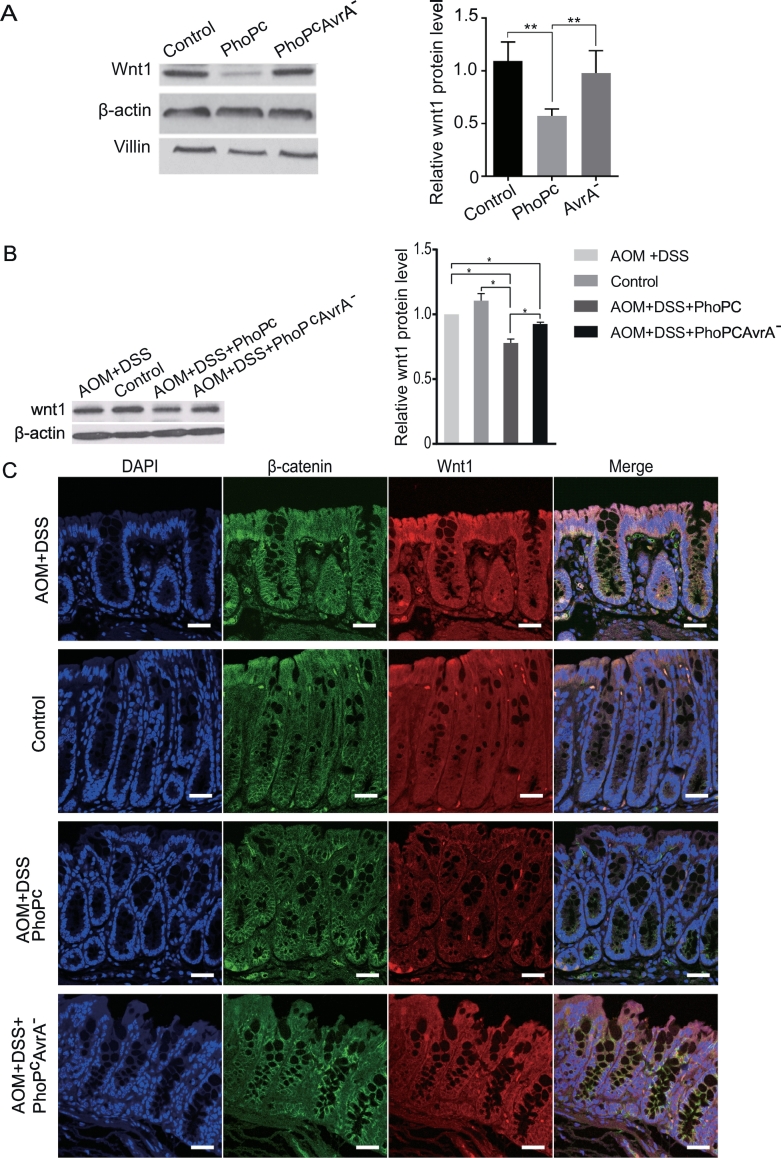
Figure 5.
Salmonella effector AvrA regulates Wnt1 expression in Salmonella-infected intestine and infection-associated tumors.
(A) Wnt1 reduction in intestinal epithelial cells in vivo after exposure to nonpathogenic Salmonella WT. Mice were infected with S. typhimurium PhoPc or PhoPc AvrA− (AvrA gene knockout). (B) Expression levels of Wnt1 as assessed by Western blotting in control mucosa. AOM/DSS-induced tumors and AOM/DSS-induced tumors exposed to Salmonella (PhoPc or PhoPc AvrA−) 49 weeks postinfection. (C) Wnt1 and β-catenin localization in control mucosa, AOM/DSS-induced mucosa, and AOM/DSS-induced mucosa exposed to Salmonella (PhoPc or PhoPc AvrA−) 49 weeks postinfection.
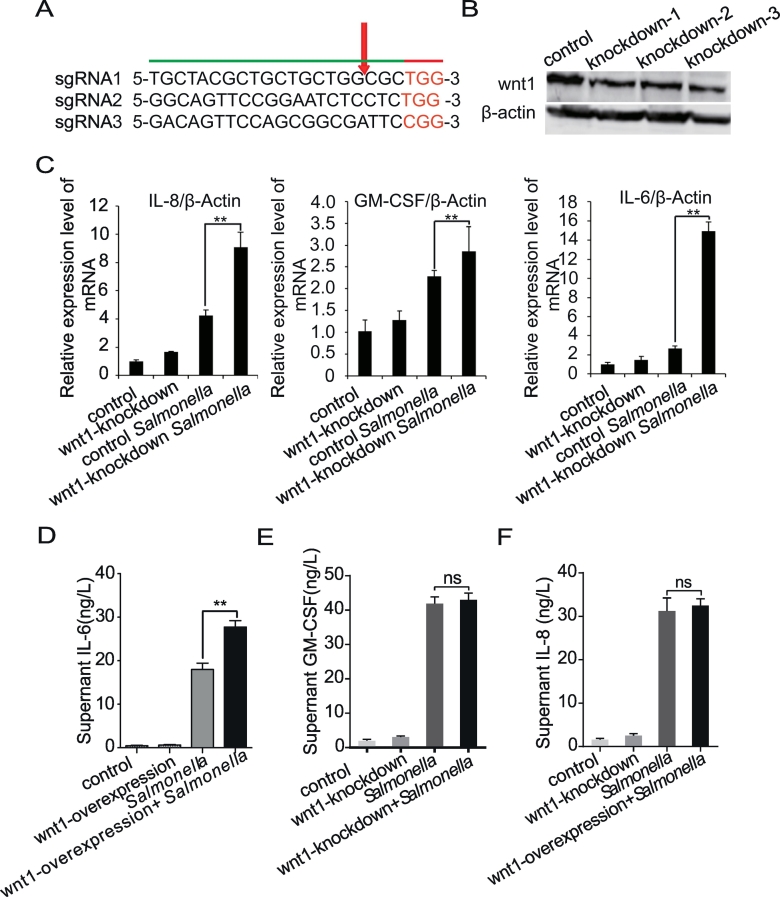
Figure 3.
Downregulation of Wnt1 expression in intestinal epithelial cells alters the inflammatory response.
(A) Design of CRISPR sgRNA sequences targeting the N-terminus of Wnt1. (B) Western blot analysis of wnt1+/− cells. A Wnt1-specific antibody was used to detect Wnt1 protein in HCT116 lysates. (C) IL-8, IL-6, and GM-CSF mRNA levels in Wnt1-knockdown HCT116 cells after Salmonella colonization. (D-F) IL-8, IL-6, and GM-CSF mRNA levels in Wnt1-overexpressing HCT116 cells after Salmonella colonization. The data are presented as the mean ± SD of three replicate experiments.
