Abstract
It is known that alcoholic fermentation is important for survival of plants under anaerobic conditions. Acetaldehyde, one of the intermediates of alcoholic fermentation, is not only reduced by alcohol dehydrogenase but also can be oxidized by aldehyde dehydrogenase (ALDH). To determine whether ALDH plays a role in anaerobic metabolism in rice (Oryza sativa L. cv Nipponbare), we characterized a cDNA clone encoding mitochondrial ALDH from rice (Aldh2a). Analysis of sub-cellular localization of ALDH2a protein using green fluorescent protein and an in vitro ALDH assay using protein extracts from Escherichia coli cells that overexpressed ALDH2a indicated that ALDH2a functions in the oxidation of acetaldehyde in mitochondria. A Southern-blot analysis indicated that mitochondrial ALDH is encoded by at least two genes in rice. We found that the Aldh2a mRNA was present at high levels in leaves of dark-grown seedlings, mature leaf sheaths, and panicles. It is interesting that expression of the rice Aldh2a gene, unlike the expression of the tobacco (Nicotiana tabacum) Aldh2a gene, was induced in rice seedlings by submergence. Experiments with ruthenium red, which is a blocker of Ca2+ fluxes in rice as well as maize (Zea mays), suggest that the induction of expression of Adh1 and Pdc1 by low oxygen stress is regulated by elevation of the cytosolic Ca2+ level. However, the induction of Aldh2a gene expression may not be controlled by the cytosolic Ca2+ level elevation. A possible involvement of ALDH2a in the submergence tolerance of rice is discussed.
Glycolysis and alcoholic fermentation are important for energy production of plants in anaerobic environments. Alcoholic fermentation is performed by two steps of reactions: the decarboxylation of pyruvate to acetaldehyde, which is catalyzed by pyruvate decarboxylase (PDC), and the following reduction of acetaldehyde to ethanol with the concomitant oxidation of NADH to NAD+, which is catalyzed by alcohol dehydrogenase (ADH) (Fig. 1; Perata and Alpi, 1993; Drew, 1997; Vartapetian and Jackson, 1997). This metabolic pathway is recognized as the principal catalytic pathway for recycling NAD+ to maintain glycolysis and the ATP level in the absence of oxygen (Perata and Alpi, 1993). It is known that expression of the genes involved in glycolysis and alcoholic fermentation (e.g. glyceraldehyde-3-P dehydrogenase, enolase, ADH, and PDC) are dramatically induced by anaerobiosis (Umeda and Uchimiya, 1994; Sachs et al., 1996). This induction is essential for anaerobic tolerance in plants. Maize (Zea mays), Arabidopsis ADH-null mutants, and rice (Oryza sativa) ADH-reduced mutants showed lower tolerance to anaerobic conditions (Schwartz, 1969; Jacobs et al., 1988; Matsumura et al., 1998).
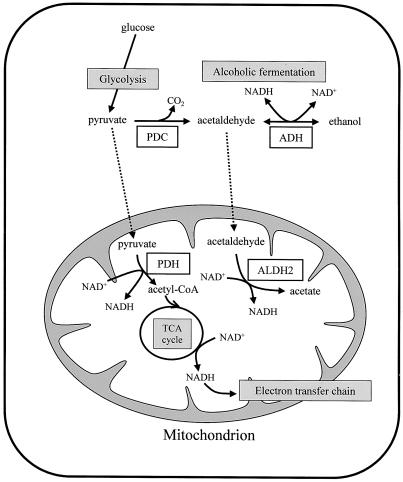
Figure 1.
Aerobic and anaerobic metabolic pathways of rice. In the aerobic pathway, pyruvate, which is produced by pathways such as glycolysis, is converted to acetyl-coenzyme A (CoA) by pyruvate dehydrogenase (PDH) and is used for the trichloroacetic acid (TCA) cycle and the electron transfer chain in mitochondria. In the anaerobic pathway, pyruvate is converted to acetaldehyde by PDC. At the same time, acetaldehyde is converted to ethanol by ADH and to acetate by aldehyde dehydrogenase (ALDH).
Aldehyde dehydrogenases [ALDHs; aldehyde:NAD(P)+ oxidoreductases, EC 1.2.1.3] are a group of enzymes catalyzing the conversion of aldehydes to the corresponding acids. In humans, the Aldh genes have been identified and characterized in detail (for review, see Yoshida et al., 1998). There are at least two isozymes of ALDH involved in ethanol metabolism (cytosolic, high-Km ALDH1 and mitochondrial, low-Km ALDH2) (Hsu et al., 1988, 1989). The mitochondrial ALDH2 is expressed in various tissues with the highest level in the liver and exhibits a high activity for oxidation of acetaldehyde, which is an intermediate of ethanol metabolism. Therefore, ALDH2 plays an important role in detoxification of acetaldehyde (Yoshida et al., 1998). In 1996, a gene for mitochondrial ALDH was identified for the first time in a plant (maize) (Cui et al., 1996). This gene, the restorer of fertility 2 (rf2) gene, was found to be a nuclear restorer gene of Texas-type cytoplasmic male sterility (Cui et al., 1996; Schnable and Wise, 1998). Two Aldh genes (Aldh2a and Aldh2b) were subsequently identified in tobacco (Nicotiana tabacum), and the tobacco Aldh2a transcript and the ALDH2a protein were present at high levels in floral tissues, especially stamens, pistils, and pollen (op den Camp and Kuhlemeier, 1997). Expression of Adh and Pdc and alcoholic fermentation also increase during pollen development in tobacco even under aerobic conditions, suggesting that alcoholic fermentation and the pathway from acetaldehyde to acetate (catalyzed by ALDH) play a role in biosynthesis and energy production during pollen development (Bucher et al., 1995; Tadege and Kuhlemeier, 1997). Under anaerobic conditions, expression of Adh, but not expression of Aldh2a, is induced in tobacco leaves. Therefore, tobacco ALDH2a seems not to function in anaerobic environments (op den Camp and Kuhlemeier, 1997).
Rice has a higher tolerance for anaerobic conditions than does tobacco. To determine whether rice ALDH functions under submergence and is involved in tolerance of anaerobic conditions, in the present study we characterized a cDNA clone encoding rice mitochondrial ALDH (Aldh2a). In contrast to the tobacco Aldh2a gene, the rice Aldh2a gene showed increased expression in rice seedlings that were submerged.
