Abstract
Circulating tumour DNA (ctDNA) is that fraction of circulating DNA that is derived from a patient’s cancer. For a number of years, patients with haematological malignancies have had their disease diagnosed or monitored using tests based on detecting specific cytological or molecular biomarkers in blood. It has long been appreciated that the more common epithelial malignancies also shed DNA into the blood and that this tumour-derived DNA generally contributes a minor percentage of the overall cell-free DNA burden in peripheral blood. The biotech revolution has transformed our ability to detect, quantify and interpret genetic events. This has led to a renewed interest in the potential of using a simple blood test to both diagnose cancer and longitudinally monitor the response to medical interventions in patients with solid organ malignancies.
In this review we provide a summary of the literature to date and describe the main attributes of the current analytical approaches to ctDNA. We then focus on the potential clinical applications. There is increasing evidence to support the routine analysis of ctDNA in clinical decision-making for certain subgroups of patients with so-called hotspot mutations, particularly in lung and colorectal cancer. With continued refinement and technological progress, non-invasive molecular biomarkers including of ctDNA may be clinically useful at all stages of cancer management from diagnosis to disease progression.
Introduction
The guiding principle of personalized medicine and its progeny—precision medicine and precision oncology is that an understanding of the molecular drivers of an individual patient’s tumour will lead to a better management strategy and outcome for that patient.1
For many years, there has been an interest in the seductive concept that cancer could be diagnosed and monitored using a simple blood test rather than repeated (and often costly) imaging investigations.2 There have been huge efforts to identify the best molecular substrate for investigation—protein, metabolites and nucleic acids. In this clinically orientated review, we will focus on the advances that have been made in the detection and analysis of circulating tumour-derived cell-free DNA (ctDNA), the technology that is used to achieve this, how this has been used clinically to date and the potential impact on cancer patients in the future.
CfDNA and ctDNA
The presence of cell-free DNA (cfDNA) in human blood was first described in 1948.3 cfDNA is normally detectable in the circulation in health. However, multiple studies have demonstrated that levels are higher in pregnancy and certain pathological states, notably inflammatory conditions, after trauma and in patients with cancer.4
The mechanism of release of cfDNA into the circulation is not well understood, although it is likely to include its release from apoptotic and necrotic cells that are not successfully phagocytosed.5 The estimated half-life of DNA in the circulation is 15 min to 1h indicating it is an active process.
A key consequence of the observation that cfDNA is detectable in health is that tumour-derived DNA accounts for only a proportion of the overall circulating DNA load. Studies have demonstrated that the tumour-derived fraction varies markedly, and critically can be very low (much less than 2%) even in patients who have advanced metastatic epithelial cancers.6 Two key concepts in the analysis of cfDNA are the mutant allele frequency (MAF)—the percentage of DNA at a particular locus that is mutated—and the detection threshold. The latter is the lowest MAF that can be reliably detected by an assay. As circulating tumour-derived DNA is often present at a very low MAF, it follows that an ideal assay would have a very low detection threshold, ideally much less than 0.1%.7,8
Approaches to CfDNA analysis
PCR
Two broad approaches are currently used for the analysis of cfDNA.8 The first comprises locus-specific assays that rely on the polymerase chain reaction (PCR). In these assays, a range of PCR-based protocols are used to detect and quantify pre-specified genetic lesions. An example may be the detection and monitoring of a specific kirsten-RAS (KRAS) mutation in plasma in patients with lung or colorectal cancer, in whom KRAS mutation testing is performed routinely in the clinic.6
PCR has a number of advantages - it is an extremely robust protocol that has been validated in multiple clinical scenarios and for which internationally respected guidelines have been established.9,10 It is a low-cost technique with no necessity for bioinformatics and has real strength in terms of accuracy and precision. It has been used successfully in a multitude of ctDNA studies in a range of common malignancies and using different PCR protocols, so is well validated.
There are however, some limitations associated with applying PCR to ctDNA analysis - the main one being that ctDNA assays are usually tailored to detect an exact DNA sequence, to discriminate a DNA variant (mutation) that is associated with a patient’s cancer but is not present in their germ-line DNA (wild type). As cancer is characterized by a myriad of mutations in many genes that vary considerably from individual to individual, it follows that most cancer associated mutations from an individual's tumour will not have a ready-made assay available.
