Abstract
Tumor metastasis is responsible for ~90% of all cancer deaths. One of the key steps of tumor metastasis is tumor cell migration and invasion. Filopodia are cell surface extensions that are critical for tumor cell migration. Fascin protein is the main actin-bundling protein in filopodia. Small-molecule fascin inhibitors block tumor cell migration, invasion and metastasis. Here we present the structural basis for the mechanism of action of these small-molecule fascin inhibitors. X-ray crystal structural analysis of a complex of fascin and a fascin inhibitor shows that binding of the fascin inhibitor to the hydrophobic cleft between the domains 1 and 2 of fascin induces a ~35° rotation of domain 1, leading to the distortion of both the actin-binding sites 1 and 2 on fascin. Furthermore, the crystal structures of an inhibitor alone indicate that the conformations of the small-molecule inhibitors are dynamic. Mutations of the inhibitor-interacting residues decrease the sensitivity of fascin to the inhibitors. Our studies provide structural insights into the molecular mechanism of fascin protein function as well as the action of small-molecule fascin inhibitors.
Keywords: crystal structure, fascin, actin cytoskeleton, small-molecule inhibitor
Graphical abstract
INTRODUCTION
Tumor metastasis is the major cause of mortality of cancer patients. Inhibition of tumor metastasis will significantly increase the survival rate of cancer patients. Metastasis is a multi-step process wherein a primary tumor spreads from its initial site to secondary tissues/organs [1–3]. Tumor cell migration and invasion are critical steps in metastasis [4]. Migration provides tumor cells the ability to leave the primary tumor bed (local invasion), enter into blood vessels, and then exit the circulation and infiltrate distant tissues/organs. For cell migration and invasion to proceed, actin cytoskeleton must be reorganized by forming polymers and bundles to cause dynamic changes in cell shapes [5–7]. Among the morphological structures supported by actin filaments, one of the most prominent and best-characterized protrusive organelles is filopodia which are fundamental to cell shape and motility events [8]. Filopodia are finger-like plasma membrane protrusions that are formed upon remodeling of the actin cytoskeleton beneath the plasma membrane [8]. They can be viewed as a sensory organ of the cells that are used to detect and assimilate signals as well as to explore and move into the surrounding microenvironment [6, 8–11]. They contain long actin filaments crosslinked into parallel bundles by the fascin protein. Metastatic tumor cells are rich in filopodia, and the numbers of filopodia correlate with their invasiveness [6, 8, 12, 13]. Filopodia-like protrusions have also been shown to be critical for metastatic tumor cells to interact with the metastatic microenvironment and to grow at the secondary tissues [14].
Fascin is the main actin cross-linker in filopodia and shows no amino acid sequence homology with other actin-binding proteins [15–20]. It has a molecular mass of ~55 kDa and functions as a monomer [17]. It fastens 10-30 parallel actin filaments together into straight, compact, and rigid bundles, to form filopodia and to impart distinct mechanical stiffness to actin bundles [8, 21, 22]. When ectopically expressed in tumor cells, fascin promotes tumor cell migration, invasion and metastasis [23]. It has also been suggested that up-regulation of fascin is part of the program of epithelial-to-mesenchymal transition that confers motility and invasion properties on tumor cells [24].
Studies on samples from human cancer patients demonstrate that fascin is a biomarker of metastases [25–29]. Elevated levels of fascin have been found in many types of metastatic tumors and are correlated with clinically aggressive phenotypes, poor prognosis, and shorter survival [23, 24, 30]. Human fascin expression is low or absent in normal adult epithelial cells, but highly expressed in metastatic tumors [31, 32]. A systematic review and meta-analysis revealed that high fascin levels are associated with increased risk of mortality, lymph node metastasis, distant metastasis, and disease progression, and may provide a novel biomarker for early identification of aggressive and metastatic tumors [30]. Furthermore, studies from pancreatic cancer patients showed that higher levels of fascin correlate with poor outcome, time to recurrence and decreased overall survival [33]. Taken together, these data from human cancer patients strongly suggest a role for fascin in cancer progression and metastasis.
Mouse genetic studies have shown that fascin gene-knockout mice are normal, likely due to the functional compensation of other actin-bundling proteins during embryonic development [34]. However, deletion of fascin gene delayed tumor development, slowed the tumor growth, reduced metastatic colonization, and increased overall survival in a spontaneous mouse model of pancreatic cancer [33]. Moreover, transgenic expression of fascin in mouse intestinal epithelium increased the tumor incidence, promoted tumor progression, and decreased the overall survival in a spontaneous mouse model of intestinal cancer [35]. These mouse genetic studies provide evidence for fascin in tumor initiation (tumor burden), tumor progression, tumor metastasis and overall survival.
We screened chemical libraries and identified small-molecule compounds that specifically inhibit the biochemical function of fascin to bundle actin filaments [36]. We optimized one of the initial fascin inhibitor hits, and showed that the improved fascin inhibitors block the actin-binding and actin-bundling activities of fascin, tumor cell migration, invasion and metastasis in mouse models [37]. However, the molecular mechanism of these fascin inhibitors is not clear. In this study, we determined the X-ray crystal structure of fascin complexed with one of these optimized fascin inhibitors, and the X-ray crystal structure of a fascin inhibitor alone. Our results reveal an induced-fit inhibitory mechanism by which a fascin inhibitor causes a major conformational change in fascin leading to the disruption of the actin-binding sites on fascin, thus impairing the actin-bundling function of fascin.
RESULTS
X-ray crystal structure of a complex of fascin and a small-molecule fascin inhibitor
From the chemical library screening for small-molecule compounds that inhibit the actin-bundling function of fascin, we identified several fascin inhibitor hits; one of them was G2 [36] (Fig. 1a). Further medicinal chemical optimization generated improved fascin inhibitors, including NP-G2-029 and NP-G2-044 [37] (Fig. 1 b and c). NP-G2-029 inhibited the actin-bundling function of fascin with an IC50 value of ~0.18 μM (Fig. 1 d and e). It also decreased the migration of mouse and human breast tumor cells (Fig. 1 f and g).
Figure 1.
