Abstract
Background
Alcohol use (both quantity and dependence) is moderately heritable, and genomewide association studies (GWAS) have identified risk genes in European, African, and Asian populations. The most reproducibly identified risk genes affect alcohol metabolism. Well-known functional variants at the gene encoding alcohol dehydrogenase B (ADH1B) and other alcohol dehydrogenases affect risk in European and African-ancestry populations. Similarly, variants mapped to these same genes and a well-known null variant that maps to the gene that encodes aldehyde dehydrogenase 2 (ALDH2) also affect risk in various Asian populations. In this study we completed the first GWAS for three traits related to alcohol use in a Thai population recruited initially for studies of methamphetamine dependence.
Methods
All subjects were evaluated with the Thai version of the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA). A total of 1045 subjects were available for analysis. Three traits were analyzed: flushing, maximum number of alcoholic beverages consumed in any lifetime 24-hour period (“MAXDRINKS”), and DSM-IV alcohol dependence criterion count. We also conducted a pleiotropy analysis with major depression, the only other psychiatric trait where summary statistics from a large-scale Asian-population GWAS are available.
Results
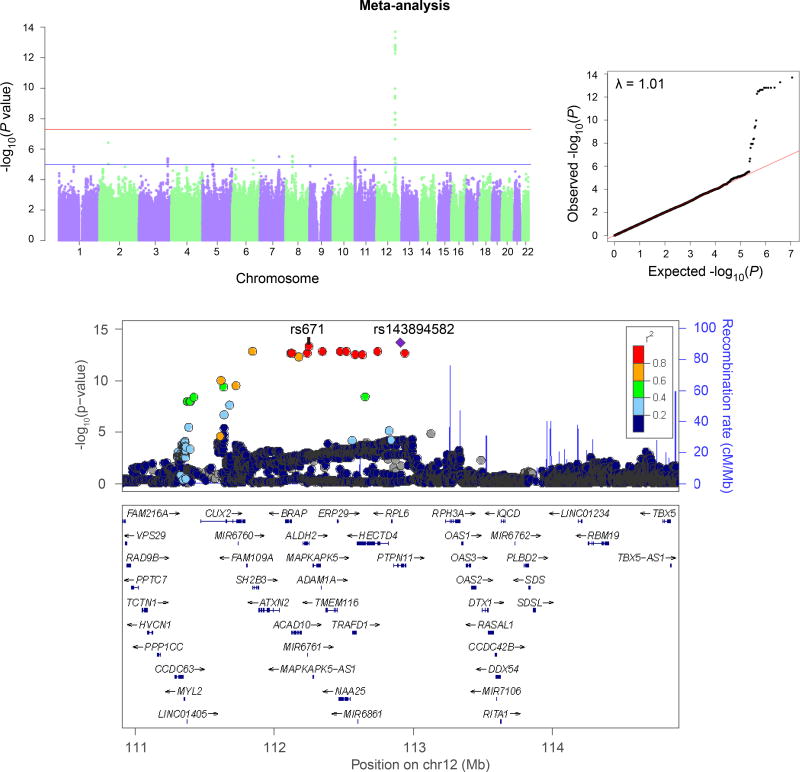
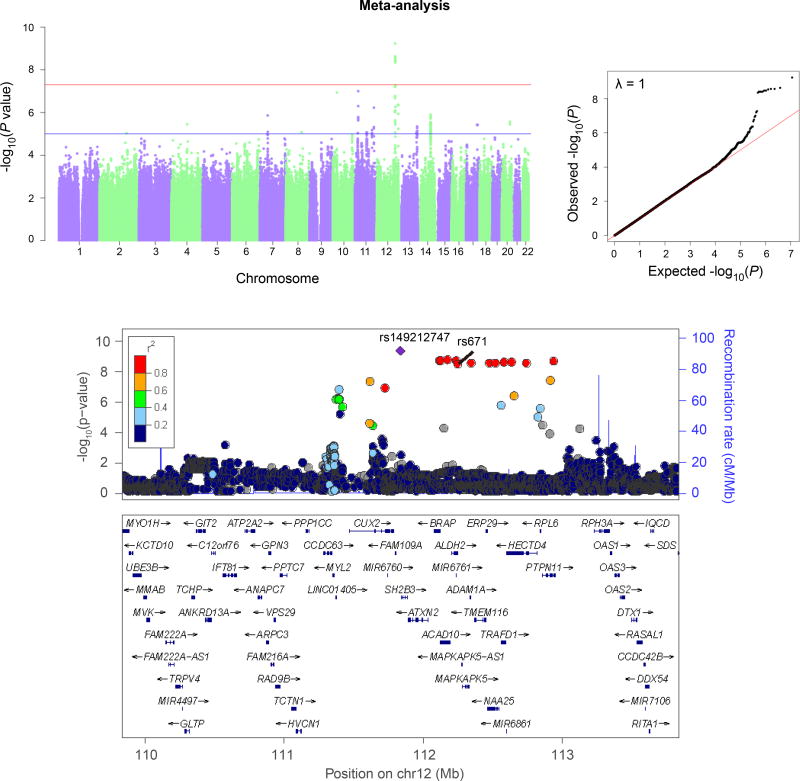
All three traits showed genomewide-significant association with variants near ALDH2, with significance ranging from 2.01×10−14 (for flushing; lead SNP PTPN11* rs143894582) to pmeta = 5.80 × 10−10 (for alcohol dependence criterion count; lead SNP rs149212747). These lead SNPs flank rs671 and span a region of over a megabase, illustrating the need for prior biological information in identifying the actual effect SNP, rs671. We also identified significant pleiotropy between major depression and flushing.