RESULTS
Characterization of Rice Aldh2a cDNA
As a first step in determining the gene for ALDH in rice, we searched the rice expressed sequence tag (EST) clone database for genes that share sequence identity with the maize rf2 gene or the tobacco Aldh2a gene. As a result, the amino acid sequences of maize RF2 protein and tobacco ALDH2a proteins were found to share sequences with the putative protein encoded by the EST clone C10151 from rice calli. The 1,855-bp insert of the cDNA clone C10151 was completely sequenced (DNA Data Bank of Japan, EMBL, and National Center for Biotechnology Information DNA accession no. AB030939). The clone C10151 contained a complete open reading frame (ORF) encoding a polypeptide of 553 amino acid residues (Fig. 2). The ORF had a significant homology with ALDH proteins of humans (Hsu et al., 1988, 1989) and yeast (Wang et al., 1998), as well as those of maize (Cui et al., 1996), tobacco (op den Camp and Kuhlemeier, 1997), and Arabidopsis (ALDH2a accession no. AB030820; M. Nakazono and A. Hirai, unpublished data) (Fig. 2). Nucleotide sequences of other copies of Arabidopsis ALDH genes (ALDH2b and ALDH1a), which were determined by the Arabidopsis genome project, have been deposited in the DNA databases (ALDH2b, accession no. AC005990; ALDH1a, accession no. AB020746). The deduced ALDH protein of rice is also homologous to the Arabidopsis ALDH2b and ALDH1a proteins (Fig. 2).
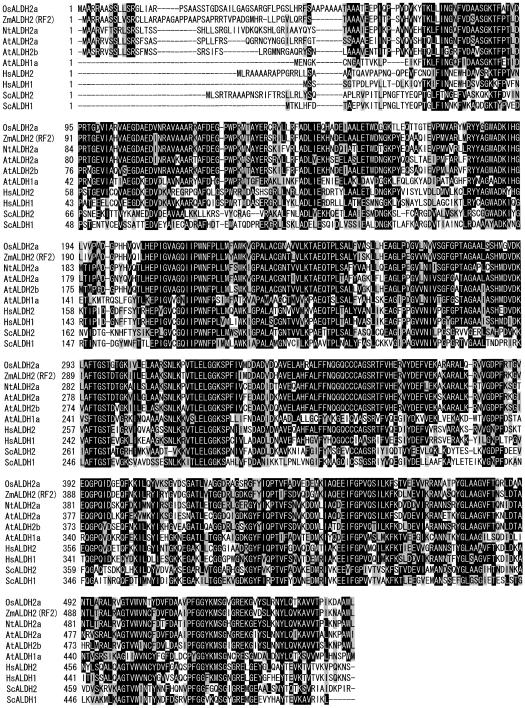
Figure 2.
Alignment of the deduced amino acid sequences of ALDH proteins from rice (OsALDH2a; this study), maize (ZmALDH2 RF2; Cui et al., 1996), tobacco (NtALDH2a; op den Camp and Kuhlemeier, 1997), Arabidopsis (AtALDH2a, accession no. AB030820; M. Nakazono and A. Hirai, unpublished data; AtALDH2b, accession no. AC005990; AtALDH1a, accession no. AB020746), human (HsALDH2 and HsALDH1; Hsu et al., 1988, 1989, respectively), and yeast (ScALDH2 [ALD5] and ScALDH1 [ALD1]; Wang et al., 1998). The alignments were generated by the Clustal W algorithm (version 1.74; Thompson et al., 1994). Black boxes indicate identical amino acids and gray boxes indicate homologous amino acids.
The amino-terminal portion of the predicted protein contains the typical mitochondrial targeting signal. To clarify whether the rice ALDH protein is imported into the mitochondria, we examined the intracellular localization of the ALDH protein in living cells using the jellyfish green fluorescent protein (GFP). Residues 1 to 95 of the ALDH protein, which contain the predicted mitochondrial targeting signal, were fused in frame to GFP, and the fusion gene was expressed transiently in suspension-cultured tobacco cv Bright Yellow-2 (BY-2) cells. As shown in Figure 3, the GFP fluorescence corresponded to the staining pattern observed using the mitochondrial-specific dye MitoTracker Red (Molecular Probes, Inc.), confirming that the rice ALDH protein is targeted to mitochondria. Therefore, the gene that encodes an ORF in the C10151 clone was designated as the rice mitochondrial Aldh2 gene (Aldh2a).
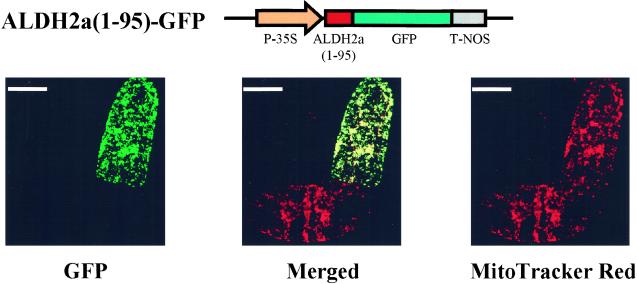
Figure 3.
Transient expression of the ALDH2a(1–95)-GFP fusion protein in suspension-cultured tobacco BY-2 cells. P-35S, A constitutive promoter from the cauliflower mosaic virus. GFP, sGFP(S65T) (Chiu et al., 1996). T-NOS, A nopaline synthase terminator. Recombinant plasmid was transformed into BY-2 cells by particle bombardment (PDS-1000, Bio-Rad, Richmond, CA). Mitochondria were stained by the mitochondrial-specific dye MitoTracker Red CMXRos (Molecular Probes, Eugene, OR) at 24 h after bombardment. Left, GFP fluorescence. Right, MitoTracker Red fluorescence. Center, Merging of the two images (GFP fluorescence and MitoTracker Red fluorescence). Each panel shows one of the optical sections through two BY-2 cells, one that is a control and the other that contains the recombinant plasmid and shows expression of the GFP fusion protein. Scale bars = 20 μm.
Expression of Recombinant ALDH2a Protein and ALDH Activity
To express recombinant mature ALDH2a protein in Escherichia coli cells, cDNA corresponding to the predicted mature protein was amplified by PCR. The modified cDNA fragment was inserted downstream of the T7 promoter in the pET-11a plasmid vector (Novagen, Madison, WI), and the resulting plasmid (termed pET-ALDH2a) was introduced into the E. coli strain BL21-CodonPlus(DE3)-RP (Stratagene, La Jolla, CA). Transformed cells were first screened for overexpression of ALDH2a and then one of these colonies was cultured. Total protein extracts were obtained by lysis of the E. coli cells that overexpressed the recombinant mature ALDH2a protein and were assayed in vitro for ALDH activity as described in “Materials and Methods.” When acetaldehyde was added as a substrate, acetaldehyde dehydrogenase activity was detected in protein extracts from the pET-ALDH2a-introduced E. coli cells (Fig. 4). By contrast, we detected no or extremely low ALDH activity in protein extracts from the E. coli cells that had been transformed with pET-11a (negative control; Fig. 4). These results indicated that rice ALDH2a protein has an activity for oxidation of acetaldehyde to acetate.