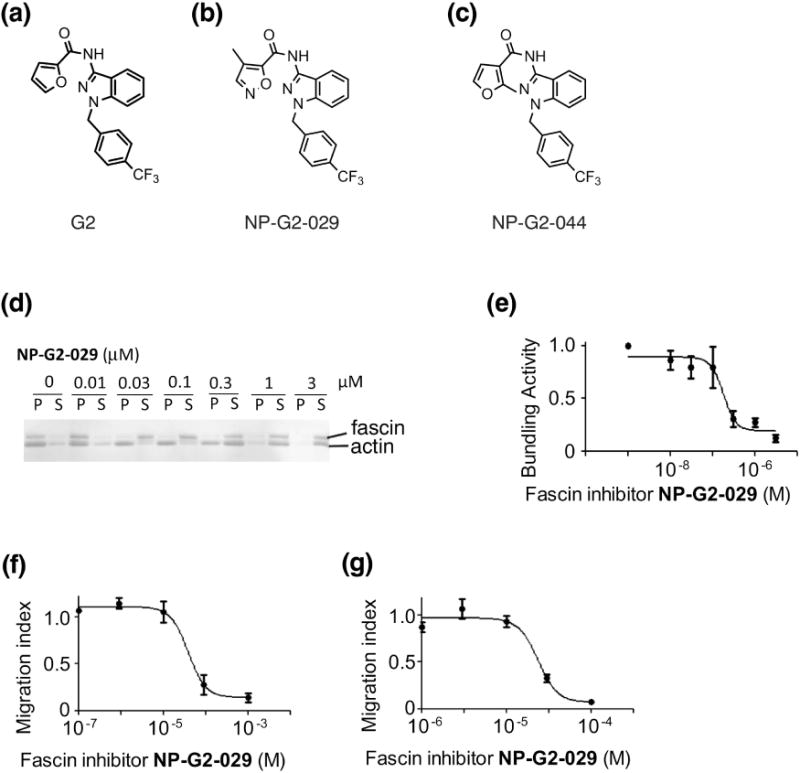
Fascin inhibitor NP-G2-029 decreases the actin-bundling activity of fascin and tumor cell migration. (a - c) The chemical structures of fascin inhibitors G2 (a), NP-G2-029 (b), and NP-G2-044 (c). (d and e) NP-G2-029 inhibits the actin-bundling activity of fascin. (d) One representative gel image of the actin-bundling assay (the low-speed sedimentation assay) is shown. Different concentrations of NP-G2-029 were added to the actin-bundling reactions. After low-speed centrifugation, the pellet (P) and supernatant (S) were separated and analyzed by SDS-PAGE. NP-G2-029 treatment shifts actin and fascin proteins from the pellet to the supernatant, indicating the inhibition of the actin-bundling function of fascin. (e) Quantification of the actin-bundling activity of fascin in the presence of different concentrations of NP-G2-029. The data are mean ± SEM. n = 3. (f and g) Boydem chamber assays show that NP-G2-029 decreases the migration of 4T1 mouse breast tumor cells (f) and MDA-MB-231 human breast tumor cells (g). Different concentrations of NP-G2-029 were added to the top of the chamber and the number of cells migrated through the membrane inserts was counted. Data are shown as mean ± SEM. n=3.
To understand the structural basis of the inhibition of fascin by NP-G2-029, we solved the X-ray crystal structure of the complex of fascin and NP-G2-029. Fascin has four β-trefoil domains (marked as domains 1 to 4) [38] (Fig. 2a). From systematic mutagenesis studies of ~100 surface residues, amino acids that are critical for actin-bundling are clustered in three regions, named as the possible actin-binding sites 1, 2, and 3 [39] (Fig. 2b). A cryo-electron tomography analysis of the filopodia from intact whole cells proposed that actin-binding sites 1 and 2 interact with two widely positioned two actin monomers on the same F-actin filament, while the actin-binding site 3 interacts with an actin monomer on a second F-actin filament [40]. In the structure of fascin and NP-G2-029, NP-G2-029 binds to a cleft between the domains 1 and 2 (the actin-binding site 2) (Fig. 2c).
Figure 2.
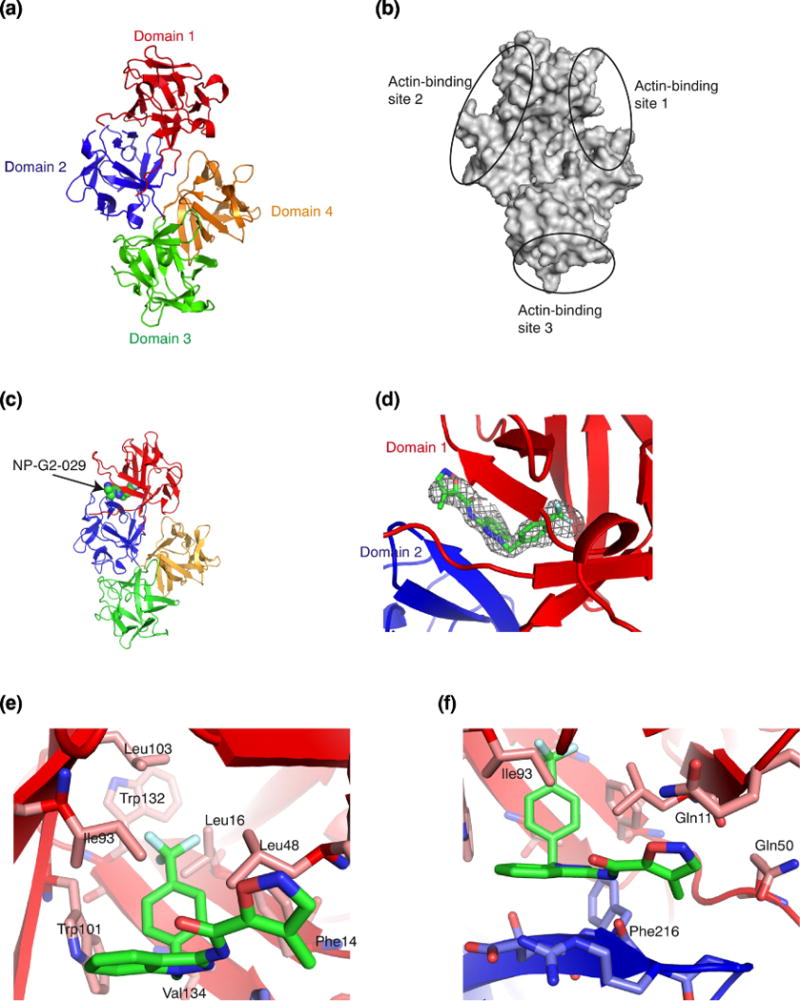
Crystal structure of the complex of fascin and the small-molecule inhibitor NP-G2-029. (a) Crystal structure of wild-type human fascin shows the relative orientation of the four β-trefoil domains (marked by 4 different colors) of one fascin molecule. (b) Diagram of fascin shows the locations of the three possible actin-binding sites based on the mutagenesis data. (c) X-ray crystal structure of the complex of fascin and NP-G2-029. NP-G2-029 occupies the actin- binding site 2. (d) The electron density map (2Fo-Fc at sigma level of 1.5) for NP-G2-029. (e and f) The binding pocket and some NP-G2-029-interacting residues on fascin are shown.
