Abstract
Attributes of alcohol sensitivity are present before alcohol use disorders (AUDs) develop, they predict those adverse alcohol outcomes, are familial in nature, and many are heritable. Whether measured by alcohol challenges or retrospective reports of numbers of drinks required for effects, alcohol sensitivity reflects multiple phenotypes, including low levels of alcohol response (low LRs) and alcohol-related stimulation. Identification of genes that contribute to alcohol sensitivity could help identify individuals carrying risks for AUDs through their alcohol responses for whom early intervention might mitigate their vulnerability. Such genes could also improve understanding of biological underpinnings of AUDs, which could lead to new treatment approaches. However, the existing literature points to a wide range of genetic mechanisms that might contribute to alcohol responses, and few such genetic findings have been widely replicated. This critical review describes the potential impact of the diverse methods used to study sensitivity on the diversity of genetic findings that have been reported, places the genetic variants mentioned in the literature into broader categories rather than isolated results, and offers suggestions regarding how to advance the field by interpreting findings in light of the methods used to select research subjects and to measure alcohol sensitivity. To date, the most promising results have been for GABA, glutamate, opioid, dopamine, serotonin, and cholinergic system genes. The more gene variants that can be identified as contributors to sensitivity the better future gene screening platforms or polygenic scores are likely to be.
Keywords: Level of Response, alcohol stimulation, alcohol, genes, research methods
Introduction
Predicting the development of complex genetically influenced disorders is complicated. Each condition (e.g., an alcohol use disorder [AUD]) is likely to encompass multiple phenotypes (e.g., for AUD: externalizing behaviors and a person’s alcohol response), each of which could explain part of the genetic contribution (e.g., Goldman et al., 2005; Reilly et al., 2017; Schuckit, 2014). Those phenotypes themselves are likely to reflect multiple genetically influenced sub-components that interrelate with the environment. The search for genes that underlie these complex genetically influenced conditions requires recognizing potential differences across phenotypes being studied. This critical review briefly addresses phenotypes related to how a person responds to alcohol, with an emphasis on specific gene variants potentially impacting alcohol sensitivity.
Alcohol challenges have identified multiple characteristics that contribute to a person’s intensity and type of alcohol responses. Such responses might differ depending on the leg of the blood alcohol concentration (BAC) curve evaluated, attributes of study participants, and alcohol administration protocols. The phenotypes include low levels of alcohol responses (low LRs), most prominent at peak and falling BACs, and high alcohol-related stimulation, typically observed at rising BACs (King et al., 2014; Quinn and Fromme, 2011). These different findings have led to questions regarding whether low LRs can stand alone in predicting later problematic drinking or if the combination of low LR with high stimulation (i.e., a Differentiator Model) is more important (e.g., Newlin and Renton, 2010). I believe both models are correct, with results differing depending on research protocols used. Therefore, this review of genetic variants potentially related to alcohol responses includes data regarding low LR, high stimulation, and their combination.
The low LR, or low sensitivity, focuses more on depressant effects of alcohol, but extends beyond sedation. This is indicated by items used to measure alcohol-induced subjective feelings in our own work (e.g., feeling high, intoxicated, or drunk) and through the first of four questions in a retrospective measure of alcohol sensitivity (i.e., standard drinks needed to first feel any alcohol effect) (Schuckit et al., 1997; Schuckit and Gold, 1988). Low LR goes beyond subjective feelings and also measures dampened alcohol-related changes in hormones, electrophysiologic measures, and patterns of functional Magnetic Resonance Imaging (Paulus et al., 2012; Schuckit et al., 2016b; Tapert et al., 2004). Enhanced alcohol stimulation might also relate to cortisol responses to alcohol. Low LR and enhanced stimulation with alcohol have sometimes been observed in the same individuals (e.g., King et al., 2011; Roche et al., 2014; Schuckit et al., 2002), although it is possible that factors that contribute to a low LR and to more intense stimulation might not be identical.
This paper reviews the current state of the search for genes that contribute to alcohol sensitivity. The goal is to help investigators prepare for future efforts to identify reliable genetic variants. Studies are described from a phenotypic perspective, but diversity in genotypic methodology is likely to be equally challenging. The relative ease of measuring aspects of alcohol sensitivity in animals has resulted in important leads from animal studies that are also referenced.
A brief review of major methods used to evaluate alcohol reaction phenotypes
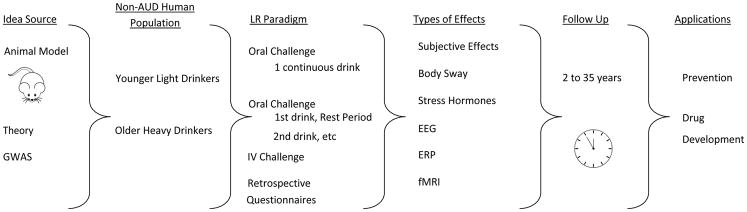
As demonstrated in Figure 1, across studies evaluations of alcohol sensitivity can flow from alcohol reaction phenomena in animals, to testing reactions in several types of non-AUD drinkers, using at least four ways to establish reactions by administering alcohol or using retrospective questionnaires, with protocols incorporating any of at least six different sensitivity measures, and at least two outcome timeframes (short term or long term). This pattern of approaches produces a wealth of information about alcohol responses, but creates hundreds of combinations of sensitivity measures that could possibly reflect different sets of genes. This suggests that it might be prudent to evaluate genetic contributors to each major method separately before combining results into a single genetic analysis.
Figure 1.
Description of Potential Pathways Across Studies Working to Identify Genes Related to Alcohol Sensitivity
Examples of measures of alcohol reactions across different populations
Our group focuses on identifying genetically-influenced characteristics that predict future binge drinking and alcohol problems in young, relatively light drinkers. We usually administer 0.75 ml ethanol/kg over 10 minutes as a single drink, with doses adjusted for height, weight, and sex, producing peak BACs of ~.06 gms/dl at about 60 minutes. Alcohol responses are measured every 15–30 minutes at rising, peak, and falling BACs using the Subjective High Assessment Scale (SHAS) and body sway. Some paradigms also include changes in prolactin, cortisol and/or adrenocorticotropin hormone (ACTH), electroencephalographic measures (EEGs), and/or functional Magnetic Resonance Imaging (fMRI) (e.g., Paulus et al., 2012; Schuckit, 1998). Other laboratories have used the Biphasic Alcohol Effects Scale (BAES) questionnaire instead of the SHAS. The BAES is a reliable 24 item self-report measure with good internal consistency that evaluates seven items each regarding sedation and stimulation during alcohol challenges
A different oral alcohol paradigm that focuses on predicting future alcohol-related problems in drinkers who have already developed alcohol binges uses a “peak and plateau” drinking schedule where subjects consume one drink, wait, and then take a second drink (e.g., Arias et al., 2013), allowing the body to react/adjust/react/adjust to alcohol. Because different gene sets might contribute to alcohol sensitivity measured by different paradigms, it may be useful to first carry out separate genetic analyses for single dose and multiple dose paradigms before combining genetic samples into a single overall analysis. Similar considerations apply to potentially different gene sets for IV vs. oral paradigms and retrospective questionnaires versus alcohol challenge-based measures. Note that oral alcohol challenges evaluate how a person responds to alcohol over several hours at a specific time of day and in a laboratory setting.
Other alcohol paradigms infuse IV alcohol at constant rates to reach peak BACs in ~20 minutes, and then maintain constant BACs (e.g., Ramshandani et al., 1999; Roh et al., 2011). This rapid BAC increase might contribute to stimulation effects of alcohol early in the experiment (Schuckit et al., 2002), and maintaining constant BACs produces intrasession tolerance.
Alcohol sensitivity can also be measured by retrospective questionnaires that record usual numbers of standard drinks required for various effects (e.g., Fleming et al., 2016; Schuckit et al., 1997, 2007, 2008). A higher number of drinks required across effects indicates a lower sensitivity per drink, and vice versa. The 12-item Self-Report of the Effects of alcohol (SRE) questionnaire asks the same four questions regarding drinks required to first feel any effect, slur speech, feel unsteady on your feet, and unwanted falling asleep for the first five times of drinking, the period of heaviest drinking, and recent 3-month drinking. The 15-item Alcohol Sensitivity Questionnaire (ASQ) includes items constructed to separately measure stimulation and sedation, asking participants if they ever experienced the effect and, if so, the minimum or maximum drinks associated with the item. The two retrospective measures compare favorably, and each generates scores relating to alcohol sensitivity (Fleming et. al., 2016). The SRE value has been adjusted for sex, age, weight, and/or the number of effects experienced, but the raw score without adjustments appears to work well. Note that, in contrast to alcohol challenges, these questionnaires ask participants to consider alcohol’s overall effects across several hours.
SRE and alcohol oral single dose challenge-based low LRs predict heavy drinking up to 35 years later in men and women in most studies in the U.S., Australia, U.K., and Germany (e.g., Daeppan et al., 2000; Ehlers et al., 1999; Eng et al., 2005; Gonçalves et al., 2017; Heath et al., 1999; Hinkers et al., 2005; Quinn and Fromme, 2011; Schuckit et al., 2007, 2008). High stimulating effects of alcohol on alcohol challenges robustly predict increases from baseline in heavy episodic (binge) drinking and alcohol problems at 2- and 6-year follow-ups (e.g., King et al., 2011, 2014, 2016).
SRE scores have validities and retest reliabilities of .70 to .80 or higher (Kalu et al., 2008; Ray et al., 2011; Schuckit et al., 1997), while repeat reliabilities of alcohol challenge based stimulation are also high (King et al., 2014, 2016). Using data from 5-year follow-ups of 66 men who had participated in both alcohol challenges and completed SRE questionnaires at baseline, a series of regression analyses predicting later alcohol quantities revealed that 60.3% of the ability of alcohol-challenge-based LR to predict alcohol outcomes overlapped with the ability of the SRE-based LR to predict the same outcome (e.g., Schuckit et al., 2009).
Two relevant LR phenotypes
Low LR
The emphasis in studies of low LR is on predicting onsets of heavy drinking and multiple alcohol problems. Therefore, subjects typically do not yet have the outcomes being predicted, and have not already passed through ages of risk for developing repetitive alcohol problems (the latter is to avoid selecting older participants who despite drinking have not developed problems and are less likely to carry the high-risk phenotype being studied). AUD risk is usually defined by an alcohol dependent relative or an ethnic group membership with high or low AUD risk (e.g., Ehlers et al., 2010; Montiero et al., 1989).
The major source of prospective data on low LR comes from the 35-year-long San Diego Prospective Study (SDPS) with single dose oral alcohol challenges in 453 drinking men (usual past consumption three drinks per occasion) at about age 22 (range 18–25). Half of participants (probands) had an alcohol dependent father and half had no relatives with AUDs, with the two groups matched on age, race, education, drinking and other substance use histories (Schuckit and Gold, 1988; Schuckit et al., 2000). Over time, SRE data were gathered from drinking spouses and offspring, generating information on ~1620 individuals. Relationships of SRE-based low LR to heavier drinking and future alcohol problems were also prospectively documented in the Avon Longitudinal Study of Parents and Children (ALSPAC), the Collaborative Study on the Genetics of Alcoholism (COGA), and other investigations (e.g., Daeppan et al., 2000; Schuckit et al., 2001, 2008). Sibling-pair, twin, and family studies indicate low LR heritabilities of 40%–60% (e.g., Heath et al., 1999; Joslyn et al., 2008; Kalu et al., 2012; Schuckit et al., 2001; Vicken et al., 2003).
Beginning in 1988, 99% of the probands from the SDPS were followed up at about age 30, when the low LR was found to relate to later heavy drinking, alcohol problems, and AUDs, but not to dependence on other substances or to major psychiatric disorders (Schuckit and Smith, 1996; Schuckit et al., 2014). Subsequent every five-year follow-ups of > 90% of original subjects documented that the relationships of low LR to future alcohol problems were partially mediated by heavy drinking friends, overly optimistic expectations of the effects of alcohol, and using alcohol to cope with stress, characteristics that became the focus of a successful program to decrease the heavy drinking risk for 500 18-year-old university students (Gonçalves et al., 2017; Savage et al., 2015; Schuckit et al., 2016a).
