Abstract
Cbl proteins are E3 ubiquitin ligases specialized for the regulation of tyrosine kinases by ubiquitylation. Human Cbl proteins are activated by tyrosine phosphorylation, thus setting up a feedback loop whereby the activation of tyrosine kinases triggers their own degradation. Cbl proteins are targeted to their substrates by a phosphotyrosine‐binding SH2 domain. Choanoflagellates, unicellular eukaryotes that are closely related to metazoans, also contain Cbl. The tyrosine kinase complement of choanoflagellates is distinct from that of metazoans, and it is unclear if choanoflagellate Cbl is regulated similarly to metazoan Cbl. Here, we performed structure‐function studies on Cbl from the choanoflagellate species Salpingoeca rosetta and found that it undergoes phosphorylation‐dependent activation. We show that S. rosetta Cbl can be phosphorylated by S. rosetta Src kinase, and that it can ubiquitylate S. rosetta Src. We also compared the substrate selectivity of human and S. rosetta Cbl by measuring ubiquitylation of Src constructs in which Cbl‐recruitment sites are placed in different contexts with respect to the kinase domain. Our results indicate that for both human and S. rosetta Cbl, ubiquitylation depends on proximity and accessibility, rather than being targeted toward specific lysine residues. Our results point to an ancient interplay between phosphotyrosine and ubiquitin signaling in the metazoan lineage.
Keywords: evolution, structure‐function, E3 ubiquitin ligase, tyrosine kinase signaling, ubiquitylation, protein–protein interactions
Short abstract
http://imolecules3d.wiley.com/imolecules3d/review/dI9tEdllk8mDutRBZipQwUhysk6T46uK2PeqDQctS3gNt4hEt2ja6O7FMx4uy5ZJ822/1617 | PDB Code(s): http://firstglance.jmol.org/fg.htm?mol=6BK5
Introduction
The ubiquitylation of proteins is carried out by the sequential action of E1, E2, and E3 enzymes that activate ubiquitin and transfer it to lysine residues in the substrate proteins. The Casitas B‐lineage Lymphoma (Cbl) proteins are well‐studied RING‐type E3 ubiquitin ligases that regulate tyrosine kinases by ubiquitylation. Following ubiquitylation by Cbl, these substrates, which are typically receptor or nonreceptor tyrosine kinases, are degraded in the lysosome.1, 2, 3 In this way, Cbl proteins play a critical role in the control of tyrosine kinase signaling pathways and thus, in the regulation of cellular processes such as cell growth and cell–cell communication.4 The human genome contains three members of the Cbl family: c‐Cbl, b‐Cbl, and Cbl‐c. All three share a tyrosine kinase binding (TKB) module, which contains a phosphotyrosine‐binding SH2 domain, and a RING domain, which is responsible for transferring ubiquitin from an E2 protein to lysine residues in the substrate [Fig. 1(A)]. The TKB‐RING domains of c‐Cbl and b‐Cbl are 86% identical, and consist of a 4‐helix domain, an EF‐hand domain, and an SH2 domain.
Figure 1.
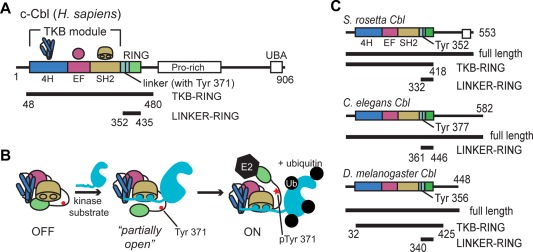
Cbl domain architecture and mechanism. (A) Domain architecture of human c‐Cbl protein. Constructs used in this study are labeled and indicated by black bars. (B) Model of the phospho‐activation mechanism of human c‐Cbl.5, 6 Domains are shown as a cartoon representation, described in (A). The kinase substrate is shown in cyan. (C) The domain architecture of Cbl homologs, S. rosetta Cbl, C. elegans Cbl, and D. melanogaster Cbl is shown along with the constructs used (black bars). http://imolecules3d.wiley.com/imolecules3d/review/dI9tEdllk8mDutRBZipQwUhysk6T46uK2PeqDQctS3gNt4hEt2ja6O7FMx4uy5ZJ822/1617.
In humans, Cbl activity is directly coupled to that of its target kinases because Cbl is activated by tyrosine phosphorylation.5, 6 In the absence of phosphorylation, Cbl exists in a “closed” inactive conformation.6 The binding of a tyrosine‐phosphorylated segment of a tyrosine kinase to the TKB module of Cbl promotes the phosphorylation of a tyrosine residue in the TKB‐RING linker region [Tyr 371 in human c‐Cbl, and see Fig. 1(A)]. Phosphorylation of this tyrosine residue converts Cbl into an “open” and active conformation [Fig. 1(B)].6, 7 In addition, the phosphorylated tyrosine residue forms hydrogen bonds with both ubiquitin, attached to the E2 active site, and the RING domain of Cbl, thus positioning ubiquitin for efficient transfer to the substrate.5, 6 Once activated, Cbl can ubiquitylate the tyrosine kinase to which it is bound, setting up a feedback loop that results, ultimately, in the degradation of the tyrosine kinase.
