Abstract
Anthrax is caused by Bacillus anthracis, a bacterium that is able to secrete the toxins protective antigen, edema factor and lethal factor. Due to the high level of secretion from the bacteria and its severe virulence, lethal factor (LF) has been sought as a biomarker for detecting bacterial infection and as an effective target to neutralize toxicity. In this study, we found three aptamers, and binding affinity was determined by fluorescently labeled aptamers. One of the aptamers exhibited high affinity, with a Kd value of 11.0 ± 2.7 nM, along with low cross reactivity relative to bovine serum albumin and protective antigen. The therapeutic functionality of the aptamer was examined by assessing the inhibition of LF protease activity against a mitogen-activated protein kinase kinase. The aptamer appears to be an effective inhibitor of LF with an IC50 value of 15 ± 1.5 µM and approximately 85% cell viability, suggesting that this aptamer provides a potential clue for not only development of both a sensitive diagnostic device of B. anthracis infection, but also the design of novel inhibitors of LF.
Introduction
Anthrax is caused by Bacillus anthracis, a Gram-positive, non-motile, spore forming bacterium. The most serious type of anthrax is pulmonary infection, where secretion of toxins in a host impairs the host immune response, allowing for fast bacterial proliferation, sepsis and death of the host within hours of symptom development [1, 2]. Three toxins are produced by B. anthracis; that is, protective antigen (PA), lethal factor (LF), and edema factor (EF) [3, 4]. These three toxins act as an AB type complex, where PA plays a role as the non-catalytic portion (B portion), and either EF or LF act as the catalytic portion (A portion) [5]. PA forms a heptameric or octameric complex capable of binding to a receptor and induce internalization of LF and EF [6]. EF is a calcium/calmodulin dependent adenylate cyclase that increases the intracellular levels of cAMP causing cell swelling [4]. LF, a zinc-dependent metalloprotease that cleaves most of the mitogen-activated protein kinase kinases (MAPKKs), interrupts multiple intracellular signaling pathways, thus resulting in the induction of apoptosis. Furthermore, it has been reported that intravenous delivery of LF toxin alone causes death in rodents [7, 8] and that the toxicity of B. anthracis strains deficient in LF was substantially reduced [9]. These reports provide strong evidence that LF plays a central role in the biological action of B. anthracis.
Proper diagnosis of anthrax has proved to be challenging because the initial symptoms of the infection are very similar to flu symptoms, and characterization of the bacterium is rather complicated due to its similarity to other members of the Bacillus group [10]. Hence, prompt diagnosis is required to treat the infection effectively with antibiotic treatments; however, antibiotics are not able to neutralize the toxins secreted from the bacteria into the body. Since LF is a secreted protein that acts as a toxin, it offers an attractive venue for a detection and therapeutic target. Additionally, it has been reported that although the PA concentration in rabbit sera was highly variable, the concentration of LF at 48 h after infection was generally higher than those of PA and EF (particularly the concentration ratio of LF and EF was 5:1) [11]. This fact also supports the notion that LF could be an attractive target for detecting the presence of B. anthracis.
Systematic evolution of ligands by exponential enrichment (SELEX) technique has been extensively used in the field of disease detection and therapeutics. SELEX was originally developed to find synthetic oligonucleotides capable of binding to a target molecule with high affinity and specificity [12–14]. The oligonucleotides can be either single stranded DNA (ssDNA) or RNA, where single stranded oligonucleotides have the ability to generate a variety of tertiary structures arising from secondary structures (reviewed in [15]) that can facilitate the interaction with a target molecule. Through the process of SELEX, tight binding ssDNAs or RNAs can be found, which are referred to as aptamers. SELEX technique has been employed to discover aptamers against B. anthracis spores [16] and PA [17, 18], and yet there are no reports of aptamers against LF. Here, we have developed ssDNA aptamers against LF using SELEX and determined their binding affinity and specificity. We also designed an aptamer-based ELISA method to visualize the presence of LF. In addition, binding sites of the aptamer and LF have been studied with the combination of experimental and computational analyses. Furthermore, the possibility of LF inhibition with the found aptamer was assessed.
1. MATERIALS AND METHODS
2.1. LF protein preparation and SELEX for LF protein
To obtain purified LF, transformed E. coli BL21(DE3) cells harboring the pET28a-LF plasmid were used for protein expression, and protein purification was performed as previously described [19].
To begin SELEX, a random 30-mer-long DNA fragment flanked by a forward primer region and a reverse primer region, 5'-GCGCGGATCCCGCGC-N(30)-CGCGCGAAGCTTGCG -3’, was purchased from Integrated DNA Technologies (Coralville, IA). Additionally, two primers were purchased with the following sequence: forward primer: 5'-GCGCGGATCCCGCGC-3' and reverse primer: 5'-CGCAAGCTTCGCGCG-3'. The template was amplified by asymmetric PCR using a thermocyler with a 10:1 forward:reverse primer ratio (0.5 µM:0.05 µM); the initial template concentration was 1.0 µM, and the template concentrations after the first round varied (approximately between 0.01 and 0.05 µM). The ssDNA was then recovered from a 6% native polyacrylamide gel using a previously described crush-and-soak method and concentrated by ethanol precipitation [20].
