Abstract
Few studies have evaluated the performance of existing breast cancer risk prediction models among women of African ancestry. In replication studies of genetic variants, a change in direction of the risk association is a common phenomenon. Termed flip-flop, it means that a variant is risk factor in one population but protective in another, affecting the performance of risk prediction models. We used data from the genome-wide association study (GWAS) of breast cancer in the African diaspora (The Root consortium), which included 3,686 participants of African ancestry from Nigeria, USA, and Barbados. Polygenic risk scores (PRSs) were constructed from the published odds ratios (ORs) of four sets of susceptibility loci for breast cancer. Discrimination capacity was measured using the area under the receiver operating characteristic curve (AUC). Flip-flop phenomenon was observed among 30%~40% of variants across studies. Using the 34 variants with consistent directionality among previous studies, we constructed a PRS with AUC of 0.531 (95% confidence interval [CI]: 0.512–0.550), which is similar to the PRS using 93 variants and ORs from European ancestry populations (AUC=0.525, 95% CI: 0.506–0.544). Additionally, we found the 34-variant PRS has good discriminative accuracy in women with family history of breast cancer (AUC=0.586, 95% CI: 0.532–0.640). In conclusion, we found that PRS based on variants identified from prior GWASs conducted in women of European and Asian ancestries did not provide a comparable degree of risk stratification for women of African ancestry. Further large-scale fine-mapping studies in African ancestry populations are desirable to discover population-specific genetic risk variants.
Keywords: breast neoplasms, polygenic risk score, women of African ancestry, flip-flop
Introduction
Breast cancer is the most common cancer in women worldwide with a total of nearly 2.2 million new cases diagnosed in 2015 [1]. To date, genome-wide association studies (GWAS) have revealed approximately 100 common single nucleotide polymorphisms (SNPs) associated with breast cancer risk [2–5]. The combined effect of these SNPs has been shown to achieve a reasonable degree of risk stratification in populations of European ancestry [6–8]. Significant SNPs can be aggregated into risk prediction models in the form of polygenic risk score (PRS), which can be used for the identification of high-risk individuals in clinical settings and for population-level screening [9,10].
To date, SNPs from GWAS have been identified almost exclusively in populations of European ancestry, and often show different association patterns among African populations [11–13]. Even among women of African ancestry, variants identified in one population are often not applicable to another population [14]. These conflicting results could be due to several reasons, including differences in allele frequencies and linkage disequilibrium (LD) blocks among different ethnicities [15], and differences in population characteristics within one ethnicity. The flip-flop phenomenon, meaning that the same allele confers risk in one population but is protective in another, has been observed as hindrance to drawing causal inference in replication studies of gene-disease associations [16]. To date, few studies have evaluated breast cancer risk prediction model with SNPs identified among women of African ancestry [17], and to our knowledge, none of the previous studies incorporated the flip-flop phenomenon to compare the model’s performance for this population.
In the present study, we had two main aims: (1) to examine the flip-flop phenomenon in approximately 100 previously identified risk variants in women of African-ancestry; (2) to construct several PRSs with aforementioned SNPs, based on consistency among ethnicity and studies, and compare their performances in women of African-ancestry.
Methods
Study participants
The study populations of the Root consortium have been described previously [11,18]. Briefly, this study included 3,686 participants of African ancestry (1,657 breast cancer cases and 2,029 controls). Ascertainment of cases and controls occurred in Ibadan, Nigeria (711 cases and 624 controls), Barbados (92 cases and 229 controls), and four sites in the United States (854 cases and 1,176 controls). The mean age of cases was 49.3 years whereas the mean age of controls was 48.4 years. Other baseline characteristics such as menopausal status and family history of breast cancer are presented in Supplemental Table 1.
SNP genotyping and imputation and ancestry estimation
Information about genotyping, quality control, imputation and principal component analysis (PCA) has been previously described [11]. Briefly, genotyping was conducted using the Illumina HumanOmni2.5-8v1 array, including approximately 2.4 million genetic variants. Genotype imputation was conducted with the IMPUTE2 software. With the 1000 Genomes Project phase 1 integrated variant set as the reference panel, 23,098,723 SNPs were imputed and passed the imputation quality filter (imputation score > 0.3). To account for population structure, the first ten principal components were computed with the smartpca program in the EIGENSOFT package [19].
Statistical analysis
Approximately 100 SNPs have been used for the creation of breast cancer PRS as of March 1, 2017, and most of those were significantly associated with breast cancer at a genome-wide level of significance (Supplemental Table 2) [17,6]. We calculated the flip-flop proportion of these SNPs, among Root consortium and three published studies, including African American Breast Cancer Consortium (AABC), the Women’s Health Initiative (WHI) and Breast Cancer Association Consortium (BCAC, with women of European ancestry) [6,2,13,12,17]. The proportion of flip-flop was calculated as the number of SNPs with the point value of odds ratios (ORs) in opposite direction between any two studies divided by the total number of SNPs tested. The point value of ORs in opposite direction means that the point value of OR for a specific SNP is greater than 1 in one study but smaller than 1 in others, and vice-versa.
PRS was calculated as the weighted sum of risk alleles [20]. We created a PRS for each individual based on the genotype at each locus, defined as , where nij is the number of risk alleles carried by individual j at the ith SNP and ORi is the per-allele odds ratio associated with the ith SNP. To remove potential bias from ancestry admixture, PRS residuals were then calculated after regression of the score on 10 principal components. This ancestry-adjusted PRS was used for further analysis. A total of 9 PRSs were generated based on different combinations of SNPs and ORs derived from BCAC with European ancestry and two consortia of women of African ancestry: WHI and AABC [6,2,13,12,17]. The Root study was taken as target sample to validate each PRS’s performance. SNPs and corresponding ORs used in the calculation of PRSs are summarized in Supplemental Table 2.
We assessed the association of these PRSs with overall breast cancer risk as well as in subgroups of ER status (ER+ or ER−), country of origin (Nigerian, African American/African Barbadian), family history of breast cancer (yes, no), and age groups (<40, 40–59, ≥60). Both continuous and categorical risk scores were examined in relation to breast cancer risk using unconditional logistic regression. Odds ratios per 1 unit standard deviation (SD) and odds ratios by percentile of PRSs, with the middle quintile (40th to 60th percentiles) as the reference group, were estimated from logistic regressions. Receiver operating characteristic (ROC) analysis was conducted and discrimination between cases and controls was measured using the area under the ROC curve (AUC). We tested the equality of ROC areas obtained from applying nine PRSs to the same sample using the method by DeLong et al [21]. Statistical analysis was conducted using Stata 15.0 (StataCorp, College Station, TX). A two-sided P-value of <0.05 was considered statistically significant.