Conclusions
These results are consistent with prior findings in Asian populations and add new information regarding alcohol use-depression pleiotropy.
Keywords: Genomewide association study, ALDH2, alcohol dependence, flushing reaction, depression
INTRODUCTION
It is well known that alcohol use disorder (AUD) is moderately heritable (Verhulst et al., 2015). Genomewide associations studies (GWAS) of alcohol dependence (AD) and habitual alcohol use have been completed in European (Gelernter et al., 2014, Mbarek et al., 2015, Clarke et al., 2017, Xu et al., 2015, Jorgenson et al., 2017) and African (Gelernter et al., 2014, Xu et al., 2015, Almli et al, 2017, Jorgenson et al., 2017) ancestry populations. Work in Asian ancestry populations is more limited, but we have completed a small GWAS in a Chinese population (Quillen et al., 2014), and other investigators have done so in Chinese (Yang et al., 2013), Koreans (Baik et al., 2011, Park et al., 2013), and Asian-Americans (Jorgenson et al., 2017). The most consistent findings have been associations with variants mapped to genes that encode alcohol metabolizing enzymes – most characteristically variants at ADH1B in European- and African-ancestry subjects (Li et al., 2011) and ALDH2*rs671 (and often ADH1B as well) in Asians (Li et al., 2012).
Alcohol consumption exacts a huge economic toll in Thailand, estimated to amount to 2% of the gross domestic product as of 2006 (Thavorncharoensap et al., 2010). However, excepting small candidate gene studies, there have been few studies on alcohol genetics in the Thai population, and to date, no GWAS. One previous study (Assanangkornchai et al., 2003) showed that ALDH2 variation is related to hazardous drinking and facial flushing in Thai males. Another study reviewed population genetics of the most relevant alcohol metabolizing enzyme variants in Asians, including Thais (Eng et al., 2007).
In the present study, we completed a GWAS of AD criterion count and the related phenotypes of “MAXDRINKS,” or maximum number of alcoholic beverages taken in any 24-hour period over a subject’s lifetime (Saccone et al., 2000), and flushing after drinking alcohol, in a Thai population collected for study of methamphetamine genetics. This was feasible because although the focus in the parent study was on methamphetamine dependence, we used the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) for assessment. The SSADDA is a comprehensive diagnostic interview designed for genetic studies of substance dependence and related phenotypes (Pierucci-Lagha et al., 2007, Pierucci-Lagha et al., 2005). We previously developed a Thai version of the SSADDA, which was translated and validated in genetic studies of opioid dependence in Northern Thailand, where it was shown to have both high inter-instrument validity and inter-rater reliability for that trait (Malison et al., 2011). There is a high level of comorbidity between methamphetamine dependence and AUDs and a high rate of AUDs in Thailand overall (9.1% in males and 1.0% in females in 2010 (World Health Organization, 2014)), and accordingly the sample is highly informative for study of alcohol-related traits.
MATERIALS and METHODS
Subject recruitment and assessment
We included subjects recruited in two stages for studies of the genetics of methamphetamine dependence. For both stages, subjects were recruited at the Princess Mother National Institute on Drug Abuse Treatment (PMNIDAT; formerly Thanyarak Institute) located in Bangkok using similar ascertainment methods (with the main phenotyping assessment via the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA—Thai version)). Studies were approved by The Faculty of Medicine, Chulalongkorn University Institutional Review Board (Med Chula IRB), the Ethical Review Committee for Research in Human Subjects, Thailand Ministry of Public Health, the Research Committee, Thanyarak Institute on Drug Abuse; and the Yale Human Investigations Committee. Subject characteristics are shown in Table 1. All subjects provided voluntary written informed consent prior to their research participation and were compensated (500 baht, or roughly US$15), per institutional review board (IRB) approval.
Table 1. Demographic characteristics of study subjects.
Samples with available genotype data and phenotype data for each trait are listed bellow.
| Sample 1 (“GSA”) | Sample 2 (“MEGA”) | Meta | |
|---|---|---|---|
| Sample size (female %) | 991 (47.4) | 589 (44.5) | 1,580 (46.3) |
| Genotyped subjects | 863 | 536 | 1399 |
| Age, mean (SD), y | 26.9 (7.1) | 30.7 (7.7) | 28.3 (7.6) |
| Phenotypes (included subjects) | |||
| Alcohol dependence | 535 | 510 | 1,045 |
| MAXDRINKS | 535 | 510 | 1,045 |
| Flushing (affected/unaffected) | 267/268 | 270/240 | 537/508 |
Sample 1: Global Screening Array (“GSA”)
This stage included methamphetamine users (n = 991) from across Thailand (Table 1). Subjects were hospitalized for four months of residential drug treatment at PMNIDAT from 2007 to 2011 (Kalayasiri et al., 2014). This initial collection was focused primarily on the genetics of methamphetamine-induced paranoia. Exclusion criteria included including (i) life-time use of MA < 11 instances; (ii) history of primary psychotic disorders; and (iii) brain disease (epilepsy, stroke, or brain trauma).