In general, a patient’s tumour will have been screened for mutations and a suitable mutation identified prior to a PCR-based assay being optimized and validated for use on ctDNA.6,11,12 Many assays have already been designed and validated for so-called ‘hotspot’ mutations—such as the clinically actionable recurring mutations in genes such as the epidermal growth factor receptor (EGFR) and KRAS.6 Such assays have led to multiple reports that confirm the potential clinical utility of a straightforward, cheap, robust test.6,8,12–14
In some clinical situations, for example if a patient was not fit to undergo an invasive biopsy, one may be interested in detecting mutations without a tissue sample to sequence. However , apart from “hotspots”, the mutation spectrum is so broad that achieving adequate coverage using a PCR based approach would be extremely difficult. Further the demands on patient plasma for each PCR assay mean that it is impractical to propose using multiple PCR-based assays.
Approaches to cfDNA analysis - next generation sequencing
The second approach to the detection and analysis of ctDNA uses the power of next generation sequencing (NGS) to screen multiple loci in a single assay.7,15,16 This is attractive because it does not depend on or assume particular genetic events—it is an unbiased survey of the genomic regions of interest. Depending on the protocol, the regions of interest may be selected for a particular tumour type (targeted resequencing), cover all protein-coding genes (exome sequencing) or whole genome sequencing.5,7,8 The experimental and analytic protocols for NGS are more complex, in particular the bioinformatics handling and interpretation of exome and whole genome sequencing. However, the pipelines are becoming increasingly automated and targeted resequencing of clinical biopsy specimens is now routine in many institutions.
To date, the detection threshold for NGS has not been able to provide the sensitivity afforded by PCR-based techniques, particularly digital PCR,8,17 and this fact as well as the simplicity and low cost of PCR has meant it has been the more popular approach. However, there have been significant developments in this regard and the lowest limit of detection in targeted NGS is down to a reported MAF of 0.02%.7 If reproducible by other groups this is extremely encouraging and will be suitable for most ctDNA applications.
In the clinical arena, it is likely that the choice of assay platform will be tailored to the clinical application. If targeted resequencing panels of commonly mutated genes can be established for different tumour types and validated/optimized to reproducibly deliver the levels of sensitivity required, then they will be very attractive assays. However, there is likely to be an ongoing demand for the extremely sensitive, targeted and cheap assays that PCR strategies can deliver. This is reflected by their development as co-diagnostic assays being developed for targeted therapeutics.
Clinical applications of ctDNA analysis
There is potential to use ctDNA to inform every stage of the management of solid organ cancers. The most common applications to date are monitoring treatment response and the early detection of relapse, and defining the mechanism of relapse in patients with progressive disease on targeted therapies.5
Monitoring of treatment response
The standard way of monitoring treatment response in patients with advanced solid organ tumours is by radiological criteria—RECIST1.1—which is a well validated but relatively crude approach that is of variable predictive value.18,19 Regular imaging can be expensive and necessitates patient exposure to radiation and contrast media.
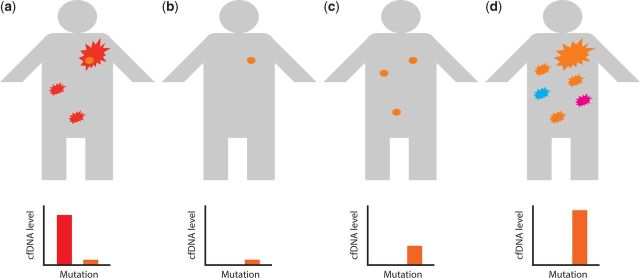
A key question is whether a simple blood test would give complementary or even more clinically useful information to interval imaging studies (Figure 1). The use of circulating biomarkers to identify minimal residual disease is well established in haematological practice, notably the myeloid leukaemias.20 Multiple studies in epithelial malignancies including lung, colorectal, breast cancer and melanoma have demonstrated that there is potential to monitor the response to treatment using ctDNA.5,8 Interestingly, in anecdotal reports of patients with lung cancer there are examples in which the radiological assessment of response and that of the circulating biomarker differ.7,21 However, there is not enough yet known about the mechanisms controlling ctDNA dynamic change, and how well radiological and ctDNA biomarkers correlate with each other or how well ctDNA predicts outcome, to understand whether there is potential for ctDNA to be used instead of rather than in addition to radiological tests. These data are beginning to emerge in larger studies. In a translational arm of a recent trial in patients with advanced EGFR mutant non-small cell lung cancer randomized to chemotherapy plus a tyrosine kinase inhibitor (TKI) or placebo, the presence or absence of the EGFR mutation in the blood after Cycle 3 of chemotherapy was a strong predictor of overall survival.22
Figure 1.