As we employed in our previously published SELEX method [18], we began to use a large quantity of protein compared to ssDNA, but as the SELEX round went on, the molar ratio of LF to ssDNA was decreased. To obtain aptamers against LF, purified protein in a binding buffer (30 mM Tris-HCl at pH 8.4, 3.0 mM KCl, 600 mM NaCl, and 0.6 mM MgCl2) was incubated at 30°C with the randomized ssDNA library for 30 min in a final volume of 50 µL, in which the amounts of LF and ssDNA used in each SELEX were 22 µg and 0.45 µg, respectively. The complex between protein and ssDNA was resolved by an electrophoretic mobility shift assay (EMSA) using a 6% polyacrylamide native gel and visualized with either ethidium bromide or SYBR gold stain. Recovery of the complexed ssDNA from the gel was achieved by a crush-and-soak method followed by ethanol precipitation and regular PCR amplification; for the normal PCR condition, the primer concentrations were 0.5 µM for both forward and reverse primers. The double stranded DNA (dsDNA) was then amplified by asymmetric PCR to produce the ssDNA for the next SELEX round. This process was repeated with the same PCR condition up to round 5, at which point the ssDNA was used to perform a counter SELEX step using Bovine Serum Albumin (BSA). The PCR condition for the counter SELEX was the same as the aforementioned asymmetric PCR condition, and 17 µg of BSA was added in the total volume of 50 µL, in which the amount of ssDNA was 0.45 µg. The incubation time was also 30 min, and the buffer used was 30 mM Tris-HCl (pH 8.0), 60 mM NaCl, 3.0 mM KCl and 0.6 mM MgCl2. After separation using EMSA, the unbound DNA was excised from the gel, and the extracted DNA using the crush-and-soak method was used for the next round 6. After one round of the counter SELEX, the selection process continued for 3 more rounds with an increasing NaCl and KCl concentration (Table 1). In the 6th round, the amounts of LF and ssDNA were 10 µg and 1 µg, respectively. The buffer condition was the same as the previous rounds. For the 7th and 8th rounds, the amounts of LF and ssDNA were the same as the round 6, but the buffer conditions were 30 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5.0 mM KCl and 0.6 mM MgCl2. The product of the final round of SELEX was amplified by PCR and used for cloning.
Table 1.
Buffer conditions of each round of SELEX*
| SELEX Round | |||
|---|---|---|---|
|
| |||
| 1st – 5th | BSA counter SELEX | 6th | 7th – 8th |
| 30 mM Tris (pH 8.0) | 30 mM Tris (pH 8.0) | 30 mM Tris (pH 8.0) | 30 mM Tris (pH 8.0) |
| 60 mM NaCl | 60 mM NaCl | 60 mM NaCl | 100 mM NaCl |
| 3.0 mM KCl | 3.0 mM KCl | 3.0 mM KCl | 5.0 mM KCl |
| 0.6 mM MgCl2 | 0.6 mM MgCl2 | 0.6 mM MgCl2 | 0.6 mM MgCl2 |
The resulting dsDNA was separated from the other PCR reaction components by running a 6% polyacrylamide gel. dsDNA was recovered by a crush-and-soak method and then inserted into pGEM-T easy vector according to the manufacturer’s instructions (Promega). E. coli BL21(DE3) cells were transformed and grown on an agar plate containing 100 µM kanamycin. Individual colonies were picked up and cultured to extract the plasmid, and the presence of the insert was confirmed by restriction enzyme digestion using EcoRI (BioLabs Inc.). Plasmids from positive colonies were subjected to sequencing.
2.2. Determination of dissociation constant (Kd) by fluorimetry
To estimate the Kd for the found aptamers binding to LF, we purchased the aptamers labeled with the fluorophore Cy3 at the 3’-end (IDT). A fixed amount of LF (0.5 µg) in 100 µL of 20 mM HEPES (pH 7.4) at room temperature was loaded onto a 96-well white opaque immunoplate (SPL, Kyounggi-do, South Korea) and was incubated for 1 h with shaking to immobilized LF on the plate. The wells containing LF were washed with 200 µL of Trisbuffered saline Tween-20, called TBST (0.5 M Tris-HCL, pH 8.4, 9% NaCl, and 0.5% Tween-20) three times. Subsequently, 100 µL of 2% BSA was added and incubated for 1 h. The equal volume of TBST was again used to wash the wells three times. Varied concentrations of ssDNA aptamers (0 – 50 nM) was added to the wells. For binding to LF, 1 h incubation with shaking was allowed. Excess ssDNA was eliminated by washing with 200 µL of 20 mM HEPES (pH 7.4) three times. The fluorescent intensities of the complex between ssDNA and LF were measured by a Molecular Devices (Sunnyvale, CA) Gemini EM fluorescence microplate reader (excitation: 545 nm; emission: 572 nm).
2.3. Aptamer-based ELISA method
LF protein (0.5 µg/well) was immobilized onto a 96 polystyrene transparent immnunoplate (SPL, Kyounggi-do, South Korea) as described in the preceding section. Varied concentrations of the aptamer ML12 labeled with biotin at the 3’-end (purchased from IDT) were added to the wells possessing immobilized LF and then incubated for 1 h with shaking. Excess ssDNA was removed by washing with 200 µL TBST three times. Subsequently, 100 µL of streptavidin-attached horseradish peroxidase (purchased from BD Biosciences, Franklin Lakes, NJ) was added to the wells with 1:1000 dilution with TBST. Incubation was carried out for 1 h with shaking. Subsequent to washing the wells with 200 µL of TBST three times, blue color was developed by 90 µL of TMB substrate solution (3,3’,5,5’-tetramethyl benzidine; R&D Systems, Minneapolis, MN). After waiting 15 min, the reaction was terminated by the addition of 90 µL of 1M H2SO4 to yield a yellow color. The color intensities in each well were measured with the Biotrak multiwall plate reader (Amersham Biosciences, Piscataway, NJ) at 450 nm.
2.4. CD analysis
To determine possible secondary structure characteristics, aptamers (5 µM) were resupended in 20 mM Na3PO4 (pH 7.5) that contained 100 mM KCl or 100 mM NaCl. Samples were heated at 90°C for 5 min, followed by gradual cooling to room temperature. CD spectra were collected by a JASCO J-810 spectropolarimeter from 320 – 220 nm, with a 1 cm path length cuvette.