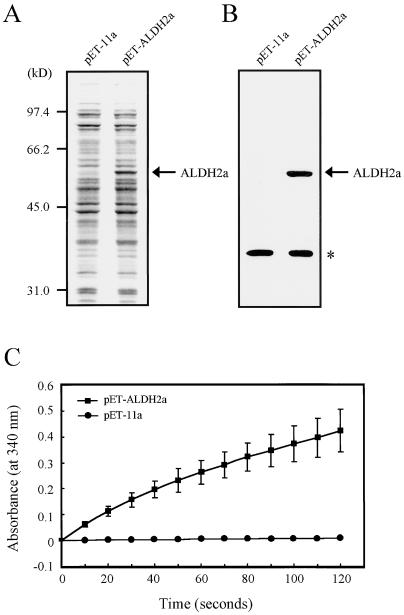
Figure 4.
In vitro assay of acetaldehyde dehydrogenase activity of E. coli cells overexpressing recombinant ALDH2a protein. A and B, E. coli cells transformed with pET-ALDH2a or pET-11a (negative control) were incubated with isopropylthio-β-galactoside to induce gene expression. Total proteins were extracted, denatured, and separated by SDS-PAGE. The gel was stained with SYPRO Orange Protein Stain (Bio-Rad) (A) and was subjected to an immunoblot analysis using an antibody raised against ALDH2 (B). The positions of the ALDH2a protein are indicated by arrows. An asterisk indicates a non-specific band. C, An in vitro ALDH assay. Immediately after acetaldehyde (final concentration 100 μm) was added to the reaction mixture, the A340 was measured at 10-s intervals for up to 120 s. Results are expressed as mean values ± se of five separate experiments.
Genomic Southern Hybridization
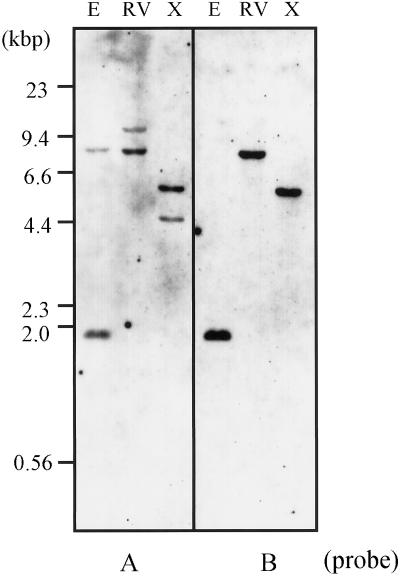
Genomic Southern hybridization was carried out using probe A, whose sequence corresponds to the coding region of Aldh2a. Probe A hybridized to two bands in the EcoRI-digest, in the EcoRV-digest, and in the XbaI-digest of rice total DNA (Fig. 5), indicating that in rice mitochondrial ALDH is actually encoded by at least two genes.
Figure 5.
Genomic Southern hybridization analysis of rice total DNA digested with EcoRI (E), EcoRV (RV), and XbaI (X). Hybridization was performed with probe A, which is specific for the coding region of Aldh2a, and probe B, which is specific for the 3′-UTR region of Aldh2a. The numbers given on the left indicate sizes of fragments in kb (kbp).
Expression of Aldh2a in Various Organs of Rice
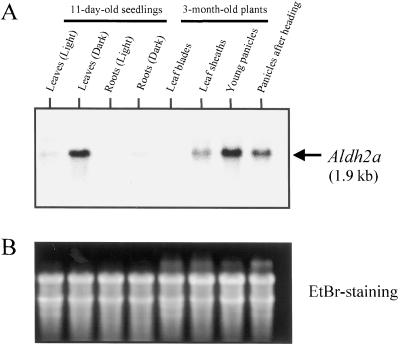
To examine the specific expression of the Aldh2a gene, we constructed a probe that is specific to the Aldh2a gene (probe B, which corresponds to the 3′-untranslated region [UTR] of Aldh2a). Genomic Southern hybridization showed that probe B hybridized to only a single band in each lane and the respective signals corresponded to one of the two bands that were detected with probe A (Fig. 5). It was confirmed that probe B is specific for the Aldh2a gene. The expression of the Aldh2a gene was examined by northern hybridization by determining the relative steady-state mRNA amounts in different organs of rice. Using total RNAs extracted from young leaves and young roots (of 11-d-old seedlings grown under light or dark conditions), mature leaf blades, mature leaf sheaths, young panicles (lengths of 7–8 cm), and panicles after heading, a single transcript of approximately 1.9 kb was observed (Fig. 6). The steady-state levels of the Aldh2a transcript in leaves of 11-d-old seedlings were higher than those levels in roots. Furthermore, the amounts of mRNA in the tissues grown under darkness were higher than those amounts in tissues grown in the light. In mature rice plants, high relative amounts of the Aldh2a mRNA were detected in young panicles, panicles after heading, and leaf sheaths (Fig. 6).
Figure 6.
A, Northern hybridization analysis of transcripts of the Aldh2a gene in various organs. Each lane was loaded with 5 μg of total RNA extracted from young leaves and young roots (of 11-d-old seedlings under light or dark conditions), mature leaf blades, mature leaf sheaths, young panicles (whose lengths were 7–8 cm), and panicles after heading. The size of the transcript of Aldh2a (1.9 kb) is shown by the arrow at the right. B, Equal loadings of total RNA were checked by ethidium bromide staining (EtBr-staining).
Increase of the Steady-State Levels of Aldh2a mRNA and ALDH2a Protein by Submergence Treatment
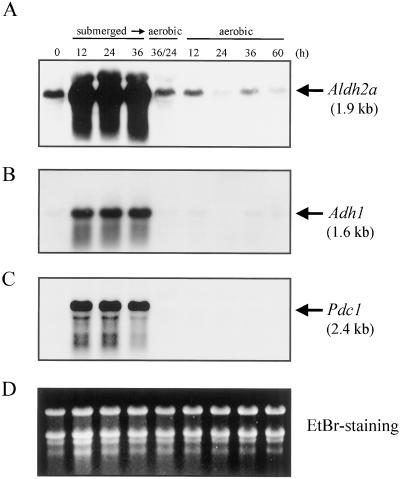
To determine whether the expression of Aldh2a, like the expression of Adh1 (Xie and Wu, 1989) and Pdc1 (Hossain et al., 1996), is induced in an anaerobic environment, 7-d-old seedlings grown under aerobic conditions were submerged for 12, 24, and 36 h and then subjected to a northern hybridization analysis. It is interesting that the steady-state level of Aldh2a mRNA dramatically increased by the submergence treatment, as did the expressions of the Adh1 and Pdc1 genes (Fig. 7, A–C). When the submerged seedlings were transferred to an aerobic environment, the amount of the Aldh2a transcript decreased (Fig. 7A).
Figure 7.
Steady-state transcript levels of Aldh2a (A), Adh1 (B), and Pdc1 (C) increase under submerged conditions. Seven-day-old rice seedlings grown in an aerobic environment in the light were submerged in the dark for 12, 24, and 36 h. After 36 h, the seedlings were returned to aerobic conditions in the darkness for 24 h (36/24). As a control, 7-d-old seedlings were kept in the dark under aerobic conditions for 12, 24, 36, and 60 h. The sizes of the transcripts of Aldh2a (1.9 kb), Adh1 (1.6 kb), and Pdc1 (2.4 kb) are shown by the arrows at the right. D, Equal loadings of total RNA were checked by ethidium bromide staining (EtBr-staining).