Schematic of potential application of ctDNA in clinical practice. A person may present with an advanced cancer bearing a mutation (red) that means the cancer is sensitive to a particular drug. However the tumour may harbour a subclonal population (orange) with a separate mutation conferring resistance to the drug. Therefore during initiation (b) and maintenance (c) therapy the dominant clone will respond while the subclone will be resistant and can metastasise (c), but may not initially cause symptomatic disease. Eventually the patient will become unwell with a high tumour load (d) with the now dominant clone (orange) and potentially other subclonal populations (blue, magenta). It should be possible to use cfDNA analysis at all stages of the patient’s management. At diagnosis the original dominant clone (red) should be readily detectable in cfDNA. The resistant subclone may be detectable with more sensitive technologies at diagnosis and following initial treatment, as well as on clinical progression (a–c). The earlier detection of the resistant subclone in plasma could influence therapeutic decision-making e.g. instituting treatment with a second-line drug with activity against both clones, if available. Irrespective of treatment modality the rise in cfDNA titre could predict subsequent clinical relapse (d) and prompt imaging investigations and a therapeutic re-evaluation.
There is clear potential for the results of ctDNA analysis to inform clinical decision-making. One obvious application would be for those patients with progressive disease to be offered alternative chemotherapy early or for potentially toxic chemotherapy to be discontinued on the grounds of futility. Another is the potential to non-invasively define the molecular characteristics that are driving the progressive disease.
Molecular mechanism of relapse
An attractive aspect of using ctDNA to detect progressive disease is that there is potential to infer the mechanism of relapse. Patients with solid organ malignancies who are treated with targeted therapies often develop resistance in a predictable way. Patients with colorectal cancer treated with anti-EGFR monoclonal antibodies will often relapse with KRAS mutant subclones.23 However, the most obvious example that has been well studied is the propensity of patients with EGFR mutant lung cancer to relapse with the so-called gatekeeper mutation—EGFR T790M—that confers resistance to first-line EGFR TKIs.12,13,16,21 It is now clear that T790M can readily be detected in the blood on relapse, and, in data presented at a recent meeting it was demonstrated that blood-based detection of T790M was as predictive of response to a second-generation TKI drug as a tissue-based detection of T790M through re-biopsy.24
Further, a recent publication has emphasised the potential of using ctDNA to define novel modes of resistance to new targeted treatments without the need for tissue samples. The authors demonstrated a new mechanism of resistance to the second-line TKIs that have activity against EGFR double mutants (L858R/exon 19 deletion plus T790M). The C797S mutation conferring this resistance was first detected in the blood of a patient who had become resistant to AZD9291, a second-line TKI. This mutation was subsequently shown to be the mechanism of resistance in other patients.25
Snapshot of subclonal structure of a patient’s cancer burden
The illustration that resistant subclones are selected under therapeutic pressure to become the dominant clone reflects another theoretical benefit of cfDNA analysis in advanced disease—the potential ability to survey the totality of the cancer burden at any given point.26 There is increasing evidence from multiple tumours that metastatic disease can comprise a number of different subclonal populations at different sites.27 This is of profound importance for designing therapeutic strategies to cope with the almost inevitable resistance to targeted therapeutics. The direct re-biopsy of only one site may not reflect the complex subclonal architecture of a patient's cancer, whereas blood ctDNA could theoretically be derived from all subclones. In certain scenarios, now and in the future, this may allow combination therapy or drug holidays to be recommended on the basis of a blood test.
Non-invasive/early diagnosis
Finally, there is great interest in the potential to use cfDNA for non-invasive diagnosis in the absence of tissue. There are two broad clinical scenarios—the first being early detection. At present, there is little published evidence to suggest this will be possible. The best study published to date in lung cancer reported on only two patients with Stage 1 disease and the authors were able to detect only one of these.7 It may be that the signal to noise of tumour-derived DNA versus DNA from normal cells will just be too low for this group of patients.28 The second group is those patients whose comorbidities preclude them from being candidates for invasive diagnostic biopsies. Although there are insufficient data as yet available, this is an obvious group for further research, as they may benefit tremendously from having a diagnosis inferred from a blood test alone, especially if they can be stratified as being candidates for a targeted therapy.
Conclusion
Circulating tumour DNA is a viable source of molecular biomarkers that can aid the management of patients with advanced solid organ malignancies. There is already sufficient evidence to justify the routine use of ctDNA in patients with EGFR mutant lung cancer.
The challenges for the future are to define what standardized protocols should be used to assay ctDNA and to define what clinical scenarios that ctDNA can reproducibly add clinically important, actionable information. For example, will ctDNA analysis benefit the vast majority of patients for whom targeted therapeutics are not yet available? Will ctDNA inform the use or monitoring of the new immune checkpoint inhibitors? What is the relationship between imaging and circulating biomarkers?
It is an exciting time in oncology at the present and it is likely that ctDNA analysis will play a major role in future clinical trials and ultimately in improving patient care.