The core components of Cbl proteins, the TKB module followed by a RING domain, can be readily recognized in the sequences of Cbl proteins distributed throughout metazoan evolution. The TKB module and the RING domain are highly conserved in metazoans, shown in Figure S1(A), Supporting Information. For example, the TKB modules of D. melanogaster and C. elegans are respectively 75% and 55% identical in sequence to the corresponding elements in human c‐Cbl. The conservation of the sequence of the TKB module is notable because the SH2 domain within it is distantly related to canonical SH2 domains and was only recognized as one after the structure was determined.8, 9 This supports the idea that the TKB module is an element that arose early in evolution and stayed together as a unit.
Cbl is found in some nonmetazoans, such as slime molds (Dictyostelium species), as well as in choanoflagellates. The choanoflagellates are unicellular aquatic species that are among the closest known relatives to metazoans.10 Choanoflagellates contain both receptor and nonreceptor tyrosine kinases, including homologs of the Src‐family kinases, as well as the adaptor protein Grb2. While the Dictyostelium Cbl proteins share low sequence identity with human Cbl (∼30% for the TKB module), the choanoflagellate Cbl is more closely related to metazoan Cbl, sharing 45–50% sequence identity across the TKB and RING domains [Fig. S1(A), Supporting Information].
Choanoflagellates contain many of the components of tyrosine kinase pathways seen in later‐branching metazoans, although it is unclear if the detailed workings of these pathways are conserved. For example, choanoflagellates contain more receptor tyrosine kinases than humans, but the extracellular domains of the choanoflagellate receptor tyrosine kinases are different from those found in vertebrates.11 The important cytoplasmic tyrosine kinase Src appears to be regulated differently in choanoflagellates than in humans. Vertebrate Src proteins are regulated by inhibitory phosphorylation on a C‐terminal tail. Previous work is conflicting on whether or not this mechanism is conserved in choanoflagellates, suggesting a number of questions remain.11, 12, 13, 14, 15 Investigating whether regulatory components of signaling pathways are conserved in nonmetazoans like choanoflagellates is intriguing because it allows us to understand if these elements were fixed before the appearance of metazoan multicellularity.
The tyrosine residue that is critical for activation of human c‐Cbl is conserved in choanoflagellate Cbl [Fig. S1(B), Supporting Information], but it has not been determined if choanoflagellate Cbl proteins are activated by phosphorylation. Receptor tyrosine kinases are the most important targets of human Cbl, but most of these proteins do not have direct counterparts in choanoflagellates. In this article, we address two main questions: First, is choanoflagellate Cbl activated by tyrosine phosphorylation? Second, given the differences in the tyrosine kinase complement of choanoflagellates and humans, are there differences in how S. rosetta and human c‐Cbl recognize their substrates?
We determined the crystal structure of the TKB module of S. rosetta Cbl. As expected from the sequence conservation, the structure resembles that of human Cbl closely.8 In particular, the conformation of the N‐terminal portion of the TKB‐RING linker is similar to that seen in the autoinhibited form of human Cbl proteins. In addition, in vitro ubiquitylation assays show preservation of the autoinhibitory mechanism. We found that Src kinases can phosphorylate S. rosetta Cbl and that this phosphorylation potentiates the ubiquitylation activity of S. rosetta Cbl. These results indicate that the feedback loop that is characteristic of the action of human Cbl on tyrosine kinases emerged before the choanoflagellates split off from the true metazoans.
To address the issue of substrate specificity, we made model Cbl substrates by fusing a segment bearing the phosphotyrosine recognized by Cbl SH2 (referred to as the “linker peptide”) to the Src kinase domain, either N‐terminal or C‐terminal to the kinase domain. We analyzed the ubiquitylation of these model substrates by human and S. rosetta Cbl. Our results suggest that Cbl ubiquitylates lysine residues in its targets based on proximity and accessibility, rather than through the recognition of sequence or structure. We also tested Cbl protein from two invertebrate organisms, C. elegans and D. melanogaster, with similar results.
Results and Discussion
The structure of the TKB module of S. rosetta Cbl
To investigate the Cbl proteins, we used a number of different constructs, defined in Figure 1. Specifically, we refer to the segment spanning residues 48–480 in human c‐Cbl as the TKB‐RING segment. S. rosetta Cbl contains 553 residues, and we define the TKB‐RING segment as residues 1–418 [Fig. 1(C)]. The phosphotyrosine residue that activates human c‐Cbl, when phosphorylated, is Tyr 371, and the corresponding residue in S. rosetta Cbl is Tyr 352 [Fig. S1(B), Supporting Information].