The crystals of the complex of fascin and NP-G2-029 diffracted to ~2.8Å. The structure was solved by molecular replacement using individual domains of the apo structure of fascin (PDB ID 3LLP) as the search model [38] (Table 1). After initial rounds of rigid body refinement, electron density matching the shape of NP-G2-029 was clearly visible at the actin-binding site 2 (Fig. 2d), and was confirmed by phenix.polder map. In this complex, the benzene ring of NP-G2-029 is surrounded by a number of hydrophobic residues including Phe14, Leu16, Trp101, Leu103, Trp132, and Val134 (Fig. 2e). This benzene ring also forms edge-to-face pi-pi-stacking with Trp101 and Phe14 (Fig. 2e). The planar indazole moiety (formed by the fusion of a pyrazole ring with a benzene ring) constitutes the base plane of NP-G2-029. The benzene ring is oriented ~90° with respect to this base plane (Fig. 2f). The backbone of Phe216 forms hydrogen bonds with the pyrazole and the amide of NP-G2-029 (Fig. 2f). In one asymmetric crystallographic unit, there were six fascin and NP-G2-029 molecules; the overall structures of these molecules were similar. The terminal isoxazole ring of NP-G2-029 displayed some flexibility among the six molecules (Fig. 3). This isoxazole ring is in a hydrophilic environment surrounded by Gln11 and Gln50 (Fig. 2f).
Table 1.
Data collection and refinement statistics
| Structure of fascin bound with NP- G2-029 a | |
|---|---|
| Native PDB 6B0T | |
| Data collection | |
| Space group | P2 |
| Cell dimensions | |
| a, b, c (Å) | 102.58, 59.25, 293.65 |
| α β γ (°) | 90, 90.02, 90 |
| Resolution (Å) | 45.44-2.8 (2.9-2.8) b |
| Number of reflections measured | 364035 (36079) |
| Number of unique reflections | 87042 (8516) |
| Rmerge | 0.09 (0.44) |
| I / σ (I) | 13.59 (3.46) |
| Completeness (%) | 99.0 (98.0) |
| Redundancy | 4.2 (4.2) |
| Refinement | |
| Resolution (Å) | 45.44-2.8 |
| No. reflections (test set) | 87025 (4264) |
| Rwork / Rfree (%) | 26.51/30.28 |
| Number of non-hydrogen atoms | |
| Protein | 22818 |
| Ligand | 174 |
| Water | 344 |
| Mean overall B factor (Å2) | 67.4 |
| Wilson B | 38.2 |
| Protein | 67.7 |
| Ligand | 76.6 |
| Water | 43.8 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.59 |
| Ramachandran plot statisticsc (%) | |
| Favored regions | 91.3 |
| Allowed regions | 8.1 |
| Disallowed regions | 0.6 |
One single crystal was used for data collection and refinement.
Values in parentheses are for highest-resolution shell.
As defined in MolProbity.
Figure 3.
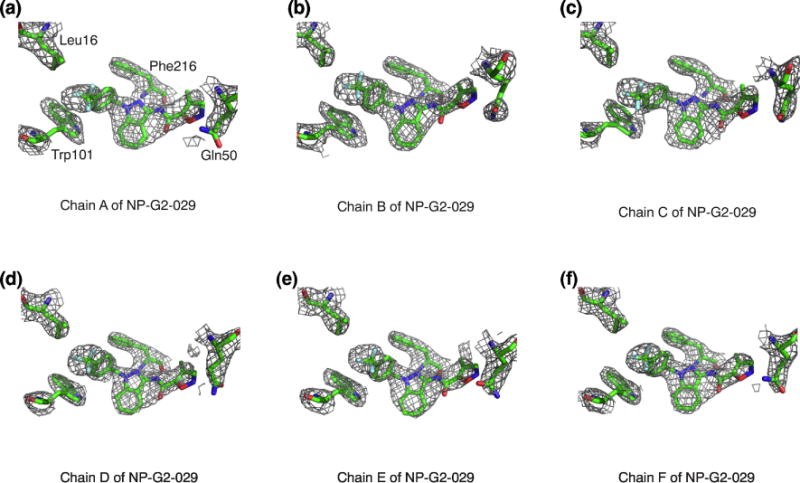
Electron density maps of the six NP-G2-029 molecules in one asymmetric crystallographic unit of the complex of fascin and NP-G2-029. 2Fo-Fc maps are shown. NP-G2-029 is shown at 1 sigma level and the side chains of nearby amino acid residues are displayed at 1.5 sigma level.
Inhibitor-induced conformational change of fascin
The binding pocket for NP-G2-029 was not present in the apo-structure of fascin (Fig. 4a). It was induced by the binding of NP-G2-029 (Fig. 4b). Structural superposition of fascin in the absence or presence of NP-G2-029 shows that the domains 2, 3, and 4 in the two crystal structures overlapped well (Fig. 4c). Structural changes of individual domains were minor. Domain-wise superposition reveals RMSDs ranging from 0.29Å (for domain 2) to 0.43Å (for domain 1). Markedly, the domain 1 rotates ~35° along with an axial shift of 2.68 Å (Fig. 4c and d). This leads to the widening of the actin-binding site 2 and the closing of the actin-binding site 1, likely disrupting the two actin-binding sites (Fig. 4c). The rotating axle centers around the binding pocket for the CF3 group of NP-G2-029 near the N-terminal region of fascin (Fig. 4d-f). The N-terminal of fascin protein is known to be essential for its actin-bundling activity [41]. This N-terminal links the actin-binding site 2 to the actin-binding site 1 (Fig. 4e). Residues Gln11, Phe14, Leu48 and Gln50 (the actin-binding site 2) form part of the binding pocket for NP-G2-029 (Fig. 4f). On the other hand, phosphorylation of Ser39 is known to decrease the actin-bundling activity of fascin is in the actin-binding site 1 (Fig. 4f). Therefore, this critical N-terminal of fascin not only couples the two actin-bundling sites, but also contributes to the inhibitory action of NP-G2-029.
Figure 4.