Diagnostic assessments were performed during each subject's rehabilitation by interviewers certified for SSADDA use based on a standard training protocol. Interviews were subjected to a rigorous quality control process, including editing and cross-editing by interviewers and final review by the Thai principal investigator (R.K.).
Sample 2: Multi-Ethnic Global Array (“MEGA”)
This stage focused both on severely affected methamphetamine dependent subjects and subjects who were methamphetamine-exposed but not dependent (n=589, Table 1). Subjects were hospitalized for four months of residential drug treatment at PMNIDAT from 2015 to 2017. The primary focus of this ongoing study is the genetics of methamphetamine dependence and related traits (including AD). Interviewers were retrained in the SSADDA in 2015 and either certified or re-certified as needed. Additional interviewers hired over the course of the study also underwent standard training. Subjects were administered the SSADDA, and SSADDA quality control procedures were performed, as for Sample 1.
Phenotypes
Only alcohol-exposed subjects were included. We studied AD (by DSM-IV criterion count), maximum number of alcoholic beverages taken in any 24-hour period over the subject’s lifetime (MAXDRINKS), and flushing. Phenotype definitions were as follows:
DSM-IV criterion count was defined as the sum of DSM-IV AD symptoms, up to 7 (AD-CrC). Subjects with missing values for AD criteria were excluded.
MAXDRINKS was defined by Thai SSADDA item E4_TotDrnk. Values >100 were set to 100 (to account for inflated and potentially spurious MAXDRINKS estimates) and were log transformed for analyses.
Flushing was defined by Thai SSADDA item E2A_1_BlushA, which translates to: ‘While drinking, has one or two drinks of alcohol ever caused you to flush or blush--that is, your face and hands felt hot and your face turned red?’ Subjects who answered ‘yes’ were classified as affected, and those answered ‘no’ were classified as unaffected.
The correlations among these traits are as follows: [AD-CrC]-MAXDRINKS, 0.537 (p<0.001); [AD-CrC]-flushing, 0.053 (p<0.05); flushing-MAXDRINKS, −0.045 (NS).
Genotyping and Quality Control
Sample 1 (GSA sample) was genotyped on the Illumina (San Diego, CA, USA) Global Screening Array (GSA). This is a 642,824 marker array with content added to provide good genomic coverage for several global populations, including East Asians. For Sample 1, there was insufficient DNA to allow use of the MEGA array (see below), which has higher DNA input requirements.
Sample 2 (MEGA sample) was genotyped on the Illumina (San Diego, CA, USA) Multi-Ethnic Global Array (MEGA). This is a 1,779,819 marker array with content added to provide excellent genomic coverage for several global populations, including East Asians.
For Sample 1 (GSA), 991 Thai subjects were phenotyped initially. Some DNA was available for 863 of these. Two hundred fifty-seven subjects were removed due to low genotype call rate (< 0.9). (A quality control (QC) call rate of 0.95 is preferred, but the quantity and quality of DNA from Sample 1 was poor, so we used a lesser standard; had we used a threshold of 0.95, more than 100 additional subjects would have been removed.) Two subjects were removed due to mismatched genotype and phenotypic sex; six subjects were removed due to high heterozygosity rate; seven were removed because they were duplicates; and ten were removed because the same subjects were also present in the Sample 2, for which better-quality DNA was available. 581 subjects remained for analysis. Only autosomal SNPs were retained. SNPs with low genotype call rate (< 0.95), MAF < 0.01, or HWE p < 10−6 were removed. 334,631 SNPs remained for imputation.
For Sample 2 (MEGA), the study is ongoing; 589 Thai subjects have been phenotyped to date; 536 of these were genotyped by MEGA array. One subject was removed due to mismatched genotype and phenotypic sex; one subject was removed due to low genotype call rate (< 0.95); four subjects were removed because they were duplicates. Five hundred and thirty subjects remained for analysis. Only autosomal SNPs were retained; SNPs with low genotype call rate (< 0.95) MAF < 0.01 or HWE P < 10−6 were removed. 728,129 SNPs remained for imputation.
Principal component analyses
To identify genetic outliers and ascertain concordance with self-reported race, we completed a principal component (PC) analysis on SNPs common to the individual genotyping arrays (pruning by linkage disequilibrium (LD) r2 > 0.2) and the 1000 Genome phase 3 reference panels (African populations, AFR; Admixed American populations, AMR; East Asian populations, EAS; European populations, EUR; and South Asian populations, SAS) using EIGENSOFT (Patterson et al., 2006, Price et al., 2006). Samples 1 and 2 were analyzed separately. We then completed a second PC analysis using Asian populations from the 1000 Genome panels (EAS and SAS), now categorized by specific region, to investigate the fine-scale population structure of the included Thai samples (Supplemental Figure 1). A third PC analysis was done within the Thai samples, and the first 3 PCs were then used to correct for population stratification.