High stimulation with alcohol
Studies of high stimulation effects of alcohol often begin with non-alcohol dependent heavy drinkers and light drinkers, attempting to predict escalations in preexisting heavy episodic drinking and increases in alcohol problems. Consistent with those goals, some subjects are in their 30s and include individuals already engaged in heavy drinking. The stimulation effects have included higher subjective feelings of stimulation, especially in early phases of the rising BAC, greater liking and wanting more alcohol during challenges, and lower sedation and lower salivary cortisol later in the alcohol challenge. The more recent work indicated that both elevated stimulation and dampened sedation might be seen in heavier drinkers even earlier in alcohol administration.
The primary source of these data involved 190 subjects who consumed oral alcohol during two five-minute periods separated by five minutes rest (e.g., King et al., 2004, 2006 2011, 2014, 2016). Similar results were seen in a separate sample of 104 individuals (Roche et al., 2014). At study entry, the 190 participants were on average age 26 (range 21–35), 104 of whom habitually engaged in weekly binge drinking, consuming 5+ drinks for men and 4+ for women one to four times per week, with between 10 and 40 drinks per week for at least the prior two years (King et al., 2006, 2011, 2016). The 86 light drinkers consumed ≤ 5 drinks per week with ≤ 5 binges per year. In a 2-year follow-up of almost all the subjects, greater alcohol-induced stimulation and lower sedation predicted increases over baseline binge drinking (King et al., 2011), a finding confirmed for stimulation in a 6-year follow-up of 156 subjects (83%) (King et al., 2014). The heavy drinkers were more likely to have alcoholic relatives, and animal studies have confirmed stimulation effects of alcohol in some genetic lines of alcohol referring rodents (e.g., Cunningham et al., 1992; Masur et al., 1986).
Studies documenting alcohol-related low LR and/or high stimulation are the focus of this review of gene variations related to the intensity of response to alcohol. As suggested above, researchers should consider differences across the various paradigms before combining results into a single analysis when searching for genes related to alcohol sensitivity.
The search for gene variants related to alcohol sensitivity (See Table 1)
Table 1.
Gene Variants Potentially Related to Alcohol Sensitivity Presented in the Text by Category, Listing Human Genetic Location, Phenotype, and Number of Cited Studies
| Gene | rs (or other ID) | Phenotype/Citations | Gene | rs (or other ID) | Phenotype/Citations | Gene | rs (or other ID) | Phenotype/Citations | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alcohol Metabolizing Enzymes* | AL-O | 12 | Glutamate/NMDA | CHRNA5/CHRNB2 Cluster | |||||||
| ALDH 2,2 | 671 | AN | GRLK1 (GluR5) | 2832407 | AD | 1 | 749132306 | AL-O | 2 | ||
| ADH1B | 1229984 | SO | AN | 2229961 | SO | ||||||
| CYP2E1 | 10776687 | SRE | 55863434 | SRE | |||||||
| GRM3 | 6465084 | AD | 2 | 80087508 | |||||||
| Stress Hormones | AN | 7 | AL-O | 2072658 | |||||||
| CRH1 | 1876831 | AL-I | BS | 051730 (A/A) | |||||||
| 242938 | AL-O | 8034191 (C/C) | |||||||||
| AL-C | GAD1 | 2241165 | SRE | 1 | |||||||
| 2058725 | Potassium and Calcium Channel | ||||||||||
| GABA | 379185 | KCNMA1 | chromosome 10q22.3 | AD | 4 | ||||||
| GABRA2 | 279858 | AL-O | 9 | AN | |||||||
| 279869 | AL-I | FYN (PTK) | T137346C | AL-C | 3 | SRE | |||||
| 279837 | AN | AN | |||||||||
| 279871 | NPY | ||||||||||
| 279844 | Opioid and Dopamine | 16147 | AD | 7 | |||||||
| 279845 | OPRM1 | 1799971 | AD | 9 | Leu/7Pro | AL-C | |||||
| 279826 | 3778150 | AL-I | AN | ||||||||
| 279828 | AL-O | ||||||||||
| 279836 | SO | Additional Genes | |||||||||
| SRE | PRMT3 | 74761974 | SRE | 1 | |||||||
| ZNF699 | 7254880 | AD | 5 | ||||||||
| GABRA1 | 1037715 | AN | 4 | ||||||||
| SRE | OPRK1 | 963549 | AL-C | 5 | AN | ||||||
| 997917 | AL-I | ||||||||||
| GABRA6 | Pro385/Ser | AL-O | 4 | AN | ALK | 17004646 | AL-O | 2 | |||
| AN | AN | ||||||||||
| GW | DAT | 28363170 | AD | 3 | GPC5 | 1330469 | AL-O | 2 | |||
| AL-C | AN | ||||||||||
| GABRG1 | 1391166 | AL-C | 1 | AL-I | KLF-3/KLF-12 | AN | 3 | ||||
| 1497571 | SRE | AL-O | |||||||||
| COL6A3 | AN | 1 | |||||||||
| SLC6A11 | 10913738 | GW | 1 | Serotonin | RYR3 | AL-O | 2 | ||||
| SRE | SLC6A4 (5-HTTLPR L allele) | AD | 4 | AN | |||||||
| AL-O | DLGAP1 | 146298733 | GW | 4 | |||||||
| SRE | SRE | ||||||||||
| Per2, Per3 | AN | 3 | |||||||||
The table offers basic descriptions of gene variants identified in the literature as potentially related to alcohol sensitivity, as described in greater detail in the text of this review. Whenever possible the genes are listed as they relate to neurochemical systems with columns offering the gene name as spelled out in the text, gene variant identifiers (usually rs numbers, if known), the type of alcohol sensitivity measure involved and the number of relevant citations for that gene variant offered in the text. The latter is offered as a guide of how often the variant has been potentially linked to a sensitivity measure. The abbreviations offered for phenotypes include: AD=alcohol use disorders; AL-C= alcohol consumption measures; AL-I= IV alcohol challenges; AL-O= oral alcohol challenges; AN= animal models; BS= body sway; GW= genome wide association studies; SO= subjective alcohol response measures other than the SRE; SRE- Self Report of the Effects of Alcohol retrospective measure. Studies involve human subjects unless noted by AN.
While no receptors are dedicated specifically to alcohol, this drug has prominent effects on gamma aminobutyric acid (GABA), glutamate, opioid, dopamine, serotonin (5-HT), and acetylcholine systems and on the hypothalamic-pituitary axis (HPA), each of which could contribute to alcohol sensitivity (Koob and Volkow, 2010). Each effect relates to sets of genes and environmental forces, and, thus, evaluations of gene × gene (G × G) and gene × environment (G × E) interactions are important in understanding how genes relate to how a person responds to alcohol (Goldman, 2010). Few gene effects are likely to follow Mendelian patterns; some gene variants are rare across families, but common within relatives (Choquet et al., 2013); and most are relatively common but explain small proportions of sensitivity phenomena (Joslyn et al., 2010, 2011; Manolio et al., 2009; McCarthy and Hirschorn, 2008; Wang et al., 2005).
The following material reviews gene variants that might relate to alcohol sensitivity and are worth considering in future investigations. These text descriptions are briefly summarized in the broader overview in Table 1, including the predominant pattern of phenotypes that have been evaluated for each variant, and the number of relevant references cited in this section.
Alcohol metabolizing enzymes
Variants of the genes for ALDH2 on chromosome 12q24.12 (e.g., rs671), ADH1B on chromosome 4q23 (e.g., rs1229984) and CYP2E1 on chromosome 10q26.3 (e.g., rs10776687) (Webb et al., 2011) are associated with increased alcohol responses and decreased AUD risks (Bujarski et al., 2015; Jaime et al., 2014; Kuo et al., 2008; Sartor et al., 2015). Those actions could interfere with identification of other genetic variants related to low sensitivity. The relationships of these gene variants to alcohol sensitivity have been supported by animal and human alcohol studies (e.g., Cook et al., 2005; Dickson et al., 2006; Fischer et al., 2007; McCarthy et al., 2010; Wall et al., 2005; Wend et al., 2009).
Gene variations related to stress responses
The stress response system is of interest for drug reactions (Koob and Kreek, 2007), but has rarely been studied regarding alcohol sensitivity. Individuals with low LR or high stimulation demonstrate less intense increases in cortisol, ACTH, and/or prolactin during oral and IV alcohol challenges (King et al., 2006; Schuckit, 1998). Also, homozygotes for C-alleles of Corticotropin Releasing Hormone (CRH) receptor 1 (chromosome 8q13.1) rs1876831 and/or carriers of the rs242938 A-allele, have histories of higher maximum drinks per occasion (perhaps reflecting a lower LR per drink), more binge drinking, higher prevalence of drunkenness (e.g., Hansson et al., 2006; Hayes et al., 2005; Treutlein et al., 2006), and heavier drinking in response to negative life events (Blomeyer et al., 2008). More definitive studies of possible relationships of sensitivity to aspects of the alcohol effects on the HPA axis are needed.
GABA-related genes
Alcohol has prominent effects on GABA-A receptors (e.g., Korpi et al., 1993; Ray and Hutchinson, 2009), variants of which may relate to AUD risks (e.g., Covault et al., 2008; Kareken et al., 2010; Kosobud et al., 2015; Krustal et al., 2006), and perhaps to sensitivity. Beginning with GABRA2 (chromosome 4p12), oral and IV alcohol challenges indicate relationships to sensitivity and heavier drinking for G-alleles of rs279858 (e.g., Arias et al., 2013; Covault et al., 2004, 2008; Kosobud et al., 2015; Lappalainen et al., 2005; Pierucci-Lagha et al., 2005; Roh et al., 2011; Uhart et al., 2012); rs279869 and rs279837 (Roh et al., 2011); rs279871 AA genotype (Kareken et al., 2010); the minor allele (T) for rs279844 (Uhart et al., 2012); and for a haplotype block of minor alleles for rs279858 (C-allele), rs279844 (T-allele), rs279845 (A-allele), rs279826 (G-allele), rs279828 (C-allele), and 279836 (A-allele) (Uhart et al., 2012).
The potential relevance to alcohol responses of GABRA1 (chromosome 5q34) variants comes from animal knockout, gene expression, and between-strain animal studies (e.g., Hanchar et al., 2005; Loh and Ball, 2000), as well as a human genome-wide association study (GWAS) using single dose oral alcohol (Wilhelmsen et al., 2003). Dick et al. (2006b) reported that SRE-based low LR was related to rs1037715.
Several animal studies implicated GABRA6 (chromosome 5q34) regarding lower LRs for cerebellar and movement-related effects (Korpi et al., 1993; Sander et al., 1999). Oral alcohol challenges indicated that GABRA6 Pro385Ser related to lower LRs (Hu et al., 2005; Schuckit et al., 1999), especially in combination with L variants of the serotonin transporter gene (5-HTTLPR).
GABRG1 (chromosome 4p12) variants in rs1391166 and rs1497571 are potentially related to low SRE-based alcohol sensitivity. Subjects with rs1497571 CC genotype had lower LRs per drink, higher drinks per occasion, and more alcohol problems (Ray and Hutchison, 2009).
Finally, regarding GABA, a GWAS and meta-analysis using SREs highlighted possible sensitivity relationships for rs10913738 in a GABA transporter, SLC6A11 (chromosome 3p25.3) (Edwards et al., submitted).
