Abstract
Muscle wasting is a decline in skeletal muscle mass and function that is associated with aging, obesity, and a spectrum of pathologies including cancer. Cancer-associated wasting not only reduces quality of life, but also directly impacts cancer mortality, chemotherapeutic efficacy, and surgical outcomes. There is an incomplete understanding of the role of tumor-derived factors in muscle wasting and sparse knowledge of how these factors impact in vivo muscle regeneration. Here, we identify several cytokines/chemokines that negatively impact in vitro myogenic differentiation. We show that one of these cytokines, CXCL1, potently antagonizes in vivo muscle regeneration and interferes with in vivo muscle satellite cell homeostasis. Strikingly, CXCL1 triggers a robust and specific neutrophil/M2 macrophage response that likely underlies or exacerbates muscle repair/regeneration defects. Taken together, these data highlight the pleiotropic nature of a novel tumor-derived cytokine and underscore the importance of cytokines in muscle progenitor cell regulation.
Keywords: lung cancer, satellite cells, myogenesis, regeneration, immune cells, cytokines
INTRODUCTION
Skeletal muscle maintenance requires balancing opposing forces of muscle breakdown and muscle growth. In healthy individuals, muscle breakdown occurs continuously as a consequence of acute muscle injury, exercise, and daily activity. This breakdown is counteracted by continuous myofiber repair and regeneration. In the context of muscle wasting, however, disease-associated and/or host-derived factors interfere with both muscle breakdown and growth/regeneration processes, ultimately leading to the loss of muscle mass and function. Muscle wasting is a serious concern when present in cancer patients, where the degree of skeletal muscle atrophy is tightly linked to cancer mortality, morbidity, therapeutic prognoses, and quality of life (Tisdale, 2002). Although the molecular basis of cancer-associated muscle wasting is unclear, several key mediators are known. Pro-inflammatory cytokines, including IL-1β, IL-6, TNF-α, and IFN-γ, are produced by host immune cells in response to the tumors and/or by the tumor itself and can directly impact skeletal muscle biology in several ways (Argiles et al., 2003) (Fearon et al., 2012). One way is by suppression of protein synthesis pathways (Emery et al., 1984). This occurs through either reduced amino acid uptake or by direct suppression of mRNA transcription and/or translation. Second, wasting may also occur due to enhanced or accelerated protein breakdown. Indeed, the ubiquitin/proteasome system is one of the most rigorously studied processes associated with muscle wasting, with regulation of ubiquitin, 26S proteasome subunits, and muscle-specific E3 ligases all implicated as mediators or drivers of muscle protein breakdown (Baracos et al., 1995; Lecker et al., 1999; Mitch and Goldberg, 1996). Importantly, while much attention has focused on muscle protein disequilibrium in mature muscle/myofibers, fewer studies have examined how muscle regeneration is impaired in the wasting state (Talbert and Guttridge, 2016).
Skeletal muscle regeneration is linked to a pool of muscle stem cells, termed satellite cells (SCs), that activates, expands, and self-renews to repair and maintain tissue mass (Dumont and Rudnicki, 2017). Loss or functional suppression of this adult stem cell compartment is implicated in diverse muscle pathologies including age-associated sarcopenia (Sousa-Victor and Munoz-Canoves, 2016), some muscular dystrophies (Blau et al., 1983; Sacco et al., 2010), and cancer-associated cachexia (He et al., 2013; Marchildon et al., 2015). Furthermore, SC dysfunction/impaired muscle regeneration is observed in multiple muscle atrophy contexts including burn injury (Corrick et al., 2015; Fry et al., 2016), chronic kidney disease (Zhang et al., 2010), diabetes (Aragno et al., 2004; Krause et al., 2011), and disuse atrophy (Mozdziak et al., 1998). Therapies that boost SC regenerative capacity can improve gross muscle mass decline and rescue functional deficits, suggesting that the SC pool is a viable therapeutic target to limit muscle atrophy. In this study, we identified secreted, tumor-derived cytokines able to suppress myogenesis. In vivo evaluation of CXCL1 – a prominent tumor derived factor not previously implicated in muscle wasting – revealed impaired muscle regeneration and satellite cell fusion, defects that may, in part, be attributable to aberrant CXCL1-mediated immune cell regulation. Taken together, our data underscore the complexity of cancer-associated muscle wasting and highlight the utility of using both cell culture and animal models to identify and evaluate novel muscle wasting factors.
RESULTS
Tumor-derived factors interfere with myoblast homeostasis and differentiation
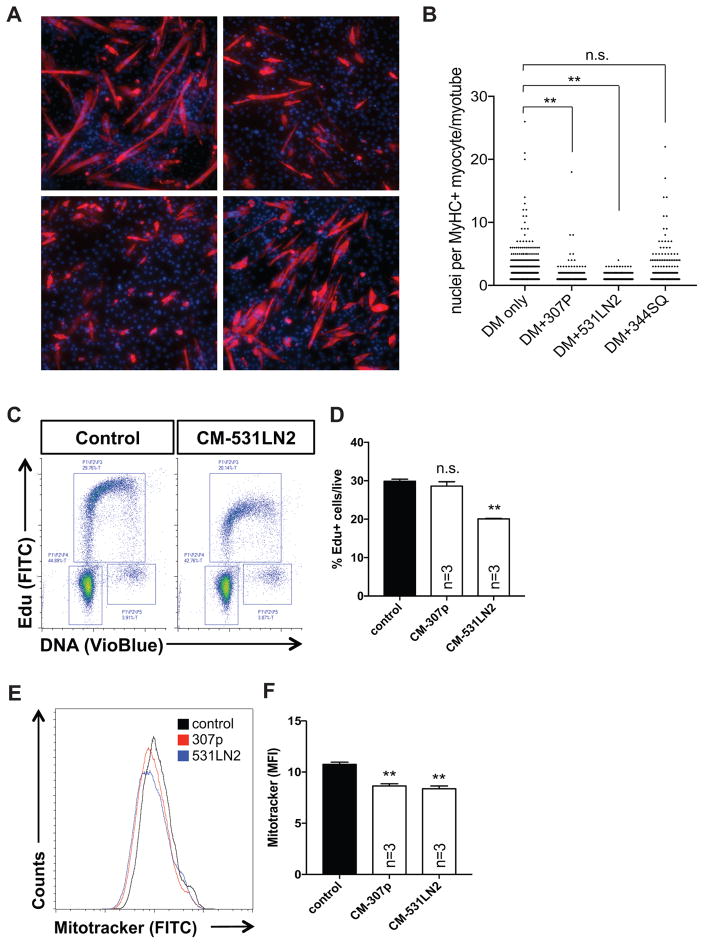
We used an established C2C12 myoblast differentiation model to screen a panel of lung cancer cell line supernatants for the ability to impair myogenic differentiation. In vitro myotube formation was assessed by exposing differentiating myoblasts to conditioned media from exponentially growing KrasLA1/+;p53R172H/Δg/+ lung adenocarcinoma cells and quantifying myotube maturation after 4 days in differentiation/conditioned media. C2C12 myoblasts cultured in the presence of control media exhibited extensive myogenic differentiation, as evidenced by the formation of Myosin Heavy Chain (MyHC)-positive, multi-nucleated myotubes. In contrast, C2C12 cells cultured using conditioned media (CM) from 307P or 531LN2 lung cancer cells formed fewer MyHC+ myotubes (control, 307P, 531LN2 mean values=2.56, 1.40, 1.15 respectively) that failed to mature into elongated, multi-nucleated structures (Figure 1A,B). Importantly, we did not observe statistically significant myogenic suppression with all lung adenocarcinoma cell lines tested (344SQ, Figure 1A,B), indicating that CM from 307P and 531LN2 lung cancer cells contains factors that actively suppress differentiation as opposed to simply depleting essential nutrients from base media. To determine whether this differentiation block was associated with defects in cell cycle progression, we performed flow cytometry based cell cycle/S-phase transit assays using 5-ethynyl-2′-deoxyuridine (EdU). We observed a significant decrease in S-phase (EdU) incorporation in 531LN2-CM C2C12 cultures (~28% in control compared to ~20% 531LN2-CM) whereas we observed slight, but insignificant deficits in 307P-CM C2C12 cultures (Figure 1C,D). Cultured primary myoblasts similarly treated with either 531LN2 or 307P CM also exhibited minor reductions in Edu incorporation compared to control cells, whereas we observed statistically significant reductions in myogenin (a marker of late myogenic differentiation) expressing cells, compared to control myoblasts (Supplementary Figure S1). To evaluate tumor-induced changes to mitochondria biogenesis in muscle cells, we measured mitochondria abundance using a Mitotracker Green dye in our lung cancer (LC)-CM C2C12 cultures. We labeled mitochondria in C2C12 cells cultured in the presence of LC-CM for 24 hours and found a ~15–20% decrease in Mitotracker Green mean fluorescence intensity (MFI), indicating a decrease in mitochondria mass in the presence of 307P and 531LN2 conditioned media (Figure 1E,F). Taken together, these data show that secreted factors in LC-CM can potently block myogenic differentiation and that these changes may, in part, result from cell line- (or cancer type-) specific defects in cell cycle progression or alterations in mitochondria biogenesis.