Imputation
Additional SNPs were imputed using IMPUTE2 (Howie et al., 2009) and 1000 Genome phase 3 (Auton et al., 2015) Asian reference panels (504 EAS and 489 SAS). GSA and MEGA samples were imputed separately. After imputation, 11,314,818 SNPs in GSA and 11,351,136 SNPs in MEGA were obtained. SNPs with imputation accuracy score ≥ 0.8, MAF ≥ 0.05 and HWE p > 10−6 were retained for association analyses. 4,683,137 SNPs in GSA and 5,477,977 SNPs in MEGA were analyzed.
Statistical analysis
We performed logistic regression for the binary trait (flushing) and linear regression for ordinal or continuous traits (AD and MAXDRINKS) on unrelated individuals (IBD < 0.05) using PLINK (Purcell et al., 2007). An additive model was used, adjusting for age, sex, and the first 3 PCs.
In instances where multiple SNPs acheived genomewide significance (GWS) in the same region, conditional analyses were performed to identify the lead SNP and to test for the presence of any additional independent signal. For each GWS SNP, we tested the association with the phenotype using age, sex, the first 3 PCs, and each of the other SNPs as covariate. The threshold for statistical significance was 5 × 10−8. Analyses were completed in each sample separately, then meta-analyzed.
Polygenic risk score
Polygenic risk scores (PRS) of related traits in discovery datasets can be used to test the polygenic effect between those traits and the study trait in the target sample. To our knowledge, there have been few large-cohort GWAS on psychiatric diseases done in East Asian populations. The CONVERGE (China, Oxford and Virginia Commonwealth University Experimental Research on Genetic Epidemiology) consortium performed the largest GWAS so far for major depressive disorder (MDD) in more than 10,000 Chinese women (Converge_Consortium, 2015). We obtained the relevant summary statistics from this study from the Psychiatric Genomics Consortium (PGC) website (http://www.med.unc.edu/pgc/results-and-downloads). Common SNPs between CONVERGE and our Thai sample were selected for downstream analysis. The summary data of the common SNPs were clumped by LD with r2 < 0.2, using a 200 kb window. As described previously (International Schizophrenia et al., 2009), MDD PRS was calculated as the sum of the risk alleles with P values less than a threshold (PT), weighted by the effect sizes. The associations between the constructed MDD PRS and alcohol-related phenotypes in our Thai cohorts were tested by linear (AD and MAXDRINKS) or logistic (flushing) regression using glm in R, adjusting for age, sex and the first 3 PCs. Samples 1 and 2 were analyzed separately and then meta-analyzed. Eight PT thresholds (0.00001, 0.0001, 0.001, 0.005, 0.01, 0.05, 0.1 and 0.5) were considered. Multiple test correction for the eight PT were considered.
RESULTS
All three traits – AD-CrC, MAXDRINKS, and flushing – showed GWS association in the total (meta-analyzed) sample with variants near ALDH2, the lead SNPs mapping to an LD block containing rs671 (Table 2). The associations differed in statistical significance, as follows: Flushing, lead SNP PTPN11*rs143894582; Sample 1 (GSA), 8.33 × 10−8; Sample 2 (MEGA), 2.76 × 10−8; pmeta=2.01 × 10−14 (Figure 1). MAXDRINKS, lead SNP rs149212747; Sample 1 (GSA), 9.03 × 10−9; Sample 2 (MEGA), 4.59 × 10−5; pmeta=1.61 × 10−12. And AD-CrC, lead SNP rs149212747; Sample 1 (GSA), 1.54 × 10−5; Sample 2 (MEGA), 6.96 × 10−6; pmeta=5.80 × 10−10 (Figure 2). The only trait without GWS in either of the separate samples taken individually was AD-CrC – although this trait reached high statistical significance in the meta-analysis. Manhattan and QQ plots for flushing (individual samples) and MAXDRINKS (individual samples and meta-analysis) are shown in Supplement Figures 2–4. Individual-sample level Manhattan and QQ plots for AD-CrC are shown in Supplemental Figure 5. (Rs671 was directly genotyped in both samples. Rs143894582 was imputed in both samples, with imputation score 0.801 in sample 1 and 0.872 in sample 2; rs149212747 was also imputed in both samples, with imputation score 0.900 in sample 1 and 0.968 in sample 2.)