Human Samples
Moderate and highly differentiated human adenocarcinoma samples were obtained from Guangdong Institute of Gastroenterology, the Sixth Affiliated Hospital, Sun Yat-Sen University, China, in accordance with approval from the institute.
Immunofluorescence
The proximal and distal portions of the colon were freshly isolated and embedded in paraffin wax after fixation with 10% neutral buffered formalin. Immunostaining was performed on paraffin-embedded sections (4 μm) of mouse colons and human colon tissue. Slides were incubated in 3% hydrogen peroxide for 20 minutes at room temperature to block endogenous peroxidase activity and then in 5% BSA in PBS for 30 minutes to reduce nonspecific background. The permeabilized tissue samples were incubated with anti-Wnt1 (1:100, Cell Signaling) and anti–β-catenin (1: 100; BD, San Jose, CA) for 10 to 12 hours at 4°C. Samples were then incubated with DAPI for 10 minutes at room temperature. Tissues were mounted with SlowFade (SlowFade AntiFade Kit, Molecular Probes) and then cover slipped. The edges were sealed to prevent drying. Specimens were examined with a Leica SP5 scanning confocal microscope.
S. typhimurium Attachment and Invasion of Human Epithelial Monolayers
HCT116 cells were infected by a previously described method [26]. For bacterial attachment, cells were stimulated with Salmonella for 0.5 hour and washed with PBS for the cell-associated bacteria, 0.9 ml LB broth was added, and each sample was mixed vigorously and quantified by plating for colony-forming units (CFUs) on MacConkey agar medium. Bacterial invasion was assessed after bacterial solutions (~20 bacteria/epithelial cell) were added at 30 minutes. Bacteria internalized in epithelial cells were released with 1% Triton X-100 after gentamicin (50 μg/ml) treatment for 20 minutes. Gentamicin does not permeate eukaryotic plasma membranes and therefore is cytolytic to only extracellular populations of bacteria; intracellular bacteria populations remain viable [27]. To quantitate internalized bacteria, 0.9 ml of LB was then added, the sample was vigorously mixed, and CFUs were quantitated by plating on MacConkey agar medium [18].
Design of the Wnt1 Single-Guide RNA Sequences
Single-guide RNA (sgRNA) sequences targeting the CDS of Wnt1 were selected using the online CRISPR design tool from the Zhang lab at the Broad Institute, and the three selected sgRNAs were predicted to have a very low probability off-target sites. The sgRNA sequences were then cloned into the Crispr-Cas9-V2-Puro vector [28]: sgRNA1 5-TGCTACGCTGCTGCTGGCGC-3, sgRNA2 5-GGCAGTTCCGGAATCTCCTC-3, and sgRNA3 5-GACAGTTCCAGCGGCGATTC-3.
Characterizing Wnt1-Knockout (KO) Lines
Western blotting confirmed that the Wnt1-KO cells expressed lower levels of Wnt1 protein. Specifically, cell lysates from WT and KO HCT116 cells were separated by SDS-PAGE, transferred to PVDF membranes, and then blotted with an anti-Wnt1 antibody (Bioworld); an anti-actin antibody (Bioworld) was used as the loading control.
Construction of the Plasmid pcDNA3.1-hWnt1
The pCDNA3.1-hWnt1 (6615 kb) plasmid was constructed by inserting an EcoRI-XbaI fragment containing the human Wnt1 complementary DNA (cDNA) (1.113 kb) into the pCDNA3.1-His plasmid (5.502 kb). PCR primers for generating a fragment EcoRI-XbaI containing the hWnt1 cDNA were as follows: EcoRI-hwnt1 forward primer: 5-CGGAATTCATGGGGCTCTGGGCGCTGTT-3; hWnt1-XbaI backward primer: 5- GCTCTAGACAGACACTCGTGCAGTACGC -3.
Co-Immunoprecipitation Assay
Cells were rinsed twice in ice-cold PBS and lysed in cold lysis buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM EGTA, pH 8.0, and 0.2 mM sodium orthovanadate) containing a protease inhibitor cocktail. Samples were precleared with protein A-agarose. Precleared lysates were incubated with 2 μg of anti-ubiquitin (Enzo Life Science, 5120 Butler Pike Plymouth Meeting, PA) or anti-IgG (Cell signaling) primary antibodies overnight at 4°C. Protein A-agarose was added to the lysate, and the mixture was incubated for 2 to 3 hours with agitation at 4°C and then pelleted by centrifugation at 3000 rpm for 5 minutes. The supernatant was removed, and the pellet was washed four times (twice with lysis buffer, once with TBS, and once with 0.05 M Tris-HCl), boiled for 5 minutes, separated by SDS-PAGE, and transferred onto a nitrocellulose membrane. Membranes were probed with an infrared-conjugated secondary antibody, and the signals were visualized and quantitated using LI-COR Odyssey v3.0 software.
Real-Time Quantitative PCR Analysis
Total RNA was extracted from epithelial cell monolayers using TRIzol reagent (Life Technology, USA). RNA integrity was verified by gel electrophoresis. RNA was reverse transcribed using the 5*All-In-One RT MasterMix (Abm, G490, CA) according to the manufacturer's directions. The RT cDNA reaction products were subjected to quantitative real-time PCR using the EvaGreen 2*qPCR MasterMix-No Dye (Abm) according to the manufacturer's directions. The following primers for β-actin, WNT1, IL-8, IL-6, and GM-CSF were used: β-actin-F 5-AGAGCAAGAGAGGCATCCTC-3, β-actin-R 5-GACGGCCGCATCTTCTTGT-3, WNT1-F 5-GAGCCACGAGTTTGGATGTT-3, WNT1-R 5-TGAGGAGAGAAGAGGGACCA-3, IL-8-F 5-TGCATAAAGACATACTCCAAACCT-3, IL-8-R 5-AATTCTCAGCCCTCTTCAAAAA-3, IL-6-F 5-AGTGGCTGCAGGACATGACAA-3, IL-6-R 5-CAATCTGAGGTGCCCATGCTA-3, GM-CSF-F 5-GGCGTCTCCTGAACCTGAGT-3, GM-CSF-R 5-GGGGATGACAAGCAGAAAGT-3, TIMP3-F 5-TCCCAGCGCAAGGGGCTGAA-3, TIMP3-R 5-GCCGGATGCAGGCGTAGTGTT-3, Nm23-F 5-ACCTGAAGGACCGTCCATTCTTTGC-3, and Nm23-R 5-GTGAAACCACAAGCCGATCTCCT-3. c-myc-F 5-TCAAGAGGCGAACACACAAC-3, c-myc-R 5-GGCCTTTTCATTGTTTTCCA-3; FlnA-F 5-GCCAGAAGAGCAGCTTCACA-3, FlnA-R 5-CCTTGAGCAGGTAGGACACG-3
Western Blot Analysis of Mouse Intestinal Epithelial Cells In Vivo
Mouse intestinal epithelial cells were lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris, pH 7.4, 1 mM EDTA, 1 mM EGTA, pH 8.0, 0.2 mM sodium orthovanadate, and protease inhibitor cocktail), and the protein concentration was measured. The cells were rinsed two times in ice-cold HBSS, lysed in protein loading buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1% bromphenol blue, and 10% glycerol), and sonicated. Equal amounts of protein were separated by SDS-PAGE, transferred to nitrocellulose membranes, and subjected to immunoblotting with primary antibodies. The following antibodies were used: anti-Wnt1 (Cell Signaling), anti-villin (Santa Cruz Biotechnology, Santa Cruz, CA), and anti–β-actin (Sigma-Aldrich, Milwaukee, WI). Bands were quantified using Kodak MI software (version 4.0.3).