Figure 1. Tumor-derived secreted factors interfere with myogenic differentiation.
(A) Representative images of C2C12 cells cultured in the presence of conditioned differentiation media from 307p, 531LN2, and 344SQ lung adenocarcinoma cell lines. Red depicts myosin heavy chain positive myocytes and maturing myotubes. Nuclei are shown in blue. Scale bar=100um. (B) Graph depicting the number of nuclei per myosin heavy chain positive structure under each experimental condition. **p<0.01 using unpaired, nonparametric, Mann-Whitney t-tests. (C) Representative flow cytometry plots of control and 531LN2-conditioned C2C12 cells subjected to an Edu incorporation/cell cycle assay. (D) Quantification of the percentage of Edu+ cells in control, 307p and 531LN2-conditioned C2C12 cells. *p<0.05, n.s.= not significant using two-tailed, unpaired t tests. (E) Representative histograms of Mitotracker fluorescence in control, 307p and 531LN2 conditioned C2C12 cells. (F) Quantification of the mean fluorescence intensity (MFI) of control, 307p and 531LN2-conditioned C2C12 cells. **p<0.01 using two-tailed, unpaired t tests.
Identification of tumor derived secreted cytokines
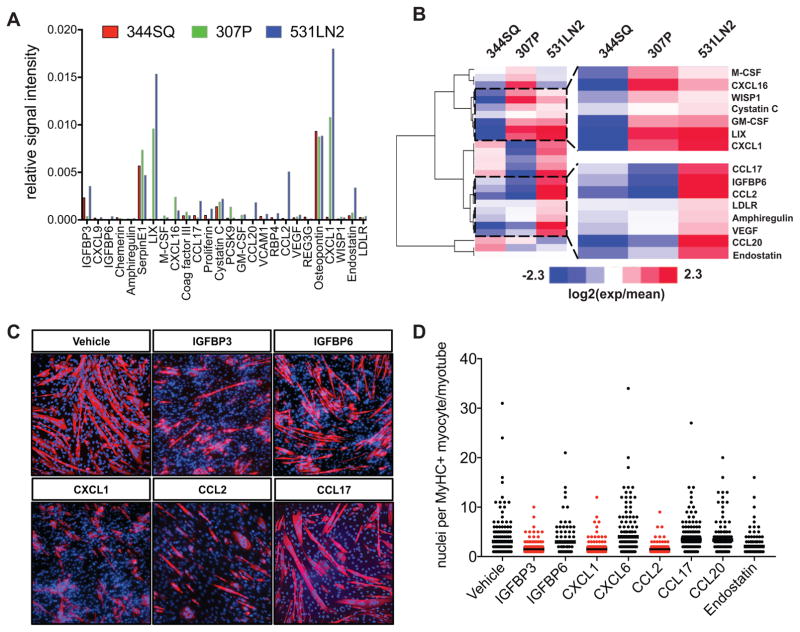
We next profiled over 100 cytokines/chemokines in 307P, 531LN2 and 344SQ lung cancer line supernatants using antibody arrays to identify secreted factors linked to suppression of myogenic differentiation. We were able to detect 26 proteins above background in at least one of three lung cancer line supernatants. Of these proteins, IGFBP3, LIX/CXCL6, SerpinE1, Osteopontin, and CXCL1/KC/GRO1 exhibited the highest relative signal intensities in our lung cancer supernatants (Figure 2A). We then performed hierarchical clustering analyses based on normalized log2 transformed relative expression values and found two cytokine clusters (344SQlow/307Plow/med/531LN2high and 344SQlow/307Pmed/high/531LN2high) that exhibited expression patterns consistent with the ability of these supernatants to suppress C2C12 differentiation (ref to Figure 2B). Based on these expression patterns, we then tested the hypothesis that individual recombinant cytokines can suppress myogenesis. Of the eight cytokines tested (IGFBP3, IGFBP6, CXCL1, CXCL6, CCL2, CCL17, CCL20, Endostatin), three – IGFBP3, CXCL1, and CCL2 – displayed a statistically significant ability to impair in vitro myogenesis, as evidenced by suppression of multinucleated MyHC+ myotube maturation (Figure 2C,D). Reverse phase protein array (RPPA) analyses of CXCL1-treated C2C12 cells revealed down-regulation of several signal transduction pathways linked to myogenic differentiation, including PI3K-AKT (Gardner et al., 2012), ErbB (Figeac et al., 2014; Golding et al., 2007), and HIF-1/AMPK (Fu et al., 2015) signaling pathways (Supplemental Figure S2). Importantly, while we did not observe suppressive activity with most cytokines tested, we cannot rule out the possibility that specific doses, lots, local concentrations, or cytokine combinations are required for these cytokines to impact myogenic differentiation. Taken together, these experiments revealed two cytokine clusters positively associated with suppression of myogenesis and provide evidence that several individual cytokines, including CXCL1, can directly impact myogenic differentiation.
Figure 2. Identification of tumor-derived secreted cytokines.