Table 2. Genomewide significant SNPs associated with different traits.
| Trait | Lead SNP and rs671 |
A1/A2 | P_GSA | P_MEGA | P_meta | Beta (se) | Freq | Gene | GWS SNPs in LD region (r2 > 0.2 with lead SNP) |
|---|---|---|---|---|---|---|---|---|---|
| Flushing | rs143894582 (chr12:112906873) | CA/C | 8.33×10−8 | 2.76×10−8 | 2.01×10−14 | 1.70 (0.22) | 0.08 | PTPN11 | chr12:111367244–112930475 (23 SNPs) |
| rs671 (chr12:112241766, r2 = 0.82 with rs143894582) | A/G | 2.94×10−8 | 1.96×10−8 | 5.15×10−14 | 1.49 (0.20) | 0.09 | ALDH2 | ||
| MAXDRINKS | rs149212747 (chr12: 111836771) | A/AC | 9.03×10−9 | 4.59×10−5 | 1.61×10−12 | 0.59 (0.08) | 0.91 | SH2B3a | chr12:111609727–112930475 (15 SNPs) |
| rs671 (r2 = 0.92 with rs149212747) | G/A | 4.02×10−8 | 6.05×10−4 | 1.25×10−10 | 0.53 (0.08) | 0.91 | ALDH2 | ||
| Alcohol dependence | rs149212747 | A/AC | 1.54×10−5 | 6.96×10−6 | 5.80×10−10 | 0.91 (0.15) | 0.91 | SH2B3a | chr12:111836771–112930475 (12 SNPs) |
| rs671 (r2 = 0.92 with rs149212747) | G/A | 1.45×10−5 | 7.99×10−5 | 4.46×10−9 | 0.84 (0.14) | 0.91 | ALDH2 |
SH2B3 is less than 10kb downstream from rs149212747.
Figure 1.
Manhattan and QQ plots (above) and regional Manhattan plot (below) for Flushing (meta-analysis). (Results for the individual samples are shown separately in Supplemental Figure 2.) Regional Manhattan plot shows locations of the lead SNP (rs143894582) and functional variant rs671.
Figure 2.
Manhattan QQ plots (above) and regional Manhattan plot (below) for Alcohol Dependence symptom criterion count (meta-analysis). (Results for the individual samples are shown separately in Supplemental Figure 5.) Regional Manhattan plot shows locations of the lead SNP (rs149212747) and functional variant rs671.
ADH1B
Rs1229984 in ADH1B was genotyped directly on both arrays. This marker passed quality control in Sample 2, however, it did not survive quality control in Sample 1. We re-imputed the 2Mb region surrounding rs1229984 in Sample 1 using either AS populations or EAS populations as references, but the imputation quality for rs1229984 did not pass the defined quality threshold (info < 0.8). We thus tested the association between rs1229984 and the alcohol-related traits only in Sample 2; there was an association with AD-CrC at nominal significance (p = 2.72 × 10−5). No association with other traits was observed.
PRS analysis
PRS results are shown in Table 3. The PRS based on the CONVERGE major depression GWAS (Converge Consortium, 2015) was positively associated with AD-CrC (PT = 0.005, NSNPS = 2410, beta = 0.151, p = 0.019) and MAXDRINKS (PT = 0.001, NSNPS = 654, beta = 0.200, p = 0.047), supporting shared genetic factors (pleiotropy) between alcohol consumption and major depression. These associations do not, however, survive correction for multiple comparisons (i.e., for the eight PT. threshholds). The major depression PRS was also negatively associated with flushing (PT = 0.001, NSNPS = 654, beta = −0.539, p = 5.00 × 10−3), with the association for flushing remaining significant after multiple-comparison correction.
Table 3. Polygenic risk analysis.
Eight p-value thresholds (PT) were used to select SNPs from the LD-clumped CONVERGE dataset for PRS construction. The PRS association labeled in bold connotes significance after correction for multiple PT comparisons.
| Alcohol dependence | MAXDRINKS | Flushing | |||||
|---|---|---|---|---|---|---|---|
| PT | #SNPs | Beta (se) | P | Beta (se) | P | Beta (se) | P |
| 0.00001 | 17 | 0.080 (0.058) | 0.173 | 0.091 (0.096) | 0.341 | −0.253 (0.180) | 0.159 |
| 0.0001 | 97 | 0.066 (0.064) | 0.298 | 0.103 (0.105) | 0.327 | −0.142 (0.197) | 0.471 |
| 0.001 | 654 | 0.065 (0.059) | 0.272 | 0.200 (0.101) | 0.047 | −0.539 (0.192) | 5.00 × 10−3 |
| 0.005 | 2,410 | 0.151 (0.065) | 0.019 | 0.136 (0.107) | 0.201 | −0.376 (0.201) | 0.061 |
| 0.01 | 4,168 | 0.071 (0.067) | 0.290 | 0.056 (0.108) | 0.608 | −0.252 (0.203) | 0.215 |
| 0.05 | 15,247 | 0.103 (0.060) | 0.087 | −0.011 (0.100) | 0.916 | −0.206 (0.188) | 0.274 |
| 0.1 | 25,780 | 0.132 (0.061) | 0.030 | 0.023 (0.101) | 0.820 | −0.190 (0.189) | 0.314 |
| 0.5 | 78,366 | 0.102 (0.060) | 0.090 | −0.005 (0.098) | 0.960 | −0.054 (0.184) | 0.768 |
Conditional analysis
After controlling for the known association at rs671, there was no independent significant association between rs143894582 and flushing or between rs149212747 and MAXDRINKS. Rs671 showed GWS for AD (p = 4.46 × 10−9), MAXDRINKS (p = 1.25 × 10−10) and flushing (p = 5.15 × 10−14). Conditional analyses did not identify any further candidates for independent associations.