Glutamate and NMDA receptors
Glutamate receptors, including NMDA, are important for alcohol intoxication, sensitivity, and withdrawal. Some aspects of this system are different in individuals with alcohol dependent relatives and drinkers with low LR (Bell et al., 2016; Joslyn et al., 2010; Krystal et al., 2003; Schumann et al., 2008). Specific variants include rs2832407 in GRLK1 (GluR5) (chromosome 21q21.3) (Kranzler et al., 2009), the homolog of which is related to low alcohol consumption and high sensitivity in rodents (Bird et al., 2008); GRM3 (chromosome 7q21.11) rs6465084 (Xia et al., 2014) which might relate to oral alcohol induced body sway and to alcohol dependence (e.g., Wilhelmsen et al., 2003); and GAD1 (chromosome 2q31.1) for SRE-based alcohol sensitivity regarding rs2241165, rs2058725, and rs379185 (Kuo et al., 2009). Sensitivity might also relate to the FYN gene (chromosome 6q21), also known as protein tyrosine kinase [PTK] fyn, especially for T137346C regarding higher maximum drinks (a possible marker for a low sensitivity per drink) (Ishiguro et al., 2000; Schuman et al., 2003). PTK fyn knockout mice have a lower sensitivity to alcohol (Miyakawa et al., 1997). Effects of PTK fyn are likely to occur through NMDA receptors NR2A and NR2B that partially mediate glutaminergic effects of alcohol (Fink and Gothert, 1996).
Opioid receptors and dopamine
Alcohol affects release of beta endorphin and impacts on ventral tegmentum and nucleus accumbens activity, with feelings of reward operating in part through mu opioid receptors (Koob and Kreek, 2007; Mague and Blendy, 2010; Otto et al., 2017). Dopamine has also been linked to craving and heavy drinking (e.g., Cloninger, 1987; Parsian and Zhang, 1997). These effects could relate to alcohol sensitivity as well.
A nonsynonymous SNP rs1799971 of OPRM1 (chromosome6q25.2) relates to IV alcohol subjective effects and cue reactivity from alcohol (Courtney et al., 2015; Ray et al., 2012; Ray and Hutchison, 2004), and might be associated with both decreased receptor glycosylation and half-life (Weerts et al., 2017). G-allele carriers of rs1799971 exhibit decreased mu opioid receptor binding compared to those with AA genotypes (Weerts et al., 2013), and the former may relate to increased responses to oral and IV alcohol and on a retrospective questionnaire, as well as lower AUD risks (Ehlers et al., 2008; Ray and Hutchison 2004; Schwantes-An et al., 2016). The same SNP might relate alcohol dependence risks and higher alcohol self-administration (Hendershot et al., 2014; Otto et al., 2017; van der Zwaluw et al., 2007, 2009). C- allele carriers of a SNP in linkage disequilibrium (LD) with rs179971, rs3778150, demonstrate decreased intensities of response to oral alcohol. (Hancock et al., 2015; Otto et al., 2017).
Kappa opioid receptor, OPRK1 (chromosome 8q11.23), its’ endogenous ligand, dynorphin, and the ligand’s precursor, prodynorphin, affect substance-related phenomena (e.g., Anderson and Becker, 2017; Gilpin et al., 2014; Walker and Koob, 2008), but have not been adequately evaluated regarding alcohol sensitivity. Acute alcohol increases dynorphin in the nucleus accumbens and frontal cortex, and a kappa receptor antagonist decreases alcohol self-administration in animals with alcohol-dependent-like syndromes (Anderson and Becker, 2017; D’Addario et al., 2011; Gilpin et al., 2014; Walker and Koob, 2008). Rs963549 and rs997917 might relate to sedating effects of alcohol as measured by drinks consumed per day and IV alcohol challenges, at least in the context of naltrexone (Ashenhurst et al., 2012; Gelernter et al., 2007).
The dopamine transporter (DAT) gene (chromosome 5q15.3) has a common variable number of tandem repeat (VNTR) polymorphism (rs28363170) with 10-repeat alleles (A10) associated with higher DAT expression in the striatum (i.e., lower synaptic dopamine). The A9 allele produces higher synaptic dopamine. Individuals with the A9 DAT VNTR and the G-allele for the OPRM1 rs1799971 have higher sensitivity with oral and IV alcohol, and lower AUD risks (Anton et al., 2012; Heinz et al., 2000; Ramchandani et al., 2011; Weerts et al., 2017).
5-HT (serotonin) systems
Low synaptic 5-HT in humans and animals is associated increased drinking; medications that increase synaptic serotonin decrease alcohol intake; 5-HT-like drugs mimic alcohol intoxication; and higher platelet 5-HT reuptake is associated with developing AUDs (Ernouf et al., 1993; George et al., 1997; LeMarquand et al., 1994; Pandey et al., 1992; Rausch et al., 1991). A variant in the promoter region (5-HTTLPR) in the serotonin transporter gene (SLC6A4; chromosome 17q11.2) might relate to a lower LR (e.g., Cope et al., 2017; Hu et al., 2005) for a long (L) repeat length polymorphism associated with faster 5-HT reuptake (Hu et al., 2005). The LA variant might be associated with single dose oral alcohol challenge- and SRE-based low LR and higher rates of future AUDs (Hinkers et al., 2006; Hu et al., 2005; Schuckit et al, 1999).
Cholinergic systems
Alcohol- and nicotine-related disorders often co-occur (e.g., Hopfer et al., 2001), the presence of either disorder relates to increased severity of the other (Ehringer et al., 2007), both alcohol and nicotine conditions are genetically influenced, and some gene variants might predispose individuals toward both disorders (Froelich et al., 2017; Hettema et al., 1999; Hopfer et al., 2001; Steensland et al., 2007; Swan et al., 1997). Nicotinic receptors might contribute to this overlap (Sherva et al., 2010; Wang et al., 2009) in that the nicotinic receptor partial agonist, varenicline, might also attenuate alcohol consumption (Froelich et al., 2017; Steensland et al., 2007). Oral alcohol challenges and SRE data have highlighted rs1051730 (A/A) and rs8034191(C/C) in the cholinergic gene cluster on chromosome 15q2 as potentially related to lower LR (Joslyn et al., 2008). A rare missense variant of CHRNA5 might relate to more intense oral alcohol challenge responses, including rs749132306, rs2229961, rs55863434, and rs80087508. Data also support possible relationships to alcohol sensitivity for rs2072658 in the CHRNB2 receptor (chromosome 1q21.3) (Ehringer et al., 2007).
Potassium and calcium channel related genes
Animal homologs of human KCNMA1 (chromosome 10q22.3) might relate to the intensity of alcohol responses in Drosophila and C. elegans (Davies et al., 2003; Wang et al., 2001). In humans, a locus on chromosome 10 near KCNMA1 related to low LRs on the SRE (Ehlers et al., 2010; Wilhelmsen et al., 2003). A similar chromosome 10 region related to smoking, and to AUDs (Agrawal et al., 2008; Gelernter et al., 2009; Li et al., 2006).
NPY
This inhibitory neuropeptide affects appetitive behaviors and emotion (Foroud et al., 2000; Hayes et al., 2005; Heilig and Widerlov, 1995; Hwang et al., 1999; Levine and Morley, 1984). Mice deficient in NPY demonstrate higher alcohol intake and lower sensitivity, rodents with high NPY have lower alcohol intake (Badia-Elder et al., 2003; Ehlers et al., 1998; Gilpen et al., 2003; Teacott and Hebelein, 1998; Thiele et al., 2000; Zhu et al., 2003), and a QTL related to alcohol consumption in rats might involve NPY (Carr et al., 1998). A relevant variant is rs16147 in the promotor region in the NPY gene (7p15.3), and another chromosomal region is a Leu7Pro missense variant in the signal peptide of human NPY where sensitivity is lower with the Leu allele (Lappalainen et al., 2002). The latter genotype is also related to heavier alcohol intake and AUDs in some studies (Hu et al., 2005; Kauhanen et al., 2000; Zhou et al., 2008).
Additional genes of potential interest
Protein Arginine Methyltransferase 3, PRMT3 (chromosome 11p15.1) variant rs74761974 (A-allele) related to SRE measures in a preliminary analysis of a recent GWAS (1.4×10−8) (Wetherell et al., 2017). A nearby gene of interest is the glycine neurotransmitter transporter, SLC6A5.
Zinc-finger gene ZNF699 (chromosome 19p13.2) is related to Drosophila gene “hang”, which is associated with increased alcohol tolerance (Scholz et al., 2005) and to QTLs involved in alcohol sedation in mice (e.g., Bennett and Johnson, 1998; Ehringer et al., 2002; Markel et al., 1997; Riley et al., 2006). In humans, several gene variants (e.g., rs7254880) might relate to alcohol dependence (Riley et al., 2006).
Homologes of human anaplastic lymphoma kinase (ALK) (also known as ALK tyrosine kinase; chromosome 2p22.3) may be associated with resistance to alcohol’s sedative effects in Drosophila, alcohol-induced ataxia in recombinant inbred mice, and with longer alcohol sedation in ALK knockout mice (Lasek et al., 2011a,b). Sequencing of human ALK indicated several variants (e.g. rs17004646) potentially associated with low sensitivity (Lasek et al. 2011b).
Glypican 5 (GPC5) (chromosome 13q31.3) modulates cellular signaling in the caudate nucleus, putamen and hippocampus. Variant rs1330469 is potentially related to alcohol-induced ataxia in mice, locomotion in Drosophila, and single dose oral alcohol induced ataxia in humans (Joslyn et al., 2011; Kong et al., 2010; Saunders et al., 1997).
Krueppel-Like Factor 12 (KLF-12) (chromosome 13q22.1) potentially relates to acute functional tolerance to alcohol in C. elegans and to effects of alcohol in the nucleus accumbens and the ventral tegementum (Adkins et al., 2017; Wolen et al., 2012). This gene has not been directly evaluated in humans regarding alcohol sensitivity.
A mouse homolog of Collagen alpha-3 (COL6A3) (chromosome 2q37.3) is of potential interest to sensitivity because of a QTL related to sensitivity to alcohol withdrawal handling-induced seizures and 2-bottle alcohol related preference in mice (Adkins et al., 2017).
Ryanodine 3 receptor-related genes (RYR, chromosome 15q13.3) potentially relate to single dose oral alcohol responses in humans, and homologs of this gene may relate to alcohol sensitivity in C. elegans (unc-68) and to tolerance development in Drosophila (Adkins et al., 2017; Joslyn et al., 2010). Ryanodine gene effects might operate, at least in part, through calcium channels and dopamine 1 receptors (Kurokawa et al., 2013).
Clock genes involved in circadian rhythms have also been reported to relate to depression and AUDs, and to glutaminergic systems (Huang et al., 2010; Kovanen et al., 2010; Spanagel et al., 2005). Most salient to the current review are animal studies that have highlighted the impact of mutations in Per2 (human chromosome 2q37.3) and Per3 (human chromosome 1p36.23) on how the time of day relates to the intensity of alcohol reactions, including alcohol sensitivity and alcohol consumption (Perreau-Lenz et al., 2009; Wang et al., 2012). No specific human gene variations have been highlighted for these effects, but the existing data support the need for genetic studies regarding these and other clock genes in human alcohol responses (Edwards et al., submitted).
Finally, a SRE-based GWAS and meta-analysis highlighted rs146298733 in DKGAP1 (DLG-Associated Protein 1) on chromosome18p11.31 as potentially related to alcohol sensitivity. Variations in this gene are also associated with obsessive-compulsive disorder and retinitis pigmentosa (Edwards et al., submitted).
Some conclusions and future directions
This review is the first to summarize gene variants potentially related to the alcohol response while emphasizing the diversity of paradigms evaluating alcohol sensitivity. There is no single best approach for evaluating alcohol sensitivity, and the existing variety of methods has the benefit of describing multiple aspects of the alcohol response. However, the information offered above highlights the complexities among the various measures of alcohol responses, and it might be important to consider differences in research methods before combining results from different approaches into a single meta-analysis or GWAS.