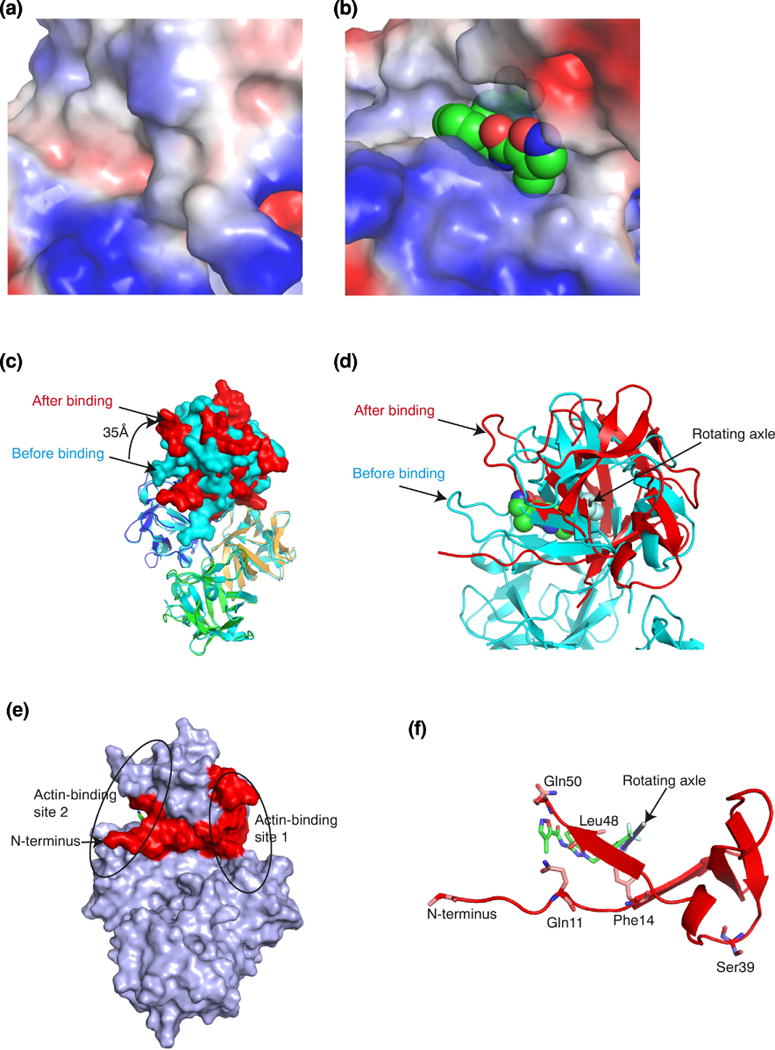
NP-G2-029 induced changes in fascin conformation. (a) Structure of the actin-binding site 2 in the absence of NP-G2-029. (b) Structure of the actin-binding site 2 with bound NP-G2-029. (c) Superposition of fascin structures in the absence or presence of NP-G2-029. The color marking of the 4 domains of fascin in the presence of NP-G2-029 is the same as in Fig. 2c. The structure of fascin in the absence of NP-G2-029 is colored in light blue. Relative to the location in the absence of NP-G2-029, domain 1 rotated ~35Å clockwise in the presence of NP-G2-029. (d) The rotating axle of domain 1 is marked by a rod. (e) The N-terminal (marked in red) of fascin couples the actin-binding sites 1 and 2. (f) The N-terminal of fascin participates in the binding of NP-G2-029.
X-ray crystal structure of a small-molecular fascin inhibitor alone
The NP-G2-029-induced conformational changes on fascin prompted us to investigate the possibility of fascin-induced conformational changes on the small-molecule inhibitors. We set up crystallization screens for various G2 analogues, and obtained the X-ray crystal structure of the small-molecular fascin inhibitor NP-G2-044 [37] (Fig. 5 a and b; Table 2). NP-G2-044 is similar to NP-G2-029 except for a furan ring replaced the isoxazole ring (Fig. 1a). In one asymmetric crystallographic unit, there were two different conformations of NP-G2-044 (Fig. 5 a and b). These two conformations were similar, with the slight orientation difference of the furan ring (Fig. 5c). When compared with the NP-G2-029 structure in the complex with fascin, the benzene ring is rotated by ~180° relative to the planar indazole moiety (Fig. 5d). Given the rotating ability of the benzene ring in relation to the indazole moiety, all these three conformations can be adopted by NP-G2-044 or NP-G2-029 (Fig. 5 a - d). Therefore, these small-molecule fascin inhibitors are flexible and sample several different conformations. It is possible that following the binding by a primary conformational selection event, optimization of side chain interactions proceeds by an induced-fit mechanism to achieve a productive inhibition. Hence the dynamic conformational ensembles of these inhibitors may play a role in the molecular recognition by fascin.
Figure 5.
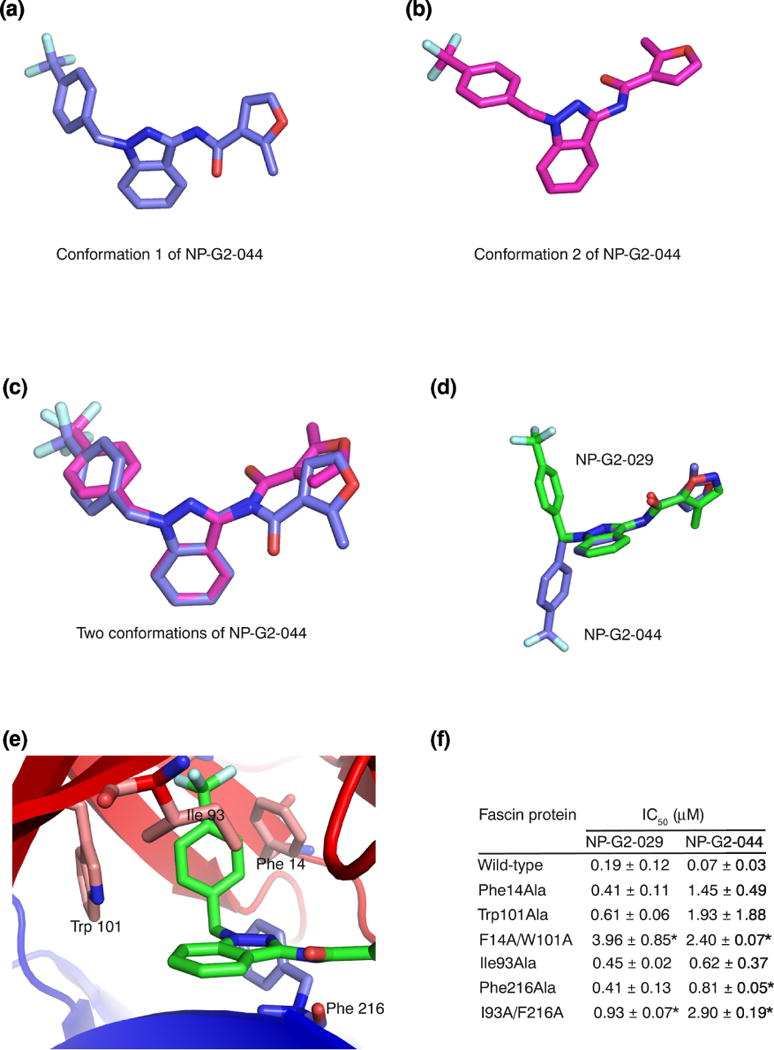
Fascin induces a conformational change on the small-molecule inhibitor. (a) X-ray crystal structure of one conformation of NP-G2-044. (b) X-ray crystal structure of the second conformation of NP-G2-044. (c) Superposition of the two conformations of NP-G2-044. (d) Superposition of one conformation of NP-G2-044 and the NP-G2-029 structure when it is bound to fascin. (e). Diagram shows the positions of Phe14, Trp101, Ile93 and Phe216 residues which are involved in the inhibitor binding. (f) Summary of the actin-bundling data of wild-type and mutant fascin proteins in the presence of various concentrations of NP-G2-029 or NP-G2-044. The IC50 values are the mean of three repeats. Data are expressed as mean ± SD and analyzed by Student’s t test with significance defined as p < 0.05. * marks the mutants, when compared with the wild-type, with p < 0.05.
Table 2.