Results
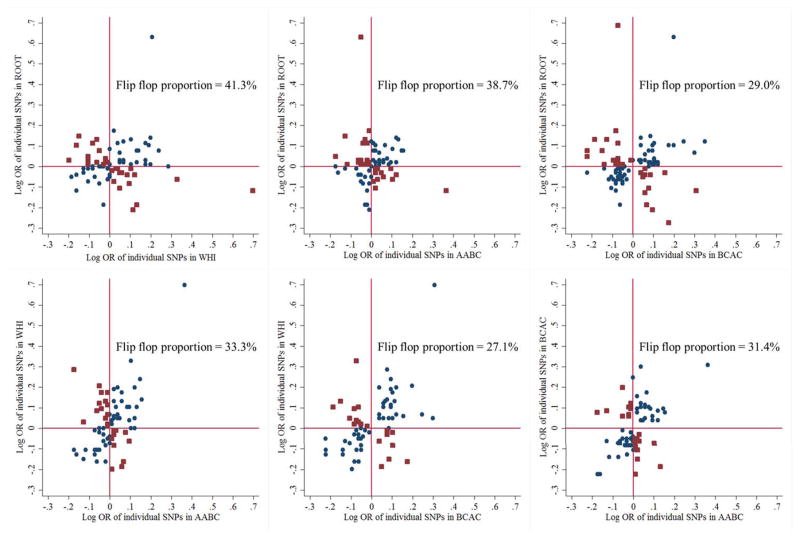
The percentages of flip-flop associations between women of European ancestry (BCAC) and women of African ancestry were 27%, 31% and 29% for WHI, AABC and ROOT, respectively (Figure 1). Among women of African ancestry, the percentages of flip-flop associations between ROOT, WHI, and AABC study ranged from 33% to 41% (Figure 1). There were 45 SNPs in the same direction between WHI and AABC (Supplemental Table 2). If further compared to the ORs from BCAC, the number of SNPs in the same direction across all three studies became 34. A total of 23 SNPs were maintained the same direction in all four datasets.
Figure 1.
Scatterplot of log odds ratio (OR) of individual SNPs in their association with breast cancer risk in ROOT, WHI, AABC and BCAC. Blue circles represent genetic variants with the same direction between the two studies, whereas red squares represent SNPs exhibiting flip-flop.
Abbreviations: WHI – Women’s Health Initiative (African American); AABC – African-American Breast Cancer GWAS study; BCAC – Breast Cancer Association Consortium (European ancestry); ROOT – GWAS of Breast Cancer in the African Diaspora.
We evaluated nine PRSs using the SNP panels from previous studies or the flip-flop analysis mentioned above (Table 1). The performance of the nine PRSs was weak in distinguishing cases and controls, and there was no statistically difference across the nine models (P=0.16). Of the four PRSs based on a single previous study, the PRS that included 93 SNPs (model 4) had the largest AUC value (0.524, 95% CI: 0.506–0.544) for overall breast cancer risk. Of the five PRSs based on results of flip-flop analysis, the PRS that included 34 SNPs with OR estimates from WHI (model 7) had the largest AUC (0.531, 95% CI: 0.512–0.550).
Table 1.
Comparison of the performance of different polygenic risk scores (PRS)
| Model | # of SNPs | SNP panel | Odds ratio source | PRS, Mean (SD) | AUC (95% CI) | |
|---|---|---|---|---|---|---|
|
| ||||||
| Case | Control | |||||
| 1 | 75 | BCAC [6] | WHI [17] | −0.017 (0.539) | 0.021 (0.530) | 0.520 (0.501–0.539) |
|
| ||||||
| 2 | 75 | BCAC [6] | AABC [13,12] | −0.012 (0.351) | 0.015 (0.352) | 0.520 (0.501–0.539) |
|
| ||||||
| 3 | 77 | BCAC [6] | BCAC [6] | −0.013 (0.425) | 0.016 (0.417) | 0.516 (0.497–0.535) |
|
| ||||||
| 4 | 93 | BCAC [6,2] | BCAC [6,2] | −0.024 (0.563) | 0.029 (0.549) | 0.525 (0.506–0.544) |
|
| ||||||
| 5 | 45 | SNPs with the effect in same direction between WHI and AABC | WHI [17] | −0.022 (0.457) | 0.027 (0.446) | 0.528 (0.509–0.547) |
| 6 | AABC [13,12] | −0.010 (0.310) | 0.012 (0.305) | 0.519 (0.500–0.538) | ||
|
| ||||||
| 7 | 34 | SNPs with the effect in same direction among WHI, AABC and BCAC | WHI [17] | −0.023 (0.423) | 0.028 (0.419) | 0.531 (0.512–0.550) |
| 8 | AABC [13,12] | −0.011 (0.292) | 0.013 (0.290) | 0.522 (0.503–0.540) | ||
| 9 | BCAC [6,2] | −0.018 (0.413) | 0.022 (0.398) | 0.524 (0.506–0.543) | ||
Abbreviation: WHI, Women’s Health Initiative (African American); AABC, African-American Breast Cancer GWAS study; BCAC, Breast Cancer Association Consortium; ROOT, GWAS of Breast Cancer in the African Diaspora; AUC, area under the receiver operating curve; CI, confidence interval.
In stratified analysis, a positive family history of breast cancer was associated with increased risk of breast cancer (OR=1.96, 95% CI: 1.59–2.41). None of the nine PRSs was correlated with family history of breast cancer, and thus there was no attenuation in odds ratio for family history after adjusting for the PRS (data not shown). The joint effects of the PRSs and family history are shown in Table 2. Interestingly, the AUCs for all the PRSs were greater in women with positive family history of breast cancer than those in women without family history (Table 2). For example, the 34-SNP PRS has a moderate discriminative capacity among women with family history of breast cancer (AUC=0.586); there was about 4.74-fold difference in breast cancer risk between women with the top 10th PRS and women with the bottom 10% of PRS (Supplemental Table 3).
Table 2.