Discussion
This study, the first GWAS of alcohol-related traits in a Thai population (n=1045 for meta-analysis), adds to the strong literature supporting ALDH2, and most likely functional variant rs671, as protective with respect to alcohol use; and adds novel genetic pleiotropy results. The association was strongest for flushing, next strongest for MAXDRINKS, then AD-CrC – consistent with the accepted mechanism of action, where rs671 inactivates ALDH2, leading to impaired clearing of acetaldehyde, the toxic first metabolic product of ethanol in its primary metabolic pathway. Presumably the aversive effect of the flushing reaction acts as a deterent to ethanol injestion. Acetaldehyde is directly responsible for the flushing reaction (Harada et al., 1981). Rs671, a glutamine to lysine substitution (E487K), results in inactivation of the enzyme subunit (Yoshida et al., 1984). Biochemistry studies – based on ALDH2 activity in liver samples – suggest that the “null” rs671 allele has a nearly dominant effect (Lai et al., 2014, Crabb et al., 1989); the heterozygote retains only about 17% enzyme activity compared to common-allele heterozygotes; minor-allele homozygotes had nearly nil measurable enzyme activity. We did not observe a significant association between ADH1B variants and any of the alcohol-related traits, but this may be attributable to lack of power: acceptable genotype data for that locus was available only for Sample 2 (510 subjects), and in that sample, we did observe a nominally-significant association (p = 2.72 × 10−5). Since acetaldehyde is implicated in various digestive tract cancers as well, these results may be relevant to a range of pathologies beyond AD per se. For example, this same variant is strongly implicated in esophageal cancer (Brooks et al., 2009) and head and neck cancers (Chang et al., 2017). We showed in a previous phenomewide association study (Polimanti et al., 2016) that ALDH2 rs671 is also nominally associated with “highest grade finished in school,” consistent with findings for ADH1B.
The observations of correlations strictly on the phenotypic level require some explanation. The correlation between AD-CrC and MAXDRINKS is expected: the larger the value for MAXDRINKS, the higher risk to have more alcohol dependence criteria. The positive correlation between AD-CrC and flushing is perhaps more surprising as it is in effect opposite to the genetic relationship between these phenotypes. The most likely explanation for this positive correlation is, we believe, simply that drinking more alcohol causes flushing directly in many subjects.
The PRS pleiotropy analyses are congruent with the previously-observed genetic overlap between AD and major depression on a population level in European-Americans (Andersen et al., 2017) and on a molecular level in African-Americans (Zhou et al., 2017). The observed positive correlation (nominally significant) between depression and MAXDRINKS and negative correlation between depression and flushing are also internally (directionally) consistent with respect to risk. Since the analysis used summary statistics from depression to predict the alcohol-related phenotypes, the outsize effect of the ALDH2 region in the latter phenotypes would not be expected to be a major confound; indeed, the most significant association between ALDH2 and major depression in the CONVERGE sample is 0.041 (and the smallest p-value in the 2Mb region is 0.0049). ALDH2*rs671 is absent from the CONVERGE dataset, again supporting that the effect is not driven by that marker. The most likely interpretation of these results is that depression and alcohol-related traits share some of their genetic risk in Asian populations, as has been previously shown in European populations (Andersen et al., 2017, Zhou et al., 2017).
For each of the three traits studied, the lead SNP is a variant other than rs671, although presumably the results are mediated by rs671, with its known functional charactersitics. Neither of the other lead variatants is itself predicted to be functional. And in fact, the lead SNPs are rather distant from rs671, although both are strongly correlated with that variant: rs143894582, lead SNP for flushing, is 665107 basepairs (bp) from rs671; and rs149212747, lead SNP for AD-CrC and MAXDRINKS, 404995 bp distant on the other side – a total span of over a megabase. This result most likely reflects random variation in the context of a large effect size in a comparatively small sample, and is illustrative of the difficulties commonly faced in identifying the true risk variant from a long associated region. Additionally, rs671 was genotyped directly, but the other lead variants were both imputed. Imperfect imputation could be an additional source of noise obfuscating the signal from rs671. In this case we know the major risk variant; it is a well-validated and well known functionally-null allele. Without that prior information, these results could have been taken to support distant SNPs mapping to different genes. For MAXDRINKS, the p-value for the lead SNP is over two orders of magnitude stronger than that for the putative functional SNP, rs671.