For example, investigations using oral alcohol challenges where alcohol is consumed in one continuous drink and paradigms giving alcohol in several servings with interspersed rest periods when alcohol plateaus, might not evaluate identical phenomena with identical genetic contributors. The same reservations apply to combining results from oral and IV alcohol paradigms, because in the latter BACs rise more rapidly, subjective responses tend to be more intense, and some IV paradigms included a phase where the BAC is maintained, with results that might reflect intrasession tolerance. Furthermore, alcohol challenges measure reactions over a relatively short laboratory session, but retrospective self-reports of drinks needed across effects relate to reactions during entire evenings of a real-life drinking. It might also be difficult to combine sensitivity results from studies of younger relatively alcohol-problem free modest drinkers (e.g., Schuckit et al., 2008) with the older heavy drinkers used to predict the escalation of baseline heavier drinking and binges (e.g., King et al., 2014). While it is possible that different methodologies to evaluate alcohol sensitivity might identify identical genes, researchers must consider that different combinations of genes might contribute to sensitivity measured through different approaches. This phenomenon might diminish the ability of meta-analyses and GWAS to consistently identify genes with small effects on how a person responds to alcohol. The optimal approach might be to first evaluate potential associated genes separately for oral alcohol administrations, IV dosing, and retrospective questionnaire-based measures before combining them into a single analysis.
Despite methodological differences, this review highlighted multiple gene variants that might contribute to alcohol responses. The most promising results are in the GABA, glutamate, opioid, dopamine, serotonin, and cholinergic systems. Finding a wide range of genetic variants likely to contribute to alcohol responses was predictable based on the characteristics of most complex genetically influenced conditions and the range of ethanol-based brain effects.
Low LR heritabilities are 40% to 60% and animal studies have confirmed alcohol stimulation in some genetic lines of alcohol referring rodents (e.g., Cunningham et al., 1992; Masur et al., 1986). Several studies confirm that lower and higher alcohol responses that may operate at different phases of the BAC curve or relate to the rapidity of rise of alcohol blood levels may be related to each other (e.g., King et al., 2016; Ray et al., 2016; Schuckit et al., 2002) and thus both types of measures have been included in this review of gene variants that potentially relate to alcohol sensitivity.
This review has several implications for my own future work. With my interest in evaluating why only some relatively problem-free lighter drinkers escalate their intake and problems, I will continue to include both single dose oral alcohol challenges and retrospective self-reports of drinks needed for effects. While higher stimulation early in the alcohol challenge has rarely been observed using our own paradigms, in future work I will add stimulation measures to our current measures of overall feelings of alcohol intoxication. Our own findings might reflect the fact that my testing paradigm involves slowly rising BACs and the major subjective measure used, the SHAS, is not as likely to pick up stimulation as the BAES that was developed years after my research began.
My collaborations with geneticists will continue to combine results across different measures of alcohol responses, but analyses will begin with preliminary evaluations of trends for variants in individual genes and gene systems (e.g., for GABA, glutamate, or clock genes) before combining results from different approaches in genetic analyses. We will work to establish whether the same or similar genes relate to stimulant, depressant, and overall intoxication effects of alcohol as risk factors for future heavy drinking and AUDs.
Improving understanding of how low LRs and alcohol-related stimulation relate to future alcohol problems has implications for prevention of AUDs. Using low LR as an example, this characteristic is relatively common in individuals from a wide range of socioeconomic strata, and relates to the AUD risk across the sexes and racial or ethnic groups (e.g., Hinkers et al., 2006; Schuckit and Smith 2017b; Schuckit et al., 2000, 2007). For low LR, several environmental and attitudinal attributes that partially mediate the risk for adverse alcohol-related outcomes have been identified. Two investigations have shown that these mediators could be addressed through relatively inexpensive internet-based education programs to decrease heavy drinking (e.g., Savage et al., 2015; Schuckit et al., 2016a). Similar programs might be used in high schools, the military, or industry to identify drinkers with high AUD risks through low alcohol LRs and to help them mitigate future alcohol-related problems. Similar results might be seen for measures of alcohol-related stimulation. Finding genes that contribute to lower LRs and higher alcohol stimulation as risk factors for future heavy drinking and alcohol problems could help with early identification and intervention in those at risk for future alcohol problems through their alcohol sensitivity. Greater understanding of the biological bases for the alcohol reaction phenotypes might also facilitate developing medications to help treat individuals who developed their AUD in the context of low LRs or higher alcohol stimulation.
It is important to consider several additional guidelines for efforts to increase our knowledge of specific gene variants that relate to alcohol sensitivity. First, studies of high stimulation or low LR need to control for the strong effects of ALDH2,2 and ADH1B genotypes, as these could obscure the effects of other genetic contributors to alcohol responses. Second, the impact of any phenotype on adverse alcohol outcomes is likely to operate through many genes and through environmental and attitudinal characteristics. Thus, whenever possible, studies should evaluate more than one gene variant at a time and search for G × G and G × E additive and mediational interrelationships (Olfson et al., 2014; Schuckit and Smith, 2017; Schuckit et al., 2017b). Third, in light of the likely small effect for any one variant when studied across families, investigators should consider evaluating gene effects vertically within families, as some variants might be seen in a third or more of members of any one family (Choquet et al., 2013) but be observed in a small proportion of the general population. Fourth, for most gene variants few, if any, specific variants will be consistently identified across almost all studies, and thought might be given to developing a standard for determining which variants are worth emphasizing in additional work (e.g., Joslyn et al., 2011). Fifth, until the national alcohol and drug institutes suggest guidelines for standardizing approaches across studies, investigators should take steps to use the same measures that are already incorporated in the recent literature in an effort to minimize the variance likely to occur when study results are combined.
There are also several caveats for this review that readers should consider. There was not sufficient space or appropriate expertise to critically review specific genetic analytic techniques. Space limitations also precluded a much-needed detailed comparison of specific phenotypic approaches, a deficiency I hope to address in the future with a review carried out jointly with researchers who study different types of participants and those who use different alcohol administration protocols.
In summary, finding gene variants that contribute to complex genetically influenced phenotypes is challenging, and alcohol sensitivity is no exception. The genes associated with such characteristics might vary depending on the population studied and test paradigms used, and such across-study differences might contribute to divergent results. This review highlighted results of studies to date, suggested options for standardizing research paradigms, and discussed issues that should be considered before combining results across studies when searching for genes that might contribute to how a person reacts to alcohol.
Acknowledgments
Supported by NIH/NIAAA grants AA008401, AA0021162, and AA005526
References
- Adkins AE, Hack LM, Bigdeli TB, Williamson VS, McMichael GO, Mamdani M, Edwards AC, Aliev F, Chan RF, Bhandari P, Raabe RC, Alaimo JT, Blackwell GMG, Moscati A, Poland RS, Rood B, Patterson DG, Walsh D, Whitfield JB, Zhu G, Montgomery GW, Henders AK, Martin NG, Heath AC, Madden PAF, Frank J, Ridinger M, Wodarz N, Soyka M, Zill P, Ising M, Nöthen MM, Klefer F, Rietschel M, Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Granzler HR, Farrer LA, Maher BS, Prescott CA, Dick DM, Bacanu SA, Mathies LD, Davies AG, Vladimirov VI, Grotewiel M, Bowers MS, Bettinger JC, Webb BT, Miles MF, Kendler KS, Riley BP German Study of the Genetics of Addiction Consortium, Collaborative Study of the Genetics of Alcoholism Consortium. Genomewide association study of alcohol dependence identifies risk loci altering ethanol-response behaviors in model organisms. Alcohol Clin Exp Res. 2017;41:911–928. doi: 10.1111/acer.13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Hinrichs AL, Dunn G, Bertelsen S, Dick DM, Saccone SF, Saccone NL, Grucza RA, Wang JC, Cloninger CR, Edenberg HJ, Foroud T, Hesselbrock V, Kramer J, Bucholz KK, Kuperman S, Nurnberger JI, Jr, Porjesz B, Schuckit MA, Goate AM, Bierut LJ. Linkage scan for quantitative traits identifies new regions of interest for substance dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Drug Alcohol Depend. 2008;93:12–20. doi: 10.1016/j.drugalcdep.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RI, Becker HC. Role of the dynorphin/kappa opioid receptor system in the motivational effects of ethanol. Alcohol Clin Exp Res. 2017;41:1402–1418. doi: 10.1111/acer.13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton RF, Voronin KK, Randall PK, Myrick H, Tiffany A. Naltrexone modification of drinking effects in a subacute treatment and bar-lab paradigm: influence of OPRM1 and dopamine transporter (SLCA3) genes. Alcohol Clin Exp Res. 2012;36:2000–2007. doi: 10.1111/j.1530-0277.2012.01807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias AJ, Covault J, Feinn R, Pond T, Yang B-Z, Ge W, Oncken C, Kranzler HR. A GABRA2 variant is associated with increased stimulation and ‘high’ following alcohol administration. Alcohol Alcohol. 2013;49:1–9. doi: 10.1093/alcalc/agt163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashenhurst JR, Bujarski S, Ray LA. Delta and kappa opioid receptor polymorphisms influence the effects of naltrexone on subjective responses to alcohol. Pharmacol Biochem Behav. 2012;103:253–259. doi: 10.1016/j.pbb.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badia-Elder NE, Stewart RB, Powrozek TA, Murphy JM, Li TK. Effects of neuropeptide Y on sucrose and ethanol intake and on anxiety-like behavior in high alcohol drinking (HAD) and low alcohol drinking (LAD) rats. Alcohol Clin Exp Res. 2003;27:894–899. doi: 10.1097/01.ALC.0000071929.17974.DA. [DOI] [PubMed] [Google Scholar]