Performance of polygenic risk scores by family history of breast cancer, and joint model of polygenic risk score and family history of breast cancer
| Model | # of SNPs | Odds ratio source | AUC (95% CI) | AUC (95% CI)* FH + PRS (N=3186) |
|
|---|---|---|---|---|---|
|
| |||||
| With FH (N=439) | No FH (N=2747) | ||||
| 1 | 75 | WHI [17] | 0.554 (0.497–0.610) | 0.516 (0.494–0.537) | 0.552 (0.532–0.572) |
|
| |||||
| 2 | 75 | AABC [13,12] | 0.558 (0.502–0.613) | 0.512 (0.490–0.533) | 0.549 (0.529–0.569) |
|
| |||||
| 3 | 77 | BCAC [6] | 0.558 (0.503–0.613) | 0.503 (0.481–0.525) | 0.543 (0.522–0.563) |
|
| |||||
| 4 | 93 | BCAC [6,2] | 0.554 (0.499–0.609) | 0.515 (0.493–0.537) | 0.552 (0.532–0.572) |
|
| |||||
| 5 | 45 | WHI [17] | 0.585 (0.530–0.640)† | 0.519 (0.498–0.541) | 0.555 (0.535–0.575) |
| 6 | AABC [13,12] | 0.570 (0.514–0.626) | 0.514 (0.492–0.536) | 0.551 (0.531–0.571) | |
|
| |||||
| 7 | 34 | WHI [17] | 0.586 (0.532–0.640)† | 0.521 (0.500–0.543) | 0.557 (0.537–0.577) |
| 8 | AABC [13,12] | 0.568 (0.513–0.623) | 0.515 (0.493–0.536) | 0.551 (0.531–0.571) | |
| 9 | BCAC [6,2] | 0.557 (0.501–0.612) | 0.520 (0.498–0.541) | 0.555 (0.535–0.575) | |
ROC analysis of joint model of family history and polygenic risk score
P<0.05 in the heterogeneity test comparing AUCs in subjects with or without family history of breast cancer.
Abbreviation: WHI, Women’s Health Initiative (African American); AABC, African-American Breast Cancer GWAS study; BCAC, Breast Cancer Association Consortium; ROOT, GWAS of Breast Cancer in the African Diaspora; AUC, area under the receiver operating curve; CI, confidence interval; FH, family history of breast cancer
We further examined the performance of the 93-SNP and 34-SNP PRSs according to demographic and clinical characteristics. The performance of the 93-SNP PRS was better in Nigerian women than in African American/African Barbadian women (Pinteraction=0.019), though this was not observed for the 34-SNP PRS (Pinteraction=0.64) (Table 3). On the other hand, we observed no significant differences across age groups for both the 93-SNP (Pinteraction=0.58) nor the 34-SNP PRSs (Pinteraction=0.74). Both the 93-SNP PRS and the 34-SNP PRS were strongly associated with ER+ breast cancer (OR per unit SD = 1.17 and 1.14, respectively), but their associations with ER− disease were weak and not statistically significant (Table 4).
Table 3.
Performance of the 93-SNP and 34-SNP polygenic risk scores by selected demographic characteristics
| 93-SNP polygenic risk score | 34-SNP polygenic risk score | |||
|---|---|---|---|---|
|
|
|
|||
| AUC (95% CI) | OR (95% CI)* | AUC (95% CI) | OR (95% CI)* | |
| Age group, years | ||||
| < 40 | 0.534 (0.495–0.573) | 1.15 (1.00–1.32) | 0.541 (0.503–0.580) | 1.23 (1.03–1.46) |
| 40–59 | 0.514 (0.486–0.542) | 1.06 (0.96–1.17) | 0.522 (0.494–0.549) | 1.13 (0.99–1.28) |
| ≥ 60 | 0.533 (0.499–0.566) | 1.12 (1.00–1.26) | 0.534 (0.500–0.568) | 1.18 (1.01–1.37) |
| P for heterogeneity | 0.58 | 0.74 | ||
|
| ||||
| Ethnicity | ||||
| Nigerian | 0.554 (0.523–0.585) | 1.23 (1.10–1.38) | 0.538 (0.507–0.569) | 1.20 (1.03–1.40) |
| African American/African Barbadian | 0.508 (0.484–0.531) | 1.04 (0.96–1.13) | 0.527 (0.503–0.550) | 1.15 (1.04–1.28) |
| P for heterogeneity | 0.019 | 0.64 | ||
|
| ||||
| Family history of breast cancer | ||||
| Yes | 0.554 (0.499–0.609) | 1.22 (1.00–1.49) | 0.586 (0.532–0.640) | 1.39 (1.14–1.70) |
| No | 0.515 (0.493–0.537) | 1.07 (0.99–1.15) | 0.521 (0.500–0.543) | 1.08 (1.00–1.17) |
| P for heterogeneity | 0.22 | 0.022 | ||
odds ratio per unit standard deviation of the polygenic risk score
Abbreviation: AUC, area under the receiver operating curve; CI, confidence interval; OR, odds ratio
Table 4.
