Abstract
Background
Transplantation of autologous mesenchymal stem cells (MSCs) may be a viable option for treatment of several diseases. MSCs efficacy depends on adequate function of their mitochondria, which might be impaired in a noxious milieu.
Objectives
We hypothesized that obesity compromises MSCs mitochondrial structure and function, possibly via micro-RNA (miRNA)-based mechanisms.
Methods
MSCs were collected from swine abdominal adipose tissue after 16 weeks of Lean or Obese diet (n=7 each). Mitochondrial structure was assessed by electron microscopy and function by membrane potential and cytochrome-c oxidase (COX)-IV activity. Oxidative stress was assessed by Mito-SOX and dihydroethidium staining. Next-generation sequencing (RNA-seq) was performed to identify miRNAs expression in MSCs, and predicted mitochondrial target genes were then identified (MitoCarta).
Results
Compared to Lean-MSCs, mitochondria from Obese-MSCs were smaller and showed cristae remodeling and loss. Mitochondrial membrane potential and COX-IV activity decreased in Obese-MSCs, associated with increased mitochondrial oxidative stress. RNA-seq generated reads for 413 miRNAs, of which 5 miRNAs were upregulated in Obese-MSCs (fold change >2, p<0.05) and found to target 43 specific mitochondrial genes.
Conclusions
Obesity impairs MSC mitochondrial structure and function, possibly mediated partly through miRNA-induced mitochondrial gene regulation, leading to increased oxidative stress. Importantly, these alterations may limit the therapeutic use of autologous MSCs in subjects with obesity.
Keywords: Mesenchymal Stem Cells, Mitochondria, Obesity, Metabolic syndrome
Introduction
Mesenchymal stem cells (MSCs) play fundamental roles in repair and self-renewal of tissues throughout life by replenishing injured cells to maintain tissue homeostasis. Cell-based therapies involving exogenous delivery of MSCs harvested from various tissue sources have become prevalent in regenerative medicine, because their potent pro-angiogenic, anti-inflammatory, and immunomodulatory properties render MSCs an attractive tool to attenuate tissue injury(Rohban and Pieber, 2017). For safety reasons and to avoid host rejection, the use of autologous MSCs is preferable compared to allogeneic transplantation, but mandates functionally robust MSCs.
Obesity is an important cardiovascular risk factor that influences almost every aspect of health and is asociated with increased cardiovascular morbidity and mortality(Han and Lean, 2016). We have recently developed a novel porcine model of obesity achieved by feeding animals a high-carbohydrate and -cholesterol diet, that mimics many features of metabolic syndrome, including abdominal obesity, hypertension, high triglycerides, and insulin resistance(Pawar et al., 2015). Furthermore, we have shown that MSCs from obese pigs have increased adipogenic differentiation and propensity for senescence(Zhu et al., 2016). Yet, the mechanisms by which obesity modulates MSC fate and function remain to be elucidated.
The viability, plasticity(Folmes et al., 2012), proliferative and differential potential, and functionality of MSCs are all affected by the function and integrity of their mitochondria(Parker, Acsadi and Brenner, 2009, Kadye et al., 2014), which are major regulators of cellular survival and senescence(Casalena et al., 2014). Furthermore, mitochondria-derived reactive oxygen species (ROS) modulate MSC fate, with low ROS levels supporting maintenance of cell plasticity(Perales-Clemente, Folmes and Terzic, 2014), whereas a higher ROS state due to stress and chronic inflammation enhances stem cell differentiation, motility, and thereby exhaustion and lost plasticity(Ludin et al., 2014). Therefore, conditions that increase mitochondrial ROS production may impair MSC function. Indeed, elucidating the molecular mechanisms that modulate MSC mitochondrial function will provide insight into the adaptive response of MSCs in the context of obesity, and will enable development of directed strategies to improve MSCs derived from patients with metabolic disorders.
Micro-RNAs (miRNAs) are small non-coding RNA fragments that act as central post-transcriptional regulators by repressing protein expression of their messenger RNA (mRNA) targets(He and Hannon, 2004). MiRNA are differentially expressed in MSCs under culture conditions that regulate important stem cell features, including proliferation and differentiation(Crabbe et al., 2016). Furthermore, miRNAs can target genes that encode proteins responsible for mitochondrial biogenesis and function(Du et al., 2016). Because miRNAs can target mitochondrial functions, and mitochondrial dysfunction is associated with metabolic disorders, we tested the hypothesis that obesity alters the activity of porcine adipose tissue-derived MSC mitochondria, which diminishes their lifespan and function, and changes expression of miRNAs that post-translationally regulate mitochondrial genes.
Materials and Methods
Experimental design
Animal studies were approved by the Institutional Animal Care and Use Committee. Three-month-old female domestic pigs were studied in 2 groups. Lean pigs (n=7) were fed a standard chow, for a total of 16 weeks, and Obese animals (n=7) a high-cholesterol/carbohydrate diet, containing (% kcal) 17% protein, 20% fructose, 20% complex carbohydrates, and 43% fat (lard, hydrogenated soybean and coconut oils), supplemented with 2% cholesterol & 0.7% sodium cholate by weight(Pawar et al., 2015).