2.5. In vitro lethal factor proteolytic activity assay
Glutathione S-transferase (GST)-tagged mitogen-activated extracellular signal-regulated kinase kinase 1(MEK1) (hereafter abbreviated as GST-MEK1) were expressed in E. coli DH5α and purified as described previously [21]. LF (0.25 ug) was pre-incubated with or without the aptamer at 37°C for 15 min. The reaction was initiated by adding substrate mixture (25 mM HEPES, pH 7.4, 1 mM DTT, 10% glycerol, 50 µM CaCl2, 5 mM MgCl2 and 0.7 µg GST-MEK1), terminated by the addition of 5 × sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer after 1 h, and the reaction was completed by boiling. Samples were assessed on a 12% SDS-polyacrylamide gel and the cleavage of GST-MEK1 was visualized by Coomassie blue staining [21]. To quantify the stained bands, ImageJ was used (freely available at http://rsbweb.nih.gov/ij/)
2.6. Cytotoxicity test
RAW 264.7 cells in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% antibiotic solution (penicillin–streptomycin) were used and counted with a hemocytometer. 2 × 105 of cells were seeded in a 96-well plate and incubated for 24 h in a 5% CO2 atmosphere at 37°C. The plate was washed with PBS (pH 7.5) and fresh media was added. Next, the cells were treated with PA for 1 h, then LF was added in the absence or presence of the ML12 concentration manner for 4 h after washing with PBS. In order to analyze cell viability, a 1 mg mL−1 of MTT solution (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was added and incubated for 4 h. Dimethyl sulfoxide (DMSO) as a detergent was added, and the absorbance was measured at 570 nM via microtiter plate reader [22, 23].
3. Results and Discussion
3.1. SELEX process
LF protein was purified for SELEX to obtain aptamers using a ssDNA library that contains a 30-mer-long randomized sequence. To increase binding affinity and specificity, after the 5th round of SELEX, BSA was added as a counter SELEX and ionic strength was increased by adding higher concentrations of NaCl and KCl. For the separation of the ssDNA:LF complex from unbound ssDNA, EMSA was performed, by which clear separation was viewed on a native gel (Supplementary Figure S1). At the 8th rounds, the PCR product bands on the gel were significantly narrower than previous round bands, and we also checked the enzyme inhibition with the pool of nucleotides at each round. There was no significant inhibition up to the 5 round, but about 20% inhibition was observed at the 6 round, and around 70% inhibition was observed at the 7 and 8 rounds. Given that there was no further inhibition improvement, the nucleotide sequence should be identified before losing a chance to obtain good candidates as inhibitors. In doing so, the dsDNA pool was inserted to the pGEM-T easy vector at the EcoRI site. Subsequently, E. coli BL21(ED3) was transformed with the cloned vector. The transformed cells appeared on an agar plate and twelve colonies were picked up. Four of the twelve colonies contained inserts of interest, and three different sequences were found among the four inserts (Table 2), where the aptamer ML12 appeared twice and other two aptamers (ML6 and ML7) appeared once. It should be noted that if one would stop at a different round, the aptamer sequences would be different. In this experiment we obtained three different sequences were obtained, and these results are not surprised because we experienced the similar observation previously [18, 20].
Table 2.
Sequence of aptamers against LF
| Aptamer | ssDNA Sequence (30-mer) | Frequency | Kd value |
|---|---|---|---|
| ML6 | 5’-GGACCAGCCGCCGCGCCTTGACCGGGGGTA-3’ | 1 | ND* |
| ML7 | 5’-GGAGAGAGGGAGACGCGCAACCTCGACCCGT-3’ | 1 | ND |
| ML12 | 5’-CGAGGGAGACGCGAACCTTCTCGCCTTGGG-3’ | 2 | 11.0 ± 2.7 nM |
Not determined within the micromolar range. The underline indicates the sequence that may contribute to a hairpin secondary structure.
3.2. Binding affinity and specificity
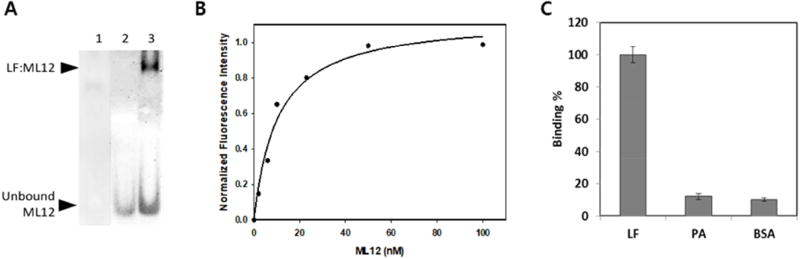
To confirm the binding of the three aptamers to LF, a native gel was loaded with samples containing LF and each aptamer. A complex successfully formed between LF (unstained) and aptamer ML12 (SYBR gold stained) as shown in Figure 1A. Also, we loaded sample containing ML12 and BSA to a 6% native polyacrylamide gel and found that no complex formation between ML12 and BSA was detected. In order to determine the binding affinity, we used a fluorimetry method with the fluorophore Cy3 attached at the 3’-end of each aptamer. Measurements were made of the Kd values for the Cy3-attached aptamers bound to LF. The results showed that only ML12 appears to have high binding affinity with a Kd value of 11.0 ± 2.7 nM, whereas the other two aptamers appear to be out of micromolar range. Figure 1B shows the results of the Kd determination of ML12 with data points fitting into a single ligand binding hyperbolic curve. In addition to binding affinity, we also tested ML12 with the flanking sequences (i.e., a 60-mer oligonucleotide) and determined the Kd value to be 17.5 ± 4.8 nM, suggesting that the 30-mer region is primarily responsible for the binding to LF. As a control experiment, we used a scramble sequence with the same length as ML12 (i.e., ssDNA 30-mer; 5’-CGCACTGAAGTGGGATACCGCCTAAACGGA-3’) and found that there was no binding observed up to 1 µM, showing that the tight binding observed with ML12 is sequence specific and is not simply a random DNA-protein interaction. Moreover, the binding of ML12 to LF was compared with the binding to BSA and PA and revealed that the binding affinity of ML12 to LF is much higher than for BSA and PA (Figure 1C). These observations suggest that the aptamer ML12 appears to bind to LF with high affinity and specificity; thus, it can be a promising detecting material for LF.
Figure 1.