Small-molecule NP-G2-044 X-ray data collection and refinement
| Empirical formula | C42 H34 F6 N6 O5 |
| Formula weight | 816.75 |
| Temperature | 100(2) K |
| Wavelength | 0.97915 (Å) |
| Crystal system | monoclinic |
| Space group | C2/c |
| Unit cell dimensions | |
| a, b, c (Å) | 35.240(7), 12.290(2), 19.970(4) |
| α β γ (°) | 90.00, 118.13, 90.00 |
| Volume | 7627.4(3) (Å3) |
| Z | 8 |
| Density (calculated) | 1.423 |
| Absorption coefficient | 0.40 mm-1 |
| F(000) | 3376 |
| Crystal size | 0.10, 0.05, 0.04 mm3 |
| Theta range for data collection | 1.81-30.32 (0) |
| Resolution range | 15.5 to 1.0 Å |
| Index range | −34 < h < 34, −12 < k < 11, −20 < l < 19 |
| Reflections collected | 12379 |
| Independent reflections | 3792 |
| Completeness to theta max = 30.32° | 86.4 % |
| Absorption correction | Empirical |
| Refinement method | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 3792/477/543 |
| Goodness-of-fit on F2 | 1.202 |
| Final R indices [I>2sigma(I)] | R1 =0.0464, wR2 =0.1212 |
| R indices (all data) | R1 = 0.0478, wR2 =0.1224 |
| Largest diff, peak and hole | 0.30 and −0.35 e.Å−3 |
Biochemical verification of inhibitor-interacting residues
To further experimentally verify the involvement of fascin residues in interacting with NP-G2-029, we mutated some specific residues and investigated the sensitivity of the mutant fascin proteins to NP-G2-029. In the co-crystal structure of fascin and NP-G2-029, NP-G2-029 forms edge-to-face pi-pi stacking with residues Phe14 and Trp101 (Fig. 5e). We generated the single (Phe14 to Ala, Trp101 to Ala) and the double (Phe14Ala/Trp101Ala) mutations on fascin. These mutant fascin proteins had lower sensitivity to the inhibition by NP-G2-029 (Fig. 5f). Furthermore, in the co-crystal structure of fascin and NP-G2-029, Phe216 forms hydrogen bonds with the pyrazole and the amide of NP-G2-029 (Fig. 5e). Ile93 forms hydrophobic interaction with the fused benzene ring of the indazole moiety of NP-G2-029 (Fig. 5e). We made the single (Ile93 to Ala, Phe216 to Ala) and the double (Ile93Ala/Phe216Ala) mutants, and observed reduced sensitivity of these mutated fascin proteins to the inhibition by NP-G2-029 (Fig. 5f). Moreover, these mutated fascin proteins had decreased sensitivity to the inhibition by NP-G2-044 as well (Fig. 5f). Together, these biochemical data support the participation of these residues in the inhibition of fascin by NP-G2-029 and NP-G2-044.
DISCUSSION
In an effort to elucidate the structural basis by which the small-molecule fascin inhibitors block the biological function of fascin, we solved the X-ray crystal structure of a complex of fascin and a small-molecule fascin inhibitor. In the structure of fascin and NP-G2-029, NP-G2-029 binds to a cleft between the domains 1 and 2 (the actin-binding site 2) of fascin. This structure of the complex also reveals that the N-terminal of fascin is critical for coupling the two actin-binding sites and for binding to NP-G2-029. Furthermore, our data indicate that the small-molecule fascin inhibitors are flexible and can adopt different conformations. This dynamics of the inhibitors might be important for the induced-fit inhibition process. Moreover, several amino acid residues on fascin participating in the interaction with the inhibitor were confirmed by mutation studies.
From the co-crystal structure of the complex of fascin and NP-G2-029, the trifluoromethyl (-CF3) group and the benzene ring of NP-G2-029 are deeply embedded into fascin. These two groups are likely the main contributors to the binding of NP-G2-029 and its analogs to fascin. This is consistent with our structure-activity relationship studies that most modifications in these positions abolished the capability of the compounds to inhibit the actin-bundling activity of fascin [37]. Furthermore, the isoxazole ring of NP-G2-029 showed some flexibility among the six NP-G2-029 molecules in the structure of the complex. This also agrees with our structure-activity relationship studies that this part of the small-molecule inhibitors could be extensively modified to fine-tune the efficacy of the inhibitors. Moreover, the NP-G2-029 binding site on fascin is formed upon the binding of NP-G2-029. In the absence of NP-G2-029, the actin-binding site 2 is fully enclosed. The rotation of the domain 1 opens up a hydrophobic and tight pocket for the binding of the -CF3 and benzene ring of NP-G2-029. This rotation results in the widening of the actin-binding site 2 and the closing of the actin-binding site 1, leading to the inhibition of the actin-bundling activity of fascin. Our data also explain that, even though we started by screening chemical libraries for protein-protein interaction inhibitors, we obtained small-molecule inhibitors capable of binding to fascin in the absence of actin proteins.
Our structural data provide a mechanistic explanation for the cooperativity among the actin binding sites. It has been shown that the actin-binding activity of fascin is cooperative [42]. Our data reveal that the N-terminal of fascin connects the actin-binding sites 1 and 2, and that conformational changes in one actin-binding site could lead to a concerted conformational change on the other actin-binding site. Hence, it is likely that binding of one F-actin filament at one actin-binding site stabilizes fascin at the active configuration and enhances the binding of actin filaments at the other actin-binding sites. It is also possible that mutations of residues in the actin-binding site 1 could affect the small-molecule inhibitor binding to the actin-binding site 2 [36, 37]. Furthermore, the unexpected flexibility of domain 1 might have implications for the role of fascin protein in the flexibility of the bundled fascin-actin filaments as well as of filopodia. Filopodia are highly dynamic cellular protrusions. They scan the local environment by turning, elongation, and retraction. These require filopodia to be flexible enough to wave about, yet rigid enough to protrude the cell surface. Previous structural studies showed that the conformations of wild-type active fascin protein and four inactive fascin mutants were similar with minor local structural variations, implying the configuration of fascin is rather rigid [38, 39, 43, 44]. Our current structural data reveal that the domain 1 could be rotated; this might underlie the flexibility of the fascin/actin bundles and of filopodia. Therefore, our results might provide insights on the roles of fascin in the stability, dynamics, and organizations of filapodia in cells. The dynamic structures of fascin and the small-molecule inhibitors offer the opportunity and the challenge in generating the next-generation of fascin inhibitors for clinical uses.
MATERIALS AND METHODS
Chemistry
Compound syntheses and analyses were done by the company ValueTek Inc (NJ, USA). All reagents and solvents were purchased from Sigma-Aldrich Chemical Co., Combi-Block Inc., Astatech Inc., Ark Pharma Inc., and Frontier Scientific Inc. and were used as received. HNMR spectra were recorded on a Bruker Fourier-400 spectrometer. Crude products were purified by silica gel chromatography. For all products, the purity was ascertained to be greater than 95% by the HPLC method using Shimadzu 2010HPLC-UV/MS system with a C-18 reverse phase column.