- Bell RL, Hauser SR, McClintick J, Rahman S, Edenberg HJ, Szumlinski KK, McBride WJ. Ethanol-associated changes in glutamate reward neurocircuitry: a minireview of clinical and preclinical genetic findings. Prog Mol Biol Transl Sci. 2016;137:41–85. doi: 10.1016/bs.pmbts.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B, Johnson TE. Development of congenics for hypnotic sensitivity to ethanol by QTL-marker-assisted counter selection. Mamm Genome. 1998;9:969–974. doi: 10.1007/s003359900908. [DOI] [PubMed] [Google Scholar]
- Bird MK, Kirchhoff J, Djouma E, Lawrence AJ. Metabotropic glutamate 5 receptors regulate sensitivity to ethanol in mice. Int J Neuropsychopharmacology. 2008;11:765–774. doi: 10.1017/S1461145708008572. [DOI] [PubMed] [Google Scholar]
- Blomeyer D, Treutlein J, Esser G, Schmidt MH, Schumann G, Laucht M. Interaction between CRHR1 gene and stressful life events predicts adolescent heavy alcohol use. Biol Psychiatry. 2008;63:146–151. doi: 10.1016/j.biopsych.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Bujarski S, Lau A, Lee S, Ray L. Genetic and environmental predictors of alcohol use in Asian American young adults. J Stud Alcohol Drugs. 2015;76:690–699. doi: 10.15288/jsad.2015.76.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, Lumeng L, Li T-K. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22:884–887. [PubMed] [Google Scholar]
- Choquet H, Joslyn G, Lee A, Kasberger J, Robertson M, Brush G, Schuckit MA, White R, Jorgenson E. Examination of rare missense variants in the CHRNA5-A3-B4 gene cluster to level of response to alcohol in the San Diego sibling pair study. Alcohol Clin Exp Res. 2013;37:1311–1316. doi: 10.1111/acer.12099. [DOI] [PubMed] [Google Scholar]
- Cloninger CR. Neurogenetic adaptive mechanisms in alcoholism. Science. 1987;236:410. doi: 10.1126/science.2882604. [DOI] [PubMed] [Google Scholar]
- Cook T, Luczak S, Shea S, Ehlers C, Carr L, Wall T. Associations of ALDH2 and ADH1B gentopyes with response to alcohol in Asian Americans. J Stud Alcohol. 2005;66:196–204. doi: 10.15288/jsa.2005.66.196. [DOI] [PubMed] [Google Scholar]
- Cope LM, Munier EC, Trucco EM, Hardee JE, Burmeister M, Zucker RA, Heitzeg MM. Effects of the serotonin transporter gene, sensitivity of response to alcohol, and parental monitoring on risk for problem alcohol use. Alcohol. 2017;59:7–16. doi: 10.1016/j.alcohol.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney KE, Ghahremani DG, Ray LA. The effect of alcohol priming on neural markers of alcohol cue-reactivity. Am J Drug Alcohol Abuse. 2015;41:300–308. doi: 10.3109/00952990.2015.1044608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33:837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Noble D. Conditioned activation induced by ethanol: role in sensitization and conditioned place preference. Pharmacol Biochem Behav. 1992;43:307–313. doi: 10.1016/0091-3057(92)90673-4. [DOI] [PubMed] [Google Scholar]
- D’Addario C, Caputi FF, Rimondini R, Gandolfi O, Del BE, Candeletti S, Romualdi P. Different alcohol exposures induce selective alterations on the expression of dynorphin and nociceptin systems related genes in rat brain. Addict Bio. 2011;18:425–433. doi: 10.1111/j.1369-1600.2011.00326.x. [DOI] [PubMed] [Google Scholar]
- Daeppen J-B, Landry U, Pécoud A, Decrey H, Yersin B. A measure of the intensity of response to alcohol to screen for alcohol use disorders in primary care. Alcohol Alcohol. 2000;35:625–627. doi: 10.1093/alcalc/35.6.625. [DOI] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- Dick DM, Plunkett J, Wetherill LF, Xuei X, Goate A, Hesselbrock V, Schuckit M, Crowe R, Edenberg HJ, Foroud T. Association between GABRA1 and drinking behaviors in the Collaborative Study on the Genetics of Alcoholism sample. Alcohol Clin Exp Res. 2006b;30:1101–1110. doi: 10.1111/j.1530-0277.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Dickson P, Heath JM, Montgomery G, Martin N, Whitfield J, Birley A. Effects of variation at the ALDH2 locus on alcohol metabolism, sensitivity, consumption, and dependence in Europeans. Alcohol Clin Exp Res. 2009;30:1093–1100. doi: 10.1111/j.1530-0277.2006.00128.x. [DOI] [PubMed] [Google Scholar]
- Edwards AC, Deak JD, Gizer IR, Chatzinakos C, Wilhelmson KP, Heron J, Hickman M, Webb BT, Bacanu A-A, Kendler KS, Dick DM, Schuckit MA. Meta-analysis of genetic influences on initial alcohol sensitivity. Addiction Biol. doi: 10.1111/acer.13896. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Garcia-Andrade C, Wall TL, Cloutier D, Phillips E. Electroencephalographic responses to alcohol challenge in Native American Mission Indians. Biol Psychiatry. 1999;45:776–787. doi: 10.1016/s0006-3223(98)00113-9. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR, Schuckit MA, Wilhelmsen KC. Genome-wide scan for self-rating of the effects of alcohol in American Indians. Psychiatr Genet. 2010;20:221–228. doi: 10.1097/YPG.0b013e32833add87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Li TK, Lumeng L, Hwang BH, Somes C, Jimenez P, Mathe AA. Neuropeptide Y levels in ethanol-naïve alcohol-preferring and -nonpreferring rats and in Wistar rats after ethanol exposure. Alcohol Clin Exp Res. 1998;22:1778–1782. [PubMed] [Google Scholar]
- Ehlers CL, Lind PA, Wilhelmsen KC. Association between single nucleotide polymorphisms in the mu opioid receptor gene (OPRM1) and self-reported responses to alcohol in American Indians. BMC Med Genet. 2008;9:35–45. doi: 10.1186/1471-2350-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH, Schlaepfer I, Smolen A, Stallings MC, Young SE, Zeiger JS. Association of the neuronal nicotinic receptor β2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet Part B (Neuropsychiatr Genet) 2007;144B:596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- Ehringer MA, Thompson J, Conroy O, Yang F, Hink R, Bennett B, Johnson TE, Sikela JM. Fine mapping of polymorphic alcohol-related quantitative trait loci candidate genes using interval-specific congenic recombinant mice. Alcohol Clin Exp Res. 2002;26:1603–1608. doi: 10.1097/01.ALC.0000036921.33411.67. [DOI] [PubMed] [Google Scholar]
- Ernouf D, Compagnon P, Lothion P, Narcisse G, Benard JR, Daoust M. Platelet 3H 5-HT uptake in descendants from alcoholic patients: a potential risk factor for alcohol dependence. Life Sci. 1993;52:989–995. doi: 10.1016/0024-3205(93)90190-e. [DOI] [PubMed] [Google Scholar]
- Fink K, Gothert M. Both ethanol and ifenprodil inhibit NMDA-evoked release of various neurotransmitters at different, yet proportional potency: potential relation to NMDA receptor subunit composition. Naunyn Schmiedebergs Arch Pharmacol. 1996;354:312–319. doi: 10.1007/BF00171062. [DOI] [PubMed] [Google Scholar]
- Fischer M, Wetherill L, Carr L, You M, Crabb D. Association of the aldehyde dehydrogenase 2 promoter polymorphism with alcohol consumption and reactions in an American Jewish population. Alcohol Clin Exp Res. 2007;31:1654–1659. doi: 10.1111/j.1530-0277.2007.00471.x. [DOI] [PubMed] [Google Scholar]
- Fleming KA, Bartholow BD, Hilgard J, McCarthy DM, O’Neill SE, Steinley D, Sher KJ. The alcohol sensitivity questionnaire: evidence for construct validity. Alcohol Clin Exp Res. 2016;40:880–888. doi: 10.1111/acer.13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroud T, Bice P, Castelluccio P, Bo R, Miller L, Ritchotte A, Lumeng L, Li TK, Carr LG. Identification of quantitative trait loci influencing alcohol consumption in the high alcohol drinking and low alcohol drinking rat lines. Behav Genet. 2000;30:131–140. doi: 10.1023/a:1001955205117. [DOI] [PubMed] [Google Scholar]
- Froehlich JC, Nicholson ER, Dilley JE, Filosa NJ, Rademacher LC, Smith TN. Varenicline reduces alcohol intake during repeated cycles of alcohol reaccess following deprivation in alcohol-preferring (P) rats. Alcohol Clin Exp Res. 2017;41:1510–1517. doi: 10.1111/acer.13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J, Farrer L. Dense genome-wide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biol Psychiatry. 2009;65:111–115. doi: 10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Gueorguieva R, Kranzler HR, k Zhang H, Cramer J, Rosenheck R, Krystal JH VA Cooperative Study #425 Study Group. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: results from the VA Cooperative Study. Alcohol Clin Exp Res. 2007;31:555–563. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- George DT, Benkelfat C, Rawlings RR, Eckardt MJ, Phillips MJ, Nutt DJ, Wynne D, Murphy DL, Linnoila M. Behavioral and neuroendocrine responses to m-chlorophenylpiperazine in subtypes of alcoholics and in healthy comparison subjects. Am J Psychiatry. 1997;154:81–87. doi: 10.1176/ajp.154.1.81. [DOI] [PubMed] [Google Scholar]
- Gilpin NW, Stewart RB, Murphy JM, Li TK, Badia-Elder NE. Neuropeptide Y reduces oral ethanol intake in alcohol-preferring (P) rats following a period of imposed ethanol abstinence. Alcohol Clin Exp Res. 2003;27:787–794. doi: 10.1097/01.ALC.0000065723.93234.1D. [DOI] [PubMed] [Google Scholar]
- Gilpin NW, Roberto M, Koob GF, Schweitzer P. Kappa opioid receptor activation decreases inhibitory transmission and antagonizes alcohol effects in rat central amygdale. Neuropharmacology. 2014;77:294–302. doi: 10.1016/j.neuropharm.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D. Gene × environment interactions in complex behavior: first, build a telescope. Biol Psychiatry. 2010;67:295–296. doi: 10.1016/j.biopsych.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Gonçalves PD, Schuckit MA, Smith T. Drinking status between ages 50 and 55 for men from the San Diego Prospective Study who developed DSM-IV alcohol abuse or dependence in prior follow-ups. J Stud Alcohol Drugs. 2017;78:512–520. doi: 10.15288/jsad.2017.78.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchar HJ, Dodson PD, Olsen RW, Otis TS, Wallner M. Alcohol-induced motor impairment caused by increased extrasynaptic GABAA receptor activity. Nature Neurosci. 2005;8:339–345. doi: 10.1038/nn1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Levy JL, Gaddis NC, Glasheen C, Saccone NL, Page GP, Hulse GK, Wildenauer D, Kelty EA, Schwab SG, Degenhardt L, Martin NG, Montgomery GW, Attia J, Holliday EG, McEvoy M, Scott RJ, Bierut LJ, Nelson EC, Kral AH, Johnson EO. Cis-expression quantitative trait loci mapping reveals replicable associations with heroin addiction in OPRM1. Biol Psychiatry. 2015;78:474–484. doi: 10.1016/j.biopsych.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson AC, Cippitelli A, Sommer WH, Fedeli A, Bjork K, Soverchia L, Terasmaa A, Massi M, Heilig M, Ciccocioppo R. Variation at the rate Crhr1 locus and sensitivity to relapse into alcohol seeking induced by environmental stress. PNAS USA. 2006;103:15236–15241. doi: 10.1073/pnas.0604419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes DM, Knapp DJ, Breese GR, Thiele TE. Comparison of basal neuropeptide Y and corticotrophin releasing factor levels between the high ethanol drinking C57BL/6J and low ethanol drinking DBA/2J inbred mouse strains. Alcohol Clin Exp Res. 2005;29:721–729. doi: 10.1097/01.ALC.0000164375.16838.F3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Madden Af, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, Rohrbaugh JW, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- Heilig M, Widerlov E. Neurobiology and clinical aspects of neuropeptide Y. Critical Rev Neurobiol. 1995;9:115–136. [PubMed] [Google Scholar]
- Heinz A, Goldman D, Jones DW, Palmour R, Hommer D, Gorey JG, Lee KS, Linnoila M, Weinberger DR. Genotype influences in vivo dopamine transporter availability in human striatum. Neuropsychopharmacology. 2000;22:133–139. doi: 10.1016/S0893-133X(99)00099-8. [DOI] [PubMed] [Google Scholar]