Association of polygenic risk scores and breast cancer risk by estrogen receptor status
| Odds ratio (95% confidence intervals) | |||
|---|---|---|---|
|
| |||
| All breast cancers | ER+ disease | ER− disease | |
| 93-SNP PRS | |||
| Per unit SD of PRS | 1.10 (1.03–1.17) | 1.17 (1.05–1.30) | 1.04 (0.93–1.16) |
| Percentile of PRS | |||
| <5 | 0.72 (0.52–1.01) | 0.68 (0.38–1.20) | 0.65 (0.37–1.16) |
| 5–10 | 0.84 (0.60–1.16) | 0.63 (0.34–1.16) | 0.74 (0.42–1.30) |
| 10–20 | 0.78 (0.60–1.00) | 0.81 (0.53–1.23) | 0.95 (0.64–1.41) |
| 20–40 | 0.83 (0.68–1.02) | 0.89 (0.63–1.24) | 0.70 (0.49–1.00) |
| 40–60 | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| 60–80 | 0.91 (0.74–1.11) | 0.94 (0.67–1.32) | 0.79 (0.56–1.11) |
| 80–90 | 0.98 (0.76–1.26) | 0.94 (0.62–1.43) | 0.91 (0.60–1.38) |
| 90–95 | 1.14 (0.82–1.57) | 1.30 (0.79–2.15) | 0.90 (0.52–1.58) |
| >95 | 1.11 (0.80–1.54) | 1.54 (0.96–2.48) | 0.94 (0.55–1.63) |
| P for trend | 0.002 | 0.002 | 0.31 |
|
| |||
| 34-SNP PRS | |||
| Per unit SD of PRS | 1.13 (1.06–1.20) | 1.14 (1.03–1.27) | 1.11 (1.00–1.24) |
| Percentile of PRS | |||
| <5 | 0.65 (0.46–0.90) | 0.67 (0.39–1.16) | 0.72 (0.38–1.36) |
| 5–10 | 0.73 (0.53–1.02) | 0.43 (0.22–0.84) | 1.69 (1.04–2.76) |
| 10–20 | 0.74 (0.58–0.96) | 0.71 (0.47–1.08) | 1.00 (0.63–1.56) |
| 20–40 | 0.99 (0.81–1.21) | 0.96 (0.69–1.33) | 1.34 (0.94–1.93) |
| 40–60 | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| 60–80 | 0.90 (0.73–1.11) | 0.77 (0.55–1.08) | 1.07 (0.73–1.55) |
| 80–90 | 1.04 (0.81–1.33) | 0.94 (0.62–1.41) | 1.42 (0.92–2.19) |
| 90–95 | 0.95 (0.69–1.32) | 0.88 (0.52–1.50) | 1.31 (0.75–2.27) |
| >95 | 1.29 (0.94–1.79) | 1.54 (0.96–2.47) | 2.13 (1.28–3.53) |
| P for trend | <0.001 | 0.009 | 0.076 |
Abbreviation: SNP, single nucleoid polymorphism; PRS, polygenic risk score; ER, estrogen receptor; SD, standard deviation
Discussion
We found that the proportion of flip-flop phenomenon was about 30%~40% regardless of whether assessing within women of African ancestry or across racial/ethnic groups. The PRS based on a subset of 34 SNPs with consistent direction among previous studies demonstrated similar risk prediction as the PRS based on 93 SNPs from European ancestry populations. However, the discriminative performance of these PRSs was still low for women of African ancestry. On the other hand, the PRSs offered good discriminative capacity for women with family history of breast cancer.
The proportion of flip-flop phenomenon was approximately 30%–40% when comparing the three studies in women of African ancestry or across different ethnic groups. One possible explanation was that the results of these three studies of African ancestry were limited by statistical power. The p-value thresholds for GWAS studies in African or African-American population were suggested to be more stringent due to the greater genetic diversity in these populations [22], and the flip-flop associations within the same ethnic group are likely to be spurious [16]. Ntzani et al evaluated the discrepancies for ancestral effect sizes for confirmed GWAS-identified associations obtained from GWAS catalog, and they reported a slight lower discrepancy proportion of 21% between European ancestry and African ancestry, while the percentages of flip-flop associations in our study ranged from 27% to 31% [23]. As discussed in their paper, the flip-flop associations across different ethnic groups have two interpretations. First, it could indicate that the heterogeneity of the same variant could be due to differences in genetic background or environmental factors across populations [16]. It is unlikely that all of the flip-flop associations could be attributed to these differences and instead indicates that some variants could be false positives, demonstrating the importance of validation studies in populations across different ancestries [24]. Second, as Wen et al proposed, the SNPs without flip-flop association across different ethnic groups are more likely to be correlated with the disease outcome and could be true causal variants [25]. In their paper, only 44 internally confirmed SNPs were applied to construct a PRS for women of Asian ancestry, and the performance of 44-SNP PRS was very close to 88-SNP PRS in their data (AUC: 0.606 vs 0.609).
In the present study, several PRSs constructed using published breast cancer susceptibility loci demonstrated inadequate predictive value for breast cancer risk assessment among women of African ancestry. The PRS of 77 SNPs developed in European ancestry populations yielded an AUC of 0.615 in a validation sample of Caucasians [6]. When this PRS was directly applied to women of African ancestry, the AUC was only 0.516. The best performed PRSs in our study (AUC≈0.53) had discriminative accuracy similar to Allman’s study (AUC=0.55, 95% CI: 0.53–0.58), which was conducted using 7,539 African American women from WHI [17]. Our study included both African Americans and women of African-ancestry living in Nigeria and Barbados. We found an interesting phenomenon that the PRSs performed better in Nigerian individuals than in African Americans and Africans in the Caribbean. A potential explanation is that spurious association may occur with an adjustment only for global ancestry or mixed ancestry, and further local ancestry adjustment might be needed to sufficiently control population stratification in populations like African Americans [26].
Furthermore, we noted that the PRS including only 34 SNPs, all with the same association direction among previous studies, performed equally well as the PRS including 93 SNPs. This result supports our hypothesis that the performance of PRS based on a subset of SNPs without flip-flop is non-inferior since it avoids the inclusion of SNPs that may be false positives. In order to apply a PRS as population-level screening tool, cost-effectiveness should also be considered [27]. Our study provides a possible strategy to identify women at increased risk for breast cancer with only 1/3 of the known index SNPs. This cost-saving may be considered when performing genetic testing among women at average breast cancer risk [9].
Another interesting finding is that the common genetic variant-based PRSs performed better for women with a positive family history of breast cancer. We estimated the AUC for the 34-SNP PRS to be 0.586 and there could be 4.74-fold risk difference between individuals in the top 10% of the PRS and individuals in the bottom 10%. This is consistent with a previous study conducted in familial cancer clinics in Australia, which demonstrated that a 22 SNPs-based polygenic risk score can distinguish familial breast cancer cases who are negative for BRCA1 or BRCA2 mutation from controls (AUC=0.654) [28]. These findings suggest that common genetic variants have potential for risk stratification in women with positive family history of breast cancer, especially those who tested negative for BRCA1 or BRCA2, among other established genetic mutations.
To the best of our knowledge, this is the first study to describe the prevalence of the flip-flop phenomenon not only across racial groups but within different ethnic groups of women of African ancestry, and confirms the non-inferiority of PRS only using SNPs showing consistent associations. However, some limitations should be considered when interpreting our study findings. We only assessed the index SNPs from GWAS among non-African Americans. Large-scale fine-mapping studies conducted in women of African ancestry may be needed to identify additional genetic variants at these loci and could be beneficial for population-specific PRS model development. Second, there are multiple reasons for the flip-flop phenomenon, including the interplay of genetic loci and environmental factors or interaction between multiple loci among populations of different ancestries, different LD architectures, sampling variations, and spurious association [16,29,23,30]. Because of our main aim of applying flip-flop information into genetic risk prediction, we could not pinpoint the exact reasons of flip-flop phenomenon in this study. Further studies on the interaction between genetic variants and environmental factors or between multiple genetic variants (epistasis) are desirable to identify biological reasons underlying the flip-flop phenomenon.