(A) Graph depicting the relative signal intensity values (a.u.) of the 26 cytokines/chemokines detected in supernatants from 307P, 531LN2, and 344SQ lung adenocarcinoma cell lines. (B) A heatmap depicting clustered, normalized, log2 transformed cytokine expression values. Blue=lower expression and red=higher expression. Areas highlighted with black dotted lines depict cytokine clusters with increased expression values across increasingly myogenesis-suppressing tumor lines. (C) Representative images of C2C12 cells cultured in the presence of differentiation media supplemented with selected recombinant cytokines at 80ng/ml. Red depicts myosin heavy chain positive myocytes and maturing myotubes. Nuclei are shown in blue. Scale bar=100um. (D) Graph depicting the number of nuclei per myosin heavy chain positive structure. Red=statistically significant (p<0.01) compared to control using unpaired, nonparametric, Mann-Whitney tests.
Tumor bearing mice exhibit weight loss and elevated CXCL1 levels
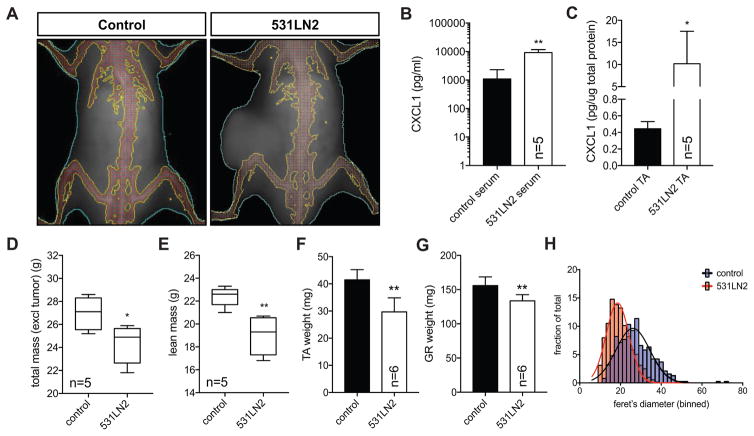
Given the ability of tumor-derived cytokines to suppress myogenic differentiation and the importance of muscle regeneration/repair in maintenance of muscle mass, we next asked 1) if we could detect elevated CXCL1, the most differentially and highly expressed cytokine identified in our cytokine arrays, in serum and skeletal muscle of tumor-bearing mice, and 2) whether tumor-bearing mice exhibited evidence of weight loss. We used a transplant-based approach previously established using related isogenic lung cancer cell lines (Gibbons et al., 2009), to initiate xenograft tumors in recipient mice. Prior to necropsy, we subjected mice to whole body dual-energy X-ray absorptiometry (DEXA) to quantify total and lean mass in the whole animal. DEXA analyses of mice bearing subcutaneous lung tumors revealed an overall reduction in total body weight excluding the tumor (mean=24.3g) compared to non-tumor control mice (mean=27.0g) (Figure 3A,D). This decline in overall weight appeared largely attributable to loss of lean mass (mean=19.0g vs. 22.4g). (Figure 3A,E). Serum analysis of 531LN2-tumor bearing mice revealed a nearly 10-fold increase in serum CXCL1 levels compared to non-tumor bearing healthy control mice (Figure 3B). Furthermore, hindlimb TA CXCL1 levels in tumor bearing mice were ~20-fold higher than control muscle samples (Figure 3C). Interestingly, we observed a striking up-regulation (>200 fold) of cxcl1 mRNA in C2C12 myoblast or myotube cultures exposed to 531LN2 media (Supplemental Figure S3), suggesting that elevated CXCL1 in the skeletal muscle of tumor bearing mice may be the combined result of both tumor-derived CXCL1 as well as tumor-induced CXCL1 production in skeletal muscle. Upon necropsy, we confirmed the DEXA-determined decline in lean mass with tibialis anterior (TA) and gastrocnemius (GR) wet weight measurements (Figure 3F,G). Morphometric evaluation of myofiber size further revealed a statistically significant downward shift in the distribution of myofiber feret diameters (Figure 3H). Taken together, our data show that lung tumor xenotransplantation results in overall weight loss, a decline in lean mass, and is associated with elevated CXCL1 levels in serum and skeletal muscle.
Figure 3. Tumor bearing mice exhibit weight loss and elevated CXCL1 levels.
(A) Representative dual-energy X-ray absorptiometry (DEXA) images of a control, non-tumor bearing mouse (left) and a mouse bearing a transplanted terminal 531LN2 lung adenocarcinoma tumor. The dotted white line and “T” depict the tumor. (B–C) Graphs quantifying CXCL1 abundance in serum (B) and hindlimb muscle (C) lysates from control, non-tumor bearing mice and terminal 531LN2 tumor mice. Serum concentrations are depicted as picograms of CXCL1 per milliliter of serum whereas muscle concentrations are shown as picograms per microgram of total muscle protein. N=5 mice in each group. *p<0.05, **p<0.01 using two-tailed, unpaired t tests. (D–E) Quantification of DEXA computed mouse weight (D) and lean mass (E), *p=0.03, **p=0.004 using two-tailed, unpaired t tests. (F–G) Bar graphs quantifying tibialis anterior (F) and gatrocnemeous (G) wet weights. **p<0.01 using two-tailed, unpaired t tests. (H) Histograms depicting binned myofiber feret diameter measurements from control and 531LN2 tumor bearing mice. Non-linear regression curves were fit (least squares method) to each sample and compared using extra sum-of-squares F tests to determine if regression curves were statistically different (p<0.0001).