Transient Transfection
Transient transfections were performed with Lipofectamine2000 (Invitrogen) in accordance with the manufacturer's instructions. At the indicated times after transfection, protein was extracted with RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) and analyzed by immunoblotting.
Western Blot Analysis of Cell Lines
Cultured infected or control cells were rinsed twice in ice-cold HBSS and lysed in protein loading buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol). Equal amounts of protein were separated by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by immunoblotting with anti-Wnt1 and anti–β-actin antibodies (Bioworld). The signals were visualized and quantitated using LI-COR Odyssey v3.0 software after probing the membranes with an infrared-conjugated secondary antibody.
Cytokine Production Assay in Culture Supernatant
HCT116 cells were cultured in DMEM followed by Salmonella-containing HBSS (1.6 × 1010 bacteria/ml) for 30 minutes, washed 3 times in HBSS, and incubated at 37°C for 6 hours. Cell supernatants were removed and assayed for IL-8 (Cat No. H008), IL-6 (Cat No. H007), and GM-CSF (Cat No. H060) by ELISA kit (Nanjing Jiancheng Company, China) in 96-well plates as described previously [18].
Cell Migration and Invasion Assay
In vitro cell migration and invasion assays were performed as described previously [29]. Cells growing in the log phase were trypsinized, resuspended in serum-free medium, and seeded into chambers (8-μm pore size in a polycarbonate membrane) (Corning, USA). The chambers were coated with Matrigel (BD Biosciences, USA) for cell invasion assays. Medium with 20% FBS (750 μl) was added to the lower chamber. After incubation for 72 hours, the cells on the top surface of the insert were removed with a cotton swab. Cells that had migrated to the bottom surface of the insert were stained in 0.1% crystal violet for 30 minutes, rinsed in PBS, and subjected to microscopic inspection. Images of four random fields (10×) were captured from each membrane, and the number of migratory or invasive cells was counted. The migration and invasion results were normalized to cell number under the same treatment conditions. Triplicate assays were performed for each experiment.
Statistical Analysis
Data are presented as the mean ± SD. All statistical tests were two-sided. P values of less than .05 indicated statistical significance. Differences between two samples were analyzed by Student's t test. Differences among three or more groups were performed using one-way ANOVA with GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA).
Result
Reduction of Wnt1 Protein in Response to Wild-Type Salmonella Infection
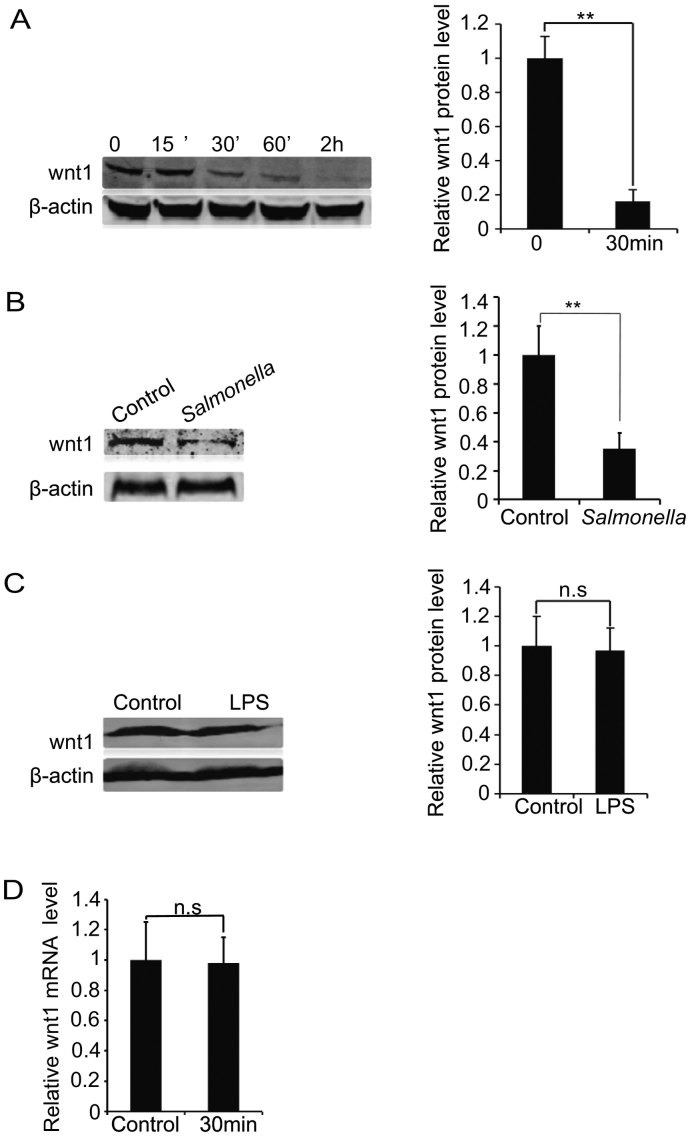
To determine whether Wnt1 protein plays a role in epithelial-Salmonella interactions, we infected human intestinal epithelial HCT116 cells with wild-type Salmonella typhimurium strain SL1314. We found that Salmonella significantly decreased the total amount of Wnt1 protein in host cells after bacterial colonization for only 30 minutes (Figure 1A). The Salmonella-induced Wnt1 reduction lasted for more than 2 hours and occurred in a time-dependent manner (Figure 1A). In the human epithelial Caco-2 cells, Salmonella colonization also significantly decreased Wnt1 protein expression (Figure 1B). To test whether the response is induced by lipopolysaccharide from pathogenic bacteria, we treated cells with lipopolysaccharide. However, we did not see a similar change in Wnt1 expression (Figure 1C), suggesting that the downregulation of Wnt1 may be specific to bacterial endotoxin.