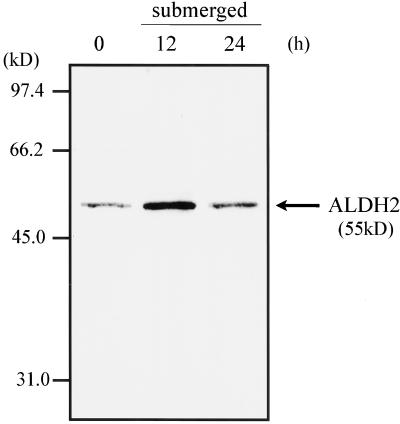
We examined whether the protein level of ALDH2a also increases under submerged conditions. An immunoblot experiment of mitochondrial total proteins, which were extracted from rice seedlings that were submerged in the dark at 28°C for 12 and 24 h, was performed using an antibody against ALDH2. The data in Figure 8 show that the amount of the ALDH2 protein, which has a molecular mass of about 55 kD, increased more in the seedlings submerged for 12 h than in the aerobically grown seedlings. However, the ALDH2 protein level in the seedlings submerged for 24 h was unexpectedly comparable to that in the control plants (0 h of submergence), suggesting that the amount of the rice ALDH2a protein transiently increases by oxygen deprivation. Although the pattern of increase of the ALDH2a protein was different from that of the Aldh2a mRNA, these results indicated that expression of the rice Aldh2a gene is induced by low oxygen and that the rice ALDH2a protein is one of the anaerobic proteins such as ADH1 and PDC1.
Figure 8.
Influence of low oxygen on the rice ALDH2 protein level. Seven-day-old aerobically grown seedlings were submerged for 12 and 24 h. Total mitochondrial proteins were extracted from leaves of the seedlings and then subjected to an immunoblot analysis using an antibody raised against ALDH2. The position of the ALDH2 protein at approximately 55 kD is indicated by an arrow.
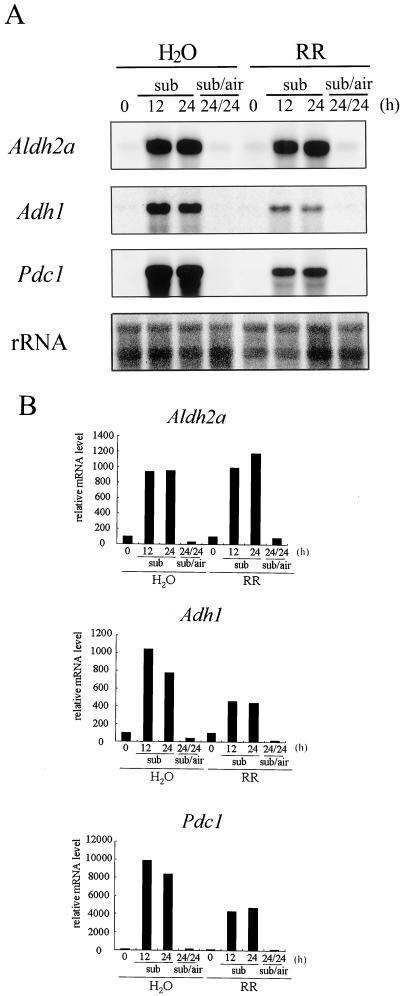
Ruthenium Red (RR) Does Not Suppress the Increase in the Steady-State Level of Aldh2a mRNA under Submerged Conditions
In maize and Arabidopsis, oxygen deprivation elevates the cytosolic Ca2+ ([Ca2+]c) level through intracellular Ca2+ flux, and RR, which is thought to be a blocker of Ca2+ fluxes from organelles, suppresses induction of expression of the anaerobic-inducible Adh gene (Subbaiah et al., 1994a, 1994b, 1998; Sedbrook et al., 1996). Here it was of interest to examine whether the increase in the amount of mRNA of the Aldh2a gene is also regulated by oxygen deprivation-induced [Ca2+]c elevation. We investigated the effect of RR on the expressions of the Aldh2a gene after 12 and 24 h of submergence. Treatment of rice seedlings with 100 μm RR under submerged conditions showed that the steady-state level of Adh1 and Pdc1 mRNAs decreased to 50% of the level induced in seedlings in water (without RR) (Fig. 9), indicating that the RR treatment affects the increase of the Adh1 and Pdc1 mRNAs by oxygen deprivation in rice, as it does in maize. On the other hand, addition of RR to rice seedlings did not affect the expressions of the Adh1 and Pdc1 genes under aerobic conditions (data not shown). Under these conditions, we examined whether the expression of the Aldh2a gene is influenced by the RR treatment under hypoxia. As shown in Figure 9, the transcript levels of Aldh2a with the RR treatment were about the same as the levels without RR. We suggest that, unlike its effect on Adh1 and Pdc1, RR does not affect the expression of Aldh2a under hypoxia.
Figure 9.
Effect of RR on the expressions of Aldh2a, Adh1, and Pdc1 genes of rice under submerged conditions. Seven-day-old aerobically grown rice seedlings were submerged in water or in 100 μm RR for 12 and 24 h (sub 12 and sub 24), and then 24-h-submerged seedlings were transferred to aerobic conditions where they were kept for 24 h (sub/air 24/24). A, Total RNA (5 μg) from each treatment was used to determine the transcript levels by northern hybridization. As a control, a probe based on the 25S/17S rRNA genes was used. B, Quantification of relative mRNA levels of Aldh2a, Adh1, and Pdc1 shown in A. All mRNA levels were normalized to the rRNA level and are presented as a percent of that observed at 0 h in water.
DISCUSSION
The predicted amino acid sequence of the rice ALDH2a protein showed high sequence identities with the sequences of the human and yeast ALDH proteins as well as the maize RF2 protein and the tobacco ALDH2a protein (Fig. 2). In contrast, the rice ALDH2a showed lower sequence conservation with other enzymes that oxidize some aldehydes (such as betaine aldehyde dehydrogenase and methylmalonate semi-aldehyde dehydrogenase; data not shown). As shown in Figure 4, rice ALDH2a has acetaldehyde dehydrogenase activity, although it might also have a role in the oxidation of other aldehydes in rice mitochondria. Analysis of intracellular localization of the rice ALDH2a protein using GFP indicated that ALDH2a functions in mitochondria (Fig. 3). Northern hybridization using total RNA extracted from several organs showed that Aldh2a is expressed mainly in leaves of dark-grown seedlings, mature leaf sheaths, and panicles (Fig. 6). This expression pattern was very similar to that of Adh1 (data not shown). Therefore, we suggest that alcoholic fermentation might occur in these organs and ALDH2a might convert the alcoholic fermentation intermediate, acetaldehyde, to acetate in the mitochondria.