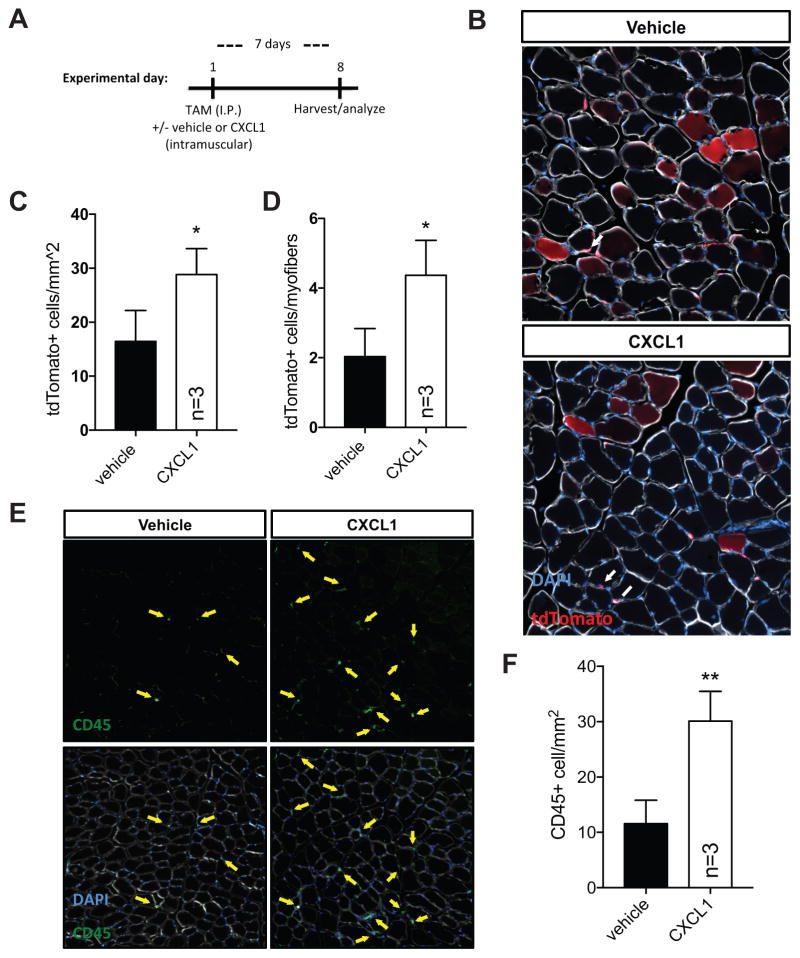
CXCL1 modulates skeletal muscle regeneration and in vivo satellite cell homeostasis
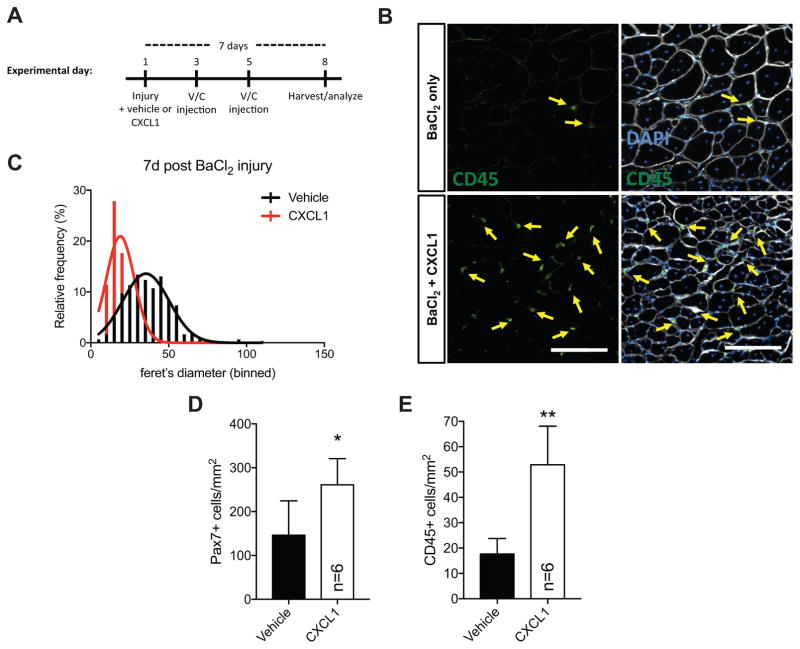
Skeletal muscle regeneration requires the activation, expansion, and differentiation of satellite cells, a resident adult muscle stem cell population. Given the antagonistic effect of CXCL1 on in vitro myogenesis, as well as increased CXCL1 abundance in lung tumor bearing mice, we next tested the hypothesis that elevated CXCL1 would limit in vivo muscle regeneration. We injured wildtype TA muscle with 1.2% barium chloride (BaCl2) in either a 0.9% saline vehicle or vehicle containing CXCL1. We chose a dose of CXCL1 (100ng) consistent with CXCL1 levels detected in cancer-associated muscle (ref to Figure 3C). Additional injections of vehicle or vehicle/CXCL1 were given 2 and 4 days following injury to maintain elevated CXCL1 levels (see experimental timeline in Figure 4A). Seven days following injury, we sacrificed experimental mice, harvested TA muscles, and performed immunohistochemical analyses. Myofiber feret diameters measured 7d post injury were significantly smaller in vehicle/CXCL1 samples compared to vehicle only controls (control mean=36um, CXCL1 mean=19um) (Figure 4B,C). We observed a ~50% increase in Pax7+ cell numbers (Figure 4D) and a nearly 3-fold increase in the number of muscle resident CD45+ immune cells (Figure 4E). Interestingly, tumor-bearing mice also exhibited elevated (~4.5 fold) CD45+ cell numbers in hindlimb muscle preparations analyzed by flow cytometry (Supplemental Figure S4). Together, these data support the hypothesis that CXCL1 impairs muscle regeneration.
Figure 4. CXCL1 disrupts skeletal muscle regeneration.
(A) Timeline of skeletal muscle injury experiment. (B) Representative images of TA muscle tissue sections from vehicle or CXCL1 injected mice 7 days post 1.2% BaCl2 injury stained with antibodies targeting CD45 (green, highlighted with yellow arrows) and laminin (white). Nuclei are labeled with DAPI (blue). (C) A histogram of binned myofiber feret measurements from experiment shown in (A) and (B). Non-linear regression curves were fit (least squares method) to each sample and compared using an extra sum-of-squares F test to determine if regression curves were statistically different (p<0.0001 between vehicle and CXCL1). Quantification of Pax7+ (D) and CD45+ cells (E) from 7d post-injury TA muscle. *p<0.05, **p<0.01 using two-tailed, unpaired t tests.
We next leveraged a recently described SC lineage tracing Pax7CreERT2;ROSA26LSL-tdTomato mouse model to directly assess the impact of CXCL1 on in vivo satellite cell function in the absence of injury. Seven days following tamoxifen-induced SC labeling and intramuscular (TA) vehicle or CXCL1 administration, tdTomato+ SCs and tdTomato+ myofibers were quantified in TA muscle sections (see experimental timeline in Figure 5A). We detected higher overall numbers of tdTomato+ SCs in CXCL1 injected muscle (~16 cells/mm2 vehicle vs. ~28 cells/mm2 CXCL1) (Figure 5B,C), indicating deficits in SC commitment and/or fusion consistent with published studies (He et al., 2013). Additionally, the tdTomato+ SC/tdTomato+ myofiber ratio was elevated ~2-fold (Figure 5D), suggesting that CXCL1 results in SC expansion or accumulation at the expense of cell cycle exit and commitment to differentiation and fusion. Strikingly, we observed evidence of elevated CD45+ immune cell numbers in the absence of overt injury (Figure 5E,F), suggesting that CXCL1 directly stimulates immune cell accumulation in skeletal muscle.
Figure 5. CXCL1 disrupts satellite cell homeostasis.
(A) Timeline of satellite cell homeostasis experiment. (B) TA muscle tissue sections from tamoxifen-treated (day 0) Pax7icreERT2;tdTomatoflox mice (to mark SCs, arrows) injected with 100ng CXCL1 (right hindlimb) or a vehicle control (left hindlimb) and harvested at day 7 to assess progenitor cell contribution to muscle (red myofibers, asterisks). N=3 mice. Bar graphs quantifying the number of tdTomato+ SCs (C) and the ratio of tdTomato+ cells versus tdTomato+ myofibers (D). *p<0.05, n.s.=not significant using two-tailed, unpaired t tests. (E) Representative images of uninjured TA muscle tissue sections from vehicle or CXCL1 injected mice 7 days post injection stained with antibodies targeting CD45 (green, highlighted with yellow arrows) and laminin (white). Nuclei are labeled with DAPI (blue). (F) Bar graph quantifying CD45+ cell abundance from (E). **p<0.01 using two-tailed, unpaired t tests.