Binding experiments between LF and ML12. (A) Gel shift assay between LF and the aptamer ML12. Lane 1 represents the gel result after loading only LF; Lane 2 represents the gel result after loading only ssDNAs; Lane 3 respresent the gel result after loading ssDNAs and LF. The top band shows the complex of LF and ML12 and the bottom band shows unbound ML12 in a 6 % native gel with 5.9 µM LF and 100 nM ML12 using SYBR gold staining. Prior to loading sample, 20 min incubation at 30 °C in buffer (30 mM Tris, pH 8.0, 100 mM NaCl, 5.0 mM KCl and 0.6 mM MgCl2) was conducted. The total sample volume was 10 µL. (B) Determination of binding affinity of aptamers. The black circles are for ML12 and the empty circles are for ML12 with flanking sequences. Binding reactions were carried out with a constant concentration of LF protein in a 96-well black plate and varied concentarions of Cy3-attached ML12. The fluorescent intensity of the bound ML12 or ML12 with flanking sequences to LF was measured after a couple of washing steps. The Kd value was determined by nonlinear fitting of the saturation binding curve. (C) Binding specificty of ML12 against BSA by the fluorescent intensity measurement of bound Cy3-ML12 to either LF or BSA. The concentration of the proteins (LF and BSA) was kept constant at 0.5 µM, and 10 nM of ML12 was used for initial incubation against the proteins.
3.3. Aptamer-based ELISA development
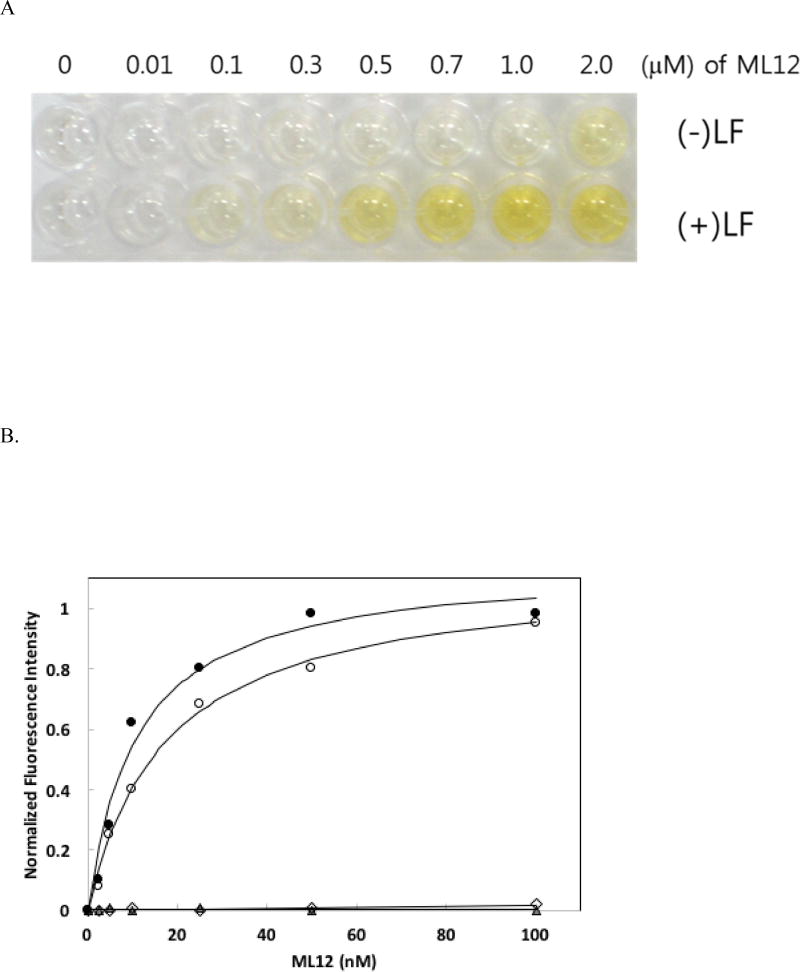
To use the ML12 aptamer for detection, we developed an aptamer-based ELISA. LF was successfully immobilize on a 96-well plate (see Method and Materials), with little to no none-specific binding of ssDNA to plate [18, 24]; ML12 aptamer, labeled with biotin at the 3’ end instead of Cy3, was added to each well in a concentration-dependent manner. After incubation and washing steps. Binding of ML12 to LF was visualize by the formation of a yellow color after addition of HRP-conjugated streptavidin, TMB, and H2SO4. This approach for an ELISA-based detecting method was successful, as shown in Figure 2. Color intensities clearly increased when the aptamer concentration increased with saturated intensities at the highest concentrations of aptamers (Figure 2A). The quantification of color intensity revealed a Kd value of 530 ± 11 nM. Meanwhile, no color development for other aptamers ML6 and ML7 was observed (Table 1). In addition, we tested ML12 with the flanking sequences (i.e. 60-mer) and the scramble sequence oligonucleotide (see the Material and Method for the sequence information) used for the preceding fluorescent measurement method and found a Kd value of 705 ± 60 nM for the 60-mer (Figure 2B) and no binding to LF for the scramble sequence using the aptamer-based ELISA method. It should be mentioned that in this aptamer-based ELISA method, the Kd value obtained was much higher than the value obtained by the Cy3 fluorescence detection method. This may be due to the requirement of multiple steps for color development in the ELISA method, including the reaction between biotin and streptavidin. In addition, the multiple washing steps require in this method increase the possibility of detachment of the aptamer from the target molecule [18, 25]. Hence, future investigation is warranted to further refine the ELISA method in an effort to reduce the apparent Kd value of the aptamer.
Figure 2.
ELISA method using biotinylated aptamers. (A) The results of ELISA using biotinylated ML12. The first row contains only BSA and the second row contains a fixed concentration of LF (0.5 µg/well) with various concentrations of biotinylated ML12 (0 – 2.0 µM). Biotinlyated aptamers interacted with horseradish peroxidase-conjugated streptavidin, followed by the addition of teramethyl benzine. To quench the reaction, sulfuric acid was added. (B) Determination of binding affinity using ELISA. The gray-color filled circles are for ML12 and the empty circles are for ML12 with flanking sequences. The absorbance (450 nm) intensity of the bound ML12 or ML12 with flanking sequences to LF was measured. The Kd value was determined by nonlinear fitting of the saturation binding curve. The gray-color filled triangles are for ML6 and the empty diamonds are for ML7.