After 16 weeks of feeding, body weight, heart rate, arterial blood pressure, total cholesterol, low-density lipoprotein (LDL), high-density lipoprotein (HDL), triglyceride (TG), and fasting glucose and insulin levels were obtained. Insulin resistance was assessed by the homeostasis model assessment of insulin resistance (HOMA-IR) index(Eirin et al., 2014a). Animals were then euthanized with intravenous sodium pentobarbital (100mg/kg, Fatal Plus, Vortech Pharmaceuticals, Dearborn, MI).
MSCs isolation, characterization, and culture
MSCs collected from swine subcutaneous abdominal fat tissue (5–10g) were digested in collagenase-H, filtered, and cultured for 3 weeks in advanced MEM medium (Gibco/Invitrogen) supplemented with 5% platelet lysate(Crespo-Diaz et al., 2011). Cellular phenotype was confirmed in both groups by cell-surface marker expression(Zhu et al., 2016), and the third passage preserved in Gibco Cell Culture Freezing Medium for subsequent studies.
Mitochondrial structure
MSC mitochondrial content was evaluated by immunofluorescence staining with the mitochondrial outer membrane marker preprotein translocases of the outer membrane (TOM)-20 (Santa CruzBiotechnology, Dallas, TX)(Eirin et al., 2016), which was quantified in 15–20 random fields using a computer-aided image analysis program (ZEN®2012 blue edition, Carl ZEISS SMT, Oberkochen, Germany). In addition, MSCs were re-suspended in Trump’s fixative solution (4% paraformaldehyde and 1% gluteraldehyde in 0.1M phosphate buffer [pH 7.3]) overnight, and mitochondrial number and structure evaluated using a digital electron microscopy (Philips CM10 Transmission Electron Microscope). In 10 randomly selected MSC the counted number of mitochondria/cell was averaged. Mitochondrial area and mitochondrial matrix density were measured using Image-J (version 1.44 for Windows)(Eirin et al., 2017a). In 5 representative mitochondria fully contained within the images. In addition, double immunofluorescence staining with 10N-nonyl-acridine-orange and TOM-20 was performed to assess the MSCs content (% area) of cardiolipin, a phospholipid exclusively located in the inner mitochondrial membrane(Eirin et al., 2012).
Mitochondrial function
Mitochondrial membrane potential was measured by staining cells with tetramethylrhodamine ethyl ester (TMRE, 50nM, ThermoFisher, CA, cat#: T669) for 20min at 37°C, which accumulates exclusively in actively respiring mitochondria with an intact membrane potential(Farrelly et al., 2001). Cytochrome-c oxidase (COX)-IV activity was assessed by fluorometric methods (Abnova Cat# KA3950). Mitochondrial ROS production was measured by staining cells with 2μM Mito-SOX red reagent (ThermoFisher, CA, cat#: M36008) for 30min at 37°C, a lipid-permeant cation that distributes across the mitochondrial membranes and rapidly oxidized by superoxide(Mukhopadhyay et al., 2007). In addition, cellular MSC oxidative stress was assessed by in-situ production of superoxide anion detected using dihydroethidium (DHE)(Eirin et al., 2015), with the percentage of area stained quantified using ZEN®.
High Throughput miRNA Sequencing and data analysis
Sequencing of miRNA was performed as previously described(Eirin et al., 2014b, Eirin et al., 2017b), and the data analyzed using the CAP-miRSeq-v1.1 workflow(Sun et al., 2014). The workflow starts with unaligned FASTQs to generate aligned BAMs, excel sheets containing both raw and normalized known mature miRNA expression counts, predicted novel miRNA, single-nucleotide variants, and various quality reports. The R-based tool from Bioconductor, edgeR2.6.2(Dudakovic et al., 2014, Robinson, McCarthy and Smyth, 2010) was used to perform the differential expression analysis. The significantly upregulated miRNAs were fed into the computational tool ComiR(Coronnello et al., 2012) to identify genes that they target, which were in turn filtered by MitoCarta 2.0, an online inventory of mammalian mitochondrial genes(Calvo, Clauser and Mootha, 2016). Subsequent functional annotation-clustering analysis utilized the DAVID 6.7 database. Primary gene ontology categories were obtained for miRNAs upregulated in Obese-MSCs, and integrated using the Cytoscape v3.4 plugin ClueGO, which allows visualization of functionally arranged gene ontology term networks and connectivity of multiple genes within a common pathway(Bindea et al., 2009).
Expression analysis of miRNAs by quantitative polymerase chain reaction (qPCR)
To validate expression of representative miRNAs, the expression of miR-196a, miR-27b, miR-212-5p, and miR-let-7c in Lean-MSCs and Obese-MSCs was measured by qPCR. Total RNA was isolated from 5×105 –1×106 MSC samples, as previously described(Saad et al., 2016). All primers were from Life Technology (mir-196a: 000495, miR-27b: 243757, miR-212-5p: 461768, and miR-let-7c: 000379). In addition, mRNA and protein expression of the mitochondrial miRNA targets Ubiquinol-Cytochrome-C Reductase Binding Protein (UQCRB) and ATP Synthase, H+ Transporting, Mitochondrial Fo Complex (ATP5s) were assessed by qPCR (Thermo Fisher Scientific, Cat# 4331348 and ss04247010) and western blotting (1:1000, ThermoFisher,PA5-48493, and 1:16000, abcam, cat# ab129774, respectively).
Statistical analysis
Statistical analysis was performed using JMP 10.0 (SAS Institute, Cary, NC). Data were expressed as mean±standard deviation. Student’s t-test was used to evaluate statistically significant differences between the Lean and Obese groups. Statistical significance was accepted if p≤0.05.