It seems likely that aerobic alcoholic fermentation is a general phenomenon in pollen and developing microspores in plants, and is controlled not by oxygen availability but by substrate (e.g. Glc) availability (Tadege and Kuhlemeier, 1997; Tadege et al., 1999). In rice, we found that Aldh2a (Fig. 6) and Adh1 (data not shown) transcripts accumulate to high levels in young panicles and panicles after heading. Xie and Wu (1989) reported that the ADH1 isozyme, but not the ADH2 isozyme, is present in rice pollen. These findings suggest that, even in an aerobic environment, alcoholic fermentation takes place in the floral tissues (the panicles) in rice as in the case of tobacco (Bucher et al., 1995; Tadege and Kuhlemeier, 1997). In fact, we detected substantial acetaldehyde and ethanol production in the developing anthers of rice by gas chromatography (data not shown).
Although the steady-state levels of Aldh2a transcripts in the leaves and roots of rice seedlings grown in darkness were higher than those in tissues grown in the light (Fig. 6), the amount of Aldh2a mRNA slightly decreased when aerobically light-grown seedlings were transferred to a dark environment under aerobic conditions (Fig. 7, aerobic). Thus it seems unlikely that the expression of Aldh2a is simply regulated by light; rather, it might be regulated by some aspect of the metabolic status in etiolated or green seedlings.
One of the findings of this study is that the steady-state levels of the rice Aldh2a mRNA dramatically increased in an anaerobic environment (Fig. 7A). When the rice plants were transferred from an anaerobic environment to an aerobic environment, the amounts of the Aldh2a transcripts decreased (Fig. 7A). This expression pattern was very similar to those of the anaerobic-inducible genes Adh1 and Pdc1 (Fig. 7, B–C). An immunoblot analysis showed that the amount of the ALDH2 protein in the mitochondria increased by submergence for 12 h and then decreased by more submergence treatment (for 24 h; Fig. 8). The reason for the difference between the patterns of increase of ALDH2a protein and Aldh2a mRNA is not known. One possible explanation for the difference is that the decrease of the ALDH2 protein after 24 h of submergence is due to a shortage of ATP and to a disappearance of the membrane potential, because both ATP and a membrane potential are required for the import of proteins into the mitochondria (for review, see Glaser et al., 1998). Under submerged conditions, efficiencies of respiration and ATP synthesis in the mitochondria are reduced because of a lack of oxygen. Thus it is possible that further submergence (more than 12 h) might cause an ATP shortage and a reduction in the membrane potential in the mitochondria and might reduce the efficiency of importing ALDH2 protein into the mitochondria.
The difference between ALDH2a protein and Aldh2a mRNA might alternatively be due to the existence of isozymes. There are at least two isozymes of mitochondrial ALDH2 in rice. Because the antibody against ALDH2 used in this study is able to recognize both mitochondrial ALDH2 isozymes, it is possible that the signals detected in the immunoblot analysis (Fig. 8) originate from both isozymes. We are now investigating the structure and expression of another copy of the rice Aldh2 gene (termed Aldh2b). It is interesting that steady-state levels of Aldh2b mRNA were relatively abundant under aerobic conditions, whereas when rice seedlings were submerged, the Aldh2b mRNA levels rapidly decreased (H. Tsuji, M. Nakazono, and A. Hirai, unpublished data). This finding suggests that changes in the expression of the rice Aldh2a and Aldh2b in response to changes in the oxygen status are antiparallel to each other. Thus the ALDH2 protein signals from rice under aerobic conditions (Fig. 8, 0 h) might have originated mostly from ALDH2b protein, and the ALDH2 signals from rice that had been submerged for 24 h might have originated mostly from an increase in ALDH2a protein. In contrast, the signal from rice that had been submerged for 12 h might have originated from both ALDH2a protein, whose amount is increasing, and ALDH2b protein, whose amount is decreasing. Further investigations will be necessary to test these possibilities.
In contrast to the rice Aldh2a transcript levels, the tobacco Aldh2a transcript levels did not increase during anaerobiosis in leaf tissue (op den Camp and Kuhlemeier, 1997). The authors proposed that a pathway involving ALDH2 is not important for normal metabolism in tobacco leaves, even in anaerobiosis. Our preliminary results showed that expression of the Arabidopsis ALDH2a gene was not enhanced under submerged conditions as in the case of tobacco Aldh2a (data not shown). We propose that rice might have a greater ability than tobacco and Arabidopsis to detoxify acetaldehyde, which is produced during alcoholic fermentation in anaerobiosis, both by ALDH and ADH. In other words, higher levels of ALDH could be one of the reasons why rice is more tolerant of submergence than other plant species such as tobacco and Arabidopsis. One means of testing this hypothesis would be to examine whether transgenic tobacco or Arabidopsis plants that overproduce ALDH2 can better tolerate anaerobiosis. In addition, in rice plants during anaerobiosis, pyruvate might be converted to acetyl-CoA by the sequential actions of PDC, ALDH, and acetyl-CoA synthetase (ACS). Kuhlemeier and his colleagues (Tadedge and Kuhlemeier, 1997; op den Camp and Kuhlemeier, 1997) proposed that acetyl-CoA produced through PDC/ALDH/ACS might be supplied as a substrate for the trichloroacetic acid cycle, lipid biosynthesis, and the glyoxylate cycle in tobacco pollen. A similar pathway might operate in rice under submerged conditions, and acetyl-CoA production via the PDC/ALDH/ACS pathway might be important for the biosynthesis of several intermediates in the process of adaptation in anaerobiosis of rice.
Under anaerobic conditions, fermentation regenerates NAD+ from NADH, allowing plant cells to continue glycolysis and maintain the ATP level (Perata and Alpi, 1993). The conversion of acetaldehyde to acetate by ALDH consumes NAD+ and this consumption could potentially block glycolysis. However, the hypoxia-inducible ALDH2a protein is localized in mitochondria and is separated from the cytosolic enzymes involved in glycolysis and alcoholic fermentation (Fig. 1). This fact suggests that reduction of NAD+ to NADH by rice ALDH2a does not influence the efficiency of glycolysis. Plants have many ALDH isozymes. We assume that at least two of these isozymes are involved in the oxidation of acetaldehyde. One isozyme is a mitochondrial enzyme (Cui et al., 1996; op den Camp and Kuhlemeier, 1997) and another is a cytosolic enzyme (Li et al., 2000). In contrast to the expression of the mitochondrial Aldh2a gene, the expression of the rice cytosolic Aldh1a gene is probably not influenced by the oxygen status (Y Li, M. Nakazono, and A. Hirai, unpublished data). Thus it seems reasonable that the mitochondrial ALDH2a protein is induced to ensure a continuation of glycolysis and that the cytosolic ALDH isozymes are not induced for this purpose.