3.4. Characterization of the aptamer ML12
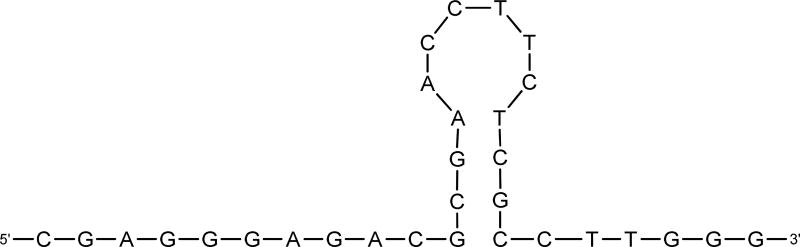
To explore potential binding sites of the aptamer, M-Fold was used to predict possible secondary structures [26]. Although our focus of the binding sites was on the complex between ML12 and LF, we examined all three aptamers to ensure that there were no structural relationships among them. As a result, MFold predicted all dissimilar secondary structures, inferring that no common binding sites would be present among the three aptamers. In the case of ML12, two different secondary structures were predicted by MFold with weak ΔG binding values of −2.66 and −1.70 kcal/mol, respectively. The secondary structure with the lowest energy value is shown in Figure 3. In this secondary structure, a 14-mer sequence forms a stem-loop secondary structure that may be a good candidate for the binding sites of ML12. When the 14-mer (5’-d(GCGAACCTTCTCGC)-3’) sequence was subject to the DNA folding prediction, the identical secondary structure was formed with a ΔG value of −2.01 kcal/mol, which is slightly lower than the value obtained for the full sequence. In an effort to obtain the experimental data of interaction between the 14-mer and LF, we attempted to determine Kd value between them. However, there was no significant fluorescent signals up to 1 µM by the 14-mer, suggesting that instead of the 14-mer, the 30-mer would be a better choice for binding LF and potentially detect its presence. It should be noted here that we also predicted the secondary structure of the ML12 with the flanking sequences (i.e., 60-mer containing ML12 30-mer sequence and two flanking sequences) and found that the predicted secondary structures are different from those of ML12. Taken together, these observations suggest that in this research, the prediction by MFold may not be essential for a binding site evaluation.
Figure 3.
Predicted secondary structure of ML12. The 14-mer (5’-GCGAACCTTCTCGC-3’) is shown as a stem-and-loop structure.
In view of aptamer composition, there are differences in the guanosine (G) content between the full sequence (30-mer) and the 14-mer. In the case of 30-mer, the G content is 36.7%, whereas the 14-mer contains only 21.4%. One might argue that due to the high G content in the 30-mer, the 30-mer may form a G-quadruplex structure instead of a stem-and-loop structure. To assess the possibility of the presence of a G-quadruplex, we used circular dichroism (CD). The CD spectra were measured in the presence of either NaCl or KCl because earlier literature reported that the presence of potassium or sodium ion and appropriate heat-and-cooling procedure would facilitate the correct formation of a G-quadruplex [27]. In most cases potassium contributes to G-quadruplex structure stabilization, but in some cases sodium plays a role in stabilizing the G-quadruplex [27–29]. We examined the CD spectra of the 30-mer in the presence of K+ or Na+ and found that no G-quadruplex spectra were observed. Although the presence of G-quadruplex typically results in a positive maximal peak near 295 nm [30, 38], the CD spectra obtained with the 30-mer in the presence and absence of K+ or Na+ showed an almost identical spectrum with a positive maximum peak near 280 nm. Thus, these results indicate that a secondary structure with B-form double helix, rather than a G-quadruplex, is present (Supplementary Figure S2) [30]. Additionally, the quadruplex-forming G-rich sequence (QGRS) mapper was employed to check the possibility of the presence of G-quadruplex, and the computational G-quadruplex prediction showed no possibility [34]. Combined, these observations suggest that there is only one possible secondary structure (14-mer) in the 30-mer and that the entire 30-mer dictates tight binding to LF.
3.5. Evaluation of an aptamer-based LF inhibitor
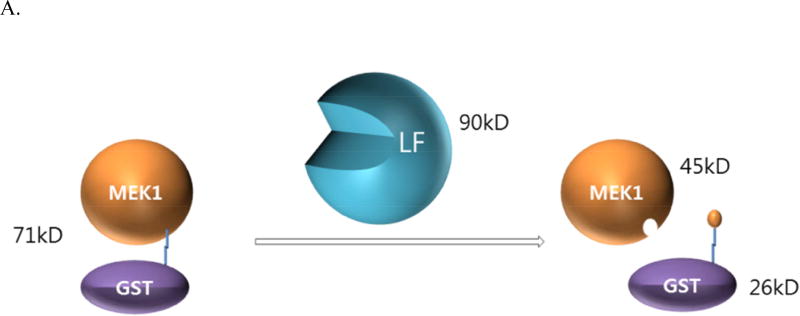
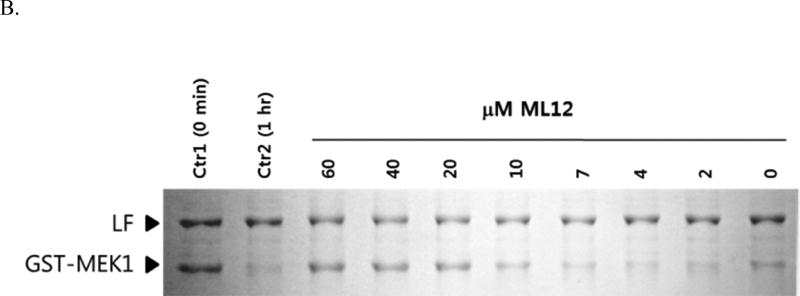
Since the aptamer ML12 tightly binds to LF, there is a possibility that ML12 would interfere with the interaction between LF and its substrate MEK1, a MAPKK that plays a key intermediary role in the mitogen-activated protein kinase pathway. LF that cleaves most of the MAPKKs interrupts the transcription of several genes including genes involved in immune response [35], and shuts down the extracellular signal-regulated kinase (ERK), the c-Jun N-terminal kinase (JNK), and p38 MAPK pathway, in which the activation of these protein is responsible for different types of responses in cell like gene expression, apoptosis and cell proliferation [36]. Thus, blocking the interaction between LF and MEK1 allows for the reduction of the toxicity of LF. Previously, we developed a reliable LF activity assay with GST-MEK1 in a native gel [21], where disappearance of the band corresponding to GST-MEK1 occurs as a result of the removal of the GST tag by the LF cleavage reaction to MEK1, as illustrated in Figure 4A.