General procedures for NP-G2-029 and NP-G2-044
A mixture of KOH (6.95 g, 124 mmol) in DMSO (165 ml) was stirred at room temperature for 5 min. 1H-indazol-3-amine (8.25 g, 62.0 mmol) was then added in one portion. The resulting mixture was stirred at room temperature for 5 min. A solution of 4-trifluoromethylbenzyl bromide (15.6 g, 65.1 mmol) in DMSO (83 ml) was then added dropwise over 30 min. When the addition was complete, the resulting mixture was stirred at room temperature for an additional 1 h. The mixture was quenched by the addition of water (200 mL). The mixture was then extracted with CH2Cl2 (3 × 100 mL). The combined extracts were washed with H2O (2 × 100 mL), brine (1 × 100 mL), then dried over MgSO4, filtered and concentrated in vacuo. Purification by flash chromatography (Silica, 200 g, 10 - 100% EtOAc/Hexanes) gave 1-(trifluoromethylbenzyl)-1H-indazol-3-amine (21.79 g, 56.6 mmol, 91.3 % yield) as an off-white crystalline solid. MS (ESI) m/z: 292 (M+H)+. To a solution of 1-(trifluoromethylbenzyl)-1H-indazol-3-amine (29.2 mg, 0.10 mmol), furan-2-carboxylic acid (12.3 mg, 0.11 mmol), and triethylamine (45.2 μl, 0.30 mmol) in dichloromethane (2 mL) was added 2,4,6-tripropyl-1,3,5,2,4,6-trioxatriphosphinane 2,4,6-trioxide (118.6 μl, 0.20 mmol). The resulting reaction mixture was stirred at room temp for 3h and then the solvent was removed. The crude product was purified by preparative HPLC (sunfire 5u 100 mm column, MeOH/H2O as solvents). 23 mg (0.0597 mmol, 59.7%) of N-(1-(4-(trifluoromethyl) benzyl)-1H-indazol-3-yl)furan-2-carboxamide (Compound G2) was obtained as an off-white solid. 1H NMR (400 MHz, chloroform-d) R 8.73 (s, 1H), 8.18 (d, J=8.2 Hz, 1H), 7.50-7.59 (m, 3H), 7.35-7.43 (m, 1H), 7.24-7.33 (m, 4H), 7.17 (ddd, J=0.88, 6.99, 8.20 Hz, 1H), 6.58 (dd, J=1.76, 3.52 Hz, 1H), 5.54 (s, 2H). MS (ESI) calcd for C20H14F3N3O2 (M+H)+ m/z 386.10, found 386.12.
Compound NP-G2-029
4-methyl-N-(1-(4-(trifluoromethyl)benzyl)-1H-indazol-3-yl)isoxazole-5-carboxamide (Compound NP-G2-029) was prepared as described for the preparation of Compound G2 with replacement of furan-2-carboxylic acid with 4-methylisisoxazole-5-carboxylic acid in 63.1% yield as an off-white solid. 1H NMR (400 MHz, chloroform-d) R 8.85 (s, 1H), 8.28 (s, 1H), 8.14 (d, J=8.25 Hz, 1H), 7.59 (d, J=7.98 Hz, 2H), 7.43 (s, 1H), 7.33-7.38 (m, 3H), 7.23 (t, J=7.57 Hz, 1H), 5.56 (s, 2H), 2.47 (s, 3H). MS (ESI) calcd for C20H15F3N4O2 (M+H)+ m/z 401.11, found 401.11.
Compound NP-G2-044
2-methyl-N-(1-(4-(trifluoromethyl)benzyl)-1H-indazol-3-yl)furan-3-carboxamide (Compound NP-G2-044) was prepared as described for the preparation of Compound G2 with replacement of furan-2-carboxylic acid with 2-methylfuran-3-carboxylic acid in 79.6% yield as an off-white solid. 1H NMR (400 MHz, chloroform-d) R 8.05-8.16 (m, 2H), 7.54 (d, J=8.14 Hz, 2H), 7.36-7.43 (m, 1H), 7.31 (d, J=1.98 Hz, 1H), 7.28 (d, J=0.66 Hz, 2H), 7.26 (s, 1H), 7.18 (ddd, J=0.88, 6.99, 8.20 Hz, 1H), 6.60 (d, J=1.98 Hz, 1H), 5.53 (s, 2H), 2.87 (s, 3H). MS (ESI) calcd for C21H16F3N3O2 (M+H)+ m/z 400.12, found 400.12.
Cell culture
Mouse 4T1 mammary tumor cells and human MDA-MB-231 breast cancer cells were obtained from American Type Culture Collection. 4T1 cells and MDA-MB-231 cells were cultured in DMEM supplemented with 10% FBS as previously described [36–38].
Human fascin-1 expression and purification
Recombinant human fascin 1 was expressed as a GST fusion protein in BL21 Escherichia coli. 1-litre of 2YT medium with ampicillin was inoculated overnight with 3 mL of BL21/DE3 culture transformed with pGEX4T-fascin 1 plasmid and grown at 37°C until attenuance at 600 nm (OD600) reached about 0.5. The culture was then transferred to 17 °C and induced by the addition of 0.1 mM isopropyl β-d-thiogalactoside (IPTG) for 16 h. Bacteria were harvested by centrifugation at 5,000 r.p.m. for 10 min. The pellets were suspended in 30 mL of Tris/HCl (20 mM Tris/HCl pH 8.0, 150 mM NaCl) supplemented with 0.2 mM PMSF, 1 mM DTT, 1 % Triton X-100 and 1 mM EDTA. After sonication, the suspension was centrifuged at 15,000 r.p.m. for 30 min to remove the cell debris. The supernatant was then incubated for 2 h with 4 mL of glutathione beads (Sigma) at 4 °C. After extensive washing with Tris/HCl (20 mM Tris/HCl pH 8.0, 150 mM NaCl), the beads were resuspended in 10 mL of thrombin cleavage buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM CaCl2, 1 mM DTT). Fascin was released from the beads by incubation overnight with 40-100 U of thrombin at 4 °C. After centrifugation, 0.2 mM PMSF was added to the supernatant to inactivate the remnant thrombin activity. The fascin protein was further concentrated with Centricon to about 50 mg/mL.
F-actin bundling assay
Actin-bundling activity was measured by low-speed centrifugation assay. Monomeric rabbit G-actin was induced to polymerize at room temperature in F-actin buffer (20 mM Tris–HCl at pH 8, 1 mM ATP, 1 mM DTT, 2 mM MgCl2 and 100 mM KCl). Recombinant fascin proteins (wide-type or mutant) were subsequently incubated with F-actin for 30 min at room temperature and centrifuged for 15 min at 10,000g in an Eppendorf 5415D tabletop centrifuge. Both supernatants and pellets were dissolved in an equivalent volume of SDS sample buffer, and the amount of fascin was determined by SDS-PAGE. We measured the intensities of fascin proteins in Coomassie-stained gels and then calculated the relative actin-bundling activity.