In summary, we present here the first GWAS of AD-CrC, MAXDRINKS, and flushing in a Thai population, confirm a strong relationship between all of these traits and ALDH2, and supply the first findings of pleiotropy between flushing and major depression (and nominally between major depression and alcohol consumption phenotypes) in an Asian population. Prior studies in other, much larger, samples, in different populations, have shown association not only with alcohol metabolizing genes, but with variants mapping elsewhere as well (Gelernter et al., 2014, Clarke et al., 2017, Schumann et al., 2016, Jorgenson et al., 2017). That we identified strong evidence for association only at and near ALDH2, most likely reflects the limited sample size available to us. When larger Asian samples characterized for AD are available, it is a strong expectation that additional risk loci will be identified. Likewise, the limited pleiotropy results reflect both the sample size limitation in the present study, and the lack of other GWAS in Asian populations for comparison purposes.
Supplementary Material
Acknowledgments
We thank Mr. Wuthichai Hasook, lead SSADDA interviewer; and staff of the Princess Mother National Institute on Drug Abuse Treatment. Technical assistance was provided by Ann Marie Lacobelle and Christa Robinson. Supported by grants from the National Institutes of Health: R01-DA 037974, R01-DA-12690, and D43-TW009087; and the VA National Center for PTSD Research.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
References
- Almli LM, Lori A, Meyers JL, Shin J, Fani N, Maihofer AX, Nievergelt CM, Smith AK, Mercer KB, Kerley K, Leveille JM, Feng H, Abu-Amara D, Flory JD, Yehuda R, Marmar CR, Baker DG, Bradley B, Koenen KC, Conneely KN, Ressler KJ. Problematic alcohol use associates with sodium channel and clathrin linker 1 (SCLT1) in trauma-exposed populations. Addiction biology. 2017 doi: 10.1111/adb.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen AM, Pietrzak RH, Kranzler HR, Ma L, Zhou H, Liu X, Kramer J, Kuperman S, Edenberg HJ, Nurnberger JI, Jr, Rice JP, Tischfield JA, Goate A, Foroud TM, Meyers JL, Porjesz B, Dick DM, Hesselbrock V, Boerwinkle E, Southwick SM, Krystal JH, Weissman MM, Levinson DF, Potash JB, Gelernter J, Han S. Polygenic Scores for Major Depressive Disorder and Risk of Alcohol Dependence. JAMA Psychiatry. 2017;74:1153–1160. doi: 10.1001/jamapsychiatry.2017.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assanangkornchai S, Noi-pha K, Saunders JB, Ratanachaiyavong S. Aldehyde dehydrogenase 2 genotypes, alcohol flushing symptoms and drinking patterns in Thai men. Psychiatry research. 2003;118:9–17. doi: 10.1016/s0165-1781(03)00043-x. [DOI] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik I, Cho NH, Kim SH, Han BG, Shin C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. The American journal of clinical nutrition. 2011;93:809–816. doi: 10.3945/ajcn.110.001776. [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Enoch M-A, Goldman D, Li T-K, Yokoyama A. The Alcohol Flushing Response: An Unrecognized Risk Factor for Esophageal Cancer from Alcohol Consumption. PLoS Medicine. 2009;6:e1000050. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, Hsiao J-R, Chen C-H. ALDH2 polymorphism and alcohol-related cancers in Asians: a public health perspective. Journal of Biomedical Science. 2017;24:19. doi: 10.1186/s12929-017-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, Porteous DJ, Deary IJ, McIntosh AM. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117) Molecular psychiatry. 2017;22:1376–1384. doi: 10.1038/mp.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converge Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. The Journal of clinical investigation. 1989;83:314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng MY, Luczak SE, Wall TL. ALDH2, ADH1B, and ADH1C Genotypes in Asians: A Literature Review. Alcohol Research & Health. 2007;30:22–27. [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Molecular psychiatry. 2014;19:41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S, Agarwal DP, Goedde HW. Aldehyde dehydrogenase deficiency as cause of facial flushing reaction to alcohol in Japanese. Lancet. 1981;2:982. doi: 10.1016/s0140-6736(81)91172-7. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia C. Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Mertens J, Weisner C, Choquet H. Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Molecular psychiatry. 2017 doi: 10.1038/mp.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalayasiri R, Verachai V, Gelernter J, Mutirangura A, Malison RT. Clinical features of methamphetamine-induced paranoia and preliminary genetic association with DBH-1021C-->T in a Thai treatment cohort. Addiction (Abingdon, England) 2014;109:965–976. doi: 10.1111/add.12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CL, Yao CT, Chau GY, Yang LF, Kuo TY, Chiang CP, Yin SJ. Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcoholism, clinical and experimental research. 2014;38 doi: 10.1111/acer.12215. [DOI] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biological psychiatry. 2011;70:504–512. doi: 10.1016/j.biopsych.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J. Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Human genetics. 2012;131:725–737. doi: 10.1007/s00439-011-1116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malison RT, Kalayasiri R, Sanichwankul K, Sughondhabirom A, Mutirangura A, Pittman B, Gueorguieva R, Kranzler HR, Gelernter J. Inter-rater reliability and concurrent validity of DSM-IV opioid dependence in a Hmong isolate using the Thai version of the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) Addictive behaviors. 2011;36:156–160. doi: 10.1016/j.addbeh.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbarek H, Milaneschi Y, Fedko IO, Hottenga JJ, de Moor MH, Jansen R, Gelernter J, Sherva R, Willemsen G, Boomsma DI, Penninx BW, Vink JM. The genetics of alcohol dependence: Twin and SNP-based heritability, and genome-wide association study based on AUDIT scores. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2015 doi: 10.1002/ajmg.b.32379. [DOI] [PubMed] [Google Scholar]
- Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, Kang TC, Nam YW, Kim GB, Shin HD, Choi IG. Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Human genetics. 2013;132:657–668. doi: 10.1007/s00439-013-1281-8. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS genetics. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, Kranzler HR. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug and alcohol dependence. 2007;91:85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug and alcohol dependence. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Polimanti R, Kranzler HR, Gelernter J. Phenome-Wide Association Study for Alcohol and Nicotine Risk Alleles in 26394 Women. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2016;41:2688–2696. doi: 10.1038/npp.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, Bakker PI, Daly MJ, Sham PC. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, Wang XY, Liu TQ, Hao W, Deng HW, Kranzler HR, Gelernter J. ALDH2 is associated to alcohol dependence and is the major genetic determinant of "daily maximum drinks" in a GWAS study of an isolated rural chinese sample. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2014;165B:103–110. doi: 10.1002/ajmg.b.32213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP. A genome screen of maximum number of drinks as an alcoholism phenotype. American journal of medical genetics. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Schumann G, Liu C, O'Reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Segura Lepe M, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke TK, Enroth S, Fischer K, Hallfors J, Harris SE, Hieber S, Hofer E, Hottenga JJ, Johansson A, Joshi PK, Kaartinen N, Laitinen J, Lemaitre R, Loukola A, Luan J, Lyytikainen LP, Mangino M, Manichaikul A, Mbarek H, Milaneschi Y, Moayyeri A, Mukamal K, Nelson C, Nettleton J, Partinen E, Rawal R, Robino A, Rose L, Sala C, Satoh T, Schmidt R, Schraut K, Scott R, Smith AV, Starr JM, Teumer A, Trompet S, Uitterlinden AG, Venturini C, Vergnaud AC, Verweij N, Vitart V, Vuckovic D, Wedenoja J, Yengo L, Yu B, Zhang W, Zhao JH, Boomsma DI, Chambers J, Chasman DI, Daniela T, de Geus E, Deary I, Eriksson JG, Esko T, Eulenburg V, Franco OH, Froguel P, Gieger C, Grabe HJ, Gudnason V, Gyllensten U, Harris TB, Hartikainen AL, Heath AC, Hocking L, Hofman A, Huth C, Jarvelin MR, Jukema JW, Kaprio J, Kooner JS, Kutalik Z, Lahti J, Langenberg C, Lehtimaki T, Liu Y, Madden PA, Martin N, Morrison A, Penninx B, Pirastu N, Psaty B, Raitakari O, Ridker P, Rose R, Rotter JI, Samani NJ, Schmidt H, Spector TD, Stott D, Strachan D, Tzoulaki I, van der Harst P, van Duijn CM, Marques-Vidal P, Vollenweider P, Wareham NJ, Whitfield JB, Wilson J, Wolffenbuttel B, Bakalkin G, Evangelou E, Liu Y, Rice KM, Desrivieres S, Kliewer SA, Mangelsdorf DJ, Muller CP, Levy D, Elliott P. KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14372–14377. doi: 10.1073/pnas.1611243113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thavorncharoensap M, Teerawattananon Y, Yothasamut J, Lertpitakpong C, Thitiboonsuwan K, Neramitpitagkul P, Chaikledkaew U. The economic costs of alcohol consumption in Thailand, 2006. BMC Public Health. 2010;10:323. doi: 10.1186/1471-2458-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychological medicine. 2015;45:1061–1072. doi: 10.1017/S0033291714002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global status report on alcohol and health – 2014 ed. WHO Press, World Health Organization; 20 Avenue Appia, 1211 Geneva 27, Switzerland: 2014. [Google Scholar]
- Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, Zhao H, Farrer LA, Gelernter J. Genomewide Association Study for Maximum Number of Alcoholic Drinks in European Americans and African Americans. Alcoholism, clinical and experimental research. 2015;39:1137–1147. doi: 10.1111/acer.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lu X, Wang L, Chen S, Li J, Cao J, Chen J, Hao Y, Li Y, Zhao L, Li H, Liu D, Wang L, Lu F, Shen C, Yu L, Wu X, Zhao Q, Ji X, Guo D, Peng X, Huang J, Gu D. Common variants at 12q24 are associated with drinking behavior in Han Chinese. The American journal of clinical nutrition. 2013;97:545–551. doi: 10.3945/ajcn.112.046482. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proceedings of the National Academy of Sciences of the United States of America. 1984;81 doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Polimanti R, Yang BZ, Wang Q, Han S, Sherva R, Nunez YZ, Zhao H, Farrer LA, Kranzler HR, Gelernter J. Genetic Risk Variants Associated With Comorbid Alcohol Dependence and Major Depression. JAMA Psychiatry. 2017;74:1234–1241. doi: 10.1001/jamapsychiatry.2017.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.