Figure 4.
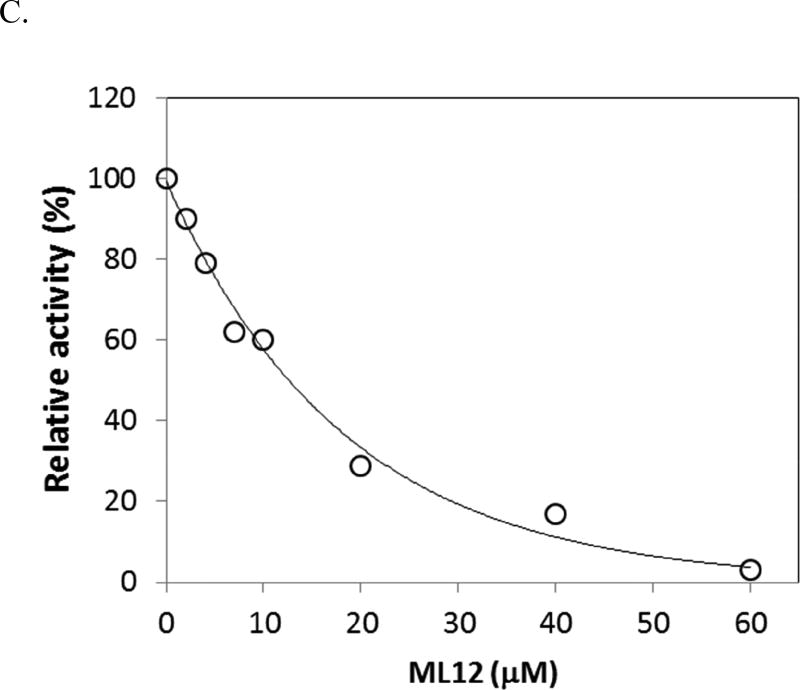
Inhibition of LF by ML12. (A) A diagram for the proteolytic assay of LF to GST-MEK1. (B) ML12 inhibits cleavage of GST-MEK1 by LF. Lane 1 (Ctr1) is a control test with no pre-incubation time for LF cleavage reaction with GST-MEK1; Lane 2 (Ctr2) is another control with l h incubation for the reaction between LF and GST-MEK.1 Subsequent lanes (third through last lanes) contains LF and GST-MEK1 with treatment of varied concentration of ML12. Except the first lane (Ctr1), the pre-incubation time for the LF cleavage reaction with GST-MEK1 was 1 h. (C) Determination of IC50 value of ML12. The intensities of the band corresponding to GST-MEK1 were plotted as a function of the concentration of ML12. The data were fitted to an exponential decay curve.
In order to ensure the validity of the designed assay, two controls were conducted. The first control experiment did not include a pre-incubation period for LF to react with GST-MEK1, and the second control experiment included a sufficient time period to complete the LF reaction. As shown in Figure 4B, the first control experiment results in intact GST-MEK1 (Lane 1), and for the second control experiment (Lane 2), the GST-MEK1 band disappears. To assess the possibility of inhibition of LF by ML12, varied concentrations of ML12 ranging from 0 to 60 µM were used in the same gel as was used for the control experiments. As illustrated in Figure 4B, as the concentration of ML12 increased, the band intensity of GST-MEK1 also increased. The result clearly provides evidence that ML12 inhibits the LF cleavage reaction, leaving GST-MEK1 intact. It would be worthwhile to note that the result by ML12 with flanking sequences was similar to that of ML12, showing that the flanking sequences do not appear to be critical to the inhibition. Furthermore, to determine the potency of inhibition, the band intensities of GST-MEK1 were quantified. The data of the band intensities gave a good fit to a single decay exponential curve with an IC50 value of 15 µM ± 1.5. This observation suggests that ML12 might block the active site of LF to prevent protease activity.
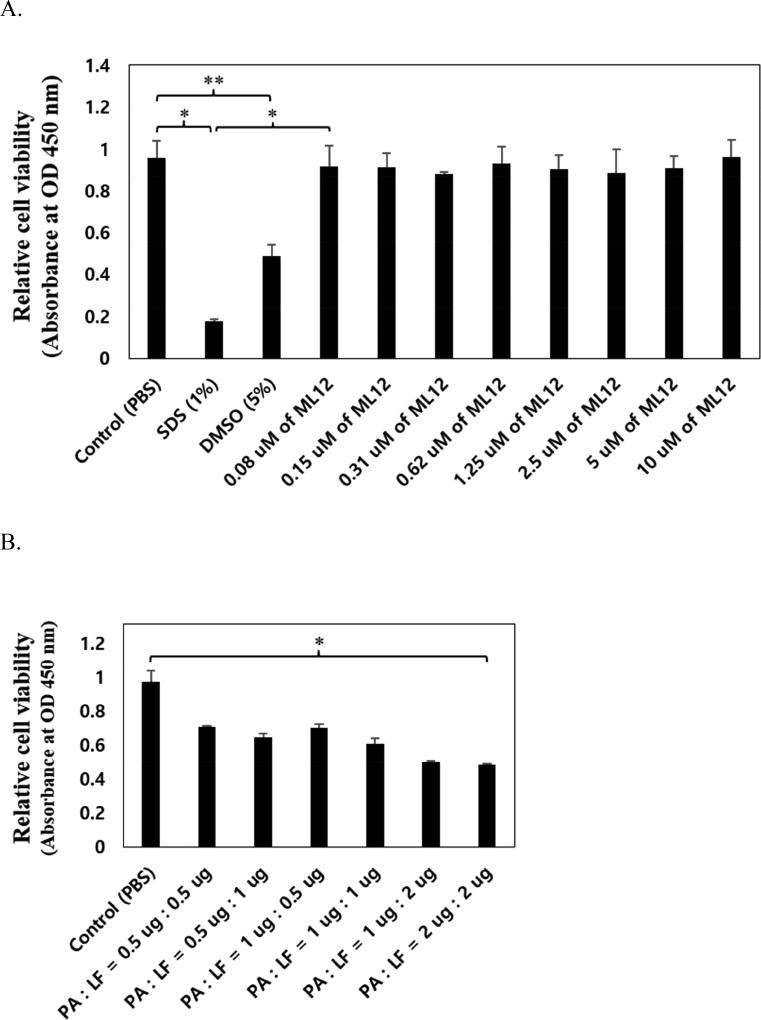
To confirm the ML12 toxicity to cell, we used RAW 264,7 macrophage cell in which anthrax toxin receptor (ATR) and anthrax toxin (PA) binding receptor proteins exist. Cytotoxicity tests were carried out via MTT assay. The ML12 with various concentration (0.08 – 10 µM) was treated in overnight cultured cell on 96-well plate. As negative control we used SDS (Sodium dodecyl sulfate) and DMSO, known to be toxic materials in cell. As shown in Figure 5A, cell viability did not change as ML12 concentration increased, that means ML12 appears to be non-toxic to the cells. Relatively, both 1% SDS and 5% DMSO induced cell toxicity with approximately 80% and 50% decreased cell viability respectively.