In conclusion, our study serves to illustrate the complexity of applying current GWAS findings across diverse racial/ethnic groups. We observed significant flip-flop in women of African ancestry using SNPs from prior GWAS conducted predominantly in women of European ancestry. Given the poor discriminative ability of currently existing PRS for women of African ancestry, we believe there are two direct implications. One is to discover additional SNPs that have true causal associations or indicate breast cancer risk among women of African ancestry by increasing the sample size coupled with fine-mapping and pathway analysis approaches [31,24]. Another implication is to develop PRS for predicting familial breast cancer, since our study showed strong predictive value among these women.
Supplementary Material
Acknowledgments
Funding sources: This work was in part supported by the American Cancer Society MRSG-13-063-01-TBG (DH) and CRP-10-119-01-CCE (OIO), National Cancer Institute CA142996 (OIO) and CA161032 (OIO), Susan G. Komen for the Cure (OIO), and Breast Cancer Research Foundation (OIO, DH). The funding agencies have no involvement in the study design, data collection, analysis, interpretation of data, writing of the manuscript, or decision to submit the manuscript for publication. SW was supported by a University of Chicago Global Health Fellowship.
This work was in part supported by the American Cancer Society (MRSG-13-063-01-TBG and CRP-10-119-01-CCE), National Cancer Institute (CA142996, CA161032), Susan G. Komen for the Cure, and Breast Cancer Research Foundation. SW was supported by a University of Chicago Global Health Fellowship.
Abbreviation
- MAF
minor allele frequency
- WHI
the Women Health Initiative
- AABC
the African-American Breast Cancer GWAS study
- BCAC
The Breast Cancer Association Consortium
- ROOT
GWAS of Breast Cancer in the African Diaspora
- OR
odds ratio
- AUC
area under the receiver operating characteristic curve
- PRS
polygenic risk score
Footnotes
Conflict of Interest Statement None declared.
References
- 1.World Health Organization IAfRoC. [Accessed 04-12 2016];GLOBOCAN2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. 2016 http://globocan.iarc.fr/Default.aspx.
- 2.Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, Maranian MJ, Bolla MK, Wang Q, Shah M, Perkins BJ, Czene K, Eriksson M, Darabi H, Brand JS, Bojesen SE, Nordestgaard BG, Flyger H, Nielsen SF, Rahman N, Turnbull C, Bocs, Fletcher O, Peto J, Gibson L, dos-Santos-Silva I, Chang-Claude J, Flesch-Janys D, Rudolph A, Eilber U, Behrens S, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Khan S, Aaltonen K, Ahsan H, Kibriya MG, Whittemore AS, John EM, Malone KE, Gammon MD, Santella RM, Ursin G, Makalic E, Schmidt DF, Casey G, Hunter DJ, Gapstur SM, Gaudet MM, Diver WR, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Berg CD, Chanock SJ, Figueroa J, Hoover RN, Lambrechts D, Neven P, Wildiers H, van Limbergen E, Schmidt MK, Broeks A, Verhoef S, Cornelissen S, Couch FJ, Olson JE, Hallberg E, Vachon C, Waisfisz Q, Meijers-Heijboer H, Adank MA, van der Luijt RB, Li J, Liu J, Humphreys K, Kang D, Choi JY, Park SK, Yoo KY, Matsuo K, Ito H, Iwata H, Tajima K, Guenel P, Truong T, Mulot C, Sanchez M, Burwinkel B, Marme F, Surowy H, Sohn C, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Gonzalez-Neira A, Benitez J, Zamora MP, Perez JI, Shu XO, Lu W, Gao YT, Cai H, Cox A, Cross SS, Reed MW, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Lindblom A, Margolin S, Teo SH, Yip CH, Taib NA, Tan GH, Hooning MJ, Hollestelle A, Martens JW, Collee JM, Blot W, Signorello LB, Cai Q, Hopper JL, Southey MC, Tsimiklis H, Apicella C, Shen CY, Hsiung CN, Wu PE, Hou MF, Kristensen VN, Nord S, Alnaes GI, Nbcs, Giles GG, Milne RL, McLean C, Canzian F, Trichopoulos D, Peeters P, Lund E, Sund M, Khaw KT, Gunter MJ, Palli D, Mortensen LM, Dossus L, Huerta JM, Meindl A, Schmutzler RK, Sutter C, Yang R, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Hartman M, Miao H, Chia KS, Chan CW, Fasching PA, Hein A, Beckmann MW, Haeberle L, Brenner H, Dieffenbach AK, Arndt V, Stegmaier C, Ashworth A, Orr N, Schoemaker MJ, Swerdlow AJ, Brinton L, Garcia-Closas M, Zheng W, Halverson SL, Shrubsole M, Long J, Goldberg MS, Labreche F, Dumont M, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Bruning T, Network G, Radice P, Peterlongo P, Manoukian S, Bernard L, Bogdanova NV, Dork T, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Devilee P, Tollenaar RA, Seynaeve C, Van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Huzarski T, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Slager S, Toland AE, Ambrosone CB, Yannoukakos D, Kabisch M, Torres D, Neuhausen SL, Anton-Culver H, Luccarini C, Baynes C, Ahmed S, Healey CS, Tessier DC, Vincent D, Bacot F, Pita G, Alonso MR, Alvarez N, Herrero D, Simard J, Pharoah PP, Kraft P, Dunning AM, Chenevix-Trench G, Hall P, Easton DF kConFab I, Group A. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nature genetics. 