Subbaiah et al. (1994a, 1994b, 1998) proposed a model for maize in which induction of Adh1 gene expression by oxygen deprivation is preceded by [Ca2+]c elevation, which might be due to Ca2+ released from the mitochondria. RR has been shown to block Ca2+ fluxes from organelles (Knight et al., 1992) and induction of ADH1 was suppressed by treatment of maize cells with RR under submerged conditions (Subbaiah et al., 1994b). In rice, treatment of seedlings by RR suppressed increase of transcripts of Adh1 and Pdc1 under anaerobiosis, as it does in maize (Fig. 9). Furthermore, we found that CaCl2, when added simultaneously with RR, prevented the effect of RR on the expressions of Adh1 (Tsuji et al., 2000) and Pdc1 (data not shown). This prevention of the effects of RR by Ca2+ supports the hypothesis that Ca2+ is involved in the expressions of the rice Adh1 and Pdc1 genes under anaerobic conditions. In contrast, treatment with RR (Fig. 9) and/or CaCl2 (data not shown) had no inhibitory effect on the anaerobic induction of the rice Aldh2a gene, suggesting that elevation of [Ca2+]c might not trigger an increase in the Aldh2a mRNA. These findings indicate that there are at least two pathways of oxygen signaling in rice: One is a signaling pathway regulated by a release of Ca2+ from intracellular stores, which is sensitive to RR, and another is a signaling pathway that is independent of [Ca2+]c elevation (Tsuji et al., 2000). In rice, it is likely that the former pathway controls the increase of the Adh1 and Pdc1 mRNAs and the latter pathway controls the increase of the Aldh2a mRNA.
In conclusion, we have demonstrated that, unlike expression of the tobacco Aldh2a gene, expression of the rice mitochondrial Aldh2a gene is induced under anaerobic conditions and that, as shown by experiments with RR, the induction of Aldh2a gene expression is not regulated by an elevation of [Ca2+]c. We propose that a high level of mitochondrial ALDH2 protein under anaerobiosis (at least for the first 12 h) might confer more tolerance to submergence in rice than in tobacco or Arabidopsis. To further test this hypothesis, we are currently attempting to produce a transgenic Arabidopsis plant that overproduces ALDH2.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Treatments
Rice (Oryza sativa L. cv Nipponbare) was grown in the light at 28°C for 10 d for extraction of total DNA. For extraction of total RNA, leaves and roots of seedlings that were grown in the light or in the dark at 28°C for 11 d were used. Mature leaf blades, mature leaf sheaths, and young panicles were prepared from 3-month-old plants and panicles after headings were prepared from 3.5-month-old plants.
For treatment of oxygen deprivation, 7-d-old aerobically grown seedlings were submerged in the dark at 28°C for 12, 24, and 36 h. After 36 h, the submerged seedlings were transferred to aerobic conditions in the dark at 28°C for 24 h. For RR treatment, seedlings were submerged in water that contained 100 μm RR (Sigma Chemical, St. Louis) in the dark at 28°C for 12 and 24 h. Leaves of the seedlings were collected after the indicated time intervals. After harvesting, the leaves were frozen in liquid nitrogen.
For isolation of mitochondrial proteins for immunoblotting, 7-d-old aerobically grown seedlings were submerged in the dark at 28°C for 12 and 24 h. Leaves of the seedlings were collected after the indicated time intervals.
EST Clone
The EST clone C10151 from rice calli was provided by the Rice Genome Research Program of the National Institute of Agrobiological Resources (Tsukuba, Japan).
Sequence Analysis
The cDNA clone was completely sequenced with an automatic DNA sequencer (model 373S; Perkin-Elmer Applied Biosystems, Foster City, CA). DNA sequencing data were analyzed with GENETYX-WIN Software (version 3, Software Development, Tokyo).
Oligonucleotides
The following 12 oligonucleotides were synthesized: ALDH-P1, 5′-TACAAGATGAGCGGCGTTGGCA-3′; ALDH-P2, 5′-ATTACTACAGCTACAACCAGGC-3′; ALDH-P3, 5′-TAGCTGTAGTAATCGATCCT-3′; ALDH-P4, 5′-TGTACAAAAGATTGCCCGAT-3′; ALDH-P5, 5′-TCACCATGGGATCCACCGTCGCGAACGTCTTC-3′; ALDH-P6, 5′-ATACATATGAGCGCTGCACCGGCCGCCGCTGCCA-3′; ALDH-P7, 5′-ATATGATCATTATTCATC GCAATCTCTGGCGTTG-3′; ADH-P1, 5′-ATTATGGTGTTGGGTAATAAGATT-3′; ADH-P2, 5′-AACTGAAACTCGTATAAATATATG-3′; PDC-P1, 5′-ATCGTCTGTGAATTAATTGT-3′; PDC-P2, 5′-CCTTCAAATCGTCCATGTTA-3′; and M13, 5′-CAGTCACGACGTTGTAAAACGACGGCCAGT-3′.
Construction and Visualization of a GFP Fusion Protein
The ALDH2a(1–95)-GFP recombinant plasmid was constructed as follows: The sequence corresponding to residues 1 to 95 of the predicted ALDH2a precursor protein was amplified by PCR from the cDNA clone using primers M13 and ALDH-P5. The primer ALDH-P5 contained an NcoI site near its 5′-end. Because the amplified fragment included an SalI site, which originated from the multicloning site of the pBluescript vector, it was digested with both SalI and NcoI and cloned in-frame into the SalI and NcoI sites of the enhanced synthetic GFP vector [CaMV35Spro::sGFP(S65T)::NOSter; a gift from Dr. Y. Niwa (University of Shizuoka, Japan); Chiu et al., 1996]. This plasmid was termed ALDH2a(1–95)-GFP (Fig. 3). Its sequence was checked before proceeding.
Ten micrograms of plasmid was precipitated onto 1.0-μm spherical gold beads. Suspension-cultured tobacco (Nicotiana tabacum) BY-2 cells were bombarded using a PDS-1000 particle delivery system (Bio-Rad) as described previously (Nakazono et al., 2000). After bombardment, the samples were placed on a benchtop for 24 h. Transformed BY-2 cells were treated with 500 nm MitoTracker Red CMXRos (Molecular Probes), a mitochondrial-specific dye, for 30 min. These cells were examined with a confocal laser-scanning microscope (Micro-Radiance MR/AG-2; Bio-Rad). The samples were illuminated with an argon ion laser (488 nm wavelength) for GFP or a green HeNe laser (543 nm) for MitoTracker Red fluorescence.