We expressed and purified full length S. rosetta Cbl using Escherichia coli and used this protein in crystallization experiments [Fig. 1(C)]. The resulting crystals did not contain the full‐length protein, and it appears that a proteolytically cleaved product containing the TKB module was crystallized instead (Material and Methods, Fig. S2, Supporting Information). We used these crystals to determine the structure of the TKB module to 2.4 Å (Material and Methods, Table SI, Supporting Information). The TKB‐RING linker can be traced to Tyr 349 in S. rosetta Cbl, just before the tyrosine residue that is phosphorylated in human c‐Cbl (Tyr 352 in S. rosetta Cbl). The structure is strikingly similar to that of the autoinhibited conformation of the TKB module of human Cbl (PDB IDs: 1B47 and 2Y1M),6, 8 with a root mean square deviation (RMSD) of 0.7 Å for 234 Cα atoms (for 1B47) or 0.9 Å for 260 Cα atoms (for 2Y1M) (Fig. 2). The TKB module consists of a 4‐helix domain, an EF hand domain, and an SH2 domain, and these form a compact structure, with a hydrophobic core at the interface of the three domains [Fig. 3(A), inset].6, 8 For human Cbl, ligand binding results in a closure of the TKB module, whereby the 4H and SH2 domains are pulled toward one another.8 S. rosetta Cbl, which was crystallized without a ligand, is open. The SH2 domain is shifted outward with respect to the 4‐helix domain, relative to the ligand‐bound human Cbl TKB (PDB ID: 2CBL),8 with a RMSD of 1.70 Å for 251 Cα atoms [Fig. S2(A), Supporting Information]. This open conformation reflects previously‐observed flexibility in the TKB module of Cbl.6
Figure 2.
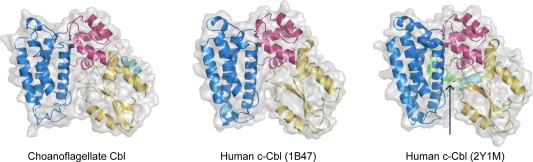
TKB module structures. Shown are the structures of the TKB modules of S. rosetta Cbl and human c‐Cbl (PDB ID: 2CBL),8 as well as the TKB‐RING segment of inactive human c‐Cbl (PDB ID: 2Y1M).6 The RING domain is indicated by an arrow.
Figure 3.
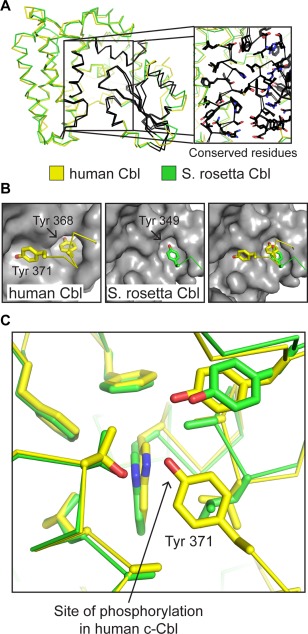
Structural similarity between S. rosetta Cbl (green) and human c‐Cbl (yellow, PDB ID: 2Y1M).6 (A) The TKB modules of S. rosetta Cbl and human c‐Cbl are shown in ribbon representation. The regions of high sequence identity are highlighted in black and are localized almost exclusively to the 4H‐SH2 interface and the SH2 domain. The inset figure is a zoomed in view of the 4H‐SH2 interface, with the side chain residues that are identical in the two structures shown in stick representation (carbon atoms are black). (B) The binding pockets for the two tyrosine residues in the linker (Tyr 368 and Tyr 371 in human c‐Cbl) are also highly conserved. Human c‐Cbl (left) and S. rosetta Cbl (right) are shown in surface representation (gray), with the region containing the two conserved tyrosine residues as ribbons and the tyrosine side chains as sticks (carbons are colored yellow for c‐Cbl and green for S. rosetta Cbl). The far‐right panel shows an overlay of the linker segments on the S. rosetta Cbl structure. (C) Although the tyrosine residue corresponding to the phosphosite in human c‐Cbl is not visualized in the S. rosetta Cbl structure, the binding pocket is highly conserved. Side chains that form this pocket are shown as sticks for both structures.
Segments with high sequence conservation between human and S. rosetta Cbls are located at the interface of the 4‐helix, EF‐hand, and SH2 domain, adjacent to the peptide‐binding cleft in the SH2 domain, or in a loop in the 4‐helix domain adjacent to the SH2 domain [highlighted in Fig. 3(A), S3, Supporting Information]. Residues in this conserved loop are involved in the recognition of residues located N‐terminal to the phosphotyrosine in substrate peptides, as shown previously for human Cbl.8, 16
Tyr 349 in S. rosetta Cbl corresponds to Tyr 368 in human c‐Cbl and lies in a pocket formed by residues in the EF‐hand and SH2 domain, stabilizing the inactive conformation [Fig. 3(B)]. In human Cbl, this interaction, in combination with others made by the tyrosine residue that is phosphorylated, Tyr 371, stabilizes the inactive conformation.6 We could not confidently model the conformation of the tyrosine residue that is potentially phosphorylated, Tyr 352 in S. rosetta Cbl, or additional C‐terminal residues, due to poor electron density. Nevertheless, sequence analysis and homology modeling of human c‐Cbl and S. rosetta Cbl, as well as almost complete conservation lining a hydrophobic pocket in which Tyr 368 of human c‐Cbl is docked, suggest that S. rosetta Cbl Tyr 352 is docked in a similar manner to human c‐Cbl Tyr 371 prior to activation [Fig. 3(B,C)].