Boyden-chamber cell migration assay
MDA-MB-231 cells (5 × 104) suspended in 100 μl starvation medium were added to the upper chamber of an insert (6.5 mm diameter, 8 μm pore size; Becton Dickson), and the insert was placed in a 24-well plate containing 700 μl starvation medium with or without 10% FBS. When used, inhibitors were added to both the upper and the lower chambers. Migration assays were performed for 6 h and cells were fixed with 3.7% formaldehyde. Cells were stained with crystal violet staining solution, and cells on the upper side of the insert were removed with a cotton swab. Three randomly selected fields (10 objectives) on the lower side of the insert were photographed, and the migrated cells were counted. Migration was expressed as average number of migrated cells in a field.
Crystallization of the complex of fascin and NP-G2-029
The concentrated fascin protein stock solution was diluted with the buffer (20 mM Tris, pH 8.0, 40 mM KBr, 0.5 mM EDTA, 1 mM DTT) to 15 mg/ml prior crystallization. The final concentration of the small-molecule inhibitors was 2 mM. The mixture was incubated at room temperature for one hour. Crystallization was performed by the vapour diffusion hanging drop method at 18°C. The complex was crystallized in 100 mM HEPES pH 7.5, 20% PEG4000, 2.5% isopropanol. Crystals were briefly transferred to the cryo-solution which consisted of the crystallization solution implemented with 15% glycerol and flash-cooled in liquid nitrogen.
Data collection, structure determination and refinement
Diffraction data were collected at 100 K using synchrotron radiation (λ = 1.3 Å) at the beamline X6A, National Synchrotron Light Source (Brookhaven, USA). The data reduction was performed with XDS, followed by merging and scaling with XSCALE [45]. The corresponding data-collection statistics are shown in Table 1, created by PHENIX (1.10.1-2155). Searching by individual β-trefoil fold of fascin (PDB 3LLP) gave the solution using PHASER molecular replacement program (version 2.5.7) in CCP4 suite (version 6.5.016) [46]. Model building, refinement and analysis were carried out using COOT (version 0.8.5) and PHENIX (1.10.1- 2155) based on σ-A weighted 2Fo-Fc and Fo-Fc maps, as well as non-crystallographic symmetry (NCS) averaged and unaveraged maps [47, 48]. The slightly high R values might be due to the tight stereochemical restraints. The model was verified with simulated annealing composite omit map created in PHENIX. Refinement statistics are also presented in Table 1, and the stereochemical quality of the refined structures was validated using MolProbity [49]. Graphical representations of structures were prepared using PyMol (DeLano Scientific, San Francisco, CA). Solvent molecules were identified from the difference Fourier map (Fo – Fc) above sigma level of 3. Six molecules per asymmetric unit have been observed with clear electron density for the small molecules at the actin-binding site 2.
Crystallization and data processing of NP-G2-044
Very thin plate like crystals of NP-G2-044 were grown at 18 °C by hanging drop method with the well solution of 24% PEG4000, 0.1M Hepes (pH 7.5), 2.5% isopropanol. Compound NP-G2-044 was dissolved in 25% PEG400, 25% PEG 4000, 0.1M Hepes (pH 7.5), 2.5% isopropanol with final concentration of 4 mM. Drops were set at 1:1 ratio of well and compound solution. X-ray diffraction data for thin plate like crystal of NP-G2-044 to 1Å resolution were collected at 100K using synchrotron radiation (l = 0.9792 Å) at NE-CAT 24ID-C beamline at APS, Argonne, using Pilatus 6M-F detector. The data were reduced using XDS. Crystals belong to the space group C2/c with cell dimensions a = 35.25 Å, b = 12.29 Å, c = 19.97 Å, and b = 118.13. Two molecules were observed in the asymmetric unit with opposite orientation of the isoxazole ring with respect to the central indazole moiety.
Acknowledgments
We thank C. Shue, G. Wu and J. Zhang for assistance with the project, and C. Hu, G. Wu, J. Zhang and S. Zheng for comments on the structures and the manuscript. This work was supported by a Sponsored Research Agreement from Novita Pharmaceuticals to Weill Cornell Medical College and by a NIH grant CA193815. Part of this work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the NIGMS from the NIH (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests: X.Y.H. is a co-founder and has equity in Novita Pharmaceuticals. The remaining authors declare no competing financial interests.
Accession codes
The atomic coordinates and structure factors for the complex of fascin and NP-G2-029 have been deposited in the Protein Data Bank under accession number 6B0T. The structure of NP-G2-044 is deposited in the Cambridge Crystallographic Data Centre (http://www.ccdc.cam.ac.uk/) under accession number 1576496.
References
- 1.Weiss L. Metastasis of cancer: a conceptual history from antiquity to the 1990s. Cancer Metastasis Rev. 2000;19(I–XI):193–383. [PubMed] [Google Scholar]
- 2.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 3.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–92. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condeelis J, Singer RH, Segall JE. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 5.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 6.Mogilner A, Rubinstein B. The physics of filopodial protrusion. Biophys J. 2005;89:782–95. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–12. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–54. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 9.Davenport RW, Dou P, Rehder V, Kater SB. A sensory role for neuronal growth cone filopodia. Nature. 1993;361:721–4. doi: 10.1038/361721a0. [DOI] [PubMed] [Google Scholar]
- 10.Bentley D, Toroian-Raymond A. Disoriented pathfinding by pioneer neurone growth cones deprived of filopodia by cytochalasin treatment. Nature. 1986;323:712–5. doi: 10.1038/323712a0. [DOI] [PubMed] [Google Scholar]
- 11.Sanders TA, Llagostera E, Barna M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature. 2013;497:628–32. doi: 10.1038/nature12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coopman PJ, Do MT, Thompson EW, Mueller SC. Phagocytosis of cross-linked gelatin matrix by human breast carcinoma cells correlates with their invasive capacity. Clin Cancer Res. 1998;4:507–15. [PubMed] [Google Scholar]
- 13.Wang W, Wyckoff JB, Frohlich VC, Oleynikov Y, Huttelmaier S, Zavadil J, et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62:6278–88. [PubMed] [Google Scholar]
- 14.Shibue T, Brooks MW, Inan MF, Reinhardt F, Weinberg RA. The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer discovery. 2012;2:706–21. doi: 10.1158/2159-8290.CD-11-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otto JJ, Kane RE, Bryan J. Formation of filopodia in coelomocytes: localization of fascin, a 58,000 dalton actin cross-linking protein. Cell. 1979;17:285–93. doi: 10.1016/0092-8674(79)90154-5. [DOI] [PubMed] [Google Scholar]