2015;47(4):373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N, Fletcher O, Peto J, Gibson L, Dos Santos Silva I, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Czene K, Irwanto A, Liu J, Waisfisz Q, Meijers-Heijboer H, Adank M, Hereditary B, van der Luijt RB, Hein R, Dahmen N, Beckman L, Meindl A, Schmutzler RK, Muller-Myhsok B, Lichtner P, Hopper JL, Southey MC, Makalic E, Schmidt DF, Uitterlinden AG, Hofman A, Hunter DJ, Chanock SJ, Vincent D, Bacot F, Tessier DC, Canisius S, Wessels LF, Haiman CA, Shah M, Luben R, Brown J, Luccarini C, Schoof N, Humphreys K, Li J, Nordestgaard BG, Nielsen SF, Flyger H, Couch FJ, Wang X, Vachon C, Stevens KN, Lambrechts D, Moisse M, Paridaens R, Christiaens MR, Rudolph A, Nickels S, Flesch-Janys D, Johnson N, Aitken Z, Aaltonen K, Heikkinen T, Broeks A, Veer LJ, van der Schoot CE, Guenel P, Truong T, Laurent-Puig P, Menegaux F, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Zamora MP, Perez JI, Pita G, Alonso MR, Cox A, Brock IW, Cross SS, Reed MW, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Henderson BE, Schumacher F, Le Marchand L, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Lindblom A, Margolin S, Hooning MJ, Hollestelle A, van den Ouweland AM, Jager A, Bui QM, Stone J, Dite GS, Apicella C, Tsimiklis H, Giles GG, Severi G, Baglietto L, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Brenner H, Muller H, Arndt V, Stegmaier C, Swerdlow A, Ashworth A, Orr N, Jones M, Figueroa J, Lissowska J, Brinton L, Goldberg MS, Labreche F, Dumont M, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Bruning T, Network G, Radice P, Peterlongo P, Manoukian S, Bonanni B, Devilee P, Tollenaar RA, Seynaeve C, van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Durda K, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Bogdanova NV, Antonenkova NN, Dork T, Kristensen VN, Anton-Culver H, Slager S, Toland AE, Edge S, Fostira F, Kang D, Yoo KY, Noh DY, Matsuo K, Ito H, Iwata H, Sueta A, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Shu XO, Lu W, Gao YT, Cai H, Teo SH, Yip CH, Phuah SY, Cornes BK, Hartman M, Miao H, Lim WY, Sng JH, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Shen CY, Hsiung CN, Wu PE, Ding SL, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Blot WJ, Signorello LB, Cai Q, Zheng W, Deming-Halverson S, Shrubsole M, Long J, Simard J, Garcia-Closas M, Pharoah PD, Chenevix-Trench G, Dunning AM, Benitez J, Easton DF kConFab I, Breast, Ovarian Cancer Susceptibility C, Ovarian Cancer Research Group N, Australian Ovarian Cancer Study G. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature genetics. 2013;45(4):353–361. 361e351–352. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, Le Marchand L, Buring JE, Eccles D, Miron P, Fasching PA, Brauch H, Chang-Claude J, Carpenter J, Godwin AK, Nevanlinna H, Giles GG, Cox A, Hopper JL, Bolla MK, Wang Q, Dennis J, Dicks E, Howat WJ, Schoof N, Bojesen SE, Lambrechts D, Broeks A, Andrulis IL, Guenel P, Burwinkel B, Sawyer EJ, Hollestelle A, Fletcher O, Winqvist R, Brenner H, Mannermaa A, Hamann U, Meindl A, Lindblom A, Zheng W, Devillee P, Goldberg MS, Lubinski J, Kristensen V, Swerdlow A, Anton-Culver H, Dork T, Muir K, Matsuo K, Wu AH, Radice P, Teo SH, Shu XO, Blot W, Kang D, Hartman M, Sangrajrang S, Shen CY, Southey MC, Park DJ, Hammet F, Stone J, Veer LJ, Rutgers EJ, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Peto J, Schrauder MG, Ekici AB, Beckmann MW, Dos Santos Silva I, Johnson N, Warren H, Tomlinson I, Kerin MJ, Miller N, Marme F, Schneeweiss A, Sohn C, Truong T, Laurent-Puig P, Kerbrat P, Nordestgaard BG, Nielsen SF, Flyger H, Milne RL, Perez JI, Menendez P, Muller H, Arndt V, Stegmaier C, Lichtner P, Lochmann M, Justenhoven C, Ko YD, Gene EI, Muranen TA, Aittomaki K, Blomqvist C, Greco D, Heikkinen T, Ito H, Iwata H, Yatabe Y, Antonenkova NN, Margolin S, Kataja V, Kosma VM, Hartikainen JM, Balleine R, Tseng CC, Berg DV, Stram DO, Neven P, Dieudonne AS, Leunen K, Rudolph A, Nickels S, Flesch-Janys D, Peterlongo P, Peissel B, Bernard L, Olson JE, Wang X, Stevens K, Severi G, Baglietto L, McLean C, Coetzee GA, Feng Y, Henderson BE, Schumacher F, Bogdanova NV, Labreche F, Dumont M, Yip CH, Taib NA, Cheng CY, Shrubsole M, Long J, Pylkas K, Jukkola-Vuorinen A, Kauppila S, Knight JA, Glendon G, Mulligan AM, Tollenaar RA, Seynaeve CM, Kriege M, Hooning MJ, van den Ouweland AM, van Deurzen CH, Lu W, Gao YT, Cai H, Balasubramanian SP, Cross SS, Reed MW, Signorello L, Cai Q, Shah M, Miao H, Chan CW, Chia KS, Jakubowska A, Jaworska K, Durda K, Hsiung CN, Wu PE, Yu JC, Ashworth A, Jones M, Tessier DC, Gonzalez-Neira A, Pita G, Alonso MR, Vincent D, Bacot F, Ambrosone CB, Bandera EV, John EM, Chen GK, Hu JJ, Rodriguez-Gil JL, Bernstein L, Press MF, Ziegler RG, Millikan RM, Deming-Halverson SL, Nyante S, Ingles SA, Waisfisz Q, Tsimiklis H, Makalic E, Schmidt D, Bui M, Gibson L, Muller-Myhsok B, Schmutzler RK, Hein R, Dahmen N, Beckmann L, Aaltonen K, Czene K, Irwanto A, Liu J, Turnbull C, Rahman N, Meijers-Heijboer H, Uitterlinden AG, Rivadeneira F, Olswold C, Slager S, Pilarski R, Ademuyiwa F, Konstantopoulou I, Martin NG, Montgomery GW, Slamon DJ, Rauh C, Lux MP, Jud SM, Bruning T, Weaver J, Sharma P, Pathak H, Tapper W, Gerty S, Durcan L, Trichopoulos D, Tumino R, Peeters PH, Kaaks R, Campa D, Canzian F, Weiderpass E, Johansson M, Khaw KT, Travis R, Clavel-Chapelon F, Kolonel LN, Chen C, Beck A, Hankinson SE, Berg CD, Hoover RN, Lissowska J, Figueroa JD, Chasman DI, Gaudet MM, Diver WR, Willett WC, Hunter DJ, Simard J, Benitez J, Dunning AM, Sherman ME, Chenevix-Trench G, Chanock SJ, Hall P, Pharoah PD, Vachon C, Easton DF, Haiman CA, Kraft P breast CN, kConFab I, Australian Breast Cancer Tissue Bank I, Familial Breast Cancer S. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nature genetics. 2013;45(4):392–398. 398e391–392. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai Q, Zhang B, Sung H, Low SK, Kweon SS, Lu W, Shi J, Long J, Wen W, Choi JY, Noh DY, Shen CY, Matsuo K, Teo SH, Kim MK, Khoo US, Iwasaki M, Hartman M, Takahashi A, Ashikawa K, Matsuda K, Shin MH, Park MH, Zheng Y, Xiang YB, Ji BT, Park SK, Wu PE, Hsiung CN, Ito H, Kasuga Y, Kang P, Mariapun S, Ahn SH, Kang HS, Chan KY, Man EP, Iwata H, Tsugane S, Miao H, Liao J, Nakamura Y, Kubo M, Delahanty RJ, Zhang Y, Li B, Li C, Gao YT, Shu XO, Kang D, Zheng W. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nature genetics. 2014;46(8):886–890. doi: 10.1038/ng.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavaddat N, Pharoah PD, Michailidou K, Tyrer J, Brook MN, Bolla MK, Wang Q, Dennis J, Dunning AM, Shah M, Luben R, Brown J, Bojesen SE, Nordestgaard BG, Nielsen SF, Flyger H, Czene K, Darabi H, Eriksson M, Peto J, Dos-Santos-Silva I, Dudbridge F, Johnson N, Schmidt MK, Broeks A, Verhoef S, Rutgers EJ, Swerdlow A, Ashworth A, Orr N, Schoemaker MJ, Figueroa J, Chanock SJ, Brinton L, Lissowska J, Couch FJ, Olson JE, Vachon C, Pankratz VS, Lambrechts D, Wildiers H, Van Ongeval C, van Limbergen E, Kristensen V, Grenaker Alnaes G, Nord S, Borresen-Dale AL, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Chang-Claude J, Rudolph A, Seibold P, Flesch-Janys D, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Burwinkel B, Marme F, Schneeweiss A, Sohn C, Trentham-Dietz A, Newcomb P, Titus L, Egan KM, Hunter DJ, Lindstrom S, Tamimi RM, Kraft P, Rahman N, Turnbull C, Renwick A, Seal S, Li J, Liu J, Humphreys K, Benitez J, Pilar Zamora M, Arias Perez JI, Menendez P, Jakubowska A, Lubinski J, Jaworska-Bieniek K, Durda K, Bogdanova NV, Antonenkova NN, Dork T, Anton-Culver H, Neuhausen SL, Ziogas A, Bernstein L, Devilee P, Tollenaar RA, Seynaeve C, van Asperen CJ, Cox A, Cross SS, Reed MW, Khusnutdinova E, Bermisheva M, Prokofyeva D, Takhirova Z, Meindl A, Schmutzler RK, Sutter C, Yang R, Schurmann P, Bremer M, Christiansen H, Park-Simon TW, Hillemanns P, Guenel P, Truong T, Menegaux F, Sanchez M, Radice P, Peterlongo P, Manoukian S, Pensotti V, Hopper JL, Tsimiklis H, Apicella C, Southey MC, Brauch H, Bruning T, Ko YD, Sigurdson AJ, Doody MM, Hamann U, Torres D, Ulmer HU, Forsti A, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Andrulis IL, Knight JA, Glendon G, Marie Mulligan A, Chenevix-Trench G, Balleine R, Giles GG, Milne RL, McLean C, Lindblom A, Margolin S, Haiman CA, Henderson BE, Schumacher F, Le Marchand L, Eilber U, Wang-Gohrke S, Hooning MJ, Hollestelle A, van den Ouweland AM, Koppert LB, Carpenter J, Clarke C, Scott R, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Brenner H, Arndt V, Stegmaier C, Karina Dieffenbach A, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Offit K, Vijai J, Robson M, Rau-Murthy R, Dwek M, Swann R, Annie Perkins K, Goldberg MS, Labreche F, Dumont M, Eccles DM, Tapper WJ, Rafiq S, John EM, Whittemore AS, Slager S, Yannoukakos D, Toland AE, Yao S, Zheng W, Halverson SL, Gonzalez-Neira A, Pita G, Rosario Alonso M, Alvarez N, Herrero D, Tessier DC, Vincent D, Bacot F, Luccarini C, Baynes C, Ahmed S, Maranian M, Healey CS, Simard J, Hall P, Easton DF, Garcia-Closas M. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107(5) doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. The New England journal of medicine. 2008;358(26):2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 8.Burton H, Chowdhury S, Dent T, Hall A, Pashayan N, Pharoah P. Public health implications from COGS and potential for risk stratification and screening. Nature genetics. 2013;45(4):349–351. doi: 10.1038/ng.2582. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee N, Shi J, Garcia-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nature reviews Genetics. 2016 doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoury MJ, Iademarco MF, Riley WT. Precision Public Health for the Era of Precision Medicine. American journal of preventive medicine. 2016;50(3):398–401. doi: 10.1016/j.amepre.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huo D, Feng Y, Haddad S, Zheng Y, Yao S, Han YJ, Ogundiran TO, Adebamowo C, Ojengbede O, Falusi AG, Zheng W, Blot W, Cai Q, Signorello L, John EM, Bernstein L, Hu JJ, Ziegler RG, Nyante S, Bandera EV, Ingles SA, Press MF, Deming SL, Rodriguez-Gil JL, Nathanson KL, Domchek SM, Rebbeck TR, Ruiz-Narvaez EA, Sucheston-Campbell LE, Bensen JT, Simon MS, Hennis A, Nemesure B, Leske MC, Ambs S, Chen LS, Qian F, Gamazon ER, Lunetta KL, Cox NJ, Chanock SJ, Kolonel LN, Olshan AF, Ambrosone CB, Olopade OI, Palmer JR, Haiman CA. Genome-wide Association Studies in Women of African Ancestry Identified 3q26.21 as a Novel Susceptibility Locus for Estrogen Receptor Negative Breast Cancer. Human molecular genetics. 2016;25(21):4835–4846. doi: 10.1093/hmg/ddw305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer JR, Ruiz-Narvaez EA, Rotimi CN, Cupples LA, Cozier YC, Adams-Campbell LL, Rosenberg L. Genetic susceptibility loci for subtypes of breast cancer in an African American population. Cancer Epidemiology Biomarkers & Prevention. 