- Hendershot CS, Claus ED, Ramchandani VA. Associations of OPRM1 A118G and alcohol sensitivity with intravenous alcohol self-administration in young adults. Addiction Biol. 2014;21:125–135. doi: 10.1111/adb.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend. 1999;57:69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- Hinckers AS, Laucht M, Schmidt MH, Mann KF, Schumann G, Schuckit MA, Heinz A. Low level of response to alcohol as associated with serotonin transporter genotype and high alcohol intake in adolescents. Biol Psychiatry. 2006;60:282–287. doi: 10.1016/j.biopsych.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Hopfer CJ, Stallings MC, Hewitt JK. Common genetic and environmental vulnerability for alcohol and tobacco use in a volunteer sample of older female twins. J Stud Alcohol. 2001;62:717–723. doi: 10.15288/jsa.2001.62.717. [DOI] [PubMed] [Google Scholar]
- Hu X, Oroszi G, Chun J, Smith TL, Goldman D, Schuckit MA. An expanded evaluation of the relationship of four alleles to the level of response to alcohol and the alcoholism risk. Alcohol Clin Exp Res. 2005;29:8–16. doi: 10.1097/01.alc.0000150008.68473.62. [DOI] [PubMed] [Google Scholar]
- Huang M-C, Ho C-W, Chen C-H, Liu S-C, Chen C-C, Leu S-J. Reduced expression of circadian clock genes in male alcoholic patients. Alcohol Clin Exp Res. 2010;34:1899–1904. doi: 10.1111/j.1530-0277.2010.01278.x. [DOI] [PubMed] [Google Scholar]
- Hwang BH, Zhang JK, Ehlers CL, Lumeng L, Li TK. Innate differences of neuropeptide Y (NPY) in hypothalamic nuclei and central nucleus of the amygdale between selectively bred rats with high and low alcohol preferences. Alcohol Clin Exp Res. 1999;23:1023–1030. [PubMed] [Google Scholar]
- Ishiguro H, Saito T, Shibuya H, Toru M, Arinami T. Mutation and association analysis of the Fyn kinase gene with alcoholism and schizophrenia. Am J Med Jenet. 2000;96:716–720. [PubMed] [Google Scholar]
- Jaime L, Shafe S, Liang T, Wills D, Berg G, Ehlers C. Subjective response to alcohol and ADH polymorphisms in a select sample of young adult male East Indians and Africans in Trinidad and Tobago. J Stud Alcohol Drugs. 2014;75:827–838. doi: 10.15288/jsad.2014.75.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslyn G, Brush G, Robertson M, Smith TL, Kalmijn J, Schuckit M, White RL. Chromosome 15q25.1 genetic markers associated with level of response to alcohol in humans. PNAS. 2008;105:20368–20373. doi: 10.1073/pnas.0810970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslyn G, Ravindranathan A, Brush G, Schuckit M, White RL. Human variation in alcohol response is influenced by variation in neuronal signaling genes. Alcohol Clin Exp Res. 2010;34:800–812. doi: 10.1111/j.1530-0277.2010.01152.x. [DOI] [PubMed] [Google Scholar]
- Joslyn G, Wolf FW, Brush G, Wu L, Schuckit M, White RL. Glypican gene GPC5 participates in the behavioral response to alcohol: evidence from humans, mice and fruit flies. Genes Genome Genet. 2011;1:627–635. doi: 10.1534/g3.111.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalu N, Ramchandani VA, Marshall V, Scott D, Ferguson C, Cain G, Taylor R. Heritability of level of response and association with recent drinking history in non-alcohol dependent drinkers. Alcohol Clin Exp Res. 2012;36:1034–1041. doi: 10.1111/j.1530-0277.2011.01699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalu NN, Ramchandani VA, Marshall VJ, Scott DM, Taylor RE. Correlation of self-reported effects of alcohol (SRE) with recent drinking history in non-dependent drinkers. Alcohol Clin Exp Res. 2008;32:265A. doi: 10.1111/j.1530-0277.2011.01699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareken DA, Liang T, Wetherill L, Dzemidzic M, Bragulat V, Cox C, Talavage T, O’Connor SJ, Foroud T. A polymorphism in GABRA2 is associated with the medial frontal response to alcohol cues in an fMRI study. Alcohol Clin Exp Res. 2010;34:2169–2178. doi: 10.1111/j.1530-0277.2010.01293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovanen L, Saarikoski S, Haukka J, Pirkola S, Aromaa A, Lonnqvist J, Partonen T. Circadian clock gene polymorphisms in alcohol use disorders and alcohol consumption. Alcohol Alcohol. 2010;45:303–311. doi: 10.1093/alcalc/agq035. [DOI] [PubMed] [Google Scholar]
- Kuhanen J, Karvonen MK, Pesonen U, Koulu M, Tuomainen TP, Uusitupa MI, Salonen JZZZT. Neuropeptide Y polymorphism and alcohol consumption in middle-aged men. Am J Med Genet. 2000;93:117–121. doi: 10.1002/1096-8628(20000717)93:2<117::aid-ajmg7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- King A, Munisamy G, deWit H, Lin S. Attenuated cortisol response to alcohol in heavy social drinkers. Int J Psychophysiol. 2006;59:203–209. doi: 10.1016/j.ijpsycho.2005.10.008. [DOI] [PubMed] [Google Scholar]
- King AC, Byars JA. Alcohol-induced performance impairment in heavy episodic and light social drinkers. J Stud Alcohol. 2004;65:27–36. doi: 10.15288/jsa.2004.65.27. [DOI] [PubMed] [Google Scholar]
- King AC, deWit H, McNamara PJ, Cao D. Rewarding, stimulant, and sedative alcohol responses and relationship to future binge drinking. Arch Gen Psychiatry. 2011;68:389–399. doi: 10.1001/archgenpsychiatry.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AC, Hasin D, O’Connor SJ, McNamara PJ, Cao D. A prospective 5-year re-examination of alcohol response in heavy drinkers progressing in alcohol use disorder. Biol Psychiatry. 2016;79:489–498. doi: 10.1016/j.biopsych.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AC, McNamara PJ, Hasin DS, Cao D. Alcohol challenge responses predict future alcohol use disorder symptoms: a 6-year prospective study. Biol Psychiatry. 2014;75:798–806. doi: 10.1016/j.biopsych.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong EC, Allouche L, Chapot PA, Vranizan K, Moore MS, Heberlein U, Wolf FW. Ethanol-regulated genes that contribute to ethanol sensitivity and rapid tolerance in Drosophila. Alcohol Clin Exp Res. 2010;34:302–316. doi: 10.1111/j.1530-0277.2009.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. Am J Psychiatry. 2007;164:1149–1159. doi: 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpi ER, Kleingoor C, Kettenmann H, Seeburg PH. Benzodiazepine-induced motor impairment linked to point mutation in cellular GABAA receptor. Nature. 1993;36:356–359. doi: 10.1038/361356a0. [DOI] [PubMed] [Google Scholar]
- Kosobud AEK, Wetherill L, Plawecki MH, Kareken DA, Liang T, Nurnberger JL, Windisch K, Xuei X, Edenberg HJ, Foroud TM, O’Connor SJ. Adaptation of subjective responses to alcohol is affected by an interaction of GABRA2 genotype and recent drinking. Alcohol Clin Exp Res. 2015;39:1148–1157. doi: 10.1111/acer.12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Gelernter J, Anton RF, Arias AJ, Herman A, Zhao H, Burian L, Covault J. Association of markers in the 3′ region of the GluR5 kainate receptor subunit gene to alcohol dependence. Alcohol Clin Exp Res. 2009;33:925–030. doi: 10.1111/j.1530-0277.2009.00913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal JH, Petrakis IL, Krupitsky E, Schütz C, Trevisan L, D’Souza DC. NMDA receptor antagonism and the ethanol intoxication signal: from alcoholism risk to pharmacotherapy. Ann NY Acad Sci. 2003;1003:176–184. doi: 10.1196/annals.1300.010. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Staley J, Mason G, Petrakis IL, Kaufman J, Harris RA, Gelernter J, Lappalainen J. Gamma-aminobutyric acid type A receptors and alcoholism: intoxication, dependence, vulnerability, and treatment. Arch Gen Psychiatry. 2006;63:957–968. doi: 10.1001/archpsyc.63.9.957. [DOI] [PubMed] [Google Scholar]
- Kuo P, Kalsi G, Prescott C, Hodgkinson C, Goldman D, Alexander J, van den Oord EJ, Chen X, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Associations of glutamate decarboxylase genes with initial sensitivity and age-at-onset of alcohol dependence in the Irish affected sib pair study of alcohol dependence. Drug Alcohol Depend. 2009;101:80–87. doi: 10.1016/j.drugalcdep.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo P, Kalsi G, Prescott C, Hodgkinson C, Goldman D, van den Oord E, Alexander J, Jiang C, Sullivan P, Patterson G, Walsh D, Kendler K, Riley B. Association of ADH and ALDH genes with alcohol dependence in the Irish affected sib pair study of alcohol dependence (IASPSAD) sample. Alcohol Clin Exp Res. 2008;32:785–795. doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Mizuno K, Ohkuma S. Dopamine D1 receptor signaling system regulates ryanodine receptor expression in ethanol physical dependence. Alcohol Clin Exp Res. 2013;37:771–783. doi: 10.1111/acer.12036. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Kranzler HR, Malison R, Price LH, Van Dyck C, Rosenheck RA, Cramer J, Southwick S, Charney D, Krystal J, Gelernter J. A functional neuropeptide Y Leu7Pro polymorphism associated with alcohol dependence in a large population sample from the United States. Arch Gen Psychiatry. 2002;59:825–831. doi: 10.1001/archpsyc.59.9.825. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Pchemlina S, Taraskina A, Zvartau E, Somberg LK, Covault J, Kranzler HR, Krystal JH, Gelernter J. Association between alcoholism and γ-amino butyric acid α2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Lasek AW, Giorgetti F, Berger KH, Taylor S, Heberlein U. Lmo genes regulate behavioral responses to ethanol in Drosophila melanogaster and the mouse. Alcohol Clin Exp Res. 2011a;35:1600–1608. doi: 10.1111/j.1530-0277.2011.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek AW, Lim J, Kliethermes CL, Berger KH, Joslyn G, Brush G, Xue L, Robertson M, Moore MS, Vranizan K, Morris SW, Schuckit MA, White RL, Heberlein U. An evolutionary conserved role for anaplastic lymphoma kinase in behavioral responses to ethanol. PlosOne. 2011b;6:e22636–e22648. doi: 10.1371/journal.pone.0022636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman RF, Ralevski E, Limoncelli D, Pittman B, O’Malley SS, Petrakis IL. Relationships between impulsivity and subjective response in an IV ethanol paradigm. Psychopharmacology. 2014;231:2867–2876. doi: 10.1007/s00213-014-3458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMarquand D, Pihl RO, Benkelfat C. Serotonin and alcohol intake, abuse, and dependence: clinical evidence. Biol Psychiatry. 1994;36:326–337. doi: 10.1016/0006-3223(94)90630-0. [DOI] [PubMed] [Google Scholar]
- Levine AS, Morley JE. Neuropeptide Y: a potent inducer of consummatory behavior in rats. Peptides. 1984;5:1025–1029. doi: 10.1016/0196-9781(84)90165-7. [DOI] [PubMed] [Google Scholar]
- Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, Crews KM, Somes G, Williams NJ, Elston RC. A genome-wide search finds major susceptibility loci for nicotine dependence on chromosome 10 in African Americans. Am J Hum Genet. 2006;79:745–751. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh EW, Ball D. Role of the GABAAalpha 1, GABAAgamma2 receptor subunit gene cluster in drug response and the development of alcohol dependence. Neurochem Int. 2000;37:413–423. doi: 10.1016/s0197-0186(00)00054-1. [DOI] [PubMed] [Google Scholar]
- Mague SD, Blendy JA. OPRM1 SNP (A118G): involvement in disease development, treatment response, and animal models. Drug Alcohol Depend. 2010;108:172–182. doi: 10.1016/j.drugalcdep.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Tuggmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Tibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel PD, Bennett B, Beeson M, Gordon L, Johnson TE. Confirmation of quantitative trait loci for ethanol sensitivity in long-sleep and short-sleep mice. Genome Res. 1997;7:92–99. doi: 10.1101/gr.7.2.92. [DOI] [PubMed] [Google Scholar]
- Masur J, Oliveira de Souza ML, Zwicker AP. The excitatory effect of ethanol: absence in rats, no tolerance and increased sensitivity in mice. Pharmacol Biochem Behav. 1986;24:1225–1228. doi: 10.1016/0091-3057(86)90175-9. [DOI] [PubMed] [Google Scholar]