Expression of the ALDH2a Protein in Escherichia coli
The cDNA clone C10151 was used as a template for PCR of a 1.68-kb fragment encoding the mature ALDH2a protein. The oligonucleotides used were the 5′-end primer ALDH-P6 and the 3′-end primer ALDH-P7. An ATG initiation codon was introduced by changing Phe-47 (the last amino acid of the mitochondrial targeting signal; Fig. 2) to Met in the 5′-end primer ALDH-P6. The primers ALDH-P6 and ALDH-P7 contained an NdeI site and a BclI site, respectively, near their 5′-ends. PCR was performed using KOD-plus, a high-fidelity thermophilic DNA polymerase (Toyobo, Tokyo). The amplified fragment was digested with both NdeI and BclI and cloned into the NdeI and BamHI sites of the pET-11a vector (Novagen). This plasmid, termed pET-ALDH2a, and pET-11a, which was used as a control, were introduced into the E. coli strain BL21-CodonPlus(DE3)-RP (Stratagene). Transformed E. coli cells were incubated at 37°C overnight in Luria-Bertani medium with 50 μg/mL ampicillin. The overnight cultures (1 mL) were added into 50 mL of fresh Luria-Bertani-ampicillin medium and incubated with shaking at 37°C until OD600 reached 0.6. Isopropyl β-d-thiogalactopyranoside, an inducer of gene expression, was added to a final concentration of 1 mm and the incubation was continued at 30°C for 3 h. Cells were washed with 100 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-NaOH (pH 7.4) and then sonicated in 100 mm HEPES-NaOH (pH7.4), 1 mm EDTA, 10% (v/v) glycerol, and 0.1% (v/v) Triton X-100 on ice. The lysate was centrifuged at 15,000g for 5 min. The supernatant was used as a crude enzyme extract for the assay of ALDH activity.
Assay of ALDH Activity against Acetaldehyde
ALDH activity was assayed according to the method of op den Camp and Kuhlemeier (1997). The reaction mixture contained 100 mm sodium-pyrophosphate (pH 9.5), 1.3 mm NAD+, 100 μm acetaldehyde, and 1 volume of E. coli extract containing 200 μg of protein in a total volume of 100 μL. The reaction was initiated by addition of acetaldehyde. ALDH activity was evaluated from the rate of increase in A340 due to the conversion of NAD+ to NADH.
Probe Labeling
The fragments that correspond to the coding region and the 3′-UTR of Aldh2a and the 3′-UTRs of Adh1 and Pdc1 were amplified from cDNA clones by PCR using the primer sets ALDH-P1/ALDH-P2, ALDH-P3/ALDH-P4, ADH-P1/ADH-P2, and PDC-P1/PDC-P2, respectively. A 7.4-kb EcoRI- fragment containing the rice 25S/17S rRNA genes was obtained from the rRNA genomic clone pRR217. These fragments were used for probes by labeling them with the DIG DNA Labeling and Detection Kit (Roche Diagnostics, Mannheim, Germany).
Extraction of Total DNA and Genomic Southern Hybridization
Total DNA was extracted using the method of Shure et al. (1983) from 10-d-old plants. Total DNA (5 μg) was digested by EcoRI, EcoRV, and XbaI, and then subjected to electrophoresis on a 0.7% (w/v) agarose gel. Southern hybridization was carried out using the method of Ohtsu et al. (1999).
Extraction of Total RNA and Northern Hybridization
Rice total RNA was extracted by the standard guanidine thiocyanate/CsCl method (Kingston, 1991). Total RNA was denatured by treatment with formaldehyde and fractionated in a 1% (w/v) denaturing agarose gel. The gel was subsequently stained with ethidium bromide and blotted onto a nylon membrane (Magna Pure Nylon, Micron Separations, Westborough, MA). Northern-blot hybridization was performed with the DIG DNA Labeling and Detection Kit (Roche Diagnostics).
Antibody Preparation
Two oligopeptides corresponding to internal sequences of rice ALDH2a and Arabidopsis ALDH2a were synthesized and purified by Biologica (Nagoya, Japan). The sequences of the synthetic peptides were as follows: NH4-(G) CAGSRTFVHERVYDEFVEK-COOH (rice ALDH2a) and NH4-(G) CAGSRTFVHEKVYDEFVEK-COOH (Arabidopsis ALDH2a). A mixture of the two oligopeptides was injected into rabbits to raise ALDH2-specific antiserum by Biologica. To purify the ALDH2-specific antibody, the antiserum was loaded onto a synthetic peptide-coupled N-hydroxysuccinimide-activated Sepharose 4 Fast Flow column (Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's instructions. Antibody was eluted from the column using 100 mm Gly-HCl (pH 2.5) and 150 mm NaCl and was neutralized with 1 m Tris [tris(hydroxymethyl)aminomethane]-HCl (pH 8.0). The purified antibody was stored at −80°C.
Preparation of Mitochondrial Protein Extracts
Mitochondria were isolated from leaves of aerobically grown rice seedlings or submergence-treated seedlings by Suc step-gradient centrifugation according to the method of Newton (1994). Mitochondrial protein extracts were obtained by lysis of isolated mitochondria with the lysis buffer (5% [w/v] sodium N-lauroyl sarcosinate, 0.25% [v/v] Triton X-100, 25 mm Tris-HCl [pH 7.5], 1 m KCl, 20 mm EDTA, and 40% [v/v] glycerol).
Immunoblotting
E. coli protein extract or mitochondrial protein extract was denatured and separated by SDS-PAGE. Each lane was loaded with 20 μg of protein. Acrylamide concentrations were 4.75% (w/v) and 10% (w/v) in the stacking and separation gels, respectively. Proteins were electroblotted onto a nitrocellulose membrane (Hybond-C extra; Amersham, Buckinghamshire, UK). The membrane was initially incubated for 1 h in 5% (w/v) skim milk (in 20 mm Tris-buffered saline plus Tween 20 [TBST: 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% {v/v}Tween 20]), and reacted with the ALDH2-specific antibody (1:200 dilution) at 4°C for 12 h. The blot was washed three times for 10 min each in 20 mm TBST, and then was incubated with 1:5,000 diluted donkey antirabbit antibody coupled to peroxidase (Amersham) at 25°C for 1 h. After three washes in 20 mm TBST for 10 min, the reaction was visualized using Lumi-Light (Roche Diagnostics) and a Lumino-image analyzer LAS-1000 (Fuji Photo Film, Tokyo).
ACKNOWLEDGMENTS
The authors thank Dr. Y. Niwa for the generous gift of the enhanced GFP expression vector and the Rice Genome Research Program of the National Institute of Agrobiological Resources for providing EST clone C10151. The authors express their appreciation to Drs. P.S. Schnable and M. Sugiura for their critical readings of the manuscript and stimulating discussions. The authors express their appreciation to Drs. J.H. Weil, Y. Yamasue, Y. Suzuki, H. Matsumura, and N. Kubo for their valuable suggestions. The authors thank Drs. T. Mikami and M. Yamamoto for their helpful assistance in the purification of the anti-ALDH2 antibody and K. Nakazono for her technical assistance.
Footnotes
This work was supported in part by grants-in-aid from the Ministry of Science, Education and Culture of Japan (grant nos. 10556001 to M.N. and 09556002 to A.H.) and by grants from the Program for Promotion of Basic Research Activities for Innovative Biosciences of Japan (grant no. 0007 to A.H.).