Figure 1.
Pathogenic Salmonella decreases Wnt1 protein expression in host cells.
(A) Wnt1 protein expression in human HCT116 cells. Cells were incubated with wild-type S. typhimurium (SL1314) for 30 minutes, washed, and incubated in fresh DMEM with 10% FBS for 15, 30, or 60 minutes or 2 hours. Total cell lysates were analyzed for total Wnt1 levels by immunoblot. *P < .05, n = 3 separate experiments. “Control” indicates “Control group without bacterial treatment”. (C) Wnt1 protein expression in Caco-2 cells. Cells were incubated with SL1314 for 30 minutes, washed, and incubated in fresh DMEM with 10% FBS for 30 minutes. Total cell lysates were analyzed for total Wnt1 levels by immunoblot. *P < .05, n = 3 separate experiments. (D) Salmonella treatment did not alter Wnt1 mRNA levels in HCT116 cells.
To determine whether Wnt1 was reduced by regulation at the transcription level, we investigated Wnt1 mRNA expression in intestinal epithelial cells treated with Salmonella. However, RT-qPCR analysis showed that the Wnt1 mRNA level was not changed by Salmonella treatment in vitro (Figure 1D). Overall, our data showed that pathogenic Salmonella reduces Wnt1 at the protein level but not the mRNA level.
Wnt1 Is Regulated by Salmonella at the Posttranslational Level
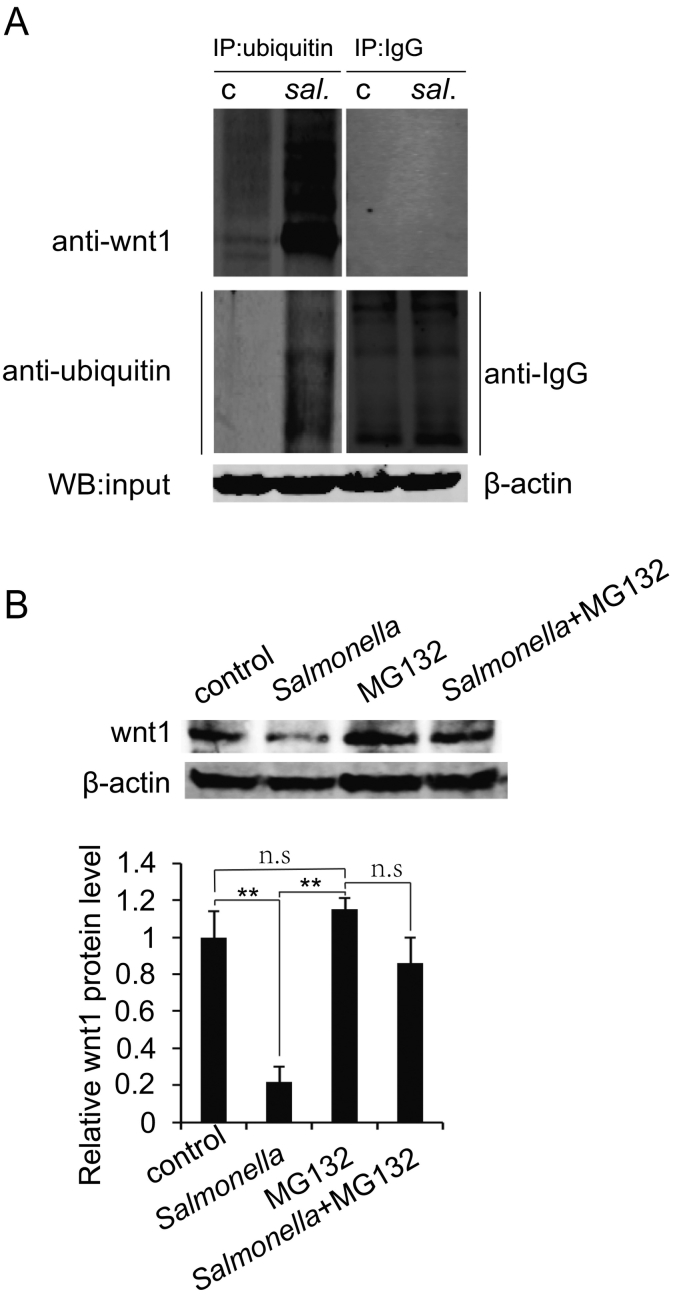
Because Wnt1 protein destabilization occurred in the early stage of Salmonella invasion, we hypothesized that Wnt1 is regulated at the posttranslational level upon Salmonella infection. Hence, we investigated whether Salmonella reduced Wnt1 protein through increased ubiquitination. To evaluate ubiquitination of Wnt1, we did immunoprecipitation using anti-ubiquitin, anti-Wnt1, or anti-IgG (negative control) antibodies, respectively. Our data indicated that Salmonella treatment induced more Wnt1 ubiquitination than did control treatment (Figure 2A, left panel). Anti-Wnt1 was not immunoprecipitated by anti-IgG (Figure 2A, right panel). Wnt1 ubiquitination was enhanced after Salmonella colonization for 30 minutes.
Figure 2.
Wnt1 reduction by Salmonella occurs through ubiquitination.
(A) Ubiquitination of Wnt1 increased in Salmonella-colonized cells by using immunoprecipitation of anti-ubiquitin or anti-IgG (negative control) and Wnt1. (B) The proteasome inhibitor MG132 stabilized Wnt1 protein after Salmonella colonization. Cells treated with MG132 (40 μM) for 4 hours had increased levels of Wnt1 protein. The data are presented as the mean ± SD of three replicate experiments.
To determine whether Wnt1 level was reduced due to increased proteasomal degradation, cells were treated with the proteasome inhibitor MG132. In the presence of MG132, Wnt1 protein level was stabilized after Salmonella infection to a level comparable to that of control cells without treatment (Figure 2B). Taken together, these data showed that increased ubiquitination and proteasomal degradation of Wnt1 occur during Salmonella infection.
Wnt1 Protein Is Directly Involved in Salmonella-Induced Inflammation
To further investigate the biological role of Wnt1 in Salmonella-induced inflammation, we designed three separate CRISPR sgRNA sequences to target the N-terminus of Wnt1, thus generating truncating mutations (Figure 3A). After puromycin selection, three clones were identified, and Western blotting analysis confirmed the heterozygous knockout status of these clones (Figure 3B). We then analyzed the inflammatory response by evaluating the proinflammatory cytokines IL-8, IL-6, and GM-CSF in Wnt1-knockdown cells. Upon Salmonella colonization, the mRNA expression levels of IL-8, IL-6, and GM-CSF were significantly increased in Wnt1-knockdown cells compared to control HCT116 cells (Figure 3C). Consistent with mRNA level, ELISA showed the IL-6 production in culture supernatant was significantly increased in Wnt1-knockdown supernatant compared to that of control group (Figure 3D). However, we did not find any changes in IL-8 and GM-SF (Figure 3, E-F). These data implicated that different cytokines may have different secretion times or are being regulated differently.