- McCarthy D, Pedersen S, Lobos E, Todd R, Wall T. ADH1B*3 and response to alcohol in African-Americans. Alcohol Clin Exp Res. 2010;34:1274–1281. doi: 10.1111/j.1530-0277.2010.01205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17(R2):R156–R165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T, Yagi T, Kitazawa H, Yasuda M, Kawai N, Tsuboi, Niki H. Fyn-kinase as a determinant of ethanol sensitivity: relation to NMDA-receptor function. Science. 1997;278:698–701. doi: 10.1126/science.278.5338.698. [DOI] [PubMed] [Google Scholar]
- Monteiro MG, Klein JL, Schuckit MA. High levels of sensitivity to alcohol in young adult Jewish men: a pilot study. J Stud Alcohol. 1991;52:464–469. doi: 10.15288/jsa.1991.52.464. [DOI] [PubMed] [Google Scholar]
- Morzorati SL, Ramshandani VA, Glury L, O’Connors Self-reported subjective perception of intoxication reflects family history of alcoholism when breath alcohol levels are constant. Alcohol Clin Exp Res. 2002;26:1299–1306. doi: 10.1097/01.ALC.0000025886.41927.83. [DOI] [PubMed] [Google Scholar]
- Newlin DB, Renton RM. High risk groups often have higher levels of alcohol response than low risk: the other side of the coin. Alcohol Clin Exp Res. 2010;34:199–202. doi: 10.1111/j.1530-0277.2009.01081.x. [DOI] [PubMed] [Google Scholar]
- Olfson E, Edenberg HJ, Nurnberger J, Jr, Agrawal A, Bucholz KK, Almasy LA, Chorlian D, Dick DM, Hesselbrock VM, Kramer JR, Kuperman S, Porjesz B, Schuckit MA, Tischfield JA, Wang J-C, Wetherill L, Foroud TM, Rice J, Goate A, Bierut L. An ADH1B variant and peer drinking in progression to adolescent drinking milestones: evidence of a gene-by-environment interaction. Alcohol Clin Exp Res. 2014;38:2541–2549. doi: 10.1111/acer.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto JM, Gizer IR, Deak JD, Fleming KA, Bartholow BD. A cis-eQTL in OPRM1 is associated with subjective response to alcohol and alcohol use. Alcohol Clin Exp Res. 2017;41:929–938. doi: 10.1111/acer.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandy SC, Piano M, Schwertz DW, David JM, Pandey GN. Effect of ethanol dependence and withdrawal on serotonin2 receptors and phosphoinositide system in rat brain. Alcohol Clin Exp Res. 1992;16:392. doi: 10.1111/j.1530-0277.1992.tb00706.x. [DOI] [PubMed] [Google Scholar]
- Parsian A, Zhang ZH. Human dopamine transporter gene polymorphism (VNTR) and alcoholism. Am J Med Genet Part A. 1997;74:480–482. doi: 10.1002/(sici)1096-8628(19970919)74:5<480::aid-ajmg4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Paulus MP, Schuckit MA, Tapert SF, Tolentino NJ, Matthews SC, Smith TL, Trim RS, Hall SA, Simmons AN. High versus low level of response to alcohol: evidence of differential reactivity to emotional stimuli. Biol Psychiatry. 2012;72:848–855. doi: 10.1016/j.biopsych.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau-Lenz S, Zghoul T, de Fonseca FR, Spanagel R, Bilbao A. Circadian regulation of central ethanol sensitivity by the mPer2 gene. Addict Biol. 2009;14:253–259. doi: 10.1111/j.1369-1600.2009.00165.x. [DOI] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Covault J, Feinn R, Nellissery M, Hernandez-Avila C, Oncken C, Morrow AL, Kranzler HR. GABRA2 alleles moderate the subjective effects of alcohol, which are attenuated by finasteride. Neuropsychopharmacology. 2005;30:1193–1203. doi: 10.1038/sj.npp.1300688. [DOI] [PubMed] [Google Scholar]
- Quinn PD, Fromme K. Subjective response to alcohol challenge: a quantitative review. Alcohol Clin Exp Res. 2011;35:1759–1770. doi: 10.1111/j.1530-0277.2011.01521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandani VA, Bolane J, Li T-K, O’Connor S. A physiologically-based pharmacokinetic (PBPK) model for alcohol facilitates rapid BrAC clamping. Alcohol Clin Exp Res. 1999;23:617–623. [PubMed] [Google Scholar]
- Ramchandani VA, Umhau J, Pavon FJ, Ruiz-Velasco V, Margas W, Sun H, Damadzic R, Eskay R, Schoor M, Thorsell A, Schwandt ML, Sommer WH, George DT, Parsons LH, Herscovitch P, Hommer D, Heilig M. A genetic determinant of the striatal dopamine response to alcohol in men. Mol Psychiatry. 2011;16:809–817. doi: 10.1038/mp.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch JL, Monteiro MG, Schuckit MA. Platelet serotonin uptake in men with family histories of alcoholism. Neuropsychopharmacology. 1991;4:83–86. [PubMed] [Google Scholar]
- Ray LA, Barr CS, Blendy JA, Oslin D, Goldman D, Anton RF. The role of the Asn40Asp polymorphism of the mu opioid receptor gene (OPRM1) on alcoholism etiology and treatment: a critical review. Alcohol Clin Exp Res. 2012;36:385–394. doi: 10.1111/j.1530-0277.2011.01633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Bujarski S, Roche DJO. Subjective response to Alcohol as a Research Domaine Criterion. Alcohol Clin Exp Res. 2016;40:6–17. doi: 10.1111/acer.12927. [DOI] [PubMed] [Google Scholar]
- Ray LA, Courtney KE, Hutchison KE, Mackillop J, Galvan A, Ghanhremani DG. Initial evidence that OPRM1 genotype moderates ventral and dorsal striatum functional connectivity during alcohol cues. Alcohol Clin Exp Res. 2014;38:78–89. doi: 10.1111/acer.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Hart EJ, Chin PF. Self-Rating of the Effects of Alcohol (SRE): predictive utility and reliability across interview and self-report administrations. Addict Behav. 2011;36:241–243. doi: 10.1016/j.addbeh.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. A polymorphism of the μ-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28:1789–1795. doi: 10.1097/01.alc.0000148114.34000.b9. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. Associations among GABRG1, level of response to alcohol, and drinking behaviors. Alcohol Clin Exp Res. 2009;33:1382–1390. doi: 10.1111/j.1530-0277.2009.00968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MT, Noronha A, Goldman D, Koob GF. Genetic studies of alcohol dependence in the context of the addiction cycle. Neuropharmacology. 2017;122:3–21. doi: 10.1016/j.neuropharm.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BP, Kalsi G, Kuo P-H, Vladimirov V, Thiselton DL, Vittum J, Wormley B, Grotewiel MS, Patterson DG, Sullivan PF, van den Oord E, Walsh D, Kendler KS, Prescott CA. Alcohol dependence is associated with the ZNF699 gene, a human locus related to Drosophila hangover, in the Irish affected sib pair study of alcohol dependence (IASPSAD) sample. Mol Psychiatry. 2006;11:1025–1031. doi: 10.1038/sj.mp.4001891. [DOI] [PubMed] [Google Scholar]
- Roche DJO, Palmeir MD, King AC. Acute alcohol response phenotype in heavy social drinkers is robust and reproducible. Alcohol Clin Exp Res. 2014;38:844–852. doi: 10.1111/acer.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh S, Matsushita S, Hara S, Maesato H, Matsui T, Suzuki G, Miyakawa T, Ramchandani VA, Li T-K, Higuchi S. Role of GABRA2 in moderating subjective responses to alcohol. Alcohol Clin Exp Res. 2011;35:400–407. doi: 10.1111/j.1530-0277.2010.01357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueger SY, King AC. Validation of the Brief Biphasic Alcohol Effects Scale (B-BAES) Alcohol Clin Exp Res. 2013;37:470–476. doi: 10.1111/j.1530-0277.2012.01941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander T, Ball D, Murray R, Patel J, Samochowiec J, Winterer G, Rommelspacher H, Schmidt LG. Association analysis of sequence variants of the GABAA alpha6, beta2, and gamma2 gene cluster and alcohol dependence. Alcohol Clin Exp Res. 1999;23:427–431. [PubMed] [Google Scholar]
- Sartor CE, Wang Z, Xu K, Kranzler HR, Gelernter J. The joint effects of ADH1B variants and childhood adversity on alcohol related phenotypes in African-American and Eorpean-American women and men. Alcohol Clin Exp Res. 2015;38:2907–2914. doi: 10.1111/acer.12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders S, Paine-Saunders S, Lander AD. Expression of the cell surface proteoglycan glypican-5 is developmentally regulated in kidney, limb, and brain. Dev Biol. 1997;190:78–93. doi: 10.1006/dbio.1997.8690. [DOI] [PubMed] [Google Scholar]
- Savage JE, Neale Z, CVho SB, Hancock L, Kalmijn JA, Smith TL, Schuckit MA, Donovan KK, Dick DM. Level of response to alcohol as a factor for targeted prevention in college students. Alcohol Clin Exp Res. 2015;39:2215–2223. doi: 10.1111/acer.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz H, Franz M, Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. doi: 10.1038/nature03864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. Biological, psychological and environmental predictors of the alcoholism risk: a longitudinal study. J Stud Alcohol. 1998;59:485–494. doi: 10.15288/jsa.1998.59.485. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. A brief history of research on the genetics of alcohol and other drug use disorders. J Stud Alcohol Drugs Suppl. 2014;17:59–67. doi: 10.15288/jsads.2014.s17.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Edenberg HJ, Kalmijn J, Flury L, Smith TL, Reich T, Bierut L, Goate A, Foroud T. A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol Clin Exp Res. 2001;25:323–329. [PubMed] [Google Scholar]
- Schuckit MA, Gold EO. A simultaneous evaluation of multiple markers of ethanol/placebo challenges in sons of alcoholics and controls. Arch Gen Psychiatry. 1988;45:211–216. doi: 10.1001/archpsyc.1988.01800270019002. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Mazanti C, Smith TL, Ahmed U, Radel M, Iwata N, Goldman D. Selective genotyping for the role of 5-HT2A, 5-HT2C, and GABAα6 receptors and the serotonin transporter in the level of response to alcohol: a pilot study. Biol Psychiatry. 1999;45:647–651. doi: 10.1016/s0006-3223(98)00248-0. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Rayses V. Ethanol ingestion: differences in blood acetaldehyde concentrations in relatives of alcoholics and controls. Science. 1978;203:54–55. doi: 10.1126/science.758678. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. An 8-year follow-up of 450 sons of alcoholic and control subjects. Arch Gen Psychiatry. 1996;53:202–210. doi: 10.1001/archpsyc.1996.01830030020005. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. Mediation of effects of the level of response to alcohol and impulsivity 15 years later in 36-year-old men: implications for prevention efforts. Drug Alcohol Depend. 2017;180:356–362. doi: 10.1016/j.drugalcdep.2017.08.030. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Beltran I, Waylen A, Horwood J, Davis JM The ALSPAC Study Team. Performance of a self-report measure of the level of response to alcohol in 12- to 13-year-old adolescents. J Stud Alcohol. 2005;66:452–458. doi: 10.15288/jsa.2005.66.452. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Clausen P, Fromme K, Skidmore J, Shafir A, Kalmijn J. The low level of response to alcohol-based heavy drinking prevention program: one-year follow-up. J Stud Alcohol Drugs. 2016a;77:25–37. doi: 10.15288/jsad.2016.77.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Danko G, Anthenelli R, Schoen L, Kawamura M, Kramer J, Dick DM, Neale Z, Kuperman S, McCutcheon V, Anokhin AP, Hesselbrock V, Hesselbrock M, Bucholz K. A prospective comparison of how the level of response to alcohol and impulsivity relate to future DSM-IV alcohol problems in the COGA youth panel. Alcohol Clin Exp Res. 2017b;41:1329–1339. doi: 10.1111/acer.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Danko GP, Pierson J, Hesselbrock V, Bucholz KK, Kramer J, Kuperman S, Dietiker C, Brandon R, Chan G. The ability of the Self-Rating of the Effects of Alcohol (SRE) Scale to predict alcohol-related outcomes five years later. J Stud Alcohol Drugs. 2007;68:371–378. doi: 10.15288/jsad.2007.68.371. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Kalmijn JA. The patterns of drug and alcohol use and associated problems over 30 years in 397 men. Alcohol Clin Exp Res. 2014;38:227–234. doi: 10.1111/acer.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Kalmijn JA, Danko GP, Buckman KR, Eng MY. Rapid vs. slow drinking: the effect on the level of response to alcohol. Alcohol Clin Exp Res. 2002;26(Suppl):159A. [Google Scholar]