2013;22(1):127–134. doi: 10.1158/1055-9965.EPI-12-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y, Stram DO, Rhie SK, Millikan RC, Ambrosone CB, John EM, Bernstein L, Zheng W, Olshan AF, Hu JJ, Ziegler RG, Nyante S, Bandera EV, Ingles SA, Press MF, Deming SL, Rodriguez-Gil JL, Palmer JR, Olopade OI, Huo D, Adebamowo CA, Ogundiran T, Chen GK, Stram A, Park K, Rand KA, Chanock SJ, Le Marchand L, Kolonel LN, Conti DV, Easton D, Henderson BE, Haiman CA. A comprehensive examination of breast cancer risk loci in African American women. Human molecular genetics. 2014;23(20):5518–5526. doi: 10.1093/hmg/ddu252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F, Chen GK, Stram DO, Millikan RC, Ambrosone CB, John EM, Bernstein L, Zheng W, Palmer JR, Hu JJ, Rebbeck TR, Ziegler RG, Nyante S, Bandera EV, Ingles SA, Press MF, Ruiz-Narvaez EA, Deming SL, Rodriguez-Gil JL, Demichele A, Chanock SJ, Blot W, Signorello L, Cai Q, Li G, Long J, Huo D, Zheng Y, Cox NJ, Olopade OI, Ogundiran TO, Adebamowo C, Nathanson KL, Domchek SM, Simon MS, Hennis A, Nemesure B, Wu SY, Leske MC, Ambs S, Hutter CM, Young A, Kooperberg C, Peters U, Rhie SK, Wan P, Sheng X, Pooler LC, Van Den Berg DJ, Le Marchand L, Kolonel LN, Henderson BE, Haiman CA. A genome-wide association study of breast cancer in women of African ancestry. Hum Genet. 2013;132(1):39–48. doi: 10.1007/s00439-012-1214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wall JD, Pritchard JK. Haplotype blocks and linkage disequilibrium in the human genome. Nature reviews Genetics. 2003;4(8):587–597. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- 16.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. American journal of human genetics. 2007;80(3):531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allman R, Dite GS, Hopper JL, Gordon O, Starlard-Davenport A, Chlebowski R, Kooperberg C. SNPs and breast cancer risk prediction for African American and Hispanic women. Breast cancer research and treatment. 2015;154(3):583–589. doi: 10.1007/s10549-015-3641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian F, Feng Y, Zheng Y, Ogundiran TO, Ojengbede O, Zheng W, Blot W, Ambrosone CB, John EM, Bernstein L, Hu JJ, Ziegler RG, Nyante S, Bandera EV, Ingles SA, Press MF, Nathanson KL, Hennis A, Nemesure B, Ambs S, Kolonel LN, Olopade OI, Haiman CA, Huo D. Genetic variants in microRNA and microRNA biogenesis pathway genes and breast cancer risk among women of African ancestry. Hum Genet. 2016 doi: 10.1007/s00439-016-1707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS genetics. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong SW, Lee IH, Leshchiner I, Krier J, Kraft P, Rehm HL, Green RC, Kohane IS, MacRae CA. Summarizing polygenic risks for complex diseases in a clinical whole-genome report. Genetics in medicine: official journal of the American College of Medical Genetics. 2015;17(7):536–544. doi: 10.1038/gim.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 22.Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Stat Sci. 2009;24(4):561–573. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ntzani EE, Liberopoulos G, Manolio TA, Ioannidis JP. Consistency of genome-wide associations across major ancestral groups. Human genetics. 2012;131(7):1057–1071. doi: 10.1007/s00439-011-1124-4. [DOI] [PubMed] [Google Scholar]
- 24.Asimit JL, Hatzikotoulas K, McCarthy M, Morris AP, Zeggini E. Trans-ethnic study design approaches for fine-mapping. European journal of human genetics: EJHG. 2016;24(9):1330–1336. doi: 10.1038/ejhg.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen W, Shu XO, Guo X, Cai Q, Long J, Bolla MK, Michailidou K, Dennis J, Wang Q, Gao YT, Zheng Y, Dunning AM, Garcia-Closas M, Brennan P, Chen ST, Choi JY, Hartman M, Ito H, Lophatananon A, Matsuo K, Miao H, Muir K, Sangrajrang S, Shen CY, Teo SH, Tseng CC, Wu AH, Yip CH, Simard J, Pharoah PD, Hall P, Kang D, Xiang Y, Easton DF, Zheng W. Prediction of breast cancer risk based on common genetic variants in women of East Asian ancestry. Breast cancer research: BCR. 2016;18(1):124. doi: 10.1186/s13058-016-0786-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Zhu X, Qin H, Cooper RS, Ewens WJ, Li C, Li M. Adjustment for local ancestry in genetic association analysis of admixed populations. Bioinformatics (Oxford, England) 2011;27(5):670–677. doi: 10.1093/bioinformatics/btq709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen KD, Dukhovny D, Siebert U, Green RC. Assessing the Costs and Cost-Effectiveness of Genomic Sequencing. J Pers Med. 2015;5(4):470–486. doi: 10.3390/jpm5040470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawyer S, Mitchell G, McKinley J, Chenevix-Trench G, Beesley J, Chen XQ, Bowtell D, Trainer AH, Harris M, Lindeman GJ, James PA. A role for common genomic variants in the assessment of familial breast cancer. J Clin Oncol. 2012;30(35):4330–4336. doi: 10.1200/JCO.2012.41.7469. [DOI] [PubMed] [Google Scholar]
- 29.Zaykin DV, Shibata K. Genetic Flip-Flop without an Accompanying Change in Linkage Disequilibrium. American journal of human genetics. 2008;82(3):794–796. doi: 10.1016/j.ajhg.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet (London, England) 2003;361(9360):865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 31.Fachal L, Dunning AM. From candidate gene studies to GWAS and post-GWAS analyses in breast cancer. Current opinion in genetics & development. 2015;30:32–41. doi: 10.1016/j.gde.2015.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.