Based on above effects of downregulation Wnt1, we further hypothesized that overexpression of Wnt1 would induce less inflammation in response to Salmonella infection. However, contrary to expectation, overexpression of Wnt1 inhibited inflammation response like that of downregulation of Wnt1 (Figure S1); therefore, we further infer that there may be coordination effects of different Wnts in response to bacterial infection. Our previously published data showed that overexpression of both Wnt2 and Wnt11 inhibited IL-8 production, and downregulation of Wnt2 promoted IL-8 production [17], [18]; therefore, we checked the Wnt2 and Wnt11 expression upon deregulation of Wnt1. As shown in Figure S2, Wnt2 and Wnt11 showed upregulation with knockdown Wnt1; in contrast, Wnt2 showed downregulation with overexpression of wnt1. Hence, these data implicated that host cells may have complicated coordination effects between the three wnts in response to pathogenic bacteria infection. Taken together, our data indicated that the reduction in Wnt1 levels associated with Wnt2 and Wnt 11 may play a protective role in bacteria-induced intestinal inflammation.
Wnt1 Reduction Protects Cells from Salmonella Invasion
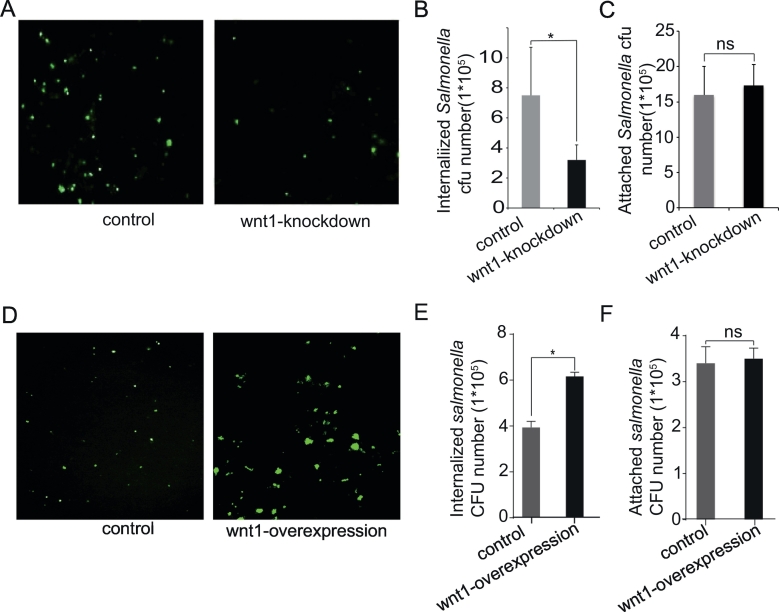
To study the physiological relevance of Wnt1 in Salmonella-host interactions, a green fluorescence-tagged Salmonella strain was used to detect bacterial invasion in human intestinal epithelial HCT116 cells. There were fewer green Salmonella invasion in Wnt1-knockdown cells than those in wild-type cells (Figure 4A). We counted the number of Salmonella that invaded HCT116 cells with normal or reduced Wnt1 protein levels. Wnt1-knockdown epithelial cells had fewer internalized Salmonella than did control cells with basal Wnt1 expression (Figure 4B). However, there was no difference of the number of cells with attached Salmonella between the control and Wnt1-knockdown groups (Figure 4C). In the gain-functional study, overexpression of Wnt1 led to more internalized Salmonella than control cells (Figure 4, D-E). Regarding the attached Salmonella, there was no difference between the control and Wnt1-overexpression groups (Figure 4F). Hence, these data indicated that Wnt1 regulated Salmonella invasion but not the attachment during the bacterial-intestinal epithelial cells interactions.
Figure 4.
Wnt1 expression in intestinal epithelial cells changed bacterial invasion.
(A) Cells with Wnt1 knockdown were resistant to invasion by GFP-Salmonella (green). (B) Number of invaded Salmonella in HCT116 cells with downregulation of Wnt1 expression compared to control group. (C) Number of Salmonella associated with HCT116 cells with/without knockdown of Wnt1. HCT116 cells with control RNAi and Wnt1knockdown cells were stimulated with Salmonella for 0.5 hour, respectively, and washed for the cell-associated bacteria; 0.9 ml LB broth was added; and each sample was mixed vigorously and quantified by plating for CFUs on MacConkey agar medium. The mean ± SD is from three replicate experiments. (D) Wnt1-overexpressing cells were sensitive to invasion by GFP-Salmonella (green). (E) Number of invaded Salmonella in HCT116 cells with overexpression of Wnt1 expression compared to control group. (F) Number of Salmonella associated with HCT116 cells with/without overexpression of Wnt1. The data are presented as the mean ± SD of three replicate experiments.
Downregulating Wnt1 Promotes Cancer Cell Invasion and Migration
Our previous study reported the pathological changes associated with chronic Salmonella infection and differences in tumor development [19]. Wnt ligands are involved in colon cancer development [20]. Thus, we assessed the physiological relevance of Wnt1 in epithelial cells from a bacteria-infected mouse model and an infection-associated colon cancer model. AvrA is a Salmonella effector protein known to activate the Wnt signaling pathway [17], [18]. We find that Wnt1 expression levels were downregulated by AvrA-expressing Salmonella but stabilized by AvrA-deficient Salmonella in the intestine of Salmonella-infected mice (Figure 5A). We found that Wnt1 protein levels decreased significantly in tumors of cancer group infected with the AvrA-sufficient Salmonella strain (PhoPc) and increased significantly in tumors from mice infected with the AvrA-deficient Salmonella strain PhoPc AvrA− (Figure 5B). However, Wnt1 protein levels did not change significantly in tumors after AOM-DSS treatment, just like that of control group. By immunostaining, we confirmed the Wnt1 distribution in colon of mice with or without tumor (Figure 5C, red color). We used β-catenin as a control with Wnt1 for our co-staining (Figure 5C, green color). Whereas Wnt1 was reduced in tumors and adjacent “normal” mucosa in AvrA-sufficient Salmonella PhoPc, the β-catenin was enhanced in the tumors. Activated β-catenin was also found transclocated into nucleus (Figure 5B for adjacent “normal” mucosa and Figure S3 for tumors). Taken together, we found that Wnt protein levels were lower in colon from mice infected with AvrA-expressing bacteria than in those treated with AOM/DSS alone or colonized with AvrA-deficient Salmonella.