S. rosetta Cbl can be activated by tyrosine phosphorylation
We studied whether S. rosetta Cbl has a phosphorylation requirement for activation by utilizing an in vitro assay based on standard methods,17 whereby we mix ubiquitylation enzymes (e.g., E1, E2, and E3) with substrate and necessary buffer components (e.g., ATP and MgCl2). We expressed and purified the TKB‐RING segment of human c‐Cbl using an E. coli expression system (Material and Methods section) and tested its ubiquitylation activity. Ubiquitylation is identified by a higher molecular weight “smear” on a coomassie‐stained SDS‐PAGE gel and validated by using a ubiquitin‐specific antibody and western blotting. Reactions were carried out in the presence of either catalytically inactive chicken c‐Src protein (D368N; the construct includes the SH3, SH2, and kinase domains and is referred to here as Src‐inactive) or the active kinase domain of Src (Src‐KD). Our results show protein ubiquitylation occurs only in the presence of active, but not inactive, Src [Fig. 4(B,C), S4(A), Supporting Information], which is consistent with tyrosine phosphorylation‐induced activation seen in human c‐Cbl.18, 19
Figure 4.
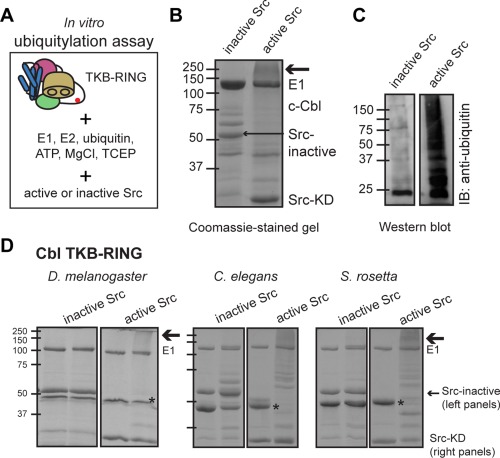
Cbl proteins require phosphorylation for ubiquitylation activity. (A) Schematic of the in vitro ubiquitylation assay, using the Cbl TKB‐RING module. (B) Coomassie‐stained protein gel of an in vitro ubiquitylation assay showing the requirement for an active kinase (here, Src‐KD) in human c‐Cbl activation. A higher molecular weight smear, indicated by an arrow, is only seen in the presence of Src‐KD (M w ∼35 kDa; right lane) and not the Src‐inactive protein (M w ∼ 52 kDa; left lane). t = 60 m for Src‐inactive and 30 m for Src‐KD. (C) An antiubiquitin western blot of a ubiquitylation assay using D. melanogaster Cbl reveals a requirement for active Src in Cbl activity. In the absence of Src‐KD, there is a build‐up of E2∼Ub species at ∼22 kDa, but no protein bands at higher molecular weight. This is in contrast to the right‐hand sample, which contains Src‐KD. t = 30 m. (D) Coomassie‐stained gels showing the results of ubiquitylation reactions using constructs containing the TKB‐RING segments of S. rosetta Cbl, C. elegans Cbl, and D. melanogaster Cbl. Two lanes are shown for each experiment, including either Src‐inactive (left gel panels) or Src‐KD (right gel panels).
We next investigated whether S. rosetta Cbl is activated by phosphorylation. For comparison, we also tested D. melanogaster and C. elegans Cbl. The constructs that we used are shown in Figure 1(C). In vitro ubiquitylation assays using active or inactive chicken Src, as described above, confirmed that S. rosetta Cbl also requires the presence of an active tyrosine kinase for activity, as do D. melanogaster and C. elegans Cbl proteins [Fig. 4(D), S4(B,C), Supporting Information]. We also tested the ability of S. rosetta Src to activate S. rosetta Cbl in our assay. S. rosetta Src, containing the SH3‐SH2‐kinase module, activates and is subsequently ubiquitylated by S. rosetta Cbl [Fig. S4(D), Supporting Information].
We also tested the ubiquitylation activity of linker‐RING constructs of all four Cbl proteins, lacking the TKB module (Fig. 5). In all cases, the linker‐RING constructs were activated by phosphorylation. As discussed earlier, Tyr 371 has two roles in human c‐Cbl regulation. First, when unphosphorylated, it stabilizes the autoinhibited conformation of Cbl, causing phosphorylation to disrupt the autoinhibited state.6 Second, in human Cbl, the phosphorylated form of Tyr 371 stabilizes the catalytically active conformation with ubiquitin bound.5 Our results with the linker‐RING constructs suggest that the role of the phosphotyrosine residue in positioning ubiquitin and the RING domain by directly interacting with ubiquitin is conserved across species.