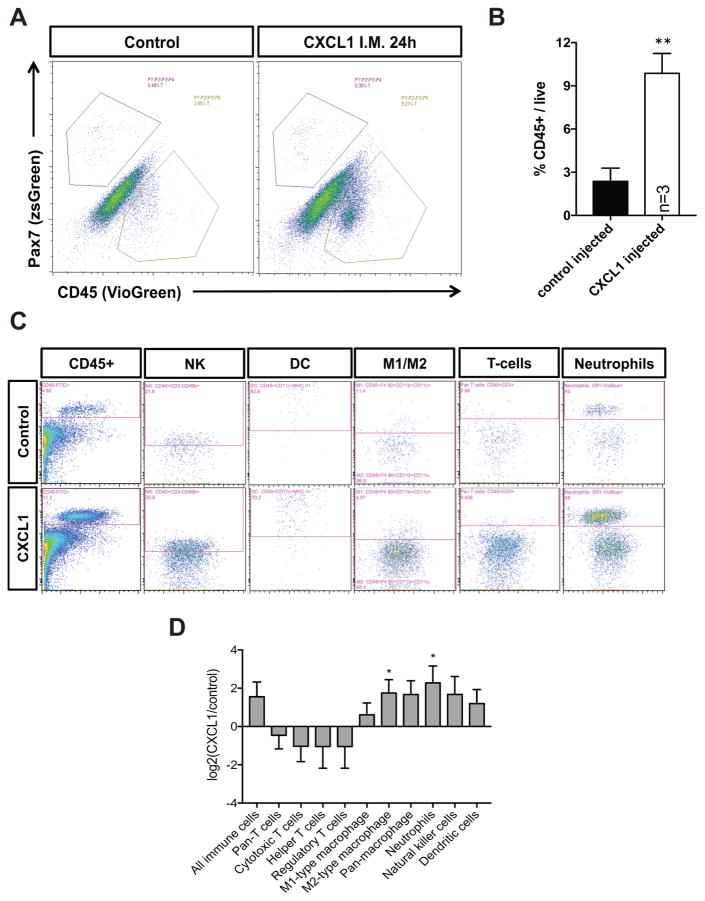
CXCL1 is an established chemokine linked to neutrophil recruitment (De Filippo et al., 2013). To identify the nature of the infiltrating CD45+ immune cell population, we used flow cytometry-based antibody panels to query the spectrum of immune cell subtypes present in skeletal muscle before and after an acute CXCL1 injection. In total, we assessed the presence of T-cells (cytotoxic, regulatory, and helper), macrophages (M1 and M2), neutrophils, dendritic cells, and natural killer (NK) cells in wildtype mice +/− CXCL1. Consistent with our immunofluorescence analyses, we detected a ~3 fold increase in CD45+ cells in the CXCL1-injected cohort after an acute dose (24h) of CXCL1 (Figure 6A,B). Most of this increase was attributable to statistically significant neutrophil and M2 macrophage expansion (Figure 6C,D). Overall, these data therefore show that CXCL1 can elicit a potent neutrophil/macrophage response, and that together with the ability to suppress myogenic differentiation, likely contributes to cancer-associated muscle wasting.
Figure 6. CXCL1 promotes a robust immune cell response in vivo.
(A) Representative flow cytometry plots of control or CXCL1 injected TA muscle isolated 24h post injection from Pax7zsGreen mice and stained with an antibody targeting CD45. (B) Bar graph depicting CD45+ immune cell abundance with respect to total live cell number in vehicle or CXCL1 injected mice. **p<0.01 using two-tailed, unpaired t tests. (C) Representative flow cytometry plots of control or CXCL1 injected TA muscle isolated 24h post injection from wildtype mice and stained with antibody cocktails designed to identify (L to R): all immune cells, natural killer cells, dendritic cells, M1/M2 type macrophages, T-cells, and neutrophils. (D) A bar graph depicting the log2 normalized CXCL1/vehicle ratio for each indicated immune cell subtype. *p<0.05 using two-tailed, one sample t tests (relative to mean=0).
DISCUSSION
We focused on identifying and characterizing tumor-derived factors that interfere with myogenic progression, in part based on accumulating evidence that impaired regeneration/repair contributes to cancer-associated wasting (He et al., 2013; Talbert and Guttridge, 2016). Consistent with loss of muscle mass, we show that tumor-derived factors interfere with myoblast differentiation and cell cycle progression. Furthermore, we observe decreased mitochondrial biogenesis which is consistent with reported tumor-mediated decrements in respiratory chain activity (Padrao et al., 2013) (Fermoselle et al., 2013) or dysregulated ATP generation (Shum et al., 2012; Tzika et al., 2013). We then implicate one of these tumor-derived factors – CXCL1 – in muscle repair/regeneration and in vivo SC homeostasis. Taken together, we propose that cytokine-mediated disruption of myoblast function is a potential mechanism to partially explain loss of lean mass observed in 531LN2 lung tumor bearing mice.
To demonstrate the modulatory effects of CXCL1 on in vivo myogenesis, we utilized a murine SC lineage tracing model that tracked tdTomato fluorescence in Pax7+ SCs and all progeny (including myofibers). We observed an accumulation of lineage marked cells and an increase in the SC/myofiber ratio in CXCL1 treated mice, implicating satellite cell differentiation defects. Several reports in stem cells (Jung et al., 2015), fibroblasts (le Rolle et al., 2015), and cancer cells (Bolitho et al., 2010) (le Rolle et al., 2015) suggest that CXCL1 can function as a mitogen. Thus, CXCL1 may inhibit differentiation by antagonizing cell cycle exit and promoting SC proliferation. Alternatively, overexpression studies showed that CXCL1 can modulate skeletal muscle metabolism (Pedersen et al., 2011). Given the dynamic metabolic changes occurring during myogenic differentiation (Ryall et al., 2015), it is possible that elevated CXCL1 levels impair SC metabolism or protein synthesis/accretion to limit muscle regeneration. Future studies more rigorously investigating the cellular and molecular impact of CXCL1 (and other tumor-associated cytokines) on SCs is needed to fully appreciate the mechanism by which myogenesis is impaired.
In addition to tumor-derived secretion of CXCL1 (Acharyya et al., 2012; Eck et al., 2003; Keane et al., 2004; Lo et al., 2013; Miyake et al., 2016), CXCL1 is secreted by several tissues which influence the physiology of skeletal muscle, including liver (Pedersen et al., 2011) and immune cells, namely neutrophils (Katsanos et al., 2008; Kobayashi, 2008). Interestingly, CXCL1 stimulated both neutrophil and M2-type macrophage expansion in skeletal muscle. During muscle regeneration, neutrophils and M2 macrophages are generally thought to drive distinct repair processes, with the former promoting cell clearance while the latter aids tissue repair and remodeling (Tidball, 2017). That CXCL1 promotes rapid expansion of these distinct immune cell subpopulations in skeletal muscle suggests that impaired regeneration may result, in part, from disruption of the ordered recruitment of immune cell subtypes to damaged muscle. Given the well-documented influence of the immune system on satellite cell function and muscle regeneration, our work underscores the importance of evaluating myogenic factors both in vitro and in relevant physiological settings to more fully reveal potential mechanisms of action.
Potentiating myogenesis has proven effective in mitigating functional deficits associated with other skeletal muscle wasting disorders such as age-associated sarcopenia and muscular dystrophy (Bernet et al., 2014; Cosgrove et al., 2014; Dumont et al., 2015). In some of these contexts, removing disease or age-associated blocks that impaired SC activity were sufficient to attenuate or even reverse muscle atrophy (Sousa-Victor et al., 2014) (Garcia-Prat et al., 2016). Importantly, enhancing muscle regeneration would likely complement other therapeutic strategies, such as those that target aberrant immune cell activity and/or hyperactive muscle breakdown pathways. Ultimately, slowing and/or reversing cancer-associated muscle wasting will likely require a therapeutic program that incorporates both pro-regeneration and anti-breakdown strategies.
In summary, we describe a role for CXCL1 in negatively regulating muscle regeneration and satellite cell homeostasis, and altering the immune cell profile in skeletal muscle. That CXCL1 can impair both myogenesis and immune cell homeostasis underscores the likelihood that other cytokines act on multiple cellular targets during muscle regeneration/repair. Thus, as anti-cytokine therapies move forward, it will be important to consider all potential target cells/signaling pathways in order to avoid or minimize off-target effects.
METHODS
Animals
Mice were bred and housed according to NIH guidelines for the ethical treatment of animals in a pathogen-free facility at the Mayo Clinic (Rochester, MN campus). Mayo Clinic’s Institutional Animal Care and Use Committee (IACUC) approved all animal protocols. The following mouse strains were used in this study: Pax7iCreERT2(Murphy et al, 2011), ROSA26LSL-tdTomato (Jackson Labs, 007909), and Pax7zsgreen (Bosnakovski et al, 2008). Wild type mice were C57BL/6J (Jackson Labs) or FVB (Charles River Labs). Pax7+ SCs were lineage marked as described previously (Pawlikowski et al., 2015) with a tdTomato fluorophore using tamoxifen/cre-mediated recombination. Using this mouse model, recombinant CXCL1 was injected in the right hindlimb TA muscle and a vehicle control injected in the contralateral limb. Cre-mediated recombination was induced via intraperitoneal injections of tamoxifen (Sigma Aldrich) in corn oil dosed at 2mg tamoxifen/20g mouse weight. Tumor transplantations were performed by subcutaneous injection of 0.5×106 KrasLA1/+;p53R172H/Δg/+ lung adenocarcinoma cells into the flank of wildtype 129S2/SvPasCrl recipient mice (Gibbons et al, 2009). Mice were monitored twice weekly and imaged at the end of the study (~6 weeks post injection).
Animal imaging
Imaging-based determination of total mass and lean mass was performed following induction of anesthesia (ketamine/xyzaline). Scanning was performed on a restrained mouse using dual-energy X-ray absorptiometry (DEXA; PIXImus; Lunar Inc.) and analyzed using manufacturer included software.
Cell culture
C2C12 cells (ATCC) (Yaffe and Saxel, 1977) were grown on tissue culture treated dishes in growth media consisting of DMEM media supplemented with 10% fetal bovine serum and antibiotics. Differentiation-inducing media replaces 10% FBS with 2% horse serum. Primary dispersed SCs were isolated and cultured as previously described (Hausburg et al, 2015). Conditioned media from lung adenocarcinoma cells was collected by culturing cells (~70–90% confluence) in serum free (DMEM) media for 8 hours, followed by centrifugation of cell debris and adding serum as needed (10% FBS for growth experiments, 2% horse serum for differentiation experiments). Differentiating cultures were maintained by alternating fresh media additions and complete media changes on a daily basis for up to 5 days. KrasLA1/+;p53R172H/Δg/+ lung adenocarcinoma cell lines (307P, 531LN2, 344SQ) were generated as previously described (Gibbons et al, 2009).
Immunostaining
Tissue sections (8–10um) were post-fixed in 4% paraformaldehyde for 5 minutes at room temperature prior to immunostaining. Once fixed, isolated SCs, myofibers or tissue sections were permeabilized with 0.5% Triton-X100 in PBS followed by blocking with 3% BSA in PBS. Primary antibody incubations occurred at RT for 90 minutes or overnight at 4 degrees followed by incubation with secondary antibody at RT for 30 minutes in 3% BSA in PBS. The following antibodies were used in this study: CD45 (BD 553076), MF-20 (Developmental Hybridoma Bank), BF-F3 (Developmental Hybridoma Bank), SC-71 (Abcam ab15580), Laminin (Sigma 4HB-2), Sdc4 (Cornelison et al., 2001), and myogenin F5D (Santa Cruz Biotech sc-52903). Secondary antibodies were all Alexa fluorescent conjugates (488, 555, or 647) from Invitrogen or Jackson ImmunoResearch. Click-IT Edu staining was performed according to manufacturer instructions (Thermo Fisher).
Flow cytometry
Cell cycle/Edu experiments were performed by incubating growth phase C2C12 myoblast cultures with 10uM 5-ethynyl-2′-deoxyuridine (EdU) for 2 hours, fixing, counterstaining DNA with DAPI, and identifying Edu+ cells using a Click-IT Edu Flow Cytometry Kit (Thermo Fisher). Mitochondria were labeled in live cells using MitoTracker Green FM (Thermo Fisher) and analyzed by flow cytometry. Immune cell profiling was performed as follows: hindlimb muscles were enzymatically digested and filtered according to standard protocols (Bernet et al, 2014), cells were labeled with fluorescently conjugated primary antibodies according to manufacturer’s instructions, and flow cytometry performed on a MACSquant 10 analyzer. The following antibodies (Miltenyi Biotec) were used in this study: anti-CD45, anti-CD3, anti-CD49b, anti-CD11c, anti-MHC class II, anti-F4/80, anti-CD11c, and anti-GR1. The following is the gating strategy used to identify individual immune cell subtypes: NK: CD45+, CD3e−, CD49b+; Dendritic cell: CD45+, CD11c+, MHC class II+; M1 macrophage: CD45+, F4/80+, CD11b+, CD11c+; M2 macrophage: CD45+, F4/80+, CD11b+, CD11c−; Neutrophil: CD45+, CD11b+, GR1+; Pan T cells: CD45+, CD3e+; Cytotoxic T cells: CD45+, CD3e+, CD8+; Helper T cells: CD45+, CD3e+, CD4+; Regulatory T cells: CD45+, CD3e+, CD4+, CD25+. Samples were analyzed using either MACSquant software or FlowJo v10.
Cytokine studies
Conditioned media from lung adenocarcinoma cells was collected as described above for C2C12 studies. Cytokine arrays were performed using Proteome Profiler Mouse XL Cytokine Arrays (R&D Systems) and visualized/quantified using a LI-COR Odyssey FC system. Ectopic cytokine studies were performed using C2C12 cells plated at a density of 100–500K/well of a 6-well tissue culture dish and differentiated as noted above for conditioned media experiments. All cytokines were purchased from Peprotech and used as noted in the text based on published IC50 ranges defined in other cell types/experimental contexts: IGFBP3 (cat#: 100-008), IGFBP6 (cat#: 350-07B), CXCL1 (cat#: 250-11), LIX/CXCL6 (cat#: 250-17), CCL2 (cat#: 250-10), CCL17 (cat#: 300-30), CCL20 (cat#: 250-27), and endostatin (cat#: 150-01). We noted substantial lot-to-lot variability with the CXCL1 preparations, so we advise recalibrating phenotypes if using different cytokine lots. CXCL1 ELISAs were performed using a Mouse CXCL1/KC DuoSet ELISA kit (R&D Systems) and serum or liquid nitrogen prepared whole muscle lysates from terminal tumor-bearing (531LN2) mice.
Supplementary Material
Acknowledgments
We wish to thank members of the Doles lab for helpful discussions and critical reading of the manuscript. J.D. is a PCAN/AACR Career Development Awardee and is supported by NIH/NIAMS R00AR66696, Mayo Clinic start-up funds, and a Development Award from the Mayo Clinic SPORE in Pancreatic Cancer (NIH/NCI CA102701). A.S. is a PREP scholar with partial support from NIH/NIGMS GM075148 and an NIH/NIAMS diversity supplement (AR66696-03S1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, Norton L, Brogi E, Massague J. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragno M, Mastrocola R, Catalano MG, Brignardello E, Danni O, Boccuzzi G. Oxidative stress impairs skeletal muscle repair in diabetic rats. Diabetes. 2004;53:1082–1088. doi: 10.2337/diabetes.53.4.1082. [DOI] [PubMed] [Google Scholar]
- Argiles JM, Busquets S, Lopez-Soriano FJ. Cytokines in the pathogenesis of cancer cachexia. Current opinion in clinical nutrition and metabolic care. 2003;6:401–406. doi: 10.1097/01.mco.0000078983.18774.cc. [DOI] [PubMed] [Google Scholar]
- Baracos VE, DeVivo C, Hoyle DH, Goldberg AL. Activation of the ATP-ubiquitin-proteasome pathway in skeletal muscle of cachectic rats bearing a hepatoma. The American journal of physiology. 1995;268:E996–1006. doi: 10.1152/ajpendo.1995.268.5.E996. [DOI] [PubMed] [Google Scholar]
- Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, Olwin BB. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nature medicine. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Webster C, Pavlath GK. Defective myoblasts identified in Duchenne muscular dystrophy. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:4856–4860. doi: 10.1073/pnas.80.15.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolitho C, Hahn MA, Baxter RC, Marsh DJ. The chemokine CXCL1 induces proliferation in epithelial ovarian cancer cells by transactivation of the epidermal growth factor receptor. Endocrine-related cancer. 2010;17:929–940. doi: 10.1677/ERC-10-0107. [DOI] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Li W, Thet S, Cleaver O, Perlingeiro RC, Kyba M. Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem cells (Dayton, Ohio) 2008;26:3194–3204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrick KL, Stec MJ, Merritt EK, Windham ST, Thomas SJ, Cross JM, Bamman MM. Serum from human burn victims impairs myogenesis and protein synthesis in primary myoblasts. Front Physiol. 2015;6:184. doi: 10.3389/fphys.2015.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, Llewellyn ME, Delp SL, Blau HM. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nature medicine. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo K, Dudeck A, Hasenberg M, Nye E, van Rooijen N, Hartmann K, Gunzer M, Roers A, Hogg N. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood. 2013;121:4930–4937. doi: 10.1182/blood-2013-02-486217. [DOI] [PubMed] [Google Scholar]
- Dumont NA, Rudnicki MA. Characterizing Satellite Cells and Myogenic Progenitors During Skeletal Muscle Regeneration. Methods in molecular biology (Clifton, NJ) 2017;1560:179–188. doi: 10.1007/978-1-4939-6788-9_12. [DOI] [PubMed] [Google Scholar]
- Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, Rudnicki MA. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nature medicine. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck M, Schmausser B, Scheller K, Brandlein S, Muller-Hermelink HK. Pleiotropic effects of CXC chemokines in gastric carcinoma: differences in CXCL8 and CXCL1 expression between diffuse and intestinal types of gastric carcinoma. Clinical and experimental immunology. 2003;134:508–515. doi: 10.1111/j.1365-2249.2003.02305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery PW, Lovell L, Rennie MJ. Protein synthesis measured in vivo in muscle and liver of cachectic tumor-bearing mice. Cancer research. 1984;44:2779–2784. [PubMed] [Google Scholar]
- Fearon KC, Glass DJ, Guttridge DC. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell metabolism. 2012;16:153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Fermoselle C, Garcia-Arumi E, Puig-Vilanova E, Andreu AL, Urtreger AJ, de Kier Joffe ED, Tejedor A, Puente-Maestu L, Barreiro E. Mitochondrial dysfunction and therapeutic approaches in respiratory and limb muscles of cancer cachectic mice. Experimental physiology. 2013;98:1349–1365. doi: 10.1113/expphysiol.2013.072496. [DOI] [PubMed] [Google Scholar]
- Figeac N, Serralbo O, Marcelle C, Zammit PS. ErbB3 binding protein-1 (Ebp1) controls proliferation and myogenic differentiation of muscle stem cells. Dev Biol. 2014;386:135–151. doi: 10.1016/j.ydbio.2013.11.017. [DOI] [PubMed] [Google Scholar]
- Fry CS, Porter C, Sidossis LS, Nieten C, Reidy PT, Hundeshagen G, Mlcak R, Rasmussen BB, Lee JO, Suman OE, Herndon DN, Finnerty CC. Satellite cell activation and apoptosis in skeletal muscle from severely burned children. J Physiol. 2016;594:5223–5236. doi: 10.1113/JP272520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Zhu MJ, Dodson MV, Du M. AMP-activated Protein Kinase Stimulates Warburg-Like Glycolysis and Activation of Satellite Cells during Muscle Regeneration. J Biol Chem. 2015 doi: 10.1074/jbc.M115.665232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S, Anguiano M, Rotwein P. Defining Akt actions in muscle differentiation. Am J Physiol Cell Physiol. 2012;303:C1292–C1300. doi: 10.1152/ajpcell.00259.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Prat L, Munoz-Canoves P, Martinez-Vicente M. Dysfunctional autophagy is a driver of muscle stem cell functional decline with aging. Autophagy. 2016;12:612–613. doi: 10.1080/15548627.2016.1143211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Lin W, Creighton CJ, Zheng S, Berel D, Yang Y, Raso MG, Liu DD, Wistuba II, Lozano G, Kurie JM. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PloS one. 2009;4:e5401. doi: 10.1371/journal.pone.0005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding JP, Calderbank E, Partridge TA, Beauchamp JR. Skeletal muscle stem cells express anti-apoptotic ErbB receptors during activation from quiescence. Exp Cell Res. 2007;313:341–356. doi: 10.1016/j.yexcr.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Hausburg MA, Doles JD, Clement SL, Cadwallader AB, Hall MN, Blackshear PJ, Lykke-Andersen J, Olwin BB. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. eLife. 2015;4:e03390. doi: 10.7554/eLife.03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WA, Berardi E, Cardillo VM, Acharyya S, Aulino P, Thomas-Ahner J, Wang J, Bloomston M, Muscarella P, Nau P, Shah N, Butchbach ME, Ladner K, Adamo S, Rudnicki MA, Keller C, Coletti D, Montanaro F, Guttridge DC. NF-kappaB-mediated Pax7 dysregulation in the muscle microenvironment promotes cancer cachexia. The Journal of clinical investigation. 2013;123:4821–4835. doi: 10.1172/JCI68523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Lee SJ, Kim J, Lee S, Sung HJ, An J, Park Y, Kim BS. CXCR2 and its related ligands play a novel role in supporting the pluripotency and proliferation of human pluripotent stem cells. Stem cells and development. 2015;24:948–961. doi: 10.1089/scd.2014.0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanos GS, Anogeianaki A, Orso C, Tete S, Salini V, Antinolfi PL, Sabatino G. Mast cells and chemokines. Journal of biological regulators and homeostatic agents. 2008;22:145–151. [PubMed] [Google Scholar]
- Keane MP, Belperio JA, Xue YY, Burdick MD, Strieter RM. Depletion of CXCR2 inhibits tumor growth and angiogenesis in a murine model of lung cancer. Journal of immunology (Baltimore, Md: 1950) 2004;172:2853–2860. doi: 10.4049/jimmunol.172.5.2853. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y. The role of chemokines in neutrophil biology. Frontiers in bioscience: a journal and virtual library. 2008;13:2400–2407. doi: 10.2741/2853. [DOI] [PubMed] [Google Scholar]
- Krause MP, Moradi J, Nissar AA, Riddell MC, Hawke TJ. Inhibition of plasminogen activator inhibitor-1 restores skeletal muscle regeneration in untreated type 1 diabetic mice. Diabetes. 2011;60:1964–1972. doi: 10.2337/db11-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Rolle AF, Chiu TK, Fara M, Shia J, Zeng Z, Weiser MR, Paty PB, Chiu VK. The prognostic significance of CXCL1 hypersecretion by human colorectal cancer epithelia and myofibroblasts. Journal of translational medicine. 2015;13:199. doi: 10.1186/s12967-015-0555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Solomon V, Mitch WE, Goldberg AL. Muscle protein breakdown and the critical role of the ubiquitin-proteasome pathway in normal and disease states. The Journal of nutrition. 1999;129:227s–237s. doi: 10.1093/jn/129.1.227S. [DOI] [PubMed] [Google Scholar]
- Lo HM, Shieh JM, Chen CL, Tsou CJ, Wu WB. Vascular endothelial growth factor induces CXCL1 chemokine release via JNK and PI-3K-dependent pathways in human lung carcinoma epithelial cells. International journal of molecular sciences. 2013;14:10090–10106. doi: 10.3390/ijms140510090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchildon F, Lamarche É, Lala-Tabbert N, St-Louis C, Wiper-Bergeron N. Expression of CCAAT/Enhancer Binding Protein Beta in Muscle Satellite Cells Inhibits Myogenesis in Cancer Cachexia. PLoS One. 2015;10:e0145583. doi: 10.1371/journal.pone.0145583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitch WE, Goldberg AL. Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. The New England journal of medicine. 1996;335:1897–1905. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- Miyake M, Hori S, Morizawa Y, Tatsumi Y, Nakai Y, Anai S, Torimoto K, Aoki K, Tanaka N, Shimada K, Konishi N, Toritsuka M, Kishimoto T, Rosser CJ, Fujimoto K. CXCL1-Mediated Interaction of Cancer Cells with Tumor-Associated Macrophages and Cancer-Associated Fibroblasts Promotes Tumor Progression in Human Bladder Cancer. Neoplasia (New York, NY) 2016;18:636–646. doi: 10.1016/j.neo.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdziak PE, Truong Q, Macius A, Schultz E. Hindlimb suspension reduces muscle regeneration. Eur J Appl Physiol Occup Physiol. 1998;78:136–140. doi: 10.1007/s004210050398. [DOI] [PubMed] [Google Scholar]
- Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development (Cambridge, England) 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrao AI, Oliveira P, Vitorino R, Colaco B, Pires MJ, Marquez M, Castellanos E, Neuparth MJ, Teixeira C, Costa C, Moreira-Goncalves D, Cabral S, Duarte JA, Santos LL, Amado F, Ferreira R. Bladder cancer-induced skeletal muscle wasting: disclosing the role of mitochondria plasticity. The international journal of biochemistry & cell biology. 2013;45:1399–1409. doi: 10.1016/j.biocel.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Pawlikowski B, Pulliam C, Betta ND, Kardon G, Olwin BB. Pervasive satellite cell contribution to uninjured adult muscle fibers. 2015;5:42. doi: 10.1186/s13395-015-0067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L, Pilegaard H, Hansen J, Brandt C, Adser H, Hidalgo J, Olesen J, Pedersen BK, Hojman P. Exercise-induced liver chemokine CXCL-1 expression is linked to muscle-derived interleukin-6 expression. The Journal of physiology. 2011;589:1409–1420. doi: 10.1113/jphysiol.2010.200733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryall JG, Dell’Orso S, Derfoul A, Juan A, Zare H, Feng X, Clermont D, Koulnis M, Gutierrez-Cruz G, Fulco M, Sartorelli V. The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell stem cell. 2015;16:171–183. doi: 10.1016/j.stem.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE, Blau HM. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum AM, Mahendradatta T, Taylor RJ, Painter AB, Moore MM, Tsoli M, Tan TC, Clarke SJ, Robertson GR, Polly P. Disruption of MEF2C signaling and loss of sarcomeric and mitochondrial integrity in cancer-induced skeletal muscle wasting. Aging. 2012;4:133–143. doi: 10.18632/aging.100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Victor P, Gutarra S, Garcia-Prat L, Rodriguez-Ubreva J, Ortet L, Ruiz-Bonilla V, Jardi M, Ballestar E, Gonzalez S, Serrano AL, Perdiguero E, Munoz-Canoves P. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- Sousa-Victor P, Munoz-Canoves P. Regenerative decline of stem cells in sarcopenia. Molecular aspects of medicine. 2016;50:109–117. doi: 10.1016/j.mam.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Talbert EE, Guttridge DC. Impaired regeneration: A role for the muscle microenvironment in cancer cachexia. Seminars in cell & developmental biology. 2016;54:82–91. doi: 10.1016/j.semcdb.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball JG. Regulation of muscle growth and regeneration by the immune system. Nature reviews Immunology. 2017;17:165–178. doi: 10.1038/nri.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale MJ. Biochemical mechanisms of cellular catabolism. Current opinion in clinical nutrition and metabolic care. 2002;5:401–405. doi: 10.1097/00075197-200207000-00009. [DOI] [PubMed] [Google Scholar]
- Tzika AA, Fontes-Oliveira CC, Shestov AA, Constantinou C, Psychogios N, Righi V, Mintzopoulos D, Busquets S, Lopez-Soriano FJ, Milot S, Lepine F, Mindrinos MN, Rahme LG, Argiles JM. Skeletal muscle mitochondrial uncoupling in a murine cancer cachexia model. International journal of oncology. 2013;43:886–894. doi: 10.3892/ijo.2013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- Zhang K, Sha J, Harter ML. Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells. J Cell Biol. 2010;188:39–48. doi: 10.1083/jcb.200904144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.