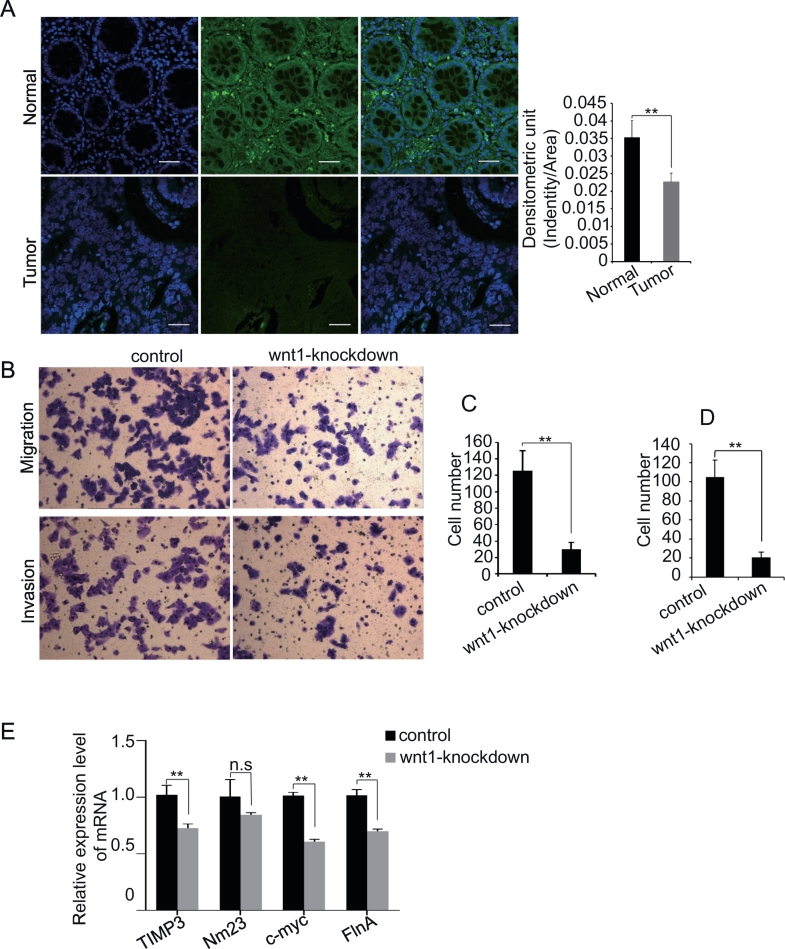
We then examined the Wnt1 protein in the colorectal tumor tissue in four human clinical samples. All of four colorectal samples showed highly and moderately differentiated adenocarcinoma. As shown in Figure 6A, Wnt1 protein level was dramatically decreased in the differentiated adenocarcinoma tissues compared to paracarcinoma tissue. Taken together, these data implicated that Wnt1 is involved in the colon cancer development.
Figure 6.
Role of Wnt1 downregulation in cancer development.
Wnt1 signaling is decreased in the tissues of differentiated adenocarcinoma (lower panel) relative to paracarinoma tissue (up-panel). The wnt1 fluorescence density from tumor tissue is significantly lower compared to paracarcinoma tissue (left panel, n = 4, moderately and highly differentiated adenocarcinoma). Four human clinical samples were collected from the Sixth Affiliated Hospital, Sun Yat-Sen University). (B) Downregulation of Wnt1 affects cancer cell migration and invasion. Cells were allowed to migrate for 24 hours in Transwell chambers (Cell Migration) or for 48 hours in chambers coated with Matrigel (Cell Invasion). Each figure is representative of three independent experiments. (C) The cell number of migration is decreased in Wnt1 knockout cells. The mean ± SD is from three replicate experiments. (D) The cell number of invasion is decreased in Wnt1 Knockout cells. The mean ± SD is from three replicate experiments. (E) The TIMP3, C-myc, FlnA, and Nim23 mRNA levels are decreased in Wnt1 knockout cells (P < .05).
To detect the role of knockdown of Wnt1 in the colon cancer development, we examined the cell migration and invasion. We found that the migration and invasion abilities of HCT116 cells with Wnt1 knockdown were dramatically decreased compared with controls (Figure 6, B-C). Accordingly, the related proliferation, migration, and invasion genes, c-myc, FlnA, TIMP3, and Nm23, are downregulated in Wnt1 knockdown cells compared to control cells (Figure 6D). These data indicated that Wnt1 participated in the development of CRC.
Discussion
In our current study, we show for the first time that Salmonella infection reduces Wnt1 protein expression in intestinal epithelial cells both in vitro and in vivo. Downregulation of Wnt1 expression increased the expression of inflammation cytokines, including IL-8, GM-CSF, and IL-6, in response to Salmonella infection and inhibited Salmonella invasion. Interestingly, we found that Wnt1 protein degradation was associated with ubiquitination. Additionally, we showed that Wnt1 protein levels were reduced in the colorectal mucosa of bacteria-infected mice. The reduction in Wnt1 protein levels promoted cancer cell migration and invasion. Hence, we have demonstrated that Wnt1 is directly involved in regulating intestinal inflammation and cancer development induced by enteric bacteria.
As summarized in Figure 7, we uncovered several novel characteristics of Wnt1: 1) Wnt1 is reduced by bacterial infection at the protein level through ubiquitination; 2) Wnt1 protects cells from bacterial invasion and inflammation; and 3) decreased Wnt1 expression in human colon cancer cells and the Salmonella-induced colon cancer model may inhibit cancer cell migration and invasion.
Figure 7.