Results
Systemic characteristics
After 16 weeks of diet, body weight, blood pressure, and cholesterol fractions were higher in Obese compared to Lean pigs (Table 1). Their fasting glucose levels were unaltered, but fasting insulin levels and HOMA-IR score were higher in Obese pigs.
Table 1.
Systemic measurements (mean ± standard deviation) in domestic pigs after 16 weeks of lean or obese diet (n=7 each)
| Parameter | Lean | Obese |
|---|---|---|
| Body Weight (Kg) | 67.6±4.6 | 93.6±1.1* |
| Systolic blood pressure (mmHg) | 129.3±7.4 | 150.4±4.6* |
| Diastolic blood pressure (mmHg) | 88.8±3.7 | 110.4±4.2* |
| Mean blood pressure (mmHg) | 100.7±4.5 | 122.4±3.4* |
| Total cholesterol (mg/dl) | 85.5±8.6 | 335.7±10.0* |
| HDL cholesterol (mg/dl) | 48.5±6.2 | 158.0±20.3* |
| LDL cholesterol (mg/dl) | 35.2±6.6 | 434.7±143.8* |
| Triglycerides (mg/dl) | 9.0±4.5 | 15.3±2.3* |
| Fasting glucose (mg/dl) | 121.2±14.9 | 123.3±13.6 |
| Fasting insulin (μU/ml) | 0.4±0.1 | 0.7±0.1* |
| HOMA-IR score | 0.7±0.1 | 1.6±0.3* |
p<0.05 vs. Lean.
HDL: High-density lipoprotein, LDL: Low-density lipoprotein, HOMA-IR: Homeostasis model assessment of insulin resistance.
Obesity induces MSC mitochondrial injury and dysfunction
Mitochondria visually assessed by electron microscopy showed a similar number, yet mitochondrial area (size) and matrix density decreased in Obese-MSCs (Figure 1A–B). MSC expression of TOM-20 was similar among the groups, in line with their unchanged mitochondrial number, yet mitochondrial cardiolipin content decreased in Obese- compared to Lean-MSCs (Figure 1C). Mitochondrial membrane potential decreased in Obese-MSCs, as did COX-IV activity (Figure 2A). Furthermore, Obese-MSCs exhibited markedly higher production of mitochondrial ROS and superoxide anion in situ (Figure 2B).
Figure 1.
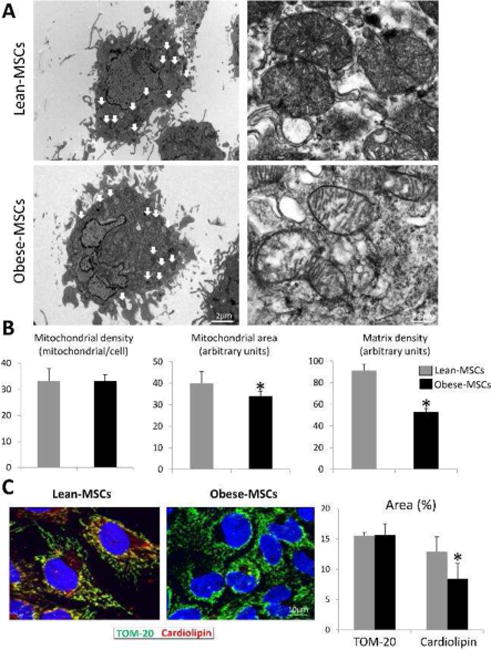
A: Representative transmission electron microscopic images of Lean-MSCs and Obese-MSCs showing decreased cristae membranes and matrix material in Obese-MSC. B: Although mitochondrial number remained unaltered, obesity decreased mitochondrial area and matrix density. C: Fluorescent staining for preprotein translocases of the outer membrane (TOM)-20 and cardiolipin (red) and its quantification, showing decreased cardiolipin content in Obese-MSCs. *p<0.05 vs. Lean-MSCs.
Figure 2.
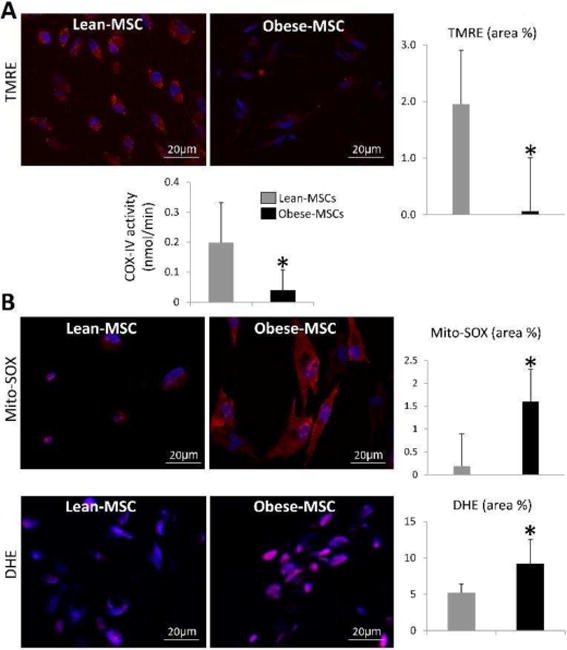
A: Tetramethylrhodamine ethyl ester (TMRE, red) immunoreactivity and cytochrome-c oxidase (COX)-IV activity decreased in Obese- compared to Lean-MSCs. B: Mito-SOX (red) and dihydroethidium (DHE, pink) staining were higher in Obese-MSCs compared to Lean-MSCs (B-D). *p<0.05 vs. Lean-MSCs.
Obesity dysregulated expression of miRNAs targeting mitochondrial genes in MSC
RNA-seq identified a total of 413 MSCs miRNAs, of which miR-196a, miR-27b, miR-504, miR-212-5p, and miR-let-7c were upregulated in Obese- compared to Lean-MSCs (Figure 3A) as confirmed by qPCR (Figure 3B). These miRNAs target 43 mitochondrial genes, including structural and functional genes (Table 2). Expression patterns of the representative mitochondrial miRNA targets UQCRB and ATP5s were confirmed by qPCR and western blot analyses (Figure 3C–D). Functional annotation clustering and pathway analysis of these 43 genes revealed their association with cellular respiration, mitochondrial translation, mitochondrial inner membrane structure, coenzyme biosynthetic process and nucleoside monophosphate biosynthetic process (Figure 4A–B).
Figure 3.
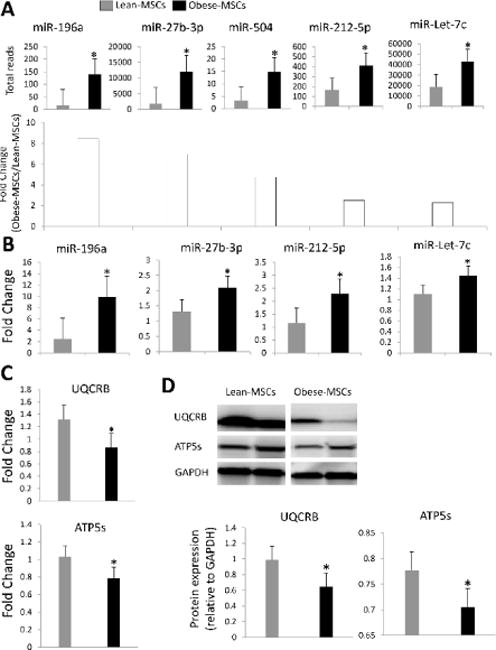
A: RNAseq-derived miRNA expression and fold-change, showing upregulated miR-196a, miR-27b, miR-504, miR-212-5p, and miR-let-7c in Obese-MSCs. B: Expression (qPCR) of miR-196a, miR-27b, miR-212-5p, and miR-let-7c was concordant with the miRNAseq findings. qPCR (C) and western blot (D) confirmed the expression patterns of the of the mitochondrial miRNA targets Ubiquinol-Cytochrome-C Reductase Binding Protein (UQCRB) and ATP Synthase, H+ Transporting, Mitochondrial Fo Complex (ATP5s). *p<0.05 vs. Lean-MSCs.
Table 2.
Forty-three mitochondrial genes targeted by miRNAs upregulated in Obese-MSCs
| Gene ID | Description | Function |
|---|---|---|
| NDUFS1 | NADH dehydrogenase (ubiquinone) Fe-S protein-1 | Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex-I) |
| MAVS | mitochondrial antiviral signaling protein | Peroxisomal and mitochondrial MAVS act sequentially to create an antiviral cellular state. |
| PDPR | pyruvate dehydrogenase phosphatase regulatory subunit | Among its related pathways are Metabolism and Respiratory electron transport |
| SPTLC2 | serine palmitoyltransferase, long chain base subunit-2 | Related pathways include Metabolism and Sphingolipid metabolism. |
| LONP2 | Lon Peptidase-2, Peroxisomal | Among its related pathways are Transport to the Golgi and subsequent modification and Protein folding |
| ALDH5A1 | Aldehyde Dehydrogenase 5 Family, Member-A1 | Encoding a mitochondrial NAD(+)-dependent succinic semialdehyde dehydrogenase |
| HEMK1 | HemK Methyltransferase Family Member-1 | Responsible for the methylation of the GGQ triplet of the mitochondrial translation release factor MTRF1L. |
| RCN2 | Reticulocalbin-2, EF-Hand Calcium Binding Domain | Encoding a protein that contains six conserved regions with similarity to a high affinity Ca (+2)-binding motif, the EF-hand. |
| SLC16A7 | Solute Carrier Family-16 (Monocarboxylate Transporter), | Catalyzes rapid transport across the plasma membrane of many monocarboxylates |
| DNAJC15 | DnaJ (Hsp40) Homolog, Subfamily-C, Member-15 | Negative regulator of the mitochondrial respiratory chain. |
| SLC25A16 | Solute Carrier Family-25 (Mitochondrial Carrier), Member-16 | Rapid transporter between the cytosol and mitochondrial matrix space. |
| METTL8 | Methyltransferase Like-8 | Related to methyltransferase and histone acetyltransferase activities |
| MRS2 | MRS2 Magnesium Transporter | Magnesium transporter that may mediate magnesium influx into the mitochondrial matrix. |
| ATP5S | ATP Synthase, H+ Transporting, Mitochondrial Fo Complex | Involved in regulation of mitochondrial membrane ATP synthase. |
| OPA3 | Optic Atrophy-3 | This protein co-purifies with the mitochondrial inner membrane. |
| PANK2 | Pantothenate Kinase-2 | Catalyzes the first committed step in the universal biosynthetic pathway leading to CoA |
| MRPS25 | Mitochondrial Ribosomal Protein-S25 | Mitochondrial ribosomes consist of a small 28S subunit and a large 39S subunit. |
| MRPL35 | Mitochondrial Ribosomal Protein-L35 | Related pathways include Organelle biogenesis and maintenance and Viral mRNA Translation |
| PCBD2 | Pterin-4 Alpha-Carbinolamine Dehydratase/Dimerization | Involved in tetrahydrobiopterin biosynthesis. |
| RFK | Riboflavin Kinase | Essential for TNF-induced reactive oxygen species (ROS) production. |
| ALDH1L2 | Aldehyde Dehydrogenase-1 Family, Member-L2 | Participates in distribution of one-carbon groups between the cytosol and mitochondria |
| NDUFA9 | NADH Dehydrogenase (Ubiquinone)-1 Alpha Subcomplex | A subunit of the hydrophobic protein fraction of NADH: ubiquinone oxidoreductase (complex-I), located in the inner mitochondrial membrane. |
| SDHC | Succinate Dehydrogenase Complex, Subunit-C | As mitochondrial complex-II, a key enzyme complex of the tricarboxylic acid cycle and aerobic respiratory chains of mitochondria. |
| GUF1 | GUF1 Homolog, GTPase | Promotes mitochondrial protein synthesis. |
| PDK1 | Pyruvate Dehydrogenase Kinase, Isozyme-1 | A mitochondrial multi-enzyme complex that catalyzes oxidative decarboxylation of pyruvate |
| SLC30A6 | Solute Carrier Family-30 (Zinc Transporter), Member-6 | Zinc-efflux transporter which allocates the cytoplasmic zinc to the trans-Golgi network and vesicular compartment |
| APOOL | Apolipoprotein O-Like | A large protein complex of the mitochondrial inner membrane; participates in crista junction formation and mitochondrial function |
| UQCRB | Ubiquinol-Cytochrome-C Reductase Binding Protein | A component of complex-III or cytochrome b–c1 complex of the mitochondrial respiratory chain. |
| AK4 | Adenylate Kinase-4 | Involved in maintaining homeostasis of cellular nucleotides by catalyzing interconversion of nucleoside phosphates |
| ACP6 | Acid Phosphatase-6, Lysophosphatidic | Hydrolyzes lysophosphatidic acid containing a medium-length fatty acid chain to the corresponding monoacylglycerol. |
| ACSL6 | Acyl-CoA Synthetase Long-Chain Family Member-6 | Activation of long-chain fatty acids for synthesis of cellular lipids and degradation via beta-oxidation. |
| SLC25A32 | Solute Carrier Family-25 (Mitochondrial Folate Carrier), | Transports folate across the mitochondrial inner membranes |
| FUNDC2 | FUN14 Domain Containing-2 | Negative genetic interaction between KRASG13D/+ and KRAS+/− |
| CA5B | Carbonic Anhydrase-VB, Mitochondrial | Reversible hydration of carbon dioxide |
| C15orf40 | Chromosome-15 Open Reading | Increased nuclear migration speed |
| PDE12 | Phosphodiesterase-12 | Enzyme degraded triphosphorylated 2–5A to produce AMP and ATP. |
| XPNPEP3 | X-Prolyl Aminopeptidase-3, Mitochondrial | Localizes to the mitochondria of renal cells; participates in ciliary function. |
| LONP1 | Lon Peptidase-1, Mitochondrial | Encods a mitochondrial matrix protein belonging to the Lon family of ATP-dependent proteases. |
| PPTC7 | PTC7 Protein Phosphatase Homolog | GO annotations related to this gene include phosphoprotein phosphatase activity |
| MRPL42 | Mitochondrial Ribosomal Protein-L42 | Related pathways include Organelle biogenesis and maintenance and Mitochondrial translation. |
| SYNJ2BP | Synaptojanin-2 Binding Protein | Related to protein C-terminus binding and type-II activin receptor binding. |
| AMACR | Alpha-Methylacyl-CoA Racemase | Racemization of 2-methyl-branched fatty acid CoA esters. |
| SLC25A53 | Solute Carrier Family-25, Member-53 | No Compound Related Data Available |
Figure 4.
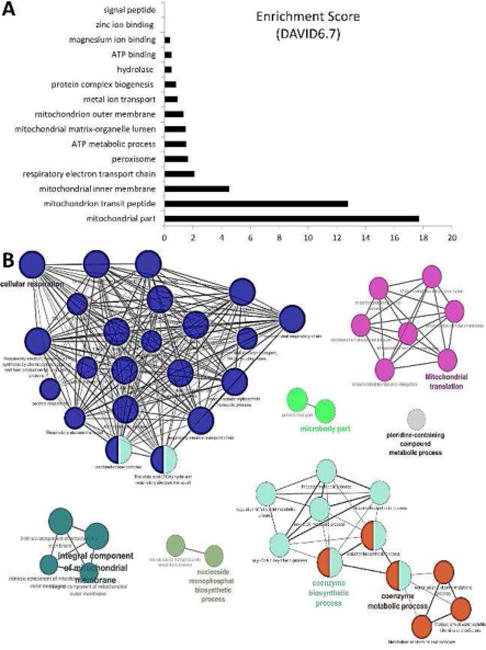
Enrichment of biological functions of miRNA-targeted 43 mitochondrial genes upregulated in Obese-MSCs compared to Lean-MSCs using DAVID6.7 (A) and ClueGo (B), a Cytoscape plugin. The size of the nodes represents the number of genes involved in this network, and the black lines the connectivity of multiple genes within a common pathway. Most miRNA-targeted genes regulate mitochondrial ATP production (blue nodes), translation (pink nodes), and coenzyme metabolic processes (orange and light blue nodes).
Discussion
The current study demonstrates that obesity impairs mitochondrial structure and function in porcine adipose tissue-derived MSCs. Obese-MSCs mitochondria were characterized by fragmentation, decreased cardiolipin content, and matrix loss. Furthermore, mitochondrial membrane potential and COX-IV activity decreased in Obese-MSCs, associated with increased oxidative stress, indicating mitochondrial dysfunction. RNA-seq revealed 5 miRNAs upregulated in Obese-MSCs that regulate gene expression related to mitochondrial structure and function, suggesting that obesity-induced mitochondrial dysfunction might be mediated partly through miRNA-induced mitochondrial gene regulation. Importantly, these alterations may interfere with the repair potency of endogenous MSCs, and should be taken into account when considering the use of autologous MSCs in subjects with obesity.
Transplantation of autologous MSCs has become increasingly popular, resulting in myriad clinical trials (www.clinicaltrials.gov) evaluating its efficacy in patients(Wei et al., 2013). Adipose tissue is a popular source of MSCs for transplantation, as they are relatively easy to harvest from the perivascular fraction of fat, with isolation success rates close to 100%(Kern et al., 2006). These pericyte-like stromal cells are highly abundant in adipose tissue, thus small fat reservoirs appear to be sufficient for MSCs isolation. However, MSCs residing in adipose tissue are exposed to the deleterious metabolic effects of obesity that creates a noxious milieu of inflammation and oxidative stress, which may lead to cellular damage. Therefore, inadequate MSCs vitality, impaired repair capacity, or loss may limit their use for cell-based therapy in obese individuals.
We have recently shown that MSCs from obese pigs exhibit increased propensity for senescence, associated with upregulated TNF-α expression in adipose tissue, suggesting that micro-environmental inflammatory changes during development of obesity affect MSC function(Zhu et al., 2016). The current study extends our previous observations, demonstrating that obesity induces in MSC mitochondrial abnormalities and dysfunction. Specifically, while mitochondrial number was preserved, we found that Obese-MSC mitochondria were smaller compared to Lean-MSCs, and lost cristae membranes and matrix material. A decrease in cardiolipin content in Obese-MSCs also indicates alterations in the inner mitochondrial membrane.
Mitochondrial function heavily relies on their structure, as changes in the matrix and inner membrane impact directly on mitochondrial bioenergetics. Indeed, we found that obesity-induced mitochondrial structural changes were associated with decreased mitochondrial membrane potential. Mitochondrial ATP generation ATP is primarily driven by their membrane potential, the electrical component of the proton motive force generated by serial reduction of electrons through the respiratory electron transport chain(Ramzan et al., 2010). Furthermore, activity of COX-IV which catalyzes the final step in this chain diminished in Obese- compared to Lean-MSCs, suggesting defective respiration and impaired energy production.
Notably, changes in obesity-induced mitochondrial dysfunction in MSC were associated with increased mitochondrial-derived oxidative stress, disclosed by increased mitochondrial as well as cellular production of superoxide anion. Previous studies have shown that cardiolipin remodeling and loss trigger opening of the mitochondrial permeability-transition pore (mPTP) allowing release of mitochondrial ROS to the cytosol, contributing to cellular oxidative stress(Niimi et al., 2012). This postulation is supported by the observation that restoration of mitochondrial cardiolipin attenuates cardiac and renal oxidative stress in porcine renovascular disease(Eirin et al., 2014a, Eirin et al., 2014c, Eirin et al., 2016, Eirin et al., 2012).
To interrogate potential mechanisms by which obesity compromised MSC mitochondrial structure and function, we used miRNA-seq to identify miRNAs upregulated in Obese-MSCs, and their predicted mitochondrial target genes. Previous studies have shown that obesity modulates cellular mechanisms by altering the expression of miRNAs(Jordan et al., 2011, Chen et al., 2014), and that differentiation of human bone marrow-derived MSCs is regulated by miRNAs(Oskowitz et al., 2008). In agreement, we found upregulated expression of miR-196a (8.5-fold), miR-27b (7-fold), miR-504 (5-fold), miR-212-5p (2.5-fold), and miR-let-7c (2-fold) in Obese- compared to Lean-MSCs. Increased miR-196a expression induces a brown adipocyte-like phenotype in white adipose tissue, favoring cellular resistance to obesity(Mori et al., 2012). Because miR-27b attenuates adipocyte differentiation and mitochondrial function, increased expression impairs mitochondrial structure and biogenesis, as well as Complex-I activity, thereby increasing mitochondrial ROS production(Kang et al., 2013). Furthermore, miR-504 modulates mitochondrial biogenesis, oxidative phosphorylation, and Complex-III activity(Zhao et al., 2015), whereas miRNA-212 in conjunction with miR-132 regulates cardiac hypertrophy and mitophagy by targeting the pro-autophagic transcription factor FoxO3(Ucar et al., 2012). Lastly, miR-let-7c is involved in regulation of mitochondrial gene expression by targeting mitochondrial mRNAs(Barrey et al., 2011).
Collectively, these miRNAs target 43 genes that are critical for preserving the structure and function of mitochondria, including genes related to mitochondrial respiration, translation, and electron transport, as well as structural outer and inner membrane genes. One of these putative targets is apolipoprotein-O Like (APOOL), a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex. Being essential for preserving cristae membrane junctions, inner membrane architecture, and formation of contact sites to the outer membrane(Weber et al., 2013, Abdoul-Azize et al., 2016), miRNA-induced APOOL silencing may contribute to matrix and cardiolipin loss, mPTP formation, as well as oxidative stress. Furthermore, dysregulation of other genes involved in mitochondrial function (e.g., NDUFA9, NDUFS1 SDHC, UQCRB, ATP5S) may account for compromised membrane potential. Taken together, our findings may implicate miRNA-induced silencing or post-transcriptional regulation of mitochondria related genes in mitochondrial injury and dysfunction in obese-MSCs. Nevertheless, additional mechanisms might have altered the gene expression profile of Obese-MSCs, including transcription factors, histone methylation or alternative splicing.
Our study is limited by the use of relatively short duration of obesity, and lack of co-morbidities that may exacerbate obesity-induced MSCs mitochondrial damage. Nevertheless, adipose tissue-derived MSCs exposure to an obesity milieu for 16 weeks was sufficient to impair their mitochondria, triggering oxidative stress and apoptosis. Nonyl-acridine-orange signal, used to assess cardiolipin content, may also be modulated by changes in mitochondrial membrane potential and respiration rate. Further studies are also needed to explore whether obesity-induced MSC mitochondrial dysfunction impairs the in-vivo MSC efficacy and regenerative capacity, and if such changes are reversible.
In summary, this study provides evidence that obesity induces mitochondrial damage and dysfunction in porcine adipose tissue-derived MSCs, which results in increased oxidative stress. Furthermore, miRNAs that target structural and functional mitochondrial genes were upregulated in Obese-MSCs, suggesting that miRNA-based mechanisms might be partly responsible for the deleterious effects of obesity on MSCs mitochondria. Therefore, our findings highlight the vulnerability of this endogenous repair system, and suggest novel therapeutic targets for development of adequate tools to preserve the regenerative potency of MSCs and their suitability for autologous transplantation in obese patients.
Acknowledgments
This study was partly supported by the NIH grant numbers: DK73608, DK104273, HL123160, and DK102325, DK106427, and the Mayo Clinic: Mary Kathryn and Michael B. Panitch Career Development Award. We also appreciate the generous philanthropic support of William and Karen Eby.
Funding: Contract grant sponsor: NIH; Contract grant numbers: DK73608, DK104273, HL123160, and DK102325, DK106427.
Footnotes
This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: [10.1002/jcp.26402]
Conflict of Interest
The authors declare no conflict of interest.
References
- Abdoul-Azize S, et al. Pyr3, a TRPC3 channel blocker, potentiates dexamethasone sensitivity and apoptosis in acute lymphoblastic leukemia cells by disturbing Ca(2+) signaling, mitochondrial membrane potential changes and reactive oxygen species production. Eur J Pharmacol. 2016;784:90–98. doi: 10.1016/j.ejphar.2016.05.014. [DOI] [PubMed] [Google Scholar]
- Barrey E, et al. Pre-microRNA and mature microRNA in human mitochondria. PLoS One. 2011;6(5):e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(D1):D1251–1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalena G, et al. Mpv17 in mitochondria protects podocytes against mitochondrial dysfunction and apoptosis in vivo and in vitro. Am J Physiol Renal Physiol. 2014;306(11):F1372–1380. doi: 10.1152/ajprenal.00608.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, et al. MiR-146b is a regulator of human visceral preadipocyte proliferation and differentiation and its expression is altered in human obesity. Mol Cell Endocrinol. 2014;393(1–2):65–74. doi: 10.1016/j.mce.2014.05.022. [DOI] [PubMed] [Google Scholar]
- Coronnello C, et al. Novel modeling of combinatorial miRNA targeting identifies SNP with potential role in bone density. PLoS Comput Biol. 2012;8(12):e1002830. doi: 10.1371/journal.pcbi.1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe MA, et al. Using miRNA-mRNA Interaction Analysis to Link Biologically Relevant miRNAs to Stem Cell Identity Testing for Next-Generation Culturing Development. Stem Cells Transl Med. 2016;5(6):709–722. doi: 10.5966/sctm.2015-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Diaz R, et al. Platelet lysate consisting of a natural repair proteome supports human mesenchymal stem cell proliferation and chromosomal stability. Cell Transplant. 2011;20(6):797–811. doi: 10.3727/096368910X543376. [DOI] [PubMed] [Google Scholar]
- Du JK, et al. Upregulation of microRNA-22 contributes to myocardial ischemia-reperfusion injury by interfering with the mitochondrial function. Free Radic Biol Med. 2016;96:406–417. doi: 10.1016/j.freeradbiomed.2016.05.006. [DOI] [PubMed] [Google Scholar]
- Dudakovic A, et al. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem. 2014;115(10):1816–1828. doi: 10.1002/jcb.24852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin A, et al. Restoration of Mitochondrial Cardiolipin Attenuates Cardiac Damage in Swine Renovascular Hypertension. J Am Heart Assoc. 2016;5(6) doi: 10.1161/JAHA.115.003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin A, et al. Mitochondrial protection restores renal function in swine atherosclerotic renovascular disease. Cardiovasc Res. 2014a;103(4):461–472. doi: 10.1093/cvr/cvu157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin A, et al. A mitochondrial permeability transition pore inhibitor improves renal outcomes after revascularization in experimental atherosclerotic renal artery stenosis. Hypertension. 2012;60(5):1242–1249. doi: 10.1161/HYPERTENSIONAHA.112.199919. [DOI] [PubMed] [Google Scholar]
- Eirin A, et al. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene. 2014b;551(1):55–64. doi: 10.1016/j.gene.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin A, et al. Mitochondrial targeted peptides attenuate residual myocardial damage after reversal of experimental renovascular hypertension. J Hypertens. 2014c;32(1):154–165. doi: 10.1097/HJH.0b013e3283658a53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin A, et al. The metabolic syndrome induces early changes in the swine renal medullary mitochondria. Transl Res. 2017a;184:45–56e49. doi: 10.1016/j.trsl.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin A, et al. Intra-renal delivery of mesenchymal stem cells attenuates myocardial injury after reversal of hypertension in porcine renovascular disease. Stem Cell Res Ther. 2015;6:7. doi: 10.1186/scrt541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin A, et al. Integrated transcriptomic and proteomic analysis of the molecular cargo of extracellular vesicles derived from porcine adipose tissue-derived mesenchymal stem cells. PLoS One. 2017b;12(3):e0174303. doi: 10.1371/journal.pone.0174303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrelly E, et al. A high-throughput assay for mitochondrial membrane potential in permeabilized yeast cells. Anal Biochem. 2001;293(2):269–276. doi: 10.1006/abio.2001.5139. [DOI] [PubMed] [Google Scholar]
- Folmes CD, et al. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11(5):596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TS, Lean ME. A clinical perspective of obesity, metabolic syndrome and cardiovascular disease. JRSM Cardiovasc Dis. 2016;5:2048004016633371. doi: 10.1177/2048004016633371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Jordan SD, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13(4):434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- Kadye R, et al. Guardian of the furnace: mitochondria, TRAP1, ROS and stem cell maintenance. IUBMB Life. 2014;66(1):42–45. doi: 10.1002/iub.1234. [DOI] [PubMed] [Google Scholar]
- Kang T, et al. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J Biol Chem. 2013;288(48):34394–34402. doi: 10.1074/jbc.M113.514372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern S, et al. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24(5):1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- Ludin A, et al. Reactive oxygen species regulate hematopoietic stem cell self-renewal, migration and development, as well as their bone marrow microenvironment. Antioxid Redox Signal. 2014;21(11):1605–1619. doi: 10.1089/ars.2014.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, et al. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2012;10(4):e1001314. doi: 10.1371/journal.pbio.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P, et al. Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem Biophys Res Commun. 2007;358(1):203–208. doi: 10.1016/j.bbrc.2007.04.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi K, et al. Mitochondrial permeability transition pore opening induces the initial process of renal calcium crystallization. Free Radic Biol Med. 2012;52(7):1207–1217. doi: 10.1016/j.freeradbiomed.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Oskowitz AZ, et al. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci U S A. 2008;105(47):18372–18377. doi: 10.1073/pnas.0809807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GC, Acsadi G, Brenner CA. Mitochondria: determinants of stem cell fate? Stem Cells Dev. 2009;18(6):803–806. doi: 10.1089/scd.2009.1806.edi. [DOI] [PubMed] [Google Scholar]
- Pawar AS, et al. Adipose tissue remodeling in a novel domestic porcine model of diet-induced obesity. Obesity (Silver Spring) 2015;23(2):399–407. doi: 10.1002/oby.20971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales-Clemente E, Folmes CD, Terzic A. Metabolic regulation of redox status in stem cells. Antioxid Redox Signal. 2014;21(11):1648–1659. doi: 10.1089/ars.2014.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramzan R, et al. Mitochondrial respiration and membrane potential are regulated by the allosteric ATP-inhibition of cytochrome c oxidase. Biochim Biophys Acta. 2010;1797(9):1672–1680. doi: 10.1016/j.bbabio.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohban R, Pieber TR. Mesenchymal Stem and Progenitor Cells in Regeneration: Tissue Specificity and Regenerative Potential. Stem Cells Int. 2017;2017:5173732. doi: 10.1155/2017/5173732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad A, et al. Adipose-derived mesenchymal stem cells from patients with atherosclerotic renovascular disease have increased DNA damage and reduced angiogenesis that can be modified by hypoxia. Stem Cell Res Ther. 2016;7(1):128. doi: 10.1186/s13287-016-0389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, et al. CAP-miRSeq: a comprehensive analysis pipeline for microRNA sequencing data. BMC Genomics. 2014;15:423. doi: 10.1186/1471-2164-15-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar A, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber TA, et al. APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PLoS One. 2013;8(5):e63683. doi: 10.1371/journal.pone.0063683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, et al. Mesenchymal stem cells: a new trend for cell therapy. Acta Pharmacol Sin. 2013;34(6):747–754. doi: 10.1038/aps.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, et al. miR-504 mediated down-regulation of nuclear respiratory factor 1 leads to radio-resistance in nasopharyngeal carcinoma. Oncotarget. 2015;6(18):15995–16018. doi: 10.18632/oncotarget.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XY, et al. Functional Plasticity of Adipose-Derived Stromal Cells During Development of Obesity. Stem Cells Transl Med. 2016;5(7):893–900. doi: 10.5966/sctm.2015-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]