LITERATURE CITED
- Bucher M, Brander KA, Sbicego S, Mandel T, Kuhlemeier C. Aerobic fermentation in tobacco pollen. Plant Mol Biol. 1995;28:739–750. doi: 10.1007/BF00021197. [DOI] [PubMed] [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Cui X, Wise RP, Schnable PS. The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science. 1996;272:1334–1336. doi: 10.1126/science.272.5266.1334. [DOI] [PubMed] [Google Scholar]
- Drew MC. Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:223–250. doi: 10.1146/annurev.arplant.48.1.223. [DOI] [PubMed] [Google Scholar]
- Glaser E, Sjöling S, Tanudji M, Whelan J. Mitochondrial protein import in plants: signals, sorting, targeting, processing and regulation. Plant Mol Biol. 1998;38:311–338. doi: 10.1023/a:1006020208140. [DOI] [PubMed] [Google Scholar]
- Hossain MA, Huq E, Grover A, Dennis ES, Peacock WJ, Hodges TK. Characterization of pyruvate decarboxylase genes from rice. Plant Mol Biol. 1996;31:761–770. doi: 10.1007/BF00019464. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Bendel RE, Yoshida A. Genomic structure of the human mitochondrial aldehyde dehydrogenase gene. Genomics. 1988;2:57–65. doi: 10.1016/0888-7543(88)90109-7. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Chang WC, Yoshida A. Genomic structure of the human cytosolic aldehyde dehydrogenase gene. Genomics. 1989;5:857–865. doi: 10.1016/0888-7543(89)90127-4. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Dolferus R, van den Bossche D. Isolation and biochemical analysis of ethyl methanesulfonate-induced alcohol dehydrogenase null mutants of Arabidopsis thaliana (L.) Heynh. Biochem Genet. 1988;26:105–122. doi: 10.1007/BF00555492. [DOI] [PubMed] [Google Scholar]
- Kingston RE. Guanidinium methods for total RNA preparation. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene Publishing Associates & Wiley-Interscience; 1991. pp. 4.2.1–4.2.8. [Google Scholar]
- Knight MR, Smith SM, Trewavas AJ. Wind-induced plant motion immediately increases cytosolic calcium. Proc Natl Acad Sci USA. 1992;89:4967–4971. doi: 10.1073/pnas.89.11.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Nakazono M, Tsutsumi N, Hirai A. Molecular and cellular characterizations of a cDNA clone encoding a novel isozyme of aldehyde dehydrogenase from rice. Gene. 2000;249:67–74. doi: 10.1016/s0378-1119(00)00152-9. [DOI] [PubMed] [Google Scholar]
- Matsumura H, Takano T, Takeda G, Uchimiya H. Adh1 is transcriptionally active but its translational product is reduced in a rad mutant of rice (Oryza sativa L.), which is vulnerable to submergence stress. Theor Appl Genet. 1998;97:1197–1203. [Google Scholar]
- Nakazono M, Imamura T, Tsutsumi N, Sasaki T, Hirai A. Characterization of two cDNA clones encoding isozymes of the F1F0-ATPase inhibitor protein of rice mitochondria. Planta. 2000;210:188–194. doi: 10.1007/pl00008125. [DOI] [PubMed] [Google Scholar]
- Newton KJ. Procedure for isolating mitochondria and mitochondrial DNA and RNA. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 549–556. [Google Scholar]
- Ohtsu K, Hamanaka S, Yamazaki K, Nakazono M, Hirai A. Characterization of a cDNA encoding a novel subunit for cytochrome c oxidase (COX6b) from rice. Breed Sci. 1999;49:211–215. [Google Scholar]
- op den Camp RGL, Kuhlemeier C. Aldehyde dehydrogenase in tobacco pollen. Plant Mol Biol. 1997;35:355–365. doi: 10.1023/a:1005889129491. [DOI] [PubMed] [Google Scholar]
- Perata P, Alpi A. Plant responses to anaerobiosis. Plant Sci. 1993;93:1–17. [Google Scholar]
- Sachs MM, Subbaiah CC, Saab IN. Anaerobic gene expression and flooding tolerance in maize. J Exp Bot. 1996;47:1–15. [Google Scholar]
- Schnable PS, Wise RP. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3:175–180. [Google Scholar]
- Schwartz D. An example of gene fixation resulting from selective advantage in suboptimal conditions. Am Nat. 1969;103:479–481. [Google Scholar]
- Sedbrook JC, Kronebusch PJ, Borisy GG, Trewavas AJ, Masson PH. Transgenic AEQUORIN reveals organ-specific cytosolic Ca2+ responses to anoxia in Arabidopsis thaliana seedlings. Plant Physiol. 1996;111:243–257. doi: 10.1104/pp.111.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M, Wessler S, Fedoroff N. Molecular identification and isolation of the Waxy locus in maize. Cell. 1983;35:225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Subbaiah CC, Bush DS, Sachs MM. Elevation of cytosolic calcium precedes anoxic gene expression in maize suspension-cultured cells. Plant Cell. 1994a;6:1747–1762. doi: 10.1105/tpc.6.12.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Bush DS, Sachs MM. Mitochondrial contribution to the anoxic Ca2+ signal in maize suspension-cultured cells. Plant Physiol. 1998;118:759–771. doi: 10.1104/pp.118.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Zhang J, Sachs MM. Involvement of intracellular calcium in anaerobic gene expression and survival of maize seedlings. Plant Physiol. 1994b;105:369–376. doi: 10.1104/pp.105.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Dupuis I, Kuhlemeier C. Ethanolic fermentation: new functions for an old pathway. Trends Plant Sci. 1999;4:320–325. doi: 10.1016/s1360-1385(99)01450-8. [DOI] [PubMed] [Google Scholar]
- Tadege M, Kuhlemeier C. Aerobic fermentation during tobacco pollen development. Plant Mol Biol. 1997;35:343–354. doi: 10.1023/a:1005837112653. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Nakazono M, Saisho D, Tsutsumi N, Hirai A. Transcript levels of the nuclear-encoded respiratory genes in rice decrease by oxygen deprivation: evidence for involvement of calcium in expression of the alternative oxidase 1a gene. FEBS Lett. 2000;471:201–204. doi: 10.1016/s0014-5793(00)01411-3. [DOI] [PubMed] [Google Scholar]
- Umeda M, Uchimiya H. Differential transcript levels of genes associated with glycolysis and alcohol fermentation in rice plants (Oryza sativa L.) under submergence stress. Plant Physiol. 1994;106:1015–1022. doi: 10.1104/pp.106.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartapetian BB, Jackson MB. Plant adaptations to anaerobic stress. Ann Bot. 1997;79:3–20. [Google Scholar]
- Wang X, Mann CJ, Bai Y, Ni L, Weiner H. Molecular cloning, characterization, and potential roles of cytosolic and mitochondrial aldehyde dehydrogenases in ethanol metabolism in Saccharomyces cerevisiae. J Bacteriol. 1998;180:822–830. doi: 10.1128/jb.180.4.822-830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wu R. Rice alcohol dehydrogenase genes: anaerobic induction, organ specific expression and characterization of cDNA clones. Plant Mol Biol. 1989;13:53–68. doi: 10.1007/BF00027335. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem. 1998;251:549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]