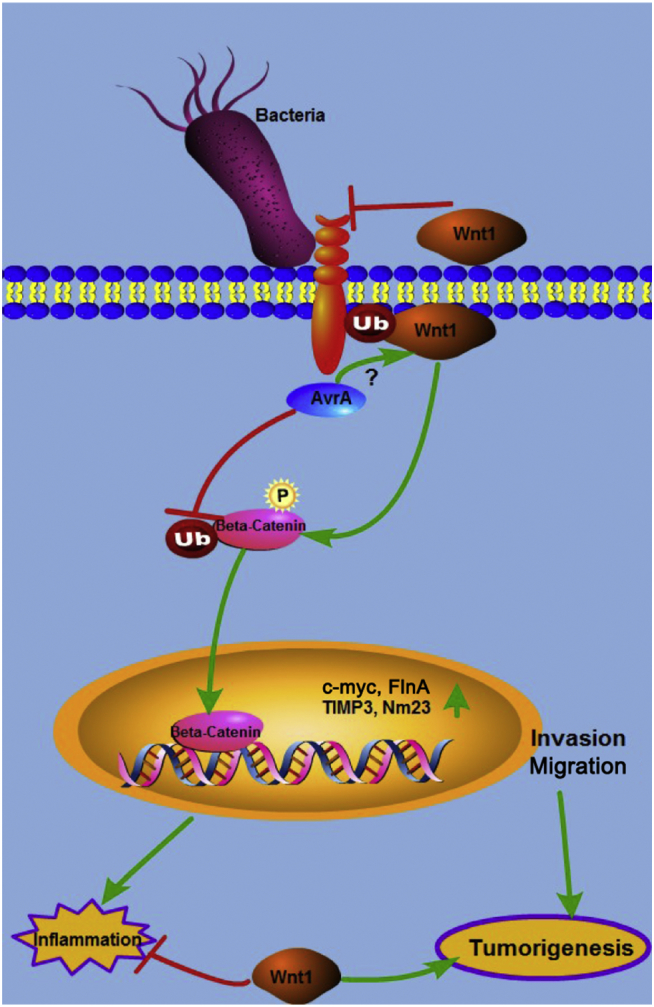
The working hypothesis on the role of Wnt1 in bacterially induced intestinal inflammation and tumorigenesis.
Wnt1 expression protects the host at different levels, including blocking pathogenic bacteria invasion, suppressing inflammation, and inhibiting cancer cell invasion and migration. The Salmonella AvrA protein reduces the expression and function of Wnt1.
Our study showed that Wnt1 protein was regulated by ubiquitination. Treatment with a proteasome inhibitor stabilized Wnt1 levels. In this study, tumors in the group infected with PhoPcAvrA+ had less Wnt1 expression than those in the group infected with the PhoPcAvrA− strain. These data were consisted with the results from the Salmonella-infected inflammation model (Figure 5A). AvrA has been reported to be a deubiquitinase [12]. Our data indicate that the Salmonella effector AvrA might regulate Wnt1 levels through an unknown mechanism.
Our study showed the presence and cellular localization of AvrA in precursor lesions using human clinical specimens [30]. Recent studies have indicated that Salmonella infection changes the composition of gut microbiota [31], [32], suggesting that other pathogenic bacteria and commensal bacteria may also regulate Wnt1 and its downstream signaling pathways. Interestingly, our data showed that tumors that developed due to AOM/DSS without Salmonella infection had no change in Wnt1 protein level; however, human CRC has deceased Wnt1. Hence, downregulation of Wnt1 in the tumor tissue of human CRC may be associated with bacterial infection and further induced ubiquitination of Wnt1.
We have demonstrated that Wnt/β-catenin is involved in Salmonella-induced intestinal inflammation [12], [15], [16]. Aberrant Wnt/β-catenin signaling is a characteristic feature of CRC development [33]. Classically, Wnt1 is viewed as a positive regulator of the Wnt/β-catenin pathway in development and cancer. However, in our study, Wnt1 levels were reduced in colon mucosa from mice with an experimental infection and in human colorectal clinical specimens, which further indicated that gut dysbiosis contributes to colon cancer development in part by deregulating the Wnt/β-catenin pathway.
Based on our data showing that Wnt1 levels were decreased by Salmonella infection and that Wnt1 levels were decreased in tumor tissue relative to normal tissue, we suggest that Wnt1 levels in intestinal tissue might determine how gut bacteria interact with host genes in colon cancer development. We have demonstrated the presence and cellular localization of AvrA in inflamed colorectal tumors and precursor lesions using both animal experimental infection models and human clinical specimens [30]. A limitation of our study is that we did not obtain fecal samples to further investigate Salmonella or other pathogenic infection associated with the downregulation of Wnt1 in human cancer specimens.
Considering the role of Wnt1 in the inflammatory response induced by Salmonella infection in cell line models, we speculate that a reduction in Wnt1 may predispose cells to tumorigenesis by limiting inflammatory responses in colitis-associated cancer. It will be interesting to investigate how Wnt1 synergistically coordinates these related pathways in inflammation and tumorigenesis.
In summary, the current study reports the protective role of Wnt1 in Salmonella infection, bacteria-induced inflammation, and colitis-associated CRC, suggesting a novel function of Wnt1 in host-pathogen interactions. This study will prompt future researchers to investigate whether decreased Wnt1 protein is associated with Salmonella or other pathogenic bacteria infection in human subjects.
Authors' Contributions
Jianwei Wang, Rong Lu, Xingyin Liu, Xinhui Fu, Zhou Dan, Xinxia Chang, and Qisha Liu: designed and performed experiments, prepared figures, and interpreted the data. Rong Lu, Yong-Guo Zhang, and Xingyin Liu: performed the animal experiment and some of the Western blots. Jianwei Wang and Rong Lu: drafted the results section of the manuscript. Yinglin Xia: verified statistical analysis and revised the manuscript. Xingyin Liu and Jun Sun: conceived and designed the study, interpreted the data, wrote and revised the manuscript, and obtained funding to support the research.
Declarations of Conflicts of Interest
No conflicts of interest.
Acknowledgements
This work was supported by NIH/NIDDK R01 DK105118, UIC Cancer Center Fund, American Cancer Society Research Scholar Award (to Jun Sun), NSFC81671983 (to Xingyin Liu), and the Jiangsu Province National Science Foundation (to Xingyin Liu: BK2016021990 and to Jianwei Wang: BK20171045).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2018.03.001.
Contributor Information
Xingyin Liu, Email: Xingyinliu@njmu.edu.cn.
Jun Sun, Email: junsun7@uic.edu.