Figure 5.
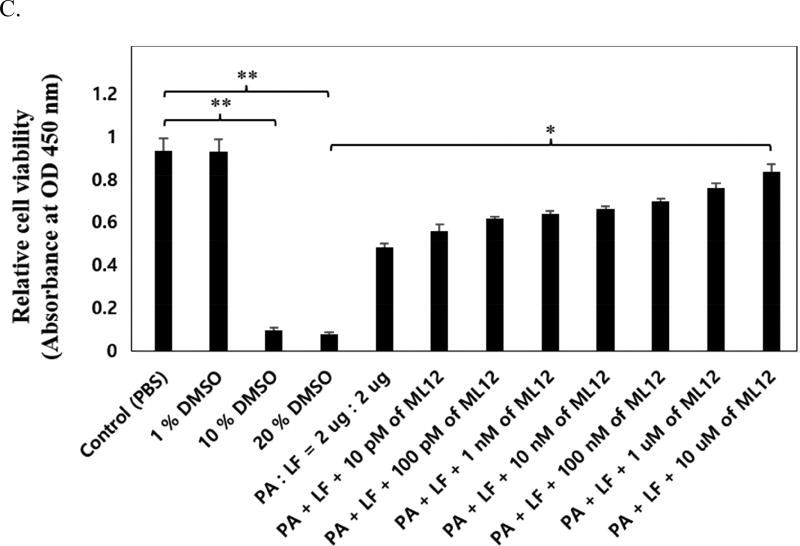
Cytotoxicity test of ML12 on Raw 264.7 cell by MTT assay. (A) Confirmation of cell toxicity for ML12. ML12 was treated with various concentrations (0.08 – 10.0 µM), and 1% SDS and 5% DMSO were used as negative controls (*p < 0.02 and **p < 0.05). (B) Optimization of anthrax toxin effect in the cell by addition of PA and LF with concentration ratio dependent (*p < 0.005). (C) Confirmation of inhibitory effect of ML12 for anthrax lethal toxin. The experiment was conducted and relative cell viability in the presence or absence of both 2 µg of PA and LF with ML12 cocentration dependent manner. The DMSO was used as a negative control. (*p < 0.002 and **p < 0.005). All measurements were conducted in triplicate.
In order to confirm an inhibitory potency of ML12 for anthrax toxin, we first optimized both PA and LF concentration that would induced maximum cytotoxicity effect. The highest concentration used to obtain low cell viability due to the activity of PA and LF was a 2 µg combination of each PA and LF (Figure 5B). After determination of optimized PA and LF concentration, we next analyzed inhibitory activity with various concentrations of ML12 and DMSO as negative control. As ML12 concentration increased, the cell viability was dramatically increased up to approximately 90% (Figure 5C). It was proved that ML12 could completely neutralize anthrax toxin by blocking a LF signaling pathway in the cell.
To date, the search to develop an effective inhibitor of LF revolves around incorporating different functional groups [31, 32, 37]. One potential focus is on the incorporation of the hydroxamate functional group, which has produced an inhibitor with an IC50 value of 60 nM, which is also in the nanomolar range of IC50 like ML12 (11 nM). However, a potential caveat is that hydroxamate functionality has the ability to reversibly bind to a number of metal ions including zinc ions, resulting in serious side effects such as musculoskeletal syndrome [33]. New types of inhibitors, such as the aptamer ML12, can be added to a repertoire of promising drug candidates for neutralization of LF toxicity. We performed SELEX against LF from B. anthracis and have demonstrated that the ssDNA aptamer ML12 binds to LF with high affinity. Visualization of binding using an aptamer-based ELISA method provides additional evidence that ML12 tightly binds to LF. Efforts to specify the binding sites of ML12 led to the conclusion that the entire sequence of ML12 is required for effective binding to LF. Although we do not yet possess sufficient data to propose exact binding sites of LF, a model might well involve the binding of ML12 to an active site of LF based on the inhibition results. The discovery of the aptamer against LF may make a contribution to developing a sensitive detecting device and drug to solve problems arising from B. anthracis infection. The found aptamers, however, would be limited when we use such a DNA-based aptamer as inhibitor in a cellular context because of aptamer degradation by nuclease activity. To enhance the aptamer’s stability against nucleases, chemical modification is necessary. These investigations are warranted for the further development of the aptamer for clinical use.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number P20GM10344. This work was also in part supported by the Korean Science and Engineering Foundation and the Ministry of Education, Science and Technology through its National Technology Program (2009-0081812) and by the Cooperative Research Program grants for Agriculture Science & Technology Development (Project No. PJ907052) funded by the Rural Development Administration, Republic of Korea.