Funding
The authors are funded/supported by the King's Health Partners Challenge Fund and by the National Institute for Health Research (NIHR) Biomedical research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. The research is also funded by the Roy Castle Lung Cancer Foundation and the Wellcome Trust. Frank McCaughan is a Wellcome Trust Intermediate Clinical Fellow (WT097143MA).
Conflict of interest: None declared.
References
- 1. Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Engl J Med 2012; 366:489–91. [DOI] [PubMed] [Google Scholar]
- 2. Sozzi G, Conte D, Mariani L, Lo Vullo S, Roz L, Lombardo C, et al. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res 2001; 61:4675–8. [PubMed] [Google Scholar]
- 3. Mandel P, Metais P. C R Seances Soc Biol Fil 1948; 142:241–3. [PubMed] [Google Scholar]
- 4. Jung K, Fleischhacker M, Rabien A. Cell-free DNA in the blood as a solid tumor biomarker–a critical appraisal of the literature. Clin Chim Acta 2010; 411:1611–24. [DOI] [PubMed] [Google Scholar]
- 5. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013; 10:472–84. [DOI] [PubMed] [Google Scholar]
- 6. Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014; 20:430–5. [DOI] [PubMed] [Google Scholar]
- 7. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014; 20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bidard FC, Weigelt B, Reis-Filho JS. Going with the flow: from circulating tumor cells to DNA. Sci Transl Med 2013; 5:207ps14. [DOI] [PubMed] [Google Scholar]
- 9. Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al. The digital MIQE guidelines: minimum information for publication of quantitative digital pcr experiments. Clin Chem 2013; 59:892–902. [DOI] [PubMed] [Google Scholar]
- 10. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55:611–22. [DOI] [PubMed] [Google Scholar]
- 11. Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2010; 2:20ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J. Ramakrishnan R, Tang Z, Fan W, Kluge A, Dowlati A, et al. Quantifying EGFR alterations in the lung cancer genome with nanofluidic digital PCR arrays. Clin Chem 2010; 56:623–32. [DOI] [PubMed] [Google Scholar]
- 13. Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O'Connell A, Messineo MM, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014; 20:1698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao F, Pfeifer E, Farah H, Karampini E, Dua D, Kamal N, et al. Microdroplet digital PCR: detection and quantitation of biomarkers in archived tissue and serial plasma samples in patients with lung cancer. J Thorac Oncol 2015; 10:212–7. [DOI] [PubMed] [Google Scholar]
- 15. Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012; 4:136ra68. [DOI] [PubMed] [Google Scholar]
- 16. Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497:108–12. [DOI] [PubMed] [Google Scholar]
- 17. Day E, Dear PH, McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods 2013; 59:101–7. [DOI] [PubMed] [Google Scholar]
- 18. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45:228–47. [DOI] [PubMed] [Google Scholar]
- 19. Toffart AC, Moro-Sibilot D, Couraud S, Merle P, Perol M, Girard N, et al. Evaluation of RECIST in chemotherapy-treated lung cancer: the Pharmacogenoscan Study. BMC Cancer 2014; 14:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grimwade D, Freeman SD. Defining minimal residual disease in acute myeloid leukemia: which platforms are ready for “Prime Time”? Blood 2014; 123:3345–55. [DOI] [PubMed] [Google Scholar]
- 21. Karampini E, Farah H, Mills G, Kamal N, Tobal K, Cane P, et al. Tumour molecular profiling and quantitative detection of circulating biomarkers in patients with lung cancer. Lung Cancer 2015; 87(Suppl. 1):S3–4. [Google Scholar]
- 22. Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and Dynamic Changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015; 21:3196–203. [DOI] [PubMed] [Google Scholar]
- 23. Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012; 486:537–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sequist LV, Goldman JW, Wakelee HA, Camidge DR, Yu HA, Varga A, et al. Efficacy of rociletinib (CO-1686) in plasma-genotyped T790M-positive non-small cell lung cancer (NSCLC) patients (pts). J Clin Oncol 2015; 33(Suppl. abstr 8001). [Google Scholar]
- 25. Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015; 21:560–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan KC, Jiang PA, Zheng YW, Liao GJ, Sun H, Wong J, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013; 59:211–24. [DOI] [PubMed] [Google Scholar]
- 27. Gerlinger M, McGranahan N, Dewhurst SM, Burrell RA, Tomlinson I, Swanton C. Cancer: evolution within a lifetime. Annu Rev Genet 2014; 48:215–36. [DOI] [PubMed] [Google Scholar]
- 28. Hori SS, Gambhir SS. Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci Transl Med 2011; 3:109ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]