Appendix A. Supplementary Data
Supplementary figures
References
- 1.Kang M, Martin A. Microbiome and colorectal cancer: Unraveling host-microbiota interactions in colitis-associated colorectal cancer development. Semin Immunol. 2017;32:3–13. doi: 10.1016/j.smim.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016;3(2):130–143. doi: 10.1016/j.gendis.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mischke M, Plosch T. The gut microbiota and their metabolites: potential implications for the host epigenome. Adv Exp Med Biol. 2016;902:33–44. doi: 10.1007/978-3-319-31248-4_3. [DOI] [PubMed] [Google Scholar]
- 4.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105(24):1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bultman SJ. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol Nutr Food Res. 2017;61(1) doi: 10.1002/mnfr.201500902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenny PA, Enver T, Ashworth A. Receptor and secreted targets of Wnt-1/beta-catenin signalling in mouse mammary epithelial cells. BMC Cancer. 2005;5:3. doi: 10.1186/1471-2407-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanczak A, Stec R, Bodnar L, Olszewski W, Cichowicz M, Kozlowski W, Szczylik C, Pietrucha T, Wieczorek M, Lamparska-Przybysz M. Prognostic significance of Wnt-1, beta-catenin and E-cadherin expression in advanced colorectal carcinoma. Pathol Oncol Res. 2011;17(4):955–963. doi: 10.1007/s12253-011-9409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You J, Nguyen AV, Gregory Albers C, Lin F, Holcombe RF. Wnt pathway-related gene expression in inflammatory bowel disease. Dig Dis Sci. 2008;53(4):1013–1019. doi: 10.1007/s10620-007-9973-3. [DOI] [PubMed] [Google Scholar]
- 9.Gradel KO, Nielsen HL, Schønheyder HC, Ejlertsen T, Kristensen B, Nielsen H. Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. 2009;137(2):495–501. doi: 10.1053/j.gastro.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Kato I, Boleij A, Kortman GAM, Roelofs R, Djuric Z, Severson RK, Tjalsma H. Partial associations of dietary iron, smoking and intestinal bacteria with colorectal cancer risk. Nutr Cancer. 2013;65(2):169–177. doi: 10.1080/01635581.2013.748922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J, Hobert ME, Rao AS, Neish AS, Madara JL. Bacterial activation of beta-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol. 2004;287(1):G220–7. doi: 10.1152/ajpgi.00498.2003. [DOI] [PubMed] [Google Scholar]
- 12.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007;171(3):882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang YG, Wu S, Xia Y, Chen D, Petrof EO, Claud EC, Hsu H, Sun J. Axin1 prevents Salmonella invasiveness and inflammatory response in intestinal epithelial cells. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0034942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahoun A, Mahajan S, Paxton E, Malterer G, Donaldson DS, Wang D, Tan A, Gillespie TL, O'Shea M, Roe AJ. Salmonella transforms follicle-associated epithelial cells into M cells to promote intestinal invasion. Cell Host Microbe. 2012;12(5):645–656. doi: 10.1016/j.chom.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Lu R, Liu X, Wu S, Xia Y, Zhang Y, Petrof EO, Claud EC, Sun J. Consistent activation of the beta-catenin pathway by Salmonella type-three secretion effector protein AvrA in chronically infected intestine. Am J Physiol Gastrointest Liver Physiol. 2012;303(10):G1113–25. doi: 10.1152/ajpgi.00453.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu R, Wu S, Liu X, Xia Y, Zhang Y, Sun J. Chronic effects of a Salmonella type III secretion effector protein AvrA in vivo. PLoS One. 2010;5(5) doi: 10.1371/journal.pone.0010505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Wu S, Xia Y, Li XE, Xia Y, Zhou ZD, Sun J. Wingless homolog Wnt11 suppresses bacterial invasion and inflammation in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2011;301(6):G992–G1003. doi: 10.1152/ajpgi.00080.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Lu R, Wu S, Zhang Y, Xia Y, Balfour Sartor R, Sun J. Wnt2 inhibits enteric bacterial-induced inflammation in intestinal epithelial cells. Inflamm Bowel Dis. 2012;18(3):418–429. doi: 10.1002/ibd.21788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu R, Wu S, Zhang YG, Xia Y, Liu X, Zheng Y, Chen H, Schaefer KL, Zhou Z, Bissonnette M. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis. 2014 doi: 10.1038/oncsis.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porfir E, Rubinfeld B, Albert I, Hovanes K, Waterman M, Polakis P. Induction of a beta-catenin-LEF-1 complex by wnt-1 and transforming mutants of beta-catenin. Oncogene. 1997;15(23):2833–2839. doi: 10.1038/sj.onc.1201462. [DOI] [PubMed] [Google Scholar]
- 21.Vider BZ, Zimber A, Chastre E, Prevot S, Gespach C, Estlein D, Wolloch Y, Tronick SR, Gazit A, Yaniv A. Evidence for the involvement of the Wnt 2 gene in human colorectal cancer. Oncogene. 1996;12(1):153–158. [PubMed] [Google Scholar]
- 22.Jeong JJ, Kim KA, Jang SE, Woo JY, Joo Han M, Kim DH. Orally administrated Lactobacillus pentosus var. plantarum C29 ameliorates age-dependent colitis by inhibiting the nuclear factor-kappa B signaling pathway via the regulation of lipopolysaccharide production by gut microbiota. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0116533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khor TO, Gul YA, Ithnin H, Seow HF. A comparative study of the expression of Wnt-1, WISP-1, survivin and cyclin-D1 in colorectal carcinoma. Int J Color Dis. 2006;21(4):291–300. doi: 10.1007/s00384-005-0002-8. [DOI] [PubMed] [Google Scholar]
- 24.Miller SI, Mekalanos JJ. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172(5):2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu R, Wu S, Zhang Y, Xia Y, Zhou Z, Kato I, Dong H, Bissonnette M, Sun J. Salmonella protein AvrA activates the STAT3 signaling pathway in colon cancer. Neoplasia. 2016;18(5):307–316. doi: 10.1016/j.neo.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick BA, Colgan SP, Delp-Archer C, Miller SI, Madara JL. Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils. J Cell Biol. 1993;123(4):895–907. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lissner CR, Swanson RN, O'Brien AD. Genetic control of the innate resistance of mice to Salmonella typhimurium: expression of the Ity gene in peritoneal and splenic macrophages isolated in vitro. J Immunol. 1983;131(6):3006–3013. [PubMed] [Google Scholar]
- 28.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habi N, Hsu PD, Wu X, Jiang W, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Zhou JM. The microbiota-gut-brain axis and its potential therapeutic role in autism spectrum disorder. Neuroscience. 2016;324:131–139. doi: 10.1016/j.neuroscience.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Lu R, Bosland M, Xia Y, Zhang Y, Kato I, Sun J. Presence of Salmonella AvrA in colorectal tumor and its precursor lesions in mouse intestine and human specimens. Oncotarget. 2017;8(33):55104–55115. doi: 10.18632/oncotarget.19052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munoz S, Guzman-Rodriguez M, Sun J, Zhang Y, Noordhof C, He SM, Allen-Vercoe E, Claud EC, Petrof EO. Rebooting the microbiome. Gut Microbes. 2016;7(4):353–363. doi: 10.1080/19490976.2016.1188248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martz SL, McDonald JAK, Sun J, Zhang Y, Gloor GB, Noordhof C, He SM, Gerbaba TK, Blennerhassett M, Hurlbut DJ. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci Rep. 2015;5:16094. doi: 10.1038/srep16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deitrick J, Pruitt WM. Wnt/beta Catenin-Mediated Signaling Commonly Altered in Colorectal Cancer. Prog Mol Biol Transl Sci. 2016;144:49–68. doi: 10.1016/bs.pmbts.2016.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures