Abstract
Upon recognition of a microbial pathogen, the innate and adaptive immune systems are linked to generate a cell-mediated immune response against the foreign invader. The culture filtrate of Mycobacterium tuberculosis contains ligands such as M. tuberculosis tRNA that activate the innate immune response and secreted antigens recognized by T cells to drive adaptive immune responses. Here, bioinformatics analysis of gene expression profiles derived from human peripheral blood mononuclear cells treated with distinct microbial ligands identified a mycobacterial tRNA-induced innate immune network resulting in the robust production of IL-12p70, a cytokine required to instruct an adaptive Th1 response for host defense against intracellular bacteria. As validated by functional studies, this pathway comprised a feed-forward loop whereby the early production of IL-18, Type I interferons (IFNs) and IL-12p70, primed NK cells to respond to IL-18 and produce IFN-γ, enhancing further production of IL-12p70. Mechanistically, tRNA activates both TLR3 and TLR8, and this synergistic induction of IL-12p70 was recapitulated by the co-addition of a specific TLR8 agonist with a TLR3 ligand to PBMC. These data indicate that M. tuberculosis tRNA activates a gene network involving the integration of multiple innate signals including both Type I and Type II IFNs, as well as distinct cell types to induce IL-12p70.
Introduction
The identification of the components of M. tuberculosis that trigger host immune responses has long been a goal of scientists studying the disease. In the 19th century, Robert Koch, after identifying M. tuberculosis as the cause of tuberculosis, went on to show that the bacilli induced protective immunity, which he ascribed to a culture filtrate preparation called “tuberculin” (1). Although tuberculin was later disproved to be a cure for tuberculosis, Koch had discovered what would become a standard diagnostic test for exposure to M. tuberculosis. The protocol for purifying tuberculin from the bacterial culture filtrate was refined by Frances Seibert, who demonstrated that the preparations confer antigenicity when injected into skin (2–7). This work led to the identification of the protein antigens from M. tuberculosis culture filtrate that are recognized by, and can elicit adaptive responses by T cells (8–10). The proteins purified from the culture filtrate elicit robust delayed type hypersensitivity responses in patients previously exposed to M. tuberculosis and individuals vaccinated with BCG. Yet protein purification largely hinders their ability to generate adaptive immune responses by themselves, suggesting that an immune adjuvant is present in the culture filtrate.
To combat the intracellular pathogen, M. tuberculosis, the innate and adaptive immune responses interact, with the innate response responsible for instructing the type of the adaptive T cell response. The innate immune system utilizes pattern recognition receptors (PRRs) to recognize a diverse array of microbial ligands known as pathogen associated molecular patterns (PAMPs). The host has deployed PRRs in a variety of subcellular locations recognizing distinct microbial ligands resulting in the activation of both common and specific innate immune responses. For example, distinct dendritic cell (DC) differentiation programs are induced by muramyl dipeptide of mycobacteria via the cytoplasmic receptor NOD2, vs. triacylated lipopeptide, a ligand for the cell surface receptor TLR2/1 (11). Furthermore, mycobacterial DNA, released from bacteria that reside in the host phagosome, gains access to the cytoplasm to trigger nucleotide sensor pathways such as STING, leading to a Type I IFN response (12–15). In chronic bacterial infections such as tuberculosis, the induction of Type I IFNs opposes the action of the Type II IFN, IFN-γ, required for an effective antimicrobial response against the causative pathogen (16, 17).
While the M. tuberculosis culture filtrate contains several protein antigens that elicit T cell responses, the culture filtrate also contains one or more microbial ligands, i.e. PAMPs, which trigger innate instruction of the adaptive T cell response, that are largely removed during the purification of the purified protein derivative. In addition to proteins, M. tuberculosis culture filtrate contains nucleic acids (7), with tRNA being an abundant form of RNA (18). Treatment of human monocytes with tRNA purified from M. tuberculosis culture filtrate induced their apoptosis, which is thought to contribute to the pathogenesis of tuberculosis (18). However, the extent and specificity of the immune response triggered by M. tuberculosis tRNA remains unknown. Therefore, we investigated whether M. tuberculosis tRNA triggers a distinct innate immune response for instruction of the adaptive T cell response.
Materials and Methods
Cell purification and culture
Whole blood was obtained from healthy donors who provided written informed consent (UCLA Institutional Review Board). PBMC were isolated by Ficoll-hypaque (GE Healthcare) density gradient centrifugation and cultured in RPMI (Gibco) supplemented with 10% FCS (Hyclone) and 1% Pen/Strep glutamine (Gibco) at a density of 2×106/ml in 96-well flat bottomed plates (Corning) at 37 °C with 4% CO2.
Reagents for Cell Stimulation
TLR2/1L, a synthetic lipopeptide derived from the 19 kDa mycobacterial lipoprotein was obtained from EMC Microcollections and used at 10 µg/ml. PolyI:C (HMW) and TLR-506 were from Invivogen and used at 2 µg/ml and 500 nM respectively. ssRNA40 (phosphothioate backbone, HPLC purified) was synthesized by IDT and used at 500 ng/ml.
Total RNA was isolated from M. tuberculosis H37Rv that was grown to OD 0.8–1 (2×108 cells/mL) and lysed using Trizol in bead-beating tubes, in the presence of antioxidants (19). The cells were agitated with 4 cycles of beating, each followed by a 5-minute rest period on ice. Chloroform (0.2 mL per mL of Trizol) was added followed by incubation at ambient temperature for 5 min. The samples were shaken and then centrifuged at 12,000 × g for 15 min at 4 °C. The aqueous phase was removed for further tRNA purifications using the Purelink miRNA Isolation Kit (Invitrogen) according to the manufacturer’s instructions. Ethanol (100%) was added to the lysate to give a 35% concentration. The mixture was loaded onto the Purelink column and centrifuged at 12,000 × g for 1 min. The flow through was then mixed with 100% ethanol to give a final concentration of 70% and the mixture was loaded onto a Purelink column and centrifuged at 12,000 × g for 1 min to yield small RNAs (<200 nt). The columns were washed using wash buffer (Invitrogen) twice. The small RNAs were eluted by adding RNase-free water and centrifuging again. tRNA was finally purified by size-exclusion chromatography with an Agilent SEC-3 column (3 µm, 300 A, 7.8 × 300 mm) eluted with 100% 8 mM ammonium acetate at 65 °C to remove contaminating miRNA and other size-resolvable RNA fragments (19). tRNA eluted between 11 and 13 minutes for each sample. Fractions containing tRNA were collected and tRNA quantity and quality were checked using an Agilent Bioanalyzer (19). The RNA purity and quality were assessed using an Agilent Bioanalyzer (19). Using endotoxin-free reagents, endotoxin levels were <2 pg per µg of tRNA.
Nucleic acid ligands (1 µg/ml) were complexed with DOTAP (Roche) according to manufacturer’s instructions to facilitate delivery to the endosome. DOTAP alone did not induce cytokine release. Reagents were determined to be endotoxin-free by Limulus amoebocyte lysate assay (Lonza).
Cytokine Quantification
Primary cells were stimulated on the same day as isolation. Cell supernatants were harvested at 24h unless otherwise noted. Cytokines measured by sandwich ELISA using antibody pairs were as follows: IL-18 (MBL Intl.), IFN-γ (BD), IL-6 and IL-12p40 (Invitrogen). IFN-α, IL-1β, IL-10, IL-12p70 and TNF-α were measured by CBA (BD, Flex Sets).
Blocking Antibodies and TLR inhibitors
PBMC were treated with the following monoclonal neutralizing antibodies compounds for 30 min before stimulation. IL-18 10 µg/ml (MBL Intl), IFN-γ 10 µg/ml (BD), IgG1 10 µg/ml from corresponding manufacturer was used as a control. The specific TLR8-inhibitor VTX-3119 and related control compound VTX-764 were gifts from VentiRx Pharmaceuticals and were used at a concentration of 100 nM which was determined by dose titration to provide optimal inhibition without off-target effects.
CD56 depletion
PBMC were depleted of CD56+ cells using CD56 MicroBeads (Miltenyi Biotech) as directed by manufacturer’s protocol. Depletion was confirmed at >99% purity by flow cytometry. CD56 depleted PBMC were cultured at 1.8×106/ml to reflect the loss of CD56+ cells, estimated to comprise 10% of PBMC.
Flow cytometry
PBMC were labeled with antibodies to CD3 (CD3-FITC Invitrogen) and CD56 (CD56-PE eBioscience), or isotype control. For intracellular detection of IFN-γ, PBMC were treated with GolgiPlug (BD) 4 h prior to harvest. Following surface staining and fixation, cells were treated with Perm/Wash Buffer (BD) and stained with IFNγ-APC (Invitrogen) or isotype control. Flow cytometry was performed on a LSRII (BD Biosciences) in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards P30 CA016042 and 5P30 AI028697. Analysis was performed in FlowJo (Tree Star Inc.).
RNA sequencing
PBMC were stimulated as described. RNA was harvested at 1, 6 and 24 h and isolated with RNeasy micro kit (Qiagen) according to manufacturer’s directions, including on-column DNase digestion. RNA was quantified by Nanodrop and quality assessed by Agilent 2100 Bioanalyzer. Libraries were created from high quality RNA using TruSeq RNA Library Prep Kit v2 (Illumina). Libraries were quantified by Qubit, pooled by donor in groups of 12, and sequenced in duplicate on a HiSeq 2500 Sequencing System (Illumina).
Differential Expression Analysis
Sequenced reads were aligned to the human reference genome (build hg19 UCSC) using TopHat and Bowtie2. Raw counts were calculated with HTseq using the hg19 Ensembl annotation. Normalization and differential expression analysis was performed using the DESeq2 package for R, which in all cases used the data for all three donors. FDR was controlled by applying the Benjamin-Hochberg correction to P-values. Differentially expressed genes were identified using the cutoffs FC > 2 vs Media and adjusted P-value < 0.05. Hierarchical clustering was performed with “hclust”, and principle component analysis via “prcomp” in R (version 3.2.4). Weighted Gene Correlation Network Analysis was performed using the WGCNA package as described (22). A network of relevant gene relationships as calculated by WGCNA was visualized using visANT.
Functional Analysis
Functional analysis of differentially expressed genes was performed using Ingenuity Pathway Analysis (Qiagen). Gene ontology term analysis was performed using the ClueGO plugin (version 2.2.5) for Cytoscape (version 3.3.0) using the GO term database files from 08.06.2016. Significantly enriched terms were identified by right-sided hypergeometric test with Bonferroni P-value correction using a cutoff of P < 0.05. A network connecting canonical pathways and biological functions relevant to induction of Th1 responses derived from IPA, GO terms, and differentially expressed genes was visualized using Gephi (beta version 0.9.1). This was accomplished by the following strategy: 1) pathways chosen from ClueGO analysis of WGCNA turquoise module genes, 2) pathways chosen from IPA analysis of both turquoise module genes as well as similar pathways identified in parallel analysis of genes induced by activation of PBMC by tRNA, 3) select IPA biological functions tRNA relevant to cell types, 4) select genes of interest from tRNA genes differentially expressed vs. TLR2/1L, 5) Pearson correlations for IL18/IL12/IFNG calculated on rLog DESeq2 results matrix; and 6) Gephi used to illustrate connections between pathways and genes connected to IL18, IL12 and IFNG as well as significantly correlated genes.
Statistics
Results are shown as mean ± SEM. Cytokine data was transformed using log10(x+1), and the Shapiro-Wilk test was used to test for normality. One-way ANOVA was performed for comparisons between three or more groups. The two-way repeated measures ANOVA was used to assess significance in experiments with multiple factors. Individual details of statistical analyses for individual experiments, such as post test for multiple comparisons, are explained in the figure legends. Statistical analyses were performed using GraphPad Prism 7 and R 3.4.1 (r-project.org).
Results
M. tuberculosis tRNA and bacterial lipopeptide induce distinct gene expression patterns
To investigate the innate immune response to M. tuberculosis tRNA, we measured the gene expression profiles induced by M. tuberculosis tRNA in human PBMC to include many of the different immune cell types that are likely to be involved. M. tuberculosis tRNA was prepared from strain H37Rv as described (19) A ssRNA 30-mer derived from the HIV-1 long terminal repeat and known to activate TLR7 and TLR8 (20) served as a positive control for detecting innate immune responses to RNA. In addition, we compared the response to a mycobacterial 19-kDa triacylated lipopeptide, which activates cell surface TLR2/1 (TLR2/1 ligand, TLR2/1L) and is known to be a major activator of transcriptional pathways in response to M. tuberculosis. The optimal concentrations of these ligands were determined by dose titration or used as previously established in our laboratory (11).
PBMC from three donors were stimulated with the various ligands, and the cells collected after 1, 6 and 24 h. The mRNAs were isolated, libraries prepared and gene expression profiles obtained by RNA-seq, which after filtering out background expression, yielded a dataset of 14,637 genes (GEO accession GSE110325; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110325). Principal component analysis (PCA) of the DESeq2 normalized counts was used to first identify samples displaying similar trends in gene expression (Fig. 1A) (5). At the 6 and 24 h timepoints, PCA indicated that gene expression for PBMC treated with M. tuberculosis tRNA and control ssRNA were similar, while gene expression for TLR2/1L-treated and untreated PBMC formed distinct groups. PCA of the gene expression data at 1 h formed a single group, with all three ligands inducing similar profiles as the control media.
Fig 1. Network analysis of M. tuberculosis tRNA, ssRNA, and TLR2/1L-induced gene expression profiles in PBMC.
(A) Principle component analysis of correlation of gene expression from RNAseq on rLog() matrix output from DESeq2. Ellipse denotes 95% confidence interval from k-means clustering of PC1 and PC2 variance. (B) Hierarchical clustering of rLog() transformed counts. Euclidean distance, complete clustering. (C) Overlap of significantly induced genes by mTB tRNA, ssRNA and TLR2/1L, defined by FC > 2 over Media and FDR < 0.05. Hypergeometric p-value calculated for overlaps excluding the 396 common genes. (D) Top Bio Functions identified by Ingenuity Pathway Analysis of significantly induced genes for mTB tRNA, ssRNA or TLR2/1L. (E) Comparison of significantly induced functional pathways induced by mTB tRNA to ssRNA or TLR2/1L.
A second unsupervised analysis, hierarchical clustering, was also performed to characterize the relationships between samples (Fig. 1B). Consistent with the PCA, gene expression profiles for both tRNA and ssRNA at the 6 and 24 h timepoints clustered together. The samples for TLR2/1L at 6 and 24 h formed their own group, as did the samples for the untreated controls at 6 and 24 h. All the 1h samples, regardless of the stimulus and including the media control, clustered on a separate branch from the 6 and 24 h samples, indicating that there was little difference in the transcriptional response at this early timepoint.
To determine the gene signatures induced in PBMC by each ligand vs. the media control, differential gene expression was calculated with DESeq2 (21). For each ligand we compared the two groups of samples (ligand vs media), with each group containing data from three donors. Differentially expressed genes for each timepoint were those that had a threshold of fold-change (FC) > 2 between the groups and a false discovery rate (FDR) < 0.05. We calculated a gene signature of significantly induced genes for each ligand by taking the union of the differentially expressed genes at each timepoint for a given ligand. Comparison of gene signatures overall showed a striking overlap [P < 10−10] of the tRNA and ssRNA signatures, with 1,465 common genes out of a total of 1,692 genes for tRNA and 1,791 for ssRNA (Fig. 1C). Of these RNA-induced genes, 396 were also induced by TLR2/1L, leaving 1,069 induced by only tRNA and ssRNA. By contrast, TLR2/1L induced a distinct signature, with 720 significantly induced genes, of which 215 were unique to TLR2/1 activation.
To investigate the immune pathways induced by each ligand, functional analysis of the gene signatures was performed using Ingenuity. Analysis using the biologic functions tool showed common pathways between the tRNA and ssRNA gene signatures, identifying roles for NK cells, DC, monocytes, and differentiation of Th1 cells (Fig. 1D). By contrast, the biologic functions of the TLR2/1L induced gene profile indicated monocytes, macrophages and neutrophil-associated pathways. Ingenuity analysis using the canonical pathways tool identified ‘DC maturation’, ‘NK-DC crosstalk’, ‘IFN signaling’, ‘PRR recognition of bacteria and viruses’, and ‘communication between innate and adaptive immune cells’, as all more significantly induced by tRNA as compared to TLR2/1L (>10−5 difference in P-value, Fig. 1E). In comparison, the significance of canonical pathways identified for the tRNA and ssRNA signatures was similar. Together, these data suggest that tRNA and ssRNA, but not TLR2/1L, induced common gene signatures in PBMC indicative of an innate pathway for induction of Th1 cells.
Identification of gene expression networks induced by M. tuberculosis tRNA
To further define the potential interaction between genes associated with tRNA/ssRNA induction and those associated with a Th1 response, we used weighted gene co-expression network analysis (WGCNA), an unbiased approach that defines modules of highly interconnected genes based on pairwise correlations (22). We tried to identify modules that were specifically induced by one of the ligands. This was accomplished by first encoding the ligands in a binary vector that was one for a specific ligand, and all its time points, and zero for all other ligands. The module expression levels were then correlated with these binary vectors to identify specific module/ligand associations.
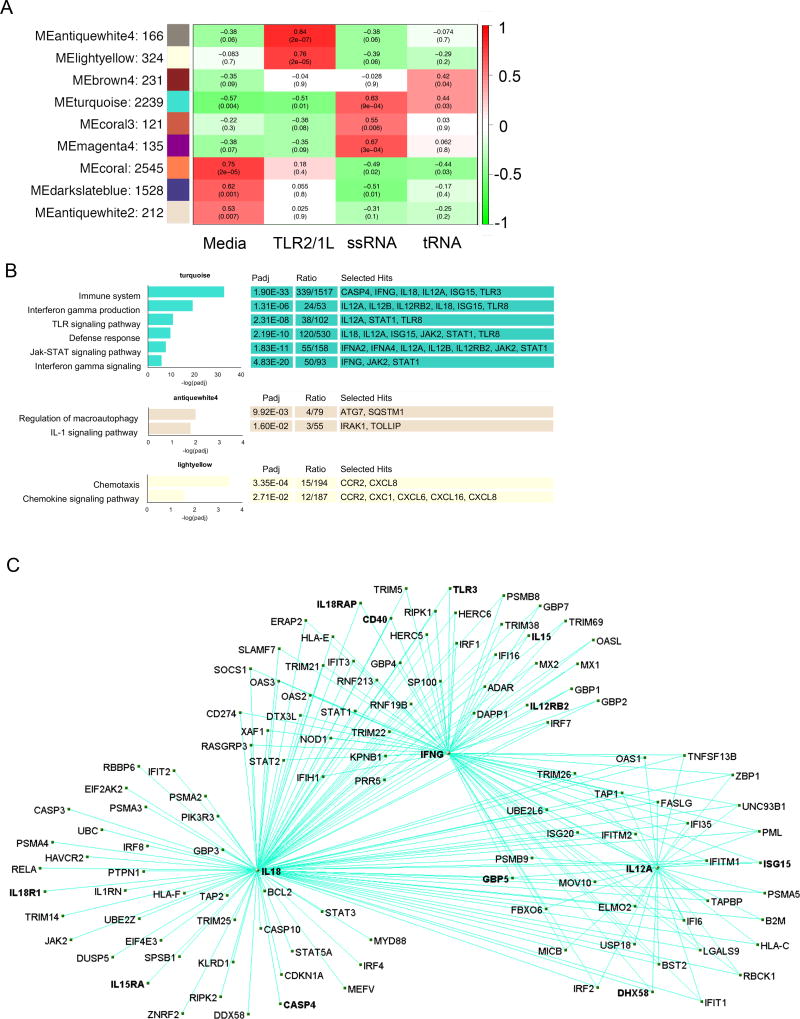
WGCNA identified 31 gene modules, of which nine were positively correlated with at least one microbial ligand (Fig. 2A). In addition to the modules associated with tRNA and ssRNA activation of PBMC, TLR2/1L activation was associated with MEantiquewhite4, indicating significant enrichment of terms including ‘regulation of macroautophagy’ and ‘IL-1 signaling pathway’ (Fig. 2B). The MElightyellow module was also associated with TLR2/1L activation, and enriched for genes associated with ‘chemotaxis’ and ‘chemokine signaling pathway’. Several modules were associated with the media-treated cells. Also, there were modules that were negatively associated with a given stimulus, but were not further studied.
Fig 2. Identification of RNA – correlated module and associated functional analysis.
(A) WGCNA eigengene modules correlated to at least one treatment condition P < 0.05. Red indicates positive correlation and green inverse correlation. (B) Top hits for functional term annotation of WGCNA modules positively correlated with mTB tRNA and ssRNA or TLR2/1L signatures. Padj is calculated by ClueGO as the P-value with Bonferroni correction for the association of the functional term with the gene expression data. Ratio represents the genes for a given functional term that are present in the module over the total number of genes for the term. (C) Visualization of the gene network derived from the WGCNA turquoise module. The module was filtered for genes exhibiting significant differential expression during tRNA treatment and annotated with the GO term “immune pathway” to select genes associated response to mTB infection.
MEturquoise was the only module that significantly correlated with both tRNA and ssRNA treatment of PBMC. Gene ontology analysis (23) of MEturquoise revealed association with the terms ‘IFN gamma production’ and ‘IFN gamma signaling’ (Fig. 2B). This module contained IL18, IFNG, and IL12A, key cytokines involved in induction of a Th1 response, thereby identifying a correlated gene network for Th1 cell differentiation as revealed by the biologic function analysis of the tRNA induced genes. The other modules that were significantly associated with either TLR2/1L or ssRNA stimulation of PBMC were not associated with immune GO terms.
To map the gene network involved in Th1 differentiation, we filtered the tRNA associated MEturquoise module using the ‘immune system’ gene ontology term, identifying 339 genes which included the key genes involved in induction of Th1 cells. This gene set was further filtered by differential expression in tRNA vs. media control (FC > 2, FDR < 0.05, calculated by DESq2 using data from all three donors) identifying 241 genes. The correlation between these individual genes as calculated by WGCNA was visualized by a connectivity map (24) (Fig. 2C), centered on IL18, IFNG and IL12A with connections of edge weight indicated (and requiring a pairwise correlation between genes ≥ 0.2). Highlighting the genes connected to IL18, IFNG and IL12A revealed a module of 120 genes, in which IFNG and IL12A were connected to each other and had multiple connections to other genes in the module. These included IL18R1, IL18RAP and IL12RB2, which encode relevant cytokine receptors, as well as STAT1, STAT2, STAT3 and STAT5A, which encode proteins involved in signaling pathways related to the induction or downstream effects of IL-18, IFNG and IL12A. Strikingly, both the Type II interferon, IFNG, and several Type I interferon genes, as well as interferon-induced downstream genes, were identified in the top 250 genes induced by tRNA (Table S1A and S1B). In fact, of the top 25 genes, 16 were Type I interferon or downstream genes, including ISG15, CASP4 and TLR3. These genes were strongly induced by tRNA and ssRNA but not by TLR2/1L. These data implicated both Type I IFN and the Type II IFN (IFNG) in the induction of a Th1 response, which was surprising to us, given that the induction of Type I IFN in tuberculosis is known to downregulate IFN-γ responses.
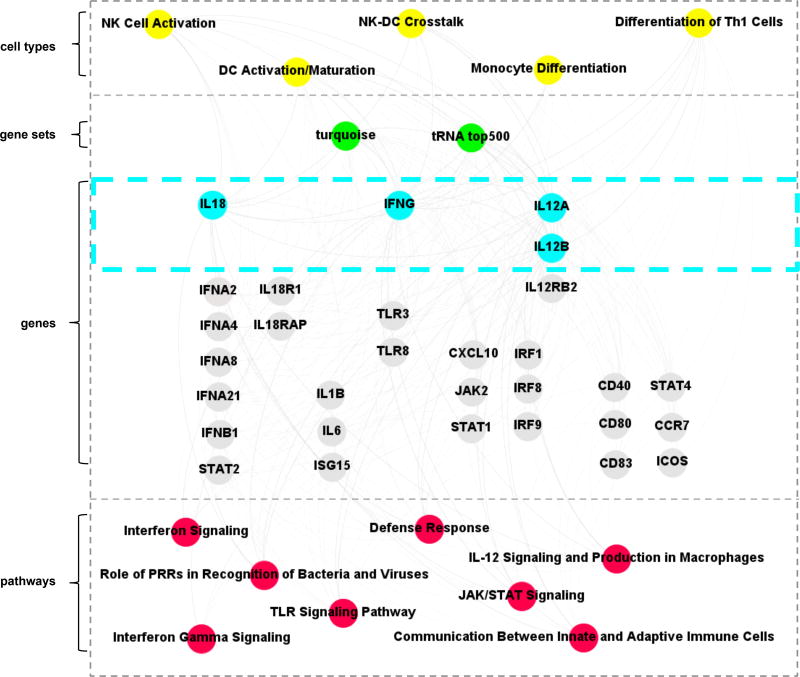
Integration of the bioinformatics analyses of the gene expression profiles for the tRNA treatment of PBMC and the WGCNA turquoise module was performed to link cell type-associated pathways with specific genes related to Th1 functional response pathways (Fig. 3). This analysis identified that M. tuberculosis tRNA directly or indirectly triggered pathways associated with monocytes, NK, DC and T cells, including Th1 cells, to induce a set of genes including IL18, IL18R1, IFNG, JAK2, STAT1, IL12A, IL12B, IL12RB2, and STAT4. These genes are contained within functional pathways including ‘Role of PRR recognition of Bacteria and Viruses’, ‘TLR signaling’, ‘IFN-γ signaling’, ‘DC maturation’, ‘T helper differentiation’ and ‘defense response’. In contrast, the top four TLR2/1L induced pathways were ‘Granulocyte Adhesion and Diapedesis’, ‘Agranulocyte Adhesion and Diapedesis, ‘Differential Regulation of Cytokine Production in Macrophages and T Helper Cells by IL-17A and IL-17F’, ‘Differential Regulation of Cytokine Production in Intestinal Epithelial Cells by IL-17A and IL-17F’. Together, these data suggest a model in which tRNA activates specific cell types that interact to establish a host-defense gene network involved in the innate instruction of an adaptive Th1 response.
Fig 3.
Integrated network of gene expression and functional analysis terms. Gephi was used to create a functional annotation network, showing connections between significant gene ontology terms, IPA canonical pathways, WGCNA modules and significantly expressed genes.
Analysis of genes characteristic of Th1 differentiation and validation in PBMC
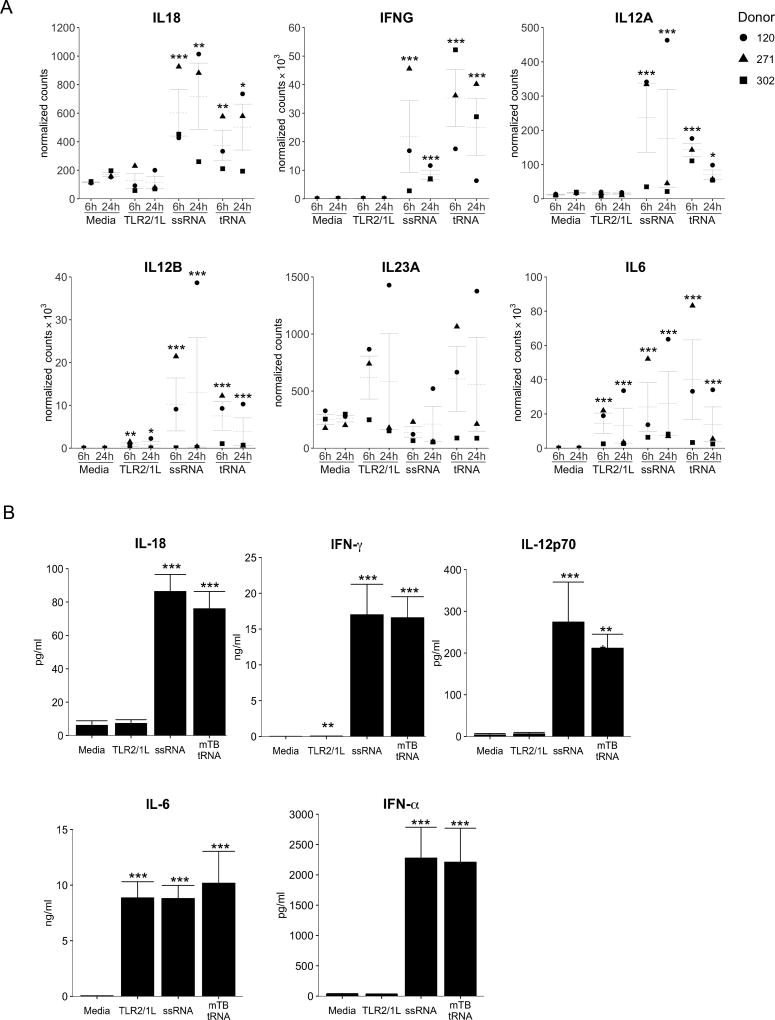
Examination of the log2 normalized counts of key genes identified by the informatics analysis as part of the M. tuberculosis tRNA-induced Th1 differentiation pathway confirmed that, both tRNA and ssRNA, but not TLR2/1L, significantly induced expression of IL18 mRNA (Fig. 4A) and its receptor heterodimer IL18R1 and IL18RAP (Fig. S1A). IFNG and IL12A mRNAs were also upregulated by both tRNA and ssRNA, but not TLR2/1L. IL12B was upregulated by all stimuli, whereas IL23Awas not significantly induced by any of the treatments. IL6 mRNA was strongly induced by all three ligands. This analysis revealed that additional genes connected to the central genes in the Th1 differentiation pathway, including Type I IFN and downstream genes, as well as genes encoding TLR3, CASP4, and ISG15, were also upregulated, but again only in response to tRNA and ssRNA, not TLR2/1L (Fig. S1B).
Fig 4. Analysis of genes characteristic of Th1 differentiation and validation in PBMC.
(A) Log-transformed, normalized counts for select genes at 6h and 24h, mean ± SEM, statistical significance of differential expression (as compared to media at each timepoint) is reported as Benjamini-Hochberg adjusted p-value. (B) PBMC were stimulated with mTB tRNA, ssRNA or TLR2/1L and cytokine secretion measured at 24h. (n=20). Statistical significance of ligand-stimulated PBMC vs. media control calculated by one-way repeated measures ANOVA followed by Dunnett’s multiple comparisons test. *p<= 0.05, ** p<=0.01, *** p<=0.001
To validate the gene expression profile, we determined whether the induction of the cytokine mRNAs leads to secretion of the encoded protein. PBMC were stimulated with M. tuberculosis tRNA, ssRNA40, and TLR2/1L, supernatants collected at 24 h and cytokine production measured. Consistent with the gene expression data, PBMC released IL-18, IFN-γ and IL-12p70 in response to tRNA (IL-18: 76±10 pg/ml; IFN-γ: 16586±2932 pg/ml; IL-12p70: 211±33 pg/ml) and ssRNA, but not media or TLR2/1L (Fig. 4B). IL-12p40 was secreted in response to all treatments, although tRNA and ssRNA induced approximately seven-times more IL-12p40 protein than TLR2/1L (not shown). IL-6 secretion served as a control to demonstrate cell activation by all treatments (Fig. 4B). Low levels of IL-23 were detected in supernatants of all three treatments (Fig. S1C). The differential production of IL-12p70 and IL-23 is consistent with the informatics analysis indicating a gene network for instruction of a Th1 immune response. In addition, IFN-α was induced by tRNA and ssRNA but not by TLR2/1L, consistent with the RNA-seq data.
Interdependence of tRNA-induced cytokines in the induction of IL-12p70
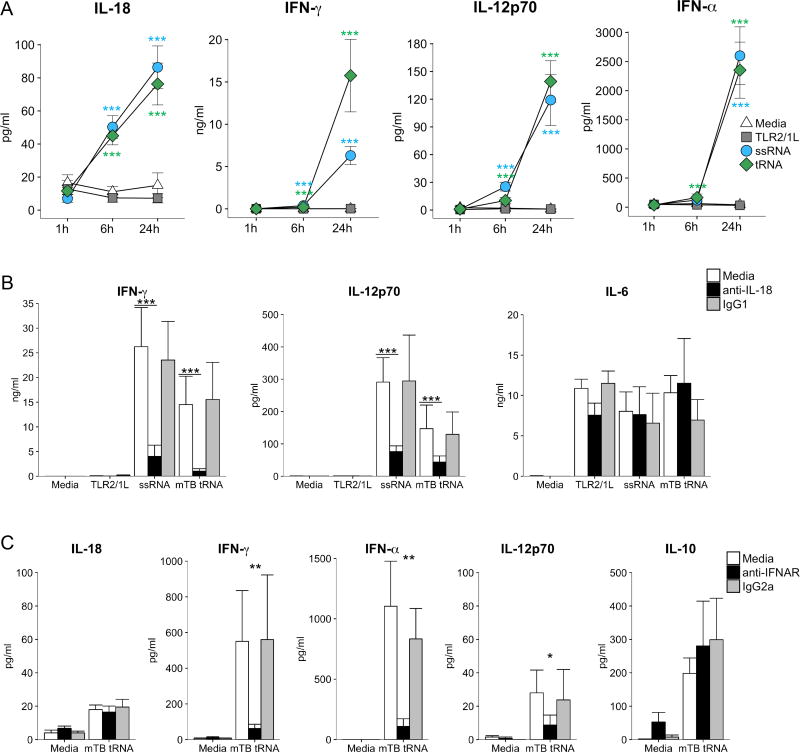
Given that M. tuberculosis tRNA induced the robust production of IL-12p70, we next investigated the immune networks that led to production of this cytokine. The kinetic sequence of cytokine induction in response to the M. tuberculosis tRNA was determined by measuring the time-course of IL-18, IFN-γ and IL-12p70 protein production (Fig. 5A). The earliest detected response was the production of IL-18 at 6 h in response to tRNA and ssRNA (tRNA: 45±5 pg/ml, P = 0.0001). IL-18 protein increased two-fold by 24 h (tRNA: 76±2 pg/ml, P= 0.0001). In contrast, little IFN-α, IFN-γ and IL-12p70 was detected at 1h or 6h, but there was significant induction by 24 h (tRNA. IFN-α: 2351±481pg/ml, P = 0.0001; IFN-γ: 15751±4287 pg/ml, P = 0.0001; IL-12p70: 139±22 pg/ml, P = 0.0001). The temporal pattern of IL-6 secretion was similar to that of IL-18; protein was detected by 6h and the concentration doubled by 24 h (Fig. S1D). These data indicate that IL-18 secretion is triggered early, followed by secretion of IFN-α, IFN-γ and IL-12p70.
Fig 5. Role of IL-18 and Type I IFN.
(A) PBMC were stimulated with mTB tRNA, ssRNA or TLR2/1L and cytokine secretion measured at 1 h, 6 h and 24 h from PBMC stimulated as above. (n=7). Statistical significance of ligand-stimulated PBMC at specific timepoints vs. media control calculated by two-way repeated measures ANOVA followed by Dunnett’s multiple comparisons test. (B) PBMC were pre-treated with monoclonal anti-IL-18 neutralizing antibody or IgG1 isotype control (n=4) or (C) anti-IFNAR neutralizing antibody or IgG2a isotype control (n=3) for 30 m before stimulation with TLR2/1L, ssRNA or mTB tRNA. Supernatants were collected at 24 h and cytokine secretion measured. Statistical significance of ligand-stimulated PBMC vs. media control in the presence of neutralizing and isotype control antibodies calculated by one-way ANOVA followed by Tukey post-hoc test. *p<= 0.05, ** p<=0.01, *** p<=0.001. Data are represented as mean ± SEM. The data within each subfigure were derived from independent donors.
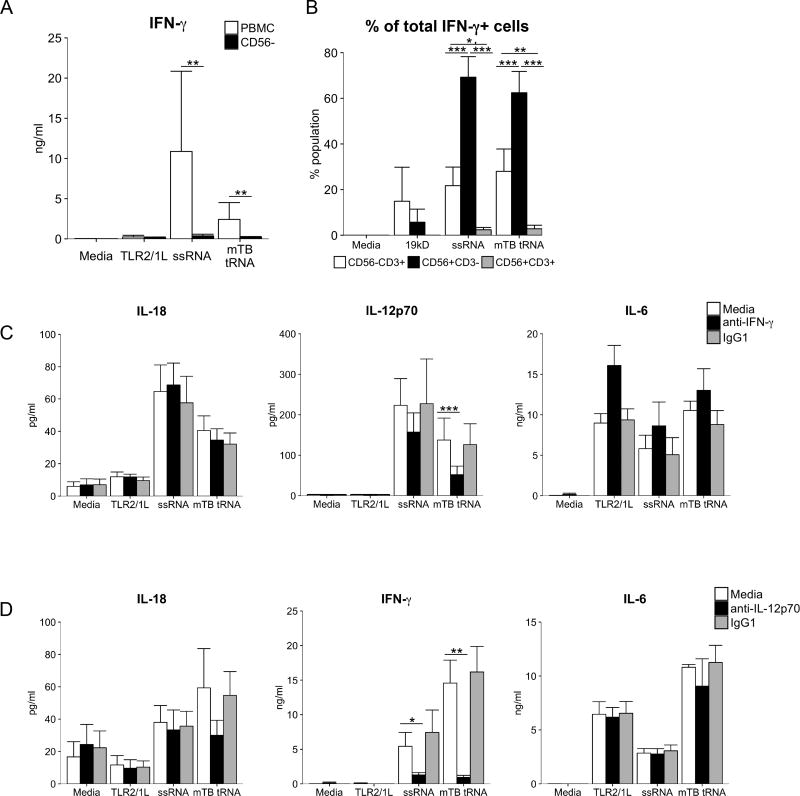
As IL-18 was detected early, i.e. at 6 h, we determined whether its secretion was required for downstream cytokine production. Pre-treatment of PBMC with anti-IL-18 neutralizing antibodies dramatically reduced tRNA-induced secretion of both IFN-γ (93% reduction, P = 0.0001) and IL-12p70 (70% reduction, P = 0.0008) (Fig. 5B). As a control, we measured induction of IL-6, which was not affected by anti-IL-18 treatment.
It has been suggested that the early production of Type I IFN can contribute to optimal induction of IL-12p70 (25). To determine the role of Type I IFN production in the M. tuberculosis tRNA induction of IL-12p70, we performed experiments using a neutralizing antibody to IFN α/β receptor A (IFNAR). Addition of anti-IFNAR mAbs had no effect on tRNA induction of IL-18, but blocked induction of IFN-γ by ∼80% (Fig. 5C). In addition, anti-IFNAR mAbs blocked tRNA induction of IFN-α, consistent with the presence of a Type I IFN-driven positive feedback loop (26, 27). Anti-IFNAR antibody significantly blocked tRNA induction of IL-12p70, while the difference between the isotype control was not significant. There was no effect of anti-IFNAR treatment on tRNA induction of IL-10.
Previous studies have shown that in PBMC, TLR7/8 agonists induce CD56+ (NK and NKT) cells to secrete IFN-γ, dependent on the production of IL-18, even with the concomitant induction of Type I IFNs (28). In order to determine if CD56+ cells were required for M. tuberculosis tRNA induction of IFN-γ, PBMC were depleted for this subset prior to stimulation, and induction of IFN-γ release was assessed. In PBMC depleted of CD56+ cells, M. tuberculosis tRNA induction of IFN-γ was almost entirely inhibited (tRNA: 141±118 pg/ml CD56 depleted vs. 4557±4157 pg/ml PBMC, 97% reduction, P = 0.0215) (Fig. 6A). Although TLR3 ligands have been reported to directly activate NK cells (29), we did not find such activation here (data not shown).
Fig 6. Role for IFN-γ and NK cells.
(A) PBMC were depleted of CD56+ cells, stimulated as shown and cytokine secretion measured at 24h (n=3). Statistical significance calculated by one-way repeated measures ANOVA and Šidák correction applied to the multiple comparison between PBMC and CD56 depleted populations. (B) PBMC were stimulated, and Brefeldin A added at 20h to arrest cytokine secretion. Cells were collected at 24h, stained with CD3-FITC, CD56-PE and IFN-γ-APC and measured by Flow cytometry. IFN-γ+ lymphocytes were divided into subpopulations determined by CD3/CD56 staining and shown as percentage of parent (IFN-γ+) population (n=3). (C) PBMC were pre-treated with monoclonal anti-IFN-γ neutralizing antibody for 30m before stimulation as previous. Cytokine secretion was measured at 24h (n=6). (D) PBMC were pre-treated with monoclonal anti-IFN-γ neutralizing antibody for 30m before stimulation as previous. Cytokine secretion was measured at 24h (n=3). Data are represented as mean ± SEM. Statistical significance of ligand-stimulated PBMC in the presence of neutralizing or isotype control antibodies for B, C, and D was calculated by two-way ANOVA followed by Tukey post-hoc test. *p<= 0.05, ** p<=0.01, *** p<=0.001
We further demonstrated that CD56+ cells were the major source of IFN-γ by intracellular cytokine labelling and flow cytometry. PBMC were stimulated as above, then stained with antibodies for IFN-γ, CD3 and CD56 to measure intracellular IFN-γ with the goal to differentiate NK, NKT and T cell populations. IFN-γ+ cells were detectable above background in tRNA- and ssRNA– treated PBMC (Fig. S2). Of IFN-γ positive lymphocytes, the majority were CD56+CD3− NK cells (tRNA: 62±9%; ssRNA 69±9%), followed by CD56−CD3+ T cells (tRNA: 28±10%; ssRNA: 22±8%), and a small number of CD56+CD3+ (tRNA: 2.8±1.5%; ssRNA: 2.4±1%) (Fig. 6B). Since the frequency of NK cells in PBMC is much lower than that of T cells (∼10% vs. ∼75%), this indicated that the populations were differentially predisposed to produce IFN-γ in response to the RNA stimuli. Although T cells are major producers of IFN-γ during the adaptive immune response, this early timepoint measures the innate immune response, in which NK cells produce the majority of IFN-γ.
Since IL-18 is known to trigger secretion of IFN-γ (30–32) and IFN-γ primes IL12A transcription (33), we hypothesized that IFN-γ contributed to the production of IL-12p70. The pre-treatment of PBMC with neutralizing anti-IFN-γ antibodies reduced the M. tuberculosis tRNA-induced secretion of IL-12p70 (62% reduction, P < 0.0001) (Fig. 6C). Secretion of IL-18 was unaffected by anti-IFN-γ treatment, consistent with the detection of IL-18 protein prior to the induction of IFN-γ. IL-6 production, which is IFN-γ independent, served a control.
The ability of TLR ligands to activate NK cells to produce IFN-γ not only involved the production of IL-18 but was also dependent on secretion of IL-12p70 (28). Purified NK cells did not produce IFN-γ in response to the same TLR ligands. We had detected the early production of IL-12p70 as soon as 6 h after stimulation with tRNA or ssRNA (Fig. 5A). Both tRNA and ssRNA induction of IFN-γ was blocked by anti-IL-12p70 neutralizing antibodies, with little effect on IL-18 or IL-6 secretion (Fig. 6D). Thus, we conclude that IL-12p70 and IL-18 are essential for the IFN-γ response to microbial RNA, and IFN-γ contributes to robust production of IL-12p70. In addition, a multicellular response to M. tuberculosis ligands involving monocytes and NK cells contributes to the induction of the Th1 pathway.
tRNA induces cytokine responses via TLR8
The secondary structure of tRNA includes both single- and double-stranded RNA regions. Given that we detected upregulation of both TLR8, a PRR for single-stranded RNA, and TLR3, a PRR for double-stranded RNA, mRNAs by M. tuberculosis tRNA, we explored the role of these PRRs in triggering the Th1 cytokine network, starting with TLR8.
In order to test the requirement of TLR8 signaling for tRNA induction of the Th1 differentiation pathway, we utilized a specific TLR8 antagonist, VTX-3119 (34). A dose titration was performed to determine the optimal concentration for the antagonist and preclude off-target effects (Fig. S3). PBMC were pre-treated with the TLR8 antagonist VTX-3119, or control molecule VTX-764, and stimulated with TL8-506, a synthetic TLR8-specific agonist (35). Pre-treatment with the TLR8 antagonist suppressed cytokine response to the TLR8 agonist vs. pre-treatment with control compound (IL-18: 60% reduction P < 0.0001; IFN-γ: 67% reduction P=0.024; IL-12p70: 47% reduction P = 0.0004). The induction of IL-6 by all three TLR ligands was not affected by the TLR8 inhibitor.
Next, PBMC were pre-incubated with the TLR8 antagonist or control compound, then stimulated with M. tuberculosis tRNA, ssRNA or TLR2/1L (Fig. 7A). Treatment with the TLR8 antagonist reduced secretion of IL-18 by 50% for activation by M. tuberculosis tRNA (mTB tRNA: 12±6 pg/ml vs. 24±10 pg/ml, P = 0.0106) and 67% for ssRNA. IFN-γ secretion was 90% lower for M. tuberculosis tRNA (mTB tRNA: 695±224 pg/ml vs. 6778±2433 pg/ml, P < 0.0001) and 58% reduced for ssRNA. Secretion of IL-12p70 was diminished by 84% for M. tuberculosis tRNA (tRNA: 42±15 pg/ml vs. 264±108 pg/ml, P < 0.0001) and 73% for ssRNA. IL-6 secretion was not significantly affected for any treatment (data not shown).
Fig 7. Role of TLR8 and synergy with TLR3.
(A) PBMC were pre-treated with TLR8 antagonist or control for 30m before treatment with TLR2/1L, ssRNA and mTB tRNA. Cytokine secretion was measured at 24h. (IL-18 and IL-6: (n=3); IL-12p70 and IFN-γ (n=5). Statistical significance calculated by two-way repeated measures ANOVA and multiple comparisons by Tukey post-hoc test. * P < 0.05, ** P < 0.01. (B) PBMC were stimulated with nucleotide oligomers complexed with Dotap (endosomal PRR) or Lipofectamine2000 (cytosolic PRR) or synthetic agonists for 24h (n>=4). Statistical significance of ligand-stimulated PBMC vs. media calculated by one-way repeated measures ANOVA followed by Dunnett’s multiple comparisons test. (C) PBMC were stimulated with poly I:C, TL8-506 or combination and cytokine secretion measured at 24h (n=3). Data summarized using a mixed-effects model with a fixed effect for ligand and a random effect for subject. A two-way interaction term was fit to each ligand to test synergy. The likelihood ratio test was used to test for statistical significance. Data are represented as mean ± SEM. *p<= 0.05, ** p<=0.01, *** p<=0.001
To define further the role of TLR8 in the induction of IL-12p70, we surveyed the ability of various oligomers and base analogues targeting endosomal and cytosolic nucleic acid PRR for ability to induce secretion of this cytokine in PBMC (Fig. 7B). As before, M. tuberculosis tRNA and ssRNA induced IL-18, with induction also observed for synthetic TLR8 agonist. IFN-γ was strongly induced by ssRNA as well as, to a lesser extent, M. tuberculosis tRNA and the TLR8-specific ligand. High levels of IL-12p70 were secreted in response to ssRNA and M. tuberculosis tRNA. Despite their induction of IL-18 and IFN-γ, only low amounts of IL-12p70 were produced in response to synthetic agonists for R848, 3M-002 (TLR7/8), and TL8-506 (TLR8). By itself, poly I:C (dsRNA, TLR3) was a weak inducer of IL-18, IFN-γ and IL-12p70. Even lower levels of these cytokines were observed in response to Imiquimod (TLR7), CpG DNA (TLR9), cyclic-di-GMP (STING), dA:dT (STING/CDS) (36–38). These data suggest that TLR8 activation alone was not sufficient to explain the strong induction of IL-12p70 by M. tuberculosis tRNA.
We next addressed whether a combination of a specific TLR8 ligand plus a second ligand activating a different PRR could lead to IL-12p70 secretion. Given that tRNA has both single- and double-stranded regions, and has previously been shown to induce type I IFN via activation of TLR3 (39), it was logical to ask whether activation of both TLR8 and TLR3 could induce IL-12p70. We found that the TLR3 agonist weakly induced IL-18 to a greater extent than the TLR8 agonist, and, when combined, the induction of IL-18 by the TLR3 and TLR8 ligands was not synergistic. In contrast, the TLR3 and TLR8 ligands by themselves weakly induced IFN-γ and IL-12p70, but the combination of the two ligands resulted in the synergistic induction of these cytokines (Fig. 7C). Both TLR3 and TLR7, but not TLR8 ligands induced IFN-α, but the combinations were not synergistic. These data suggest that the mechanism by which TLR3 synergizes with TLR8 in the induction of the Th1 pathway likely includes, but is not limited to, the production of Type I IFN. Together, these data identify an innate pathway requiring the recognition of the pathogen by distinct pattern recognition receptors and distinct cell types, involving the induction of both Type I and II IFNs as well as downstream genes, leading to the production of IL-12p70, a cytokine required for the induction of a Th1 response as part of a major host defense pathway.
Discussion
The ability of the immune system to mount a cell-mediated immune response involving Th1 cells is critical for host defense against many intracellular bacteria, including M. tuberculosis. To generate cell-mediated immunity, the innate immune system must recognize the microbial invader and instruct the adaptive T cell response. Yet the classic PAMPs found in mycobacteria, lipoproteins that activate TLR2/1, and muramyl dipeptide that activates NOD2, only weakly induce IL-12p70, a cytokine that is required for induction of a Th1 response. Here, we found that M. tuberculosis tRNA, detected in the bacterial culture filtrate, is a potent inducer IL-12p70. Exploring tRNA-induced gene expression profiles in human PBMC, we identified an integrated cellular and molecular pathway, beginning with the production of IL-18, Type I interferons, and IL-12p70, resulting in NK cell secretion of IFN-γ, and the subsequent robust induction of IL-12p70. The ability of M. tuberculosis tRNA to trigger this gene network was dependent on TLR8, yet TLR8 activation by itself was not sufficient for induction. Instead, activation of both TLR3 and TLR8 synergized for the robust induction of IFN-γ and IL-12p70, suggesting a role for the concurrent activation of nucleotide receptors in mounting a cell-mediated immune response against intracellular mycobacteria.
Initially, studies revealed that the culture filtrate of M. tuberculosis was sufficient by itself to induce T cells responses in a naïve animal, suggesting the presence of an adjuvant to activate the innate immune response along with protein/antigen signals required to stimulate an adaptive immune response (2–7). Subsequently, a purified protein derivative (PPD) fraction of the culture filtrate was isolated and shown to be useful as a diagnostic test for tuberculosis exposure, identifying individuals that had developed a delayed type hypersensitivity response because of exposure to M. tuberculosis or vaccination with BCG. The component(s) of the culture filtrate that can act as an adjuvant to stimulate the innate immune response has not been evaluated extensively, but tRNA was previously found to be a major component of the nucleotide fraction of the filtrate, which was shown to induce apoptosis in monocytes (18). Our data demonstrate a mechanism by which mycobacterial tRNA can robustly induce IL-12p70, a key cytokine in the induction of a Th1 response, and hence could serve as an adjuvant for the adaptive T cell response.
It was unexpected that a single mycobacterial ligand simultaneously induced both the Type II IFN, IFN-γ, as well as the Type I IFNs, since in chronic mycobacterial infections including tuberculosis and leprosy, the Type I IFNs inhibit the production and action of IFN-γ (40, 41). Yet as part of the pathway to induce IL-12p70, both tRNA and ssRNA were found to be potent inducers of both types of IFN, as well as IFN downstream genes. IFN-α, at low levels. has been shown to augment the ability of TLR ligands to trigger IL-12p70 production in DC (42–44), consistent with our data that Type I IFNs, as part of the acute immune response to M. tuberculosis tRNA, are involved in the production of IL-12p70. The findings presented here also demonstrate that Type I IFN was required for M. tuberculosis tRNA induction of IFN-γ by NK cells, thereby linking the production of Type I IFN to the production of Type II IFN. The induction of Type I IFNs, together with the early production of IFN-γ following tRNA activation of PBMC, is also likely to contribute to IL-12p70 production, as IFN-γ can augment the ability of other TLR ligands to induce IL-12p40 and IL-12p35 (45, 46). On the other hand, in chronic bacterial infection, Type I and Type II IFN have opposing functional roles resulting in the inhibition of IFN-γ-induced antimicrobial responses (41). The ability of Type I IFN to directly interfere with transcription of IL12B (47) leading to inhibition of IL-12p70 (48, 49) and blockade of Th1 responses (50, 51) can be overcome by the production of ISG15, which downregulates the Type I IFN response (52, 53). We note that both tRNA and ssRNA strongly induced ISG15. In viral infection, the initial induction of Type I IFN is required to clear the infection, but in chronic infections contributes to pathogenesis.
In determining the source of IFN-γ production, our data underscore the importance of NK cell activation in the robust induction of IL-12p70. We identified that M. tuberculosis tRNA, by triggering TLR8 to induce the production of IL-18, induces NK cell production of IFN-γ. The activation of NK cells to produce IFN-γ was dependent on the early production of IL-12p70, presumably through its ability to upregulate both subunits of the IL-18R on NK cells (54, 55), thereby enhancing responsiveness to IL-18. Consistent with this mechanism, tRNA upregulated IL18R1 and IL18RAP mRNA in PBMC, and the addition of neutralizing antibodies to IL-12p70 inhibited tRNA induction of IFN-γ. This early production of IFN-γ by NK cells was required for the later more robust induction of IL-12p70, since depletion of NK cells or the addition of anti-IFN-γ blocking antibodies reduced tRNA induction of IL-12p70. A role for NK cells in tuberculosis has been demonstrated in mouse models by their recruitment to the lung as early as seven days after infection (56) with a role in preventing tissue damage (57). In human tuberculosis, NK cells have been identified at the site of infection in patients, both in the pleural fluid (58) and in granulomas in pulmonary lesions (59). In addition, BCG, when used to revaccinate individuals, boosted IFN-γ producing CD56+ cells in vivo, and when added to whole blood, upregulated IFN−γ producing CD56+ cells in vitro via an IL-12 and IL-18 dependent mechanism (60).
The ability of M. tuberculosis tRNA to induce IL-12p70 is likely related to its natural location when released by M. tuberculosis in specific subcellular compartments. M. tuberculosis resides primarily in endosomes that contain the nucleotide-sensing TLRs including TLR3 (dsRNA), TLR7 (ssRNA), TLR8 (ssRNA) and TLR9 (dsDNA). TLR8 is primarily expressed in human monocytes and myeloid DC (61) and is localized to endosomes/phagosomes, allowing the innate immune system to distinguish self-RNA (nucleus, cytoplasm) from non-self RNA (endosome/phagosome). Our data indicate that the ability of M. tuberculosis tRNA to induce IL-18, IFN-γ and IL-12p70 was blocked by a specific TLR8 antagonist, consistent with previous studies (62) The mechanism by which TLR8 recognizes tRNA likely involves recognition of the ssRNA regions present in the tRNA stem loop sequences, for example in the anti-codon sequence (63). Of potential clinical relevance, a TLR8 gain-of-function polymorphism (TLR8 A1G), correlates with increased resistance to tuberculosis in humans (64–70). Presumably, the abundance of M. tuberculosis tRNA in the bacterial culture filtrate is due to a combination of accumulation through bacterial lysis and resistance to degradation (18). Although TLR8 has been shown to recognize Borrelia burgdorferi RNA in infected human macrophages (71), experiments to demonstrate a role for TLR8 during an in vitro infection of monocytes/macrophages with M. tuberculosis have not been successful, perhaps because of the slow turnover of bacteria in the short term in vitro cultures.
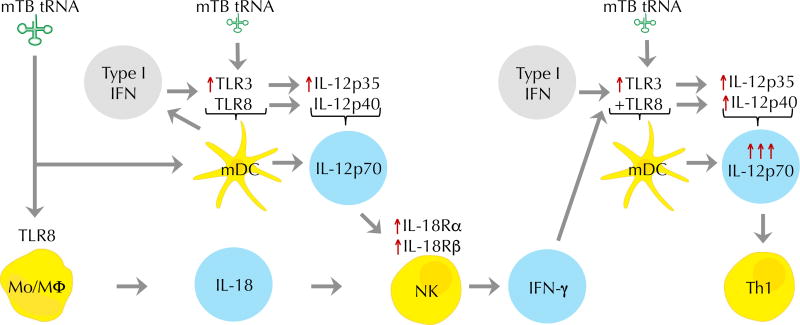
Although M. tuberculosis tRNA was a potent inducer of IL-12p70, and did so in a TLR8-dependent fashion, a TLR8 agonist alone weakly induced IL-12p70. In addition to ssRNA regions, tRNA contains dsRNA regions and has been shown to trigger TLR3 (39), prompting us to measure the response to a combination of ligands. The combination of dsRNA which activates TLR3 and a synthetic ligand that activates TLR8 induced IL-18 in an additive manner, but were synergistic in the induction of IFN-γ and IL-12p70. We therefore propose a model for the ability of tRNA to potently induce IL-12p70 (Fig. 8), involving TLR8 induction of IL-18. TLR3 activation leads to production of Type I IFN, which can enhance induction of IL-18 (72), feedback to enhance TLR3 expression (73, 74), leading to induction of IL-12p35 via an IRF3 dependent pathway (75). This in combination with early TLR8 induction of IL-12p40 leads to formation of IL-12p70, which upregulates both subunits of the IL-18R on NK cells (54, 55), thus synergizing with IL-18 to induce secretion of IFN-γ from NK cells (76). We infer that since IL-12p70 is assembled by a single cell, that myeloid DC are involved, given that they express both TLR3 and TLR8 (61) and are key producers of this cytokine. The production of IFN-γ subsequently enhances production of IL-12p70, thereby amplifying its production.
Fig. 8. M. tuberculosis tRNA triggers the induction of IL-12p70.
This model, based on the experimental data in this manuscript as well as the literature, shows that mTB tRNA induces secretion of IL-18 via TLR8. Type I IFNs may contribute to enhanced induction of IL-12p35 via upregulation of TLR3, which in combination with IL-12p40 induction leads to secretion of bioactive IL-12p70. IL-12p70 upregulates IL-18 receptor on NK cells, facilitating the ability of IL-18 and IL-12p70 to synergize to induce secretion of IFN-γ. IFN-γ in turn enhances IL-12p70 secretion. The key cell types in this process are monocytes/macrophages, NK cells and myeloid DC.
The innate pathway we describe for induction of IL-12p70 is of disease relevance as there is evidence from both murine models and from genetic studies in humans that the induction of IFN-γ is required for protective immunity against tuberculosis. Mice lacking the gene for IFN-γ died from M. tuberculosis challenge 2–3 weeks from IV challenge and within a month from aerosol challenge (77). Activation of mouse macrophages by IFN-γ and TNF-α induced killing of intracellular bacteria through the induction of nitric oxide (78, 79). Individuals with genetic disorders leading to the decreased production of, or response to IFN-γ, are highly susceptible to tuberculosis and other mycobacterial diseases (80–82). One mechanism by which IFN-γ contributes to host defense against M. tuberculosis in humans is through activation of macrophage antimicrobial activity via the vitamin-D dependent induction of the antimicrobial peptides cathelicidin and DEFB4 (83).
Our data suggest that a single microbial ligand may trigger multiple PRRs, leading to upregulation of IL-12p35 and IL-12p40 to synergize in the production of IL-12p70, thereby inducing a distinct innate immune response critical to host defense. Combinatorial analysis of how multiple innate signals trigger gene expression identified synergistic and antagonistic interactions, suggesting a functional role in the adaptation to complex stimuli (84). As such, tRNA induced a gene network that was associated with innate instruction of an adaptive T cell response. In addition to IL18, IL12A, IL12B, and IFNG, which are known to polarize towards a Th1 adaptive response, we found upregulation of multiple T-cell costimulatory molecules, including CD40 and CD80 (42, 85). Also of note were chemoattractants, such as CXCL10, which recruit T cells, NK cells, DC, and monocytes to the site of infection (86). Several of the genes in this network have been shown to be critical for host defense against mycobacterial infection in humans based on the enhanced susceptibility of individuals with genetic mutations, including IFNG, IFNGR1 and IL12RB1 (80–82). ISG15 is required to maintain IFN-γ production during mycobacterial infection (52, 53), such that genetic alteration of ISG15 increased the susceptibility of individuals to mycobacteria infection and abrogated the IFN-γ response to mycobacterial infection in vitro (87). As stated, the roles of IL-12, IL-18 and type I IFN in driving NK cell activation, as well as the ability of IFN-γ to amplify IL-12, have been established. Here, we have used both molecular and cellular approaches to identify a network by which a single microbial ligand triggers multiple PRRs leading to the production of both type I and type II IFNs, resulting in the type I and type II IFN dependent production of IL-12p70, as part of the innate immune response against many intracellular pathogens.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant (R01HL119068, R01AI022553, R01HL129887, R01AR040312, P50AR063020, R.L.M), Singapore-MIT Alliance for Research and Technology under a grant from the National Research Foundation of Singapore (P.C.D.)
References
- 1.Bloom BR, Murray CJL. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 2.Seibert FB. The isolation of a crystalline protein with tuberculin activity. Science. 1926;63:619c–620. doi: 10.1126/science.63.1642.619c. [DOI] [PubMed] [Google Scholar]
- 3.Long ER, Seibert FB. The chemical composition of the active principle of tuberculin. VII. The evidence that the active principle is a protein. Am. Rev. Tuberc. 1926;13:448. [Google Scholar]
- 4.Seibert FB. X. The isolation in crystalline form and identification of the active principle of tuberculin. Am Rev Tuberc. 1928;17:402–421. [Google Scholar]
- 5.Seibert FB. The isolation and properties of the purified protein derivative of tuberculin. Am. Rev. Tuberc. 1934;30:713–720. [Google Scholar]
- 6.Seibert FB, Glenn JT. Tuberculin Purified Protein Derivative. Preparation and Analyses of a Large Quantity for Standard. American Review of Tuberculosis and Pulmonary Diseases. 1941;44:9–25. [Google Scholar]
- 7.Seibert FB. The chemistry of tuberculin. Chem. Rev. 1944;34:107–127. [Google Scholar]
- 8.Collins FM, Lamb JR, Young DB. Biological activity of protein antigens isolated from Mycobacterium tuberculosis culture filtrate. Infect. Immun. 1988;56:1260–1266. doi: 10.1128/iai.56.5.1260-1266.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pal PG, Horwitz MA. Immunization with extracellular proteins of Mycobacterium tuberculosis induces cell-mediated immune responses and substantial protective immunity in a guinea pig model of pulmonary tuberculosis. Infect. Immun. 1992;60:4781–4792. doi: 10.1128/iai.60.11.4781-4792.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubbard RD, Flory C, Collins F. Immunization of mice with mycobacterial culture filtrate proteins. Clin. Exp. Immunol. 1992;87:94–98. doi: 10.1111/j.1365-2249.1992.tb06419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schenk M, Krutzik SR, Sieling PA, Lee DJ, Teles RM, Ochoa MT, Komisopoulou E, Sarno EN, Rea TH, Graeber TG, Kim S, Cheng G, Modlin RL. NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy. Nat. Med. 2012;18:555–563. doi: 10.1038/nm.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins AC, Cai H, Li T, Franco LH, Li XD, Nair VR, Scharn CR, Stamm CE, Levine B, Chen ZJ, Shiloh MU. Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host Microbe. 2015;17:820–828. doi: 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, Schmid-Burgk JL, Schmidt T, Hornung V, Cole ST, Ablasser A. Mycobacterium tuberculosis Differentially Activates cGAS- and Inflammasome-Dependent Intracellular Immune Responses through ESX-1. Cell Host Microbe. 2015;17:799–810. doi: 10.1016/j.chom.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE, 3rd, Freedman VH, Kaplan G. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha /beta. Proc Natl Acad Sci U S A. 2001;98:5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ottenhoff TH, Dass RH, Yang N, Zhang MM, Wong HE, Sahiratmadja E, Khor CC, Alisjahbana B, van Crevel R, Marzuki S, Seielstad M, van de Vosse E, Hibberd ML. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One. 2012;7:e45839. doi: 10.1371/journal.pone.0045839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obregon-Henao A, Duque-Correa MA, Rojas M, Garcia LF, Brennan PJ, Ortiz BL, Belisle JT. Stable extracellular RNA fragments of Mycobacterium tuberculosis induce early apoptosis in human monocytes via a caspase-8 dependent mechanism. PLoS One. 2012;7:e29970. doi: 10.1371/journal.pone.0029970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai WM, Chionh YH, Hia F, Gu C, Kellner S, McBee ME, Ng CS, Pang YL, Prestwich EG, Lim KS, Babu IR, Begley TJ, Dedon PC. A Platform for Discovery and Quantification of Modified Ribonucleosides in RNA: Application to Stress-Induced Reprogramming of tRNA Modifications. Methods Enzymol. 2015;560:29–71. doi: 10.1016/bs.mie.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautier G, Humbert M, Deauvieau F, Scuiller M, Hiscott J, Bates EE, Trinchieri G, Caux C, Garrone P. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J. Exp. Med. 2005;201:1435–1446. doi: 10.1084/jem.20041964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma F, Li B, Yu Y, Iyer SS, Sun M, Cheng G. Positive feedback regulation of type I interferon by the interferon-stimulated gene STING. EMBO Rep. 2015;16:202–212. doi: 10.15252/embr.201439366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorski KS, Waller EL, Bjornton-Severson J, Hanten JA, Riter CL, Kieper WC, Gorden KB, Miller JS, Vasilakos JP, Tomai MA, Alkan SS. Distinct indirect pathways govern human NK-cell activation by TLR-7 and TLR-8 agonists. Int. Immunol. 2006;18:1115–1126. doi: 10.1093/intimm/dxl046. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt KN, Leung B, Kwong M, Zarember KA, Satyal S, Navas TA, Wang F, Godowski PJ. APC-independent activation of NK cells by the Toll-like receptor 3 agonist double-stranded RNA. J. Immunol. 2004;172:138–143. doi: 10.4049/jimmunol.172.1.138. [DOI] [PubMed] [Google Scholar]
- 30.Ushio S, Namba M, Okura T, Hattori K, Nukada Y, Akita K, Tanabe F, Konishi K, Micallef M, Fujii M, Torigoe K, Tanimoto T, Fukuda S, Ikeda M, Okamura H, Kurimoto M. Cloning of the cDNA for human IFN-gamma-inducing factor, expression in Escherichia coli, and studies on the biologic activities of the protein. J Immunol. 1996;156:4274–4279. [PubMed] [Google Scholar]
- 31.Garcia VE, Uyemura K, Sieling PA, Ochoa MT, Morita CT, Okamura H, Kurimoto M, Rea TH, Modlin RL. IL-18 promotes type 1 cytokine production from NK cells and T cells in human intracellular infection. J. Immunol. 1999;162:6114–6121. [PubMed] [Google Scholar]
- 32.Lee HR, Yoon SY, Song SB, Park Y, Kim TS, Kim S, Hur DY, Song HK, Park H, Cho D. Interleukin-18-mediated interferon-gamma secretion is regulated by thymosin beta 4 in human NK cells. Immunobiology. 2011;216:1155–1162. doi: 10.1016/j.imbio.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Guan X, Tamura T, Ozato K, Ma X. Synergistic activation of interleukin-12 p35 gene transcription by interferon regulatory factor-1 and interferon consensus sequence-binding protein. J. Biol. Chem. 2004;279:55609–55617. doi: 10.1074/jbc.M406565200. [DOI] [PubMed] [Google Scholar]
- 34.Howbert JJ, Hershberg R, Burgess LE, Yang HW. Substituted benzoazepines as toll-like receptor modulators. Google Patents 2013 [Google Scholar]
- 35.Lu H, Dietsch GN, Matthews MA, Yang Y, Ghanekar S, Inokuma M, Suni M, Maino VC, Henderson KE, Howbert JJ, Disis ML, Hershberg RM. VTX-2337 is a novel TLR8 agonist that activates NK cells and augments ADCC. Clin. Cancer Res. 2012;18:499–509. doi: 10.1158/1078-0432.CCR-11-1625. [DOI] [PubMed] [Google Scholar]
- 36.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma F, Li B, Liu SY, Iyer SS, Yu Y, Wu A, Cheng G. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J Immunol. 2015;194:1545–1554. doi: 10.4049/jimmunol.1402066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Xiang L, Shao J, Yuan Z. The 3’ CCACCA sequence of tRNAAla(UGC) is the motif that is important in inducing Th1-like immune response, and this motif can be recognized by Toll-like receptor 3. Clin. Vaccine Immunol. 2006;13:733–739. doi: 10.1128/CVI.00019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O’Garra A. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teles RM, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, Komisopoulou E, Kelly-Scumpia K, Chun R, Iyer SS, Sarno EN, Rea TH, Hewison M, Adams JS, Popper SJ, Relman DA, Stenger S, Bloom BR, Cheng G, Modlin RL. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 2013;339:1448–1453. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hermann P, Rubio M, Nakajima T, Delespesse G, Sarfati M. IFN-alpha priming of human monocytes differentially regulates gram-positive and gram-negative bacteria-induced IL-10 release and selectively enhances IL-12p70, CD80, and MHC class I expression. J Immunol. 1998;161:2011–2018. [PubMed] [Google Scholar]
- 43.Gautier G, Humbert M, Deauvieau F, Scuiller M, Hiscott J, Bates EE, Trinchieri G, Caux C, Garrone P. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J Exp Med. 2005;201:1435–1446. doi: 10.1084/jem.20041964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heystek HC, den Drijver B, Kapsenberg ML, van Lier RA, de Jong EC. Type I IFNs differentially modulate IL-12p70 production by human dendritic cells depending on the maturation status of the cells and counteract IFN-gamma-mediated signaling. Clin Immunol. 2003;107:170–177. doi: 10.1016/s1521-6616(03)00060-3. [DOI] [PubMed] [Google Scholar]
- 45.Ma X, Chow JM, Gri G, Carra G, Gerosa F, Wolf SF, Dzialo R, Trinchieri G. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. The Journal of experimental medicine. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayes MP, Murphy FJ, Burd PR. Interferon-gamma-dependent inducible expression of the human interleukin-12 p35 gene in monocytes initiates from a TATA-containing promoter distinct from the CpG-rich promoter active in Epstein-Barr virus-transformed lymphoblastoid cells. Blood. 1998;91:4645–4651. [PubMed] [Google Scholar]
- 47.Byrnes AA, Ma X, Cuomo P, Park K, Wahl L, Wolf SF, Zhou H, Trinchieri G, Karp CL. Type I interferons and IL-12: convergence and cross-regulation among mediators of cellular immunity. European Journal of Immunology. 2001;31:2026–2034. doi: 10.1002/1521-4141(200107)31:7<2026::aid-immu2026>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 48.Manca C, Tsenova L, Freeman S, Barczak AK, Tovey M, Murray PJ, Barry C, Kaplan G. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J Interferon Cytokine Res. 2005;25:694–701. doi: 10.1089/jir.2005.25.694. [DOI] [PubMed] [Google Scholar]
- 49.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, Farlik M, Decker T, Du Pasquier RA, Romero P, Tschopp J. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 50.de Paus RA, van Wengen A, Schmidt I, Visser M, Verdegaal EM, van Dissel JT, van de Vosse E. Inhibition of the type I immune responses of human monocytes by IFN-alpha and IFN-beta. Cytokine. 2013;61:645–655. doi: 10.1016/j.cyto.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 51.Orme IM, Robinson RT, Cooper AM. The balance between protective and pathogenic immune responses in the TB-infected lung. Nat Immunol. 2015;16:57–63. doi: 10.1038/ni.3048. [DOI] [PubMed] [Google Scholar]
- 52.Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, Mansouri N, Okada S, Bryant VL, Kong XF, Kreins A, Velez MM, Boisson B, Khalilzadeh S, Ozcelik U, Darazam IA, Schoggins JW, Rice CM, Al-Muhsen S, Behr M, Vogt G, Puel A, Bustamante J, Gros P, Huibregtse JM, Abel L, Boisson-Dupuis S, Casanova JL. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, Tezcan I, Rice GI, Chen C, Mansouri N, Mahdaviani SA, Itan Y, Boisson B, Okada S, Zeng L, Wang X, Jiang H, Liu W, Han T, Liu D, Ma T, Wang B, Liu M, Liu JY, Wang QK, Yalnizoglu D, Radoshevich L, Uze G, Gros P, Rozenberg F, Zhang SY, Jouanguy E, Bustamante J, Garcia-Sastre A, Abel L, Lebon P, Notarangelo LD, Crow YJ, Boisson-Dupuis S, Casanova JL, Pellegrini S. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshimoto T, Takeda K, Tanaka T, Ohkusu K, Kashiwamura S, Okamura H, Akira S, Nakanishi K. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-gamma production. J. Immunol. 1998;161:3400–3407. [PubMed] [Google Scholar]
- 55.Fehniger TA, Shah MH, Turner MJ, VanDeusen JB, Whitman SP, Cooper MA, Suzuki K, Wechser M, Goodsaid F, Caligiuri MA. Differential cytokine and chemokine gene expression by human NK cells following activation with IL-18 or IL-15 in combination with IL-12: implications for the innate immune response. J. Immunol. 1999;162:4511–4520. [PubMed] [Google Scholar]
- 56.Junqueira-Kipnis AP, Kipnis A, Jamieson A, Juarrero MG, Diefenbach A, Raulet DH, Turner J, Orme IM. NK cells respond to pulmonary infection with Mycobacterium tuberculosis, but play a minimal role in protection. J. Immunol. 2003;171:6039–6045. doi: 10.4049/jimmunol.171.11.6039. [DOI] [PubMed] [Google Scholar]
- 57.Feng CG, Kaviratne M, Rothfuchs AG, Cheever A, Hieny S, Young HA, Wynn TA, Sher A. NK cell-derived IFN-gamma differentially regulates innate resistance and neutrophil response in T cell-deficient hosts infected with Mycobacterium tuberculosis. J. Immunol. 2006;177:7086–7093. doi: 10.4049/jimmunol.177.10.7086. [DOI] [PubMed] [Google Scholar]
- 58.Schierloh P, Aleman M, Yokobori N, Alves L, Roldan N, Abbate E, del CSM, de la Barrera S. NK cell activity in tuberculosis is associated with impaired CD11a and ICAM-1 expression: a regulatory role of monocytes in NK activation. Immunology. 2005;116:541–552. doi: 10.1111/j.1365-2567.2005.02259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Portevin D, Via LE, Eum S, Young D. Natural killer cells are recruited during pulmonary tuberculosis and their ex vivo responses to mycobacteria vary between healthy human donors in association with KIR haplotype. Cell. Microbiol. 2012;14:1734–1744. doi: 10.1111/j.1462-5822.2012.01834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suliman S, Geldenhuys H, Johnson JL, Hughes JE, Smit E, Murphy M, Toefy A, Lerumo L, Hopley C, Pienaar B, Chheng P, Nemes E, Hoft DF, Hanekom WA, Boom WH, Hatherill M, Scriba TJ. Bacillus Calmette-Guerin (BCG) Revaccination of Adults with Latent Mycobacterium tuberculosis Infection Induces Long-Lived BCG-Reactive NK Cell Responses. J Immunol. 2016;197:1100–1110. doi: 10.4049/jimmunol.1501996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J Exp Med. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sha W, Mitoma H, Hanabuchi S, Bao M, Weng L, Sugimoto N, Liu Y, Zhang Z, Zhong J, Sun B, Liu YJ. Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16059–16064. doi: 10.1073/pnas.1412487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davila S, Hibberd ML, Hari Dass R, Wong HE, Sahiratmadja E, Bonnard C, Alisjahbana B, Szeszko JS, Balabanova Y, Drobniewski F, van Crevel R, van de Vosse E, Nejentsev S, Ottenhoff TH, Seielstad M. Genetic association and expression studies indicate a role of toll-like receptor 8 in pulmonary tuberculosis. PLoS Genet. 2008;4:e1000218. doi: 10.1371/journal.pgen.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bukhari M, Aslam MA, Khan A, Iram Q, Akbar A, Naz AG, Ahmad S, Ahmad MM, Ashfaq UA, Aziz H, Ali M. TLR8 gene polymorphism and association in bacterial load in southern Punjab of Pakistan: an association study with pulmonary tuberculosis. Int. J. Immunogenet. 2015;42:46–51. doi: 10.1111/iji.12170. [DOI] [PubMed] [Google Scholar]
- 66.Daya M, van der Merwe L, van Helden PD, Moller M, Hoal EG. Investigating the Role of Gene-Gene Interactions in TB Susceptibility. PLoS One. 2014;10:e0123970. doi: 10.1371/journal.pone.0123970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai YF, Lin TM, Wang CH, Su PY, Wu JT, Lin MC, Eng HL. Functional polymorphisms of the TLR7 and TLR8 genes contribute to Mycobacterium tuberculosis infection. Tuberculosis (Edinburgh, Scotland) 2016;98:125–131. doi: 10.1016/j.tube.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 68.Salie M, Daya M, Lucas LA, Warren RM, van der Spuy GD, van Helden PD, Hoal EG, Moller M. Association of toll-like receptors with susceptibility to tuberculosis suggests sex-specific effects of TLR8 polymorphisms. Infect. Genet. Evol. 2015;34:221–229. doi: 10.1016/j.meegid.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 69.Sun Q, Zhang Q, Xiao HP, Bai C. Toll-like receptor polymorphisms and tuberculosis susceptibility: A comprehensive meta-analysis. Journal of Huazhong University of Science and Technology. Medical sciences = Hua zhong ke ji da xue xue bao. Yi xue Ying De wen ban = Huazhong keji daxue xuebao. Yixue Yingdewen ban. 2015;35:157–168. doi: 10.1007/s11596-015-1405-6. [DOI] [PubMed] [Google Scholar]
- 70.Wu L, Hu Y, Li D, Jiang W, Xu B. Screening toll-like receptor markers to predict latent tuberculosis infection and subsequent tuberculosis disease in a Chinese population. BMC Med. Genet. 2015;16:19. doi: 10.1186/s12881-015-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cervantes JL, La Vake CJ, Weinerman B, Luu S, O’Connell C, Verardi PH, Salazar JC. Human TLR8 is activated upon recognition of Borrelia burgdorferi RNA in the phagosome of human monocytes. J. Leukoc. Biol. 2013;94:1231–1241. doi: 10.1189/jlb.0413206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fang R, Hara H, Sakai S, Hernandez-Cuellar E, Mitsuyama M, Kawamura I, Tsuchiya K. Type I interferon signaling regulates activation of the absent in melanoma 2 inflammasome during Streptococcus pneumoniae infection. Infect Immun. 2014;82:2310–2317. doi: 10.1128/IAI.01572-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miettinen M, Sareneva T, Julkunen I, Matikainen S. IFNs activate toll-like receptor gene expression in viral infections. Genes Immun. 2001;2:349–355. doi: 10.1038/sj.gene.6363791. [DOI] [PubMed] [Google Scholar]
- 74.Siren J, Pirhonen J, Julkunen I, Matikainen S. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. J Immunol. 2005;174:1932–1937. doi: 10.4049/jimmunol.174.4.1932. [DOI] [PubMed] [Google Scholar]
- 75.Goriely S, Molle C, Nguyen M, Albarani V, Haddou NO, Lin R, De Wit D, Flamand V, Willems F, Goldman M. Interferon regulatory factor 3 is involved in Toll-like receptor 4 (TLR4)- and TLR3-induced IL-12p35 gene activation. Blood. 2006;107:1078–1084. doi: 10.1182/blood-2005-06-2416. [DOI] [PubMed] [Google Scholar]
- 76.Matikainen S, Paananen A, Miettinen M, Kurimoto M, Timonen T, Julkunen I, Sareneva T. IFN-alpha and IL-18 synergistically enhance IFN-gamma production in human NK cells: differential regulation of Stat4 activation and IFN-gamma gene expression by IFN-alpha and IL-12. Eur J Immunol. 2001;31:2236–2245. [PubMed] [Google Scholar]
- 77.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Newport MJ, Huxley CM, Huston S, Hawrylowicz CM, Oostra BA, Williamson R, Levin M. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 81.Jouanguy E, Lamhamedi-Cherradi S, Lammas D, Dorman SE, Fondaneche MC, Dupuis S, Doffinger R, Altare F, Girdlestone J, Emile JF, Ducoulombier H, Edgar D, Clarke J, Oxelius VA, Brai M, Novelli V, Heyne K, Fischer A, Holland SM, Kumararatne DS, Schreiber RD, Casanova JL. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat Genet. 1999;21:370–378. doi: 10.1038/7701. [DOI] [PubMed] [Google Scholar]
- 82.Altare F, Durandy A, Lammas D, Emile JF, Lamhamedi S, Le Deist F, Drysdale P, Jouanguy E, Doffinger R, Bernaudin F, Jeppsson O, Gollob JA, Meinl E, Segal AW, Fischer A, Kumararatne D, Casanova JL. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 1998;280:1432–1435. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 83.Fabri M, Stenger S, Shin DM, Yuk JM, Liu PT, Realegeno S, Lee HM, Krutzik SR, Schenk M, Sieling PA, Teles R, Montoya D, Iyer SS, Bruns H, Lewinsohn DM, Hollis BW, Hewison M, Adams JS, Steinmeyer A, Zugel U, Cheng G, Jo EK, Bloom BR, Modlin RL. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci Transl Med. 2011;3:104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cappuccio A, Zollinger R, Schenk M, Walczak A, Servant N, Barillot E, Hupe P, Modlin RL, Soumelis V. Combinatorial code governing cellular responses to complex stimuli. Nat Commun. 2015;6:6847. doi: 10.1038/ncomms7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klug-Micu GM, Stenger S, Sommer A, Liu PT, Krutzik SR, Modlin RL, Fabri M. CD40 ligand and interferon-gamma induce an antimicrobial response against Mycobacterium tuberculosis in human monocytes. Immunology. 2013;139:121–128. doi: 10.1111/imm.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lande R, Giacomini E, Grassi T, Remoli ME, Iona E, Miettinen M, Julkunen I, Coccia EM. IFN-alpha beta released by Mycobacterium tuberculosis-infected human dendritic cells induces the expression of CXCL10: selective recruitment of NK and activated T cells. J Immunol. 2003;170:1174–1182. doi: 10.4049/jimmunol.170.3.1174. [DOI] [PubMed] [Google Scholar]
- 87.Fan JB, Zhang DE. ISG15 regulates IFN-gamma immunity in human mycobacterial disease. Cell Res. 2013;23:173–175. doi: 10.1038/cr.2012.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Waddell SJ, Popper SJ, Rubins KH, Griffiths MJ, Brown PO, Levin M, Relman DA. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS One. 2010;5:e9753. doi: 10.1371/journal.pone.0009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.