Figure 5.
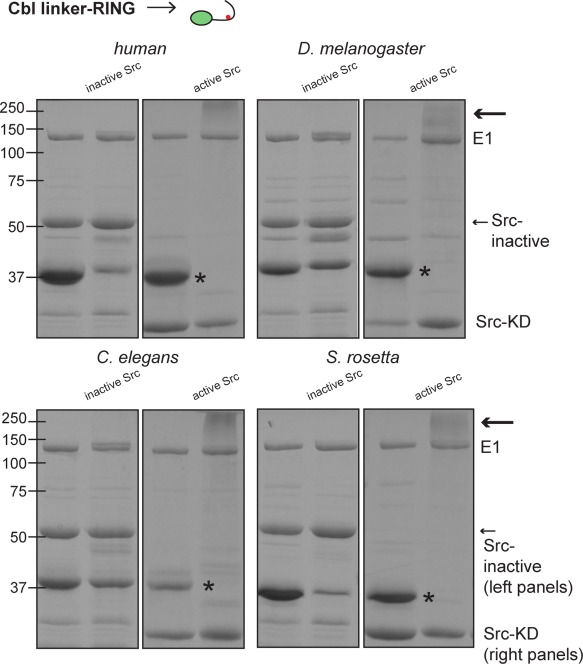
Cbl linker‐RING domains alone require phosphorylation for ubiquitylation activity. Coomassie‐stained gels showing the results of ubiquitylation assays using constructs without the TKB module, that is, the linker‐RING domains alone (see diagram of construct at top). Two lanes are shown for all experiments, for S. rosetta Cbl and C. elegans Cbl, they are t = 0 (left lane) and 15 min (right). For D. melanogaster Cbl TKB‐RING, the lanes are t = 0 (left) and t = 30 min (right). The bands corresponding to the Cbl proteins are marked with asterisks. Similar to Figure 4, a higher molecular weight smear, indicated by an arrow, is only seen in the presence of Src‐KD (M w ∼ 35 kDa; right gel panels) and not the Src‐inactive protein (M w ∼ 52 kDa; left gel panels).
In all of our experiments, we also analyzed phosphorylation of the Cbl proteins by their substrates using mass spectrometry. As discussed in detail in the Supporting Information, we were unable to resolve the tyrosine critical for regulation in all instances, with one exception. In an S. rosetta experiment with unusually high sequence coverage, we are able to identify unphosphorylated Tyr 352, as well as phosphorylated Tyr 349. Based on these results, we decided to mutagenize Tyr 352 in S. rosetta Cbl to phenylalanine. Ubiquitylation experiments with Y352F S. rosetta Cbl confirm that a tyrosine at this position is required for activation [Fig. S4(B,C), Supporting Information]. Therefore, we hypothesize that the stereochemistry of the active site is conserved between S. rosetta and human Cbl, and that Tyr 352 is the residue critical for regulation.
Proximity and accessibility govern the ubiquitylation of residues by Cbl
In humans, Cbl ubiquitylates many different tyrosine kinases, and each of these are ubiquitylated on a variety of lysine residues, as seen for example, in EGFR.20 We determined the specific lysine residues in the kinase domains of ZAP‐70 and Src that are ubiquitylated by human c‐Cbl by tandem mass spectrometry. The lysine residues that are ubiquitylated do not seem to conform to any obvious structural or sequence‐based pattern (Figs. S5, S6, Supporting Information).20
A unifying feature of Cbl substrates in humans is the location of the Cbl binding site. In human kinases, the putative Cbl SH2 binding site lies in a flexible linker segment; however, the location of this segment relative to the lysines that get ubiquitylated on the kinase domain is different, with some being N‐terminal and others being C‐terminal to the kinase domain. The sequences of the linkers in Cbl substrates are well conserved for each protein throughout vertebrate evolution, but the linkers in one kinase do not resemble those in other substrates (Fig. S7, Supporting Information). We wondered if binding of the Cbl SH2 domain to the linker of a kinase plays a role in determining which lysine residues get ubiquitylated.
The structures of phosphotyrosine‐containing linker peptides bound to human Cbl TKB domains has shown that the linkers bind similarly, with the exception of a linker in the hepatocyte growth factor receptor, c‐Met.21 The phosphorylated c‐Met linker binds to the Cbl SH2 domain in the opposite direction to canonical SH2‐phosphopeptide binding interactions. The Cbl binding site in ZAP‐70 and c‐Met is N‐terminal to the kinase domain, while the Cbl binding site in other kinases such as EGFR lies C‐terminal to the kinase domain. Given that most kinases appear to be ubiquitylated within the kinase domain, this raises the question of whether Cbl orientation with respect to the kinase domain is an important determinant of ubiquitylation efficiency. To test whether differences in binding site location relative to the kinase domain affect target lysine residue selectivity, we created a series of model substrates consisting of chimeric proteins in which the Src kinase domain is attached to ZAP‐70, EGFR, or c‐Met linkers [Fig. 6(A)]. We ubiquitylated these chimeric proteins using human, D. melanogaster, C. elegans, and S. rosetta Cbl proteins and identified targeted residues by mass spectrometry (Fig. 7).
Figure 6.
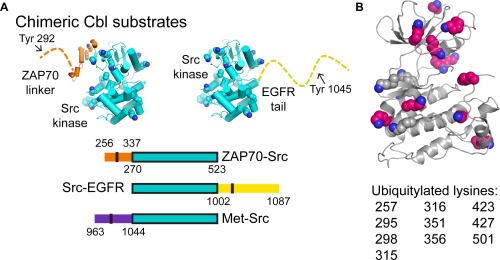
The chimeric Src kinase, linker constructs designed for ubiquitylation assays. (A) The chimeric Cbl substrates used in our ubiquitylation assays are depicted in cartoon representation for ZAP70‐Src and Src‐EGFR (above) or by domain architecture (below). The Src kinase domain (PDB ID: 1YOJ)22 is in cyan, with lysine residues highlighted as spheres by atom (carbons are green). The ZAP‐70 linker (PBD ID: 4K2R)23 is also shown in cartoon representation (orange, with lysine residues highlighted by spheres (carbons are cyan) or a dotted line. The EGFR tail is shown as a dotted line. (B) The lysine residues of chimeric proteins in the Src kinase domain that are ubiquitylated by Cbl proteins are shown as spheres (carbons are gray if not ubiquitylated, and pink if the residue is ubiquitylated). The kinase domain is shown as a gray cartoon. Ubiquitylated residues are listed below the structure.
Figure 7.
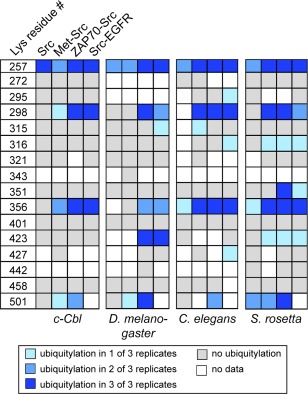
Chimeric proteins are ubiquitylated at similar residues. The results from in vitro ubiquitylation assays, followed by mass spectrometry, for Src kinase or the Src chimeras and Cbl homologs from four species are shown. The key is at the bottom.
Our results indicate that neither the position of the linker with respect to the kinase domain, nor the sequence of the linkers affect which lysine residues are ubiquitylated in the substrate. Not all of the lysine residues in Src were observed by mass spectrometry (e.g., Lys 343), but the unubiquitylated versions of the majority of the lysine‐containing peptides are detected in our experiments (Fig. 7). Overall, our data clearly suggest that certain residues are ubiquitylated (e.g., Lys 257, Lys 298, Lys 356, and Lys 501) regardless of linker identity [Figs. 6(B) and 7]. In addition, S. rosetta Cbl, C. elegans Cbl, and D. melanogaster Cbl ubiquitylate residues that we do not see in our c‐Cbl experiment (e.g., Lys 316 and Lys 423).
Conclusions
The conserved control of choanoflagellate Cbl by tyrosine phosphorylation reveals an ancient coupling of tyrosine phosphorylation and ubiquitylation. Our results show that the machinery is in place for feedback regulation of tyrosine kinase signaling, whereby activation of a tyrosine kinase substrate in turn activates its degradation by Cbl via phosphorylation, in both human and choanoflagellates. This conservation of Cbl structure and function throughout evolution, despite differences in the regulation of tyrosine kinase signaling pathways within these organisms, is remarkable because it shows that the regulation of tyrosine kinases by Cbl was in place before the tyrosine kinase signaling networks that are characteristic of metazoans arose.
Despite the importance of tyrosine phosphorylation for its activity, Cbl predates the emergence of a central role for tyrosine kinases in signaling. The slime mold Dictyostelium discoideum contains CblA, a RING‐type E3 ligase that targets the tyrosine phosphatase PTP3 for ubiquitylation.24, 25, 26 Sequence alignment suggests that CblA contains a tyrosine kinase binding module. However, CblA does not contain the conserved tyrosine for phospho‐activation, and its RING domain is more similar to that of the inhibitor of apoptosis (IAP) proteins than metazoan Cbl (Fig. S8, Supporting Information). Intriguingly, D. discoideum contains STAT proteins that are regulated by tyrosine phosphorylation, along with JAK‐ and Csk‐like kinases.27 The relationship between CblA and kinases in D. discoideum remains an open question.
The domain architecture of Cbl is unique. Analysis of the 108 annotated SH2 domains in the human proteome reveals only five SH2 domains that are associated with a coiled‐coil domain, for example, the 4‐helix domain, (Uniprot identifiers: FER, FES, STAP2, STAT1, and STAT3). Published crystal structures suggest that none of these proteins share the domain architecture of the Cbl family of proteins.28, 29, 30 It is apparent that after originating in a common ancestor of the Amoebozoa and Metazoa, the TKB binding module quickly evolved to become highly specialized for its specific activity targeting tyrosine kinases.
As our results show, the SH2‐dependent interaction between Cbl and its targets is not a determinant in specifying certain lysine residues for ubiquitylation. We conclude that SH2 binding localizes the target near the active site of Cbl, increasing ubiquitylation efficiency. Our data indicate that the role for tyrosine phosphorylation in Cbl, in promoting efficient ubiquitylation, is conserved in the choanoflagellate S. rosetta. To further confirm this, it would be interesting to see if a choanoflagellate E2:Ub:Cbl:substrate complex is structurally similar to that of the human E2:Ub:Cbl:ZAP‐70 complex.5, 6 More broadly, biochemical and structural studies on choanoflagellate tyrosine kinases and their associated signaling proteins are needed to provide a deeper understanding of the signaling pathways of nonmetazoans and how they relate to those in the metazoan lineages.
Materials and Methods
Protein expression and purification
All proteins were expressed as His‐tagged fusions in pET28a‐derived vectors (c‐Cbl, S. rosetta Cbl, and C. elegans Cbl all constructs, D. melanogaster Cbl linker‐RING, E1, E2, ubiquitin, Src‐KD, Src‐inactive, all Src chimeras) using BL21 (DE3) Escherichia coli or the pFASTBAC vector (D. melanogaster Cbl full length and TKB‐RING) using SF9 insect cells. Detailed purification procedures are in the Supporting Information.
In vitro ubiquitylation assay coupled to mass spectrometry
Ubiquitylation assays were based on standard methods17 (see Supporting Information).
Crystallization and structure determination of S. Rosetta Cbl
The crystal used for data collection crystallized at 4 mg mL−1 in well solution 2% (w/v) Tacsimate pH 7.0, 10% (w/v) polyethylene glycol 3350. Detailed data collection and structure determination procedures, as well as sequence and structural analyses methods are in Supporting Information. Coordinates and structure factors were deposited in the Protein Data Bank, with accession code 6BK5.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
JFA and JK designed the experiments. JFA, HTH, and AC expressed and purified protein. JFA, HTH, LS, MJR, and SAM performed experiments. JFA and JK wrote the paper.
Supporting information
Supporting Information Figure 1
Supporting Information Figure 2
Supporting Information Figure 3
Supporting Information Figure 4
Supporting Information Figure 5
Supporting Information Figure 6
Supporting Information Figure 7
Supporting Information Figure 8
Supporting Information
Acknowledgments
The authors would like to thank Drs. Christine Gee and Xiaoxian Cao for technical assistance, Dr. Neel Shah for critical reading of the manuscript, and the other members of the Kuriyan lab for helpful discussion. The authors would also like to thank Dr. Anthony Iavarone in the QB3/College of Chemistry Mass Spectrometry Facility for assistance in mass spectrometry sample preparation and data collection. Dr. Jeanine Amacher was supported by a Jane Coffin Childs Memorial Fund for Medical Research fellowship.
References
- 1. Thien CBF, Langdon WY (2005) c‐Cbl and Cbl‐b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem J 391:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rathinam C, Thien CBF, Flavell RA, Langdon WY (2010) Myeloid leukemia development in c‐Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell 18:341–352. [DOI] [PubMed] [Google Scholar]
- 3. Lo F‐Y, Tan Y‐HC, Cheng H‐C, Salgia R, Wang Y‐C (2011) An E3 ubiquitin ligase: c‐Cbl: a new therapeutic target of lung cancer. Cancer 117:5344–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lemmon MA, Schlessinger J (2010) Cell signaling by receptor tyrosine kinases. Cell 141:1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT (2013) Essentiality of a non‐RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat Struct Mol Biol 20:982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT (2012) Structural basis for autoinhibition and phosphorylation‐dependent activation of c‐Cbl. Nat Struct Mol Biol 19:184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kobashigawa Y, Tomitaka A, Kumeta H, Noda NN, Yamaguchi M, Inagaki F (2011) Autoinhibition and phosphorylation‐induced activation mechanisms of human cancer and autoimmune disease‐related E3 protein Cbl‐b. Proc Natl Acad Sci U S A 108:20579–20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meng W, Sawasdikosol S, Burakoff SJ, Eck MJ (1999) Structure of the amino‐terminal domain of Cbl complexed to its binding site on ZAP‐70 kinase. Nature 398:84–90. [DOI] [PubMed] [Google Scholar]
- 9. Kuriyan J, Darnell JE (1999) An SH2 domain in disguise. Nature 398:22–23. [DOI] [PubMed] [Google Scholar]
- 10. King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JGI, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D (2008) The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451:783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miller WT (2012) Tyrosine kinase signaling and the emergence of multicellularity. Biochim Biophys Acta 1823:1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li W, Young SL, King N, Miller WT (2008) Signaling properties of a non‐metazoan Src kinase and the evolutionary history of Src negative regulation. J Biol Chem 283:15491–15501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sicheri F, Moarefi I, Kuriyan J (1997) Crystal structure of the Src family tyrosine kinase Hck. Nature 385:602–609. [DOI] [PubMed] [Google Scholar]
- 14. Xu W, Harrison SC, Eck MJ (1997) Three‐dimensional structure of the tyrosine kinase c‐Src. Nature 385:595–602. [DOI] [PubMed] [Google Scholar]
- 15. Taskinen B, Ferrada E, Fowler DM (2017) Early emergence of negative regulation of the tyrosine kinase Src by the C‐terminal Src kinase. J Biol Chem 292:18518–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu J, Hubbard SR (2005) Structural characterization of a novel Cbl phosphotyrosine recognition motif in the APS family of adapter proteins. J Biol Chem 280:18943–18949. [DOI] [PubMed] [Google Scholar]
- 17. Petroski MD, Deshaies RJ, In vitro reconstitution of SCF substrate ubiquitination with purified proteins In: Deshaies RJ, Ed. (2005) Ubiquitin and protein degradation, Part A. Vol. 398. Methods in enzymology. Elsevier Academic Press; pp. 143–158. [DOI] [PubMed] [Google Scholar]
- 18. Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y (1999) Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c‐Cbl/Sli‐1. Mol Cell 4:1029–1040. [DOI] [PubMed] [Google Scholar]
- 19. Yokouchi M, Kondo T, Sanjay A, Houghton A, Yoshimura A, Komiya S, Zhang H, Baron R (2001) Src‐catalyzed phosphorylation of c‐Cbl leads to the interdependent ubiquitination of both proteins. J Biol Chem 276:35185–35193. [DOI] [PubMed] [Google Scholar]
- 20. Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell 21:737–748. [DOI] [PubMed] [Google Scholar]
- 21. Ng C, Jackson RA, Buschdorf JP, Sun Q, Guy GR, Sivaraman J (2008) Structural basis for a novel intrapeptidyl H‐bond and reverse binding of c‐Cbl‐TKB domain substrates. EMBO J 27:804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Breitenlechner CB, Kairies NA, Honold K, Scheiblich S, Koll H, Greiter E, Koch S, Schäfer W, Huber R, Engh RA (2005) Crystal structures of active SRC kinase domain complexes. J Mol Biol 353:222–231. [DOI] [PubMed] [Google Scholar]
- 23. Yan Q, Barros T, Visperas PR, Deindl S, Kadlecek TA, Weiss A, Kuriyan J (2013) Structural basis for activation of ZAP‐70 by phosphorylation of the SH2‐kinase linker. Mol Cell Biol 33:2188–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langenick J, Araki T, Yamada Y, Williams JG (2008) A Dictyostelium homologue of the metazoan Cbl proteins regulates STAT signalling. J Cell Sci 121:3524–3530. [DOI] [PubMed] [Google Scholar]
- 25. Araki T, Vu LH, Sasaki N, Kawata T, Eichinger L, Williams JG (2014) Two Dictyostelium tyrosine kinase‐like kinases function in parallel, stress‐induced STAT activation pathways. Mol Biol Cell 25:3222–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goldberg JM, Manning G, Liu A, Fey P, Pilcher KE, Xu Y, Smith JL (2006) The dictyostelium kinome–analysis of the protein kinases from a simple model organism. PLoS Genet 2:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kawata T, Shevchenko A, Fukuzawa M, Jermyn KA, Totty NF, Zhukovskaya NV, Sterling AE, Mann M, Williams JG (1997) SH2 signaling in a lower eukaryote: a STAT protein that regulates stalk cell differentiation in dictyostelium. Cell 89:909–916. [DOI] [PubMed] [Google Scholar]
- 28. Vinkemeier U, Moarefi I, Darnell JE, Kuriyan J (1998) Structure of the amino‐terminal protein interaction domain of STAT‐4. Science 279:1048–1052. [DOI] [PubMed] [Google Scholar]
- 29. Becker S, Groner B, Müller CW (1998) Three‐dimensional structure of the Stat3beta homodimer bound to DNA. Nature 394:145–151. [DOI] [PubMed] [Google Scholar]
- 30. Filippakopoulos P, Kofler M, Hantschel O, Gish GD, Grebien F, Salah E, Neudecker P, Kay LE, Turk BE, Superti‐Furga G, Pawson T, Knapp S (2008) Structural coupling of SH2‐kinase domains links Fes and Abl substrate recognition and kinase activation. Cell 134:793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1
Supporting Information Figure 2
Supporting Information Figure 3
Supporting Information Figure 4
Supporting Information Figure 5
Supporting Information Figure 6
Supporting Information Figure 7
Supporting Information Figure 8
Supporting Information