- Schuckit MA, Smith TL, Kalmijn J, Tsuang J, Hesselbrock V, Bucholz K. Response to alcohol in daughters of alcoholics: a pilot study and a comparison with sons of alcoholics. Alcohol Alcohol. 2000;35:242–248. doi: 10.1093/alcalc/35.3.242. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Paulus MP, Tapert SF, Simmons AN, Tolentino NJ, Shafir A. The ability of functional magnetic resonance imaging to predict heavy drinking and alcohol problems 5 years later. Alcohol Clin Exp Res. 2016b;40:206–213. doi: 10.1111/acer.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Shafir A, Clausen P, Danko G, Gonçalves PD, Anthenelli RM, Chan G, Kuperman S, Hesselbrock M, Hesselbrock V, Kramer J, Bucholz KK. Predictors of patterns of alcohol-related blackouts over time in youth from the Collaborative Study of the Genetics of Alcoholism: the roles of genetics and cannabis. J Stud Alcohol Drugs. 2017a;78:39–48. doi: 10.15288/jsad.2017.78.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Tipp JE. The self-rating of the effects of alcohol (SRE) form as a retrospective measure of the risk for alcoholism. Addiction. 1997;92:979–988. [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Trim R, Fukukura T, Allen R. The overlap in predicting alcohol outcome for two measures of the level of response to alcohol. Alcohol Clin Exp Res. 2009;33:563–569. doi: 10.1111/j.1530-0277.2008.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Trim RS, Heron J, Horwood J, David J, Hibbeln J ALSPAC Study Team. The self-rating of the effects of alcohol questionnaire as a predictor of alcohol-related outcomes in 12-year-old subjects. Alcohol Alcohol. 2008;43:641–646. doi: 10.1093/alcalc/agn077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Johann M, Frank J, Preuss U, Dahmen N, Laucht M, Rietschel M, Rujescu D, Lourdusamy A, Clarke T-K, Krause K, Dyer A, Depner M, Wellek S, Treutlein J, Szegedi A, Giegling I, Cichon S, Blomeyer D, Heinz A, Heath S, Lathrop M, Wodarz N, Soyka M, Spanagel R, Mann K. Systematic analysis of glutamatergic neurotransmission genes in alcohol dependence and adolescent risky drinking behavior. Arch Gen Psychiatry. 2008;65:826–838. doi: 10.1001/archpsyc.65.7.826. [DOI] [PubMed] [Google Scholar]
- Schumann G, Rujescu D, Kissling C, Soyka M, Dahmen N, Preuss UW, Wieman S, Depner M, Wellek S, Lascorz J, Bondy B, Giegling I, Anghelescu I, Cowen MS, Poustka A, Spoanagel R, Mann K, Henn FA, Szegedi A. Analysis of genetic variations of protein tyrosine kinase fyn and their association with alcohol dependence in two independent cohorts. Biol Psychiatry. 2003;54:1422–1426. doi: 10.1016/s0006-3223(03)00635-8. [DOI] [PubMed] [Google Scholar]
- Schwantes-An T-H, Zhang J, Chen L-S, Hartz SM, Culverhouse RC, Chen X, Coon H, Frank J, Kamens HM, Konte B, Kovanen L, Latvala A, Legrand LN, Maher BS, Melroy WE, Nelson EC, Reid MW, Robinson JD, Shen PH, Yang BZ, Andrews JA, Aveyard P, Beltcheva O, Brown SA, Cannon DS, Cichon S, Corley RP, Dahmen N, Degenhardt L, Foroud T, Gaebel W, Giegling I, Glatt SJ, Grucza RA, Hardin J, Hartmann AM, Heath AC, Herms S, Hodgkinson CA, Hoffmann P, Hops H, Huizinga D, Ising M, Johnson EO, Johnstone E, Kaneva RP, Kendler KS, Kiefer F, Kranzler HR, Krauter KS, Levran O, Lucae S, Lynskey MT, Maier W, Mann K, Martin NG, Mattheisen M, Montgomery GW, Müller-Myhsok B, Murphy MF, Neale MC, Nikolov MA, Nishita D, Nöthen MM, Nurnberger J, Partonen T, Pergadia ML, Reynolds M, Ridinger M, Rose RJ, Rouvinen-Lagerström N, Scherbaum N, Schmäl C, Soyka M, Stallings MC, Steffens M, Treutlein J, Tsuang M, Wall TL, Wodarz N, Yuferov V, Zill P, Bergen AW, Chen J, Cinciripini PM, Edenberg HJ, Ehringer MA, Ferrell RE, Gelernter J, Goldman D, Hewitt JK, Hopfer CJ, Iacono WG, Kaprio J, Kreek MJ, Kremensky IM, Madden PA, McGue M, Munafò MR, Philibert RA, Rietschel M, Roy A, Rujescu D, Saarikoski ST, Swan GE, Todorov AA, Vanyukov MM, Weiss RB, Bierut LJ, Saccone NL. Association of the OPRM1 variant rs1799971 (A118G) with non-specific liability to substance dependence in a collaborative de novo meta-analysis of European-ancestry cohorts. Behav Genet. 2016;46:151–169. doi: 10.1007/s10519-015-9737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, Anton RF, Oslin D, Farrer LA, Gelernter J. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Segura-Sanchise C, Magnone M, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G, Albrecht U. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nature Med. 2005;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- Steensland P, Simms JA, Holgate J, Richards JK, Bartlett SE. Varenicline, an alpha4beta2 nicotinic acetylcholine receptor partial agonist, selectively decreases ethanol consumption and seeking. PNAS USA. 2007;104:12518–12523. doi: 10.1073/pnas.0705368104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Cardon LR. Heavy consumption of cigarettes, alcohol and coffee in male twins. J Stud Alcohol. 1997;58:182–190. doi: 10.15288/jsa.1997.58.182. [DOI] [PubMed] [Google Scholar]
- Tapert SF, Pulido C, Paulus MP, Schuckit MA, Burke C. Level of response to alcohol and brain response during visual working memory. J Stud Alcohol. 2004;65:692–700. doi: 10.15288/jsa.2004.65.692. [DOI] [PubMed] [Google Scholar]
- Tecott LH, Heberlein U. Y do we drink? Cell. 1998;95:733–735. doi: 10.1016/s0092-8674(00)81695-5. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Miura GI, Marsh DJ, Bernstein IL, Palmiter RD. Neurobiological responses to ethanol in mutant mice lacking neuropeptide Y or the Y5 receptor. Pharmacol, Biochem Behav. 2000;67:683–691. doi: 10.1016/s0091-3057(00)00413-5. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Kissling C, Frank J, Wiemann S, Dong L, Depner M, Saam C, Lascorz J, Soyka M, Preuss UW, Rujescu D, Skowronek MH, Sanagel R, Heinz A, Laucht M, Mann K, Schumann G. Genetic association of the human corticotrophin release hormone receptor 1 (CRHR1) with binge drinking and alcohol intake patterns in two independent samples. Mol Psychiatry. 2006;11:594–602. doi: 10.1038/sj.mp.4001813. [DOI] [PubMed] [Google Scholar]
- Uhart M, Weerts EM, McCaul ME, Guo X, Yan X, Kranzler HR, Li N, Wand GS. GABRA2 markers moderate the subjective effects of alcohol. Addict Biol. 2012;18:357–369. doi: 10.1111/j.1369-1600.2012.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zwaluw CS, Engels RC, Buitelaar J, Verkes RJ, Franke B, Scholte RH. Polymorphisms in the dopamine transporter gene (SLC6A3/DAT1) and alcohol dependence in humans: a systematic review. Pharmacogenomics. 2009;10:853–866. doi: 10.2217/pgs.09.24. [DOI] [PubMed] [Google Scholar]
- van der Zwaluw CS, van den Wildenberg E, Wiers RW, Franke B, Buitelaar J, Scholte RHJ, Engels RCME. Polymorphisms in the mu-opioid receptor gene (OPRM1) and the implications for alcohol dependence in humans. Pharmacogenomics. 2007;8:1427–1436. doi: 10.2217/14622416.8.10.1427. [DOI] [PubMed] [Google Scholar]
- Viken RJ, Rose RJ, Morzorati SL, Christian JC, Li TK. Subjective intoxication in response to alcohol challenge: heritability and covariation with personality, breath alcohol level, and drinking history. Alcohol Clin Exp Res. 2003;27:795–803. doi: 10.1097/01.ALC.0000067974.41160.95. [DOI] [PubMed] [Google Scholar]
- Walker BM, Koob GF. Pharmacological evidence for a motivational role ofkapp-opioid systems in ethanol dependence. Neuropsychopharmacology. 2008;33:643–652. doi: 10.1038/sj.npp.1301438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall T, Shea S, Luczak S, Cook T, Carr L. Genetic associations of alcohol dehydrogenase with alcohol use disorders and endophenotypes in white college students. J Abnorm Psychol. 2005;114:456–465. doi: 10.1037/0021-843X.114.3.456. [DOI] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, Fox L, Goldstein E, Reyes O, Saccone N, Saccone S, Xuei X, Bucholz K, Kuperman S, Nurnberger J, Jr, Rice JP, Schuckit M, Tischfield J, Hesselbrock V, Porjesz B, Edenberg HJ, Bierut LJ, Goate AM. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14:501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- Wang X, Mozhul K, Li Z, Mulligan MK, Ingels JF, Zhou X, Hori RT, Chen H, Cook MN, Williams RW, Lu L. A promoter polymorphism in the Per3 gene is associated with alcohol and stress response. Transl Psychiatry. 2012;2:e73. doi: 10.1038/tp.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZW, Saifee O, Nonet ML, Salkoff L. SLO-1 potassium channels control quantal content of neurotransmitter release at the C elegans neuromuscular junction. Neuron. 2001;32:867–881. doi: 10.1016/s0896-6273(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Webb A, Lind PA, Kalmijn J, Feiler HS, Smith TL, Schuckit MA, Wilhelmsen K. The investigation into CYP2E1 in relation to the level of response to alcohol through a combination of linkage and association analysis. Alcohol Clin Exp Res. 2011;35:10–18. doi: 10.1111/j.1530-0277.2010.01317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerts EM, McCaul ME, Kuwabara H, Yang X, Xu X, Dannals RF, Frost JJ, Wong DF, Wand GS. Influence of OPRM1 Asn40Asp variant (A118G) on [11C]carfentanil binding potential: preliminary findings in human subjects. Int J Neuropsychopharmacol. 2013;16:47–53. doi: 10.1017/S146114571200017X. [DOI] [PubMed] [Google Scholar]
- Weerts EM, Wand GS, Maher B, Xu X, Stephens MA, Yang X, McCaul ME. Independent and interactive effects of OPRM1 and DAT1 polymorphisms on alcohol consumption and subjective responses in social drinkers. Alcohol Clin Exp Res. 2017;41:1093–1104. doi: 10.1111/acer.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J, Symons MSS. Ethanol-responsive genes (Crtam, Zbtb16, and Mobp) located in the alcohol –QTL region of chromosome 9 are associated with alcohol preference in mice. Alcohol Clin Exp Res. 2000;334:1409–1416. doi: 10.1111/j.1530-0277.2009.00971.x. [DOI] [PubMed] [Google Scholar]
- Wetherell L. Results of a GWAS regarding SRE measures of LR. Presented at the Collaborative Study of the Genetics of Alcoholism (COGA) annual spring meeting; March 29, 2017; Washington, D.C. 2017. [Google Scholar]
- Wilhelmsen KC, Schuckit M, Smith TL, Lee JV, Segall SK, Feiler HS, Kalmijn J. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27:1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, York TP, Williams RW, Miles MF. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. Plos One. 2012;7:e33575. doi: 10.1371/journal.pone.0033575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Wu Z, Ma D, Tang C, Liu L, Xin F, Zhu D, Hu J. Association of single-nucleotide polymorphisms in a metabotropic glutamate receptor GRM3 gene subunit to alcohol-dependent male subjects. Alcohol Alcohol. 2014;49:256–260. doi: 10.1093/alcalc/agu004. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Zhu G, Hariri AR, Enoch M-A, Scott D, Sinha R, Virkkunen M, Mash DC, Lipsky RH, Hu X-Z, Hodgkinson CA, Xu K, Buzas B, Yan Q, Shen P-H, Ferrell RE, Manuck SB, Brown SM, Hauger RL, Stohler CS, Zubieta J-K, Goldman D. Genetic variation in human NPY expression affects stress response and emotion. Nature. 2008;452:997–1001. doi: 10.1038/nature06858. [DOI] [PMC free article] [PubMed] [Google Scholar]