- 16.Bryan J, Kane RE. Separation and interaction of the major components of sea urchin actin gel. J Mol Biol. 1978;125:207–24. doi: 10.1016/0022-2836(78)90345-5. [DOI] [PubMed] [Google Scholar]
- 17.Yamashiro-Matsumura S, Matsumura F. Purification and characterization of an F-actin-bundling 55-kilodalton protein from HeLa cells. J Biol Chem. 1985;260:5087–97. [PubMed] [Google Scholar]
- 18.Vignjevic D, Yarar D, Welch MD, Peloquin J, Svitkina T, Borisy GG. Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol. 2003;160:951–62. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J Cell Biol. 2006;174:863–75. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams JC. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol. 2004;16:590–6. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Tilney LG, Connelly PS, Vranich KA, Shaw MK, Guild GM. Why are two different cross-linkers necessary for actin bundle formation in vivo and what does each cross-link contribute? J Cell Biol. 1998;143:121–33. doi: 10.1083/jcb.143.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claessens MM, Bathe M, Frey E, Bausch AR. Actin-binding proteins sensitively mediate F-actin bundle stiffness. Nat Mater. 2006;5:748–53. doi: 10.1038/nmat1718. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto Y, Kim DJ, Adams JC. The roles of fascins in health and disease. The Journal of pathology. 2011;224:289–300. doi: 10.1002/path.2894. [DOI] [PubMed] [Google Scholar]
- 24.Machesky LM, Li A. Fascin: Invasive filopodia promoting metastasis. Commun Integr Biol. 2010;3:263–70. doi: 10.4161/cib.3.3.11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darnel AD, Behmoaram E, Vollmer RT, Corcos J, Bijian K, Sircar K, et al. Fascin regulates prostate cancer cell invasion and is associated with metastasis and biochemical failure in prostate cancer. Clin Cancer Res. 2009;15:1376–83. doi: 10.1158/1078-0432.CCR-08-1789. [DOI] [PubMed] [Google Scholar]
- 26.Pelosi G, Pasini F, Fraggetta F, Pastorino U, Iannucci A, Maisonneuve P, et al. Independent value of fascin immunoreactivity for predicting lymph node metastases in typical and atypical pulmonary carcinoids. Lung cancer. 2003;42:203–13. doi: 10.1016/s0169-5002(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 27.Hashimoto Y, Shimada Y, Kawamura J, Yamasaki S, Imamura M. The prognostic relevance of fascin expression in human gastric carcinoma. Oncology. 2004;67:262–70. doi: 10.1159/000081327. [DOI] [PubMed] [Google Scholar]
- 28.Cao D, Ji H, Ronnett BM. Expression of mesothelin, fascin, and prostate stem cell antigen in primary ovarian mucinous tumors and their utility in differentiating primary ovarian mucinous tumors from metastatic pancreatic mucinous carcinomas in the ovary. Int J Gynecol Pathol. 2005;24:67–72. [PubMed] [Google Scholar]
- 29.Rodriguez-Pinilla SM, Sarrio D, Honrado E, Hardisson D, Calero F, Benitez J, et al. Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin Cancer Res. 2006;12:1533–9. doi: 10.1158/1078-0432.CCR-05-2281. [DOI] [PubMed] [Google Scholar]
- 30.Tan VY, Lewis SJ, Adams JC, Martin RM. Association of fascin-1 with mortality, disease progression and metastasis in carcinomas: a systematic review and meta-analysis. BMC Med. 2013;11:52. doi: 10.1186/1741-7015-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grothey A, Hashizume R, Sahin AA, McCrea PD. Fascin, an actin-bundling protein associated with cell motility, is upregulated in hormone receptor negative breast cancer. Br J Cancer. 2000;83:870–3. doi: 10.1054/bjoc.2000.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashimoto Y, Skacel M, Adams JC. Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? The international journal of biochemistry & cell biology. 2005;37:1787–804. doi: 10.1016/j.biocel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Li A, Morton JP, Ma Y, Karim SA, Zhou Y, Faller WJ, et al. Fascin is regulated by slug, promotes progression of pancreatic cancer in mice, and is associated with patient outcomes. Gastroenterology. 2014;146:1386–96. e1–17. doi: 10.1053/j.gastro.2014.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamakita Y, Matsumura F, Yamashiro S. Fascin1 is dispensable for mouse development but is favorable for neonatal survival. Cell Motil Cytoskeleton. 2009;66:524–34. doi: 10.1002/cm.20356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoumacher M, El-Marjou F, Lae M, Kambou N, Louvard D, Robine S, et al. Conditional expression of fascin increases tumor progression in a mouse model of intestinal cancer. European journal of cell biology. 2014;93:388–95. doi: 10.1016/j.ejcb.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Huang FK, Han S, Xing B, Huang J, Liu B, Bordeleau F, et al. Targeted inhibition of fascin function blocks tumour invasion and metastatic colonization. Nat Commun. 2015;6:7465. doi: 10.1038/ncomms8465. [DOI] [PubMed] [Google Scholar]
- 37.Han S, Huang J, Liu B, Xing B, Bordeleau F, Reinhart-King CA, et al. Improving fascin inhibitors to block tumor cell migration and metastasis. Mol Oncol. 2016;10:966–80. doi: 10.1016/j.molonc.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L, Yang S, Jakoncic J, Zhang JJ, Huang XY. Migrastatin analogues target fascin to block tumour metastasis. Nature. 2010;464:1062–6. doi: 10.1038/nature08978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang S, Huang FK, Huang J, Chen S, Jakoncic J, Leo-Macias A, et al. Molecular Mechanism of Fascin Function in Filopodial Formation. J Biol Chem. 2013;288:274–84. doi: 10.1074/jbc.M112.427971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aramaki S, Mayanagi K, Jin M, Aoyama K, Yasunaga T. Filopodia formation by crosslinking of F-actin with fascin in two different binding manners. Cytoskeleton (Hoboken) 2016;73:365–74. doi: 10.1002/cm.21309. [DOI] [PubMed] [Google Scholar]
- 41.Ono S, Yamakita Y, Yamashiro S, Matsudaira PT, Gnarra JR, Obinata T, et al. Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J Biol Chem. 1997;272:2527–33. doi: 10.1074/jbc.272.4.2527. [DOI] [PubMed] [Google Scholar]
- 42.Yamakita Y, Ono S, Matsumura F, Yamashiro S. Phosphorylation of human fascin inhibits its actin binding and bundling activities. J Biol Chem. 1996;271:12632–8. doi: 10.1074/jbc.271.21.12632. [DOI] [PubMed] [Google Scholar]
- 43.Sedeh RS, Fedorov AA, Fedorov EV, Ono S, Matsumura F, Almo SC, et al. Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein. J Mol Biol. 2010;400:589–604. doi: 10.1016/j.jmb.2010.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jansen S, Collins A, Yang C, Rebowski G, Svitkina T, Dominguez R. Mechanism of actin filament bundling by fascin. J Biol Chem. 2011;286:30087–96. doi: 10.1074/jbc.M111.251439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]