Abbreviations
- BSA
bovine serum albumin
- CD
circular dichroism
- DMEM
Dulbecco’s Modified Eagle Medium
- DMSO
dimethyl sulfoxide
- EMSA
electrophoretic mobility shift assay
- EF
Edema factor
- FBS
fetal bovine serum
- GST
Glutathione S-transferase
- LF
lethal factor
- MAPKK
mitogen-activated protein kinase kinase
- MEK
mitogen-activated extracellular signal-regulated kinase kinase kinase
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PA
protective antigen
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SELEX
systematic evolution of ligands by exponential enrichment
- TBST
Tris-buffered saline Tween-20
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baldari CT, Tonello F, Paccani SR, Montecucco C. Trends Immunol. 2006;27:434–440. doi: 10.1016/j.it.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Turk BE. Biochem. J. 2007;402:405–417. doi: 10.1042/BJ20061891. [DOI] [PubMed] [Google Scholar]
- 3.Koehler TM. Mol. Aspects Med. 2009;30:386–396. doi: 10.1016/j.mam.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young JA, Collier RJ. Annu. Rev. Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 5.Scobie HM, Young JA. Curr. Opin. Microbiol. 2005;8:106–112. doi: 10.1016/j.mib.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Kintzer AF, Thoren KL, Sterling HJ, Dong KC, Feld GK, Tang, Zhang TT, Williams ER, Berger JM, Krantz BA. J. Mol. Biol. 2009;392:614–629. doi: 10.1016/j.jmb.2009.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duesbery NS, Vande Woude GF. Cell. Mol. Life Sci. 1999;55:1599–1609. doi: 10.1007/s000180050399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moayeri M, Haines D, Young HA, Leppla SH. J. Clin. Invest. 2003;112:670–682. doi: 10.1172/JCI17991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pezard C, Berche P, Mock M. Infect. Immun. 1991;59:3472–3477. doi: 10.1128/iai.59.10.3472-3477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards KA, Clancy HA, Baeumner AJ. Anal. Bioanal. Chem. 2006;384:73–84. doi: 10.1007/s00216-005-0090-x. [DOI] [PubMed] [Google Scholar]
- 11.Molin FD, Fasanella A, Simonato M, Garofolo G, Montecucco C, Tonello F. Toxicon. 2008;52:824–828. doi: 10.1016/j.toxicon.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 13.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 14.Ellington AD, Szostak JW. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 15.Kim SK, Shin BC, Ott TA, Cha W, La IJ, Yoon MY. Biochip J. 2008;2:97–102. [Google Scholar]
- 16.Bruno JG, Kiel JL. Biosens. Bioelectron. 1999;14:457–464. doi: 10.1016/s0956-5663(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 17.Cella LN, Sanchez P, Zhong W, Myung NV, Chen W, Mulchandani A. Anal. Chem. 2010;82:2042–2047. doi: 10.1021/ac902791q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi JS, Kim SG, Lahousse M, Park HY, Park HC, Jeong B, Kim J, Kim SK, Yoon MY. J. Biomol. Screen. 16:266–271. doi: 10.1177/1087057110391787. [DOI] [PubMed] [Google Scholar]
- 19.Park S, Leppla SH. Protein Expr. Purif. 2000;18:293–302. doi: 10.1006/prep.2000.1208. [DOI] [PubMed] [Google Scholar]
- 20.Kim SK, Sims CL, Wozniak SE, Drude SH, Whitson D, Shaw RW. Chem. Biol. Drug Des. 2009;74:343–348. doi: 10.1111/j.1747-0285.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Kim YM, Koo BS, Chae YK, Yoon MY. Protein Expr. Purif. 2003;30:293–300. doi: 10.1016/s1046-5928(03)00132-3. [DOI] [PubMed] [Google Scholar]
- 22.Basha S, Rai P, Poon V, Saraph A, Gujraty K, Go MY, Sandacharan S, Frost M, Mogridge J, Kane RS. PNAS. 2006;103:13509–13513. doi: 10.1073/pnas.0509870103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SC, Gedi V, Ha NR, Cho JH, Park HC, Moon MY. Int. J. Biol. Macromol. 2015;77:293–302. doi: 10.1016/j.ijbiomac.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 24.Park HY, Park HC, Yoon MY. J. Microbiol. Methods. 2009;78:54–58. doi: 10.1016/j.mimet.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Oh BN, Lee S, Park HY, Baeg JO, Yoon MY, Kim J. Analyst. 136:3384–3388. doi: 10.1039/c0an00978d. [DOI] [PubMed] [Google Scholar]
- 26.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 27.Michalowski D, Chitima-Matsiga R, Held DM, Burke DH. Nucleic Acids Res. 2008;36:7124–7135. doi: 10.1093/nar/gkn891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonsson T. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 29.Vorlickova M, Bednarova K, Kejnovska I, Kypr J. Biopolymers. 2007;86:1–10. doi: 10.1002/bip.20672. [DOI] [PubMed] [Google Scholar]
- 30.Shum KT, Lui EL, Wong SC, Yeung P, Sam L, Wang Y, Watt RM, Tanner JA. Biochemistry. 2011;50:3261–3271. doi: 10.1021/bi2001455. [DOI] [PubMed] [Google Scholar]
- 31.Tonello F, Seveso M, Marin O, Mock M, Montecucco C. Nature. 2002;418:386. doi: 10.1038/418386a. [DOI] [PubMed] [Google Scholar]
- 32.Turk BE, Wong TY, Schwarzenbacher R, Jarrell ET, Leppla SH, Collier RJ, Liddington RC, Cantley LC. Nat. Struct. Mol. Biol. 2004;11:60–66. doi: 10.1038/nsmb708. [DOI] [PubMed] [Google Scholar]
- 33.Peterson JT. Cardiovasc. Res. 2006;69:677–687. doi: 10.1016/j.cardiores.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 34.Kikin O, D'Antonio L, Bagga PS. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young JA, Collier RJ. Annu. Rev. Biochem. 2007;76:253–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 36.Robinson MJ, Cobb MH. Curr. Opin. Cell. Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 37.Shoop WL, Xiong Y, Wiltsie J, Woods A, Guo J, Pivnichny JV, Felcetto T, Michael BF, Bansal A, Cummings RT, Cunningham BR, Friediander AM, Douglas CM, Patel SB, Wisniewski D, Scapin G, Salowe SP, Zaller DM, Chapman KT, Scolnick EM, Schmatz DM, Bartizal K, MacCoss M, Hermes JD. Proc. Natl. Acad. Sci. USA. 2005;100:7958–7963. doi: 10.1073/pnas.0502159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly JA, Feigon J, Yeates TO. J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.