Abstract
Norovirus is one of the most common causes of acute gastroenteritis among children in developing countries. No data on the prevalence and genetic variability of norovirus are available for Pakistan, where early childhood mortality due to acute gastroenteritis is common. We tested 255 fecal specimens from children under 5 years of age hospitalized between April 2006 and March 2008 with severe acute gastroenteritis in five hospitals in the four largest cities in Pakistan for norovirus by real-time RT-PCR. Positive samples were further genotyped by conventional RTPCR targeting the 5′-end of the capsid gene followed by sequencing of the positive PCR products. Overall, 41 (16.1%) samples tested positive for norovirus with an equal frequency in rotavirus-positive and rotavirus-negative samples. Nine (22%) samples were genogroup (G)I positive, 30 (73%) GII positive and two (5%) samples contained a mixture of GI and GII viruses. Sequence analyses demonstrated co-circulation of 14 norovirus genotypes including four GI genotypes (GI.3, GI.5, GI.7, GI.8) and 10 GII genotypes (GII.2, GII.3, GII.4, GII.5, GII.6, GII.7, GII.9, GII.13, GII.16, and GII.21). The most prevalent genotypes were GI.7 and GII.4 both causing 12.2% of the infections. This report confirms the presence of multiple norovirus genotypes in hospitalized children with acute gastroenteritis in Pakistan and a lack of clear predominance of GII.4 viruses.
Keywords: acute gastroenteritis, norovirus, genogroup I and II, co-infection, Pakistan
INTRODUCTION
Diarrheal disease is the second most common cause of death and accounts for 1.33 million (15%) children younger than 5 years of age worldwide [Black et al., 2010]. Pakistan is among the top five countries in diarrheal mortality, which collectively account for about 51% (or 0.676 million) of diarrhea-related deaths in young children [Black et al., 2010]. Norovirus infections are the second largest contributors of severe childhood gastroenteritis in low resource countries where they cause an estimated 1.1 million hospitalizations and 218,000 deaths in children younger than 5 years of age each year [Patel et al., 2008].
Noroviruses are non-enveloped positive stranded RNA viruses that belong to the family Caliciviridae. Genetically, they can be divided into at least six genogroups (GI-GVI) [Green, 2013] and a tentatively new genogroup (GVII) has been proposed [Vinjé, 2015]. Most infections in humans are caused by GI and GII viruses which can be further divided into 9 and 22 genotypes, respectively [Vinjé, 2015]. Noroviruses are transmitted directly from person to person or indirectly via fomites or through the consumption of contaminated food or water. For most individuals norovirus gastroenteritis is relatively mild and of short duration but children, immune-compromised individuals, and the elderly are especially vulnerable to infection with often longer and more severe symptoms [Green, 2013].
Pakistan is a densely populated country with limited access to proper health-care facilities. The country is divided into four major provinces: Sindh, Baluchistan, Khyber Pakhtunkhwa, and Punjab. Karachi, Lahore, Rawalpindi and Peshawar rank among the most populated cities in Pakistan. According to WHO estimates, Pakistan has a total population of about 173.6 million, has an under-five child mortality rate of 87 per 1,000 live births, and an estimated 465,000 deaths annually within this age group [WHO, 2012]. Mortality due to diarrhea is alarmingly high (estimated to be ~74,209 deaths) particularly when combined with pneumonia [Black et al., 2010]. Data on norovirus prevalence and distribution of genotypes are available for India [Chhabra et al., 2009; Nataraju et al., 2011; Menon et al., 2013], Bangladesh [Rahman et al., 2010; Nahar et al., 2013], Iran [Romani et al., 2012] and several other low resource countries [Yang et al., 2010; Kaplan et al., 2011; Nataraju et al., 2011; Zeng et al., 2012; Trainor et al., 2013] but lacking from Pakistan. To date, only one study on the molecular epidemiology of norovirus in hospitalized children with acute gastroenteritis in Pakistan has been published [Phan et al., 2004]. Given the importance of norovirus as a major cause of sporadic diarrhea, we determined the burden of norovirus gastroenteritis in hospitalized children younger than 5 years of age in several major cities in Pakistan.
MATERIALS AND METHODS
Study Design
This study was approved by the Ethical Review Committee of the Aga Khan University Hospital (AKU). Stool specimens were collected from children under 5 years of age from Karachi, Rawalpindi, Lahore, and Peshawar as part of a hospital-based surveillance study of the Burden of Rotavirus Gastroenteritis in Children in Pakistan that was conducted from April 2006–March 2008 [Kazi et al., 2014]. Briefly, children included in the study were hospitalized for acute gastroenteritis as the primary illness and had watery diarrhea of less than 7 days duration with severe dehydration classified as WHO’s Integrated Management of Childhood Illnesses (IMCI). All children were provided with oral or intravenous rehydration therapy. Stool samples were collected on the day of admission and tested for group A rotavirus using the IDEIA Rotavirus kit (Oxoid, Cambridge, UK) and stored at −70°C [Kazi et al., 2014]. A subset of 255 samples (51 per hospital), including 143 rotavirus positive and 112 rotavirus negative samples, were selected after randomization using Microsoft Excel for norovirus testing.
RNA Extraction
Viral RNA was extracted from clarified 10% (w/v) stool specimens in phosphate-buffered saline PBS (Gibco, Carlsbad, CA) using a guanidine hydrochloride lysis buffer and silica-based spin column. Briefly, 100 µl of lysis buffer was added to 100 µl of clarified stool suspension in a microcentrifuge tube and the mixture was incubated at room temperature for 10 min. After adding 200µl of absolute ethanol (Sigma–Aldrich, St. Louis, MO), the mixture was vortexed and loaded onto a HiBind RNA minicolumn RNA COL-02, (Omega Bio-tek, Norcross, GA), and centrifuged for 1 min at 14,000 rpm. After washing the column twice with 500 µl of 75% ethanol, viral RNA was eluted with 50 µl of TE buffer (10 mM Tris-HCl; pH 8.0, 1 mM EDTA) by centrifugation at 14,000 rpm for 1 min. Purified RNA was stored at −70°C until use.
Norovirus Real-Time RT-PCR
TaqMan real-time reverse transcription-polymerase chain reaction (RT-qPCR) was employed for detecting GI and GII norovirus strains using AgPath-ID One-Step kit (Applied Biosystem, Foster city, CA) and oligonucleotide primers and probes as described previously [Kageyama et al., 2003]. Briefly, 3 µl of extracted RNA was reverse transcribed into cDNA and amplified on a Chromo4 Real-Time PCR detection system (Bio-Rad, Hercules, CA) using the following thermocycling conditions: reverse transcription for 10 min at 45°C, denaturation for 10 min at 95°C, followed by 45 cycles of 95°C for 15 sec and 60°C for 1 min [Gentry et al., 2009].
Norovirus Conventional RT-PCR
Samples that tested positive by TaqMan real-time RT-PCR were genotyped by amplifying region C at the 5′-end of the VP1 gene by conventional RT-PCR using the OneStep RT-PCR kit (QIAGEN, Valencia, CA) using GI and GII oligonucleotide primers as described previously [Kojima et al., 2002]. Briefly, 5 µl of extracted RNA was reverse transcribed into cDNA for 30 min at 42°C followed by heat inactivation of reverse transcriptase at 95°C for 15 min and then amplified for GI using 0.5 µM of each oligonucleotide primer in a final reaction volume of 25 µl. PCR was carried out for 40 cycles using the following thermocycling conditions: 94°C for 30 sec, 50°C for 30 sec, and 72°C for 60 sec followed by a final extension for 10 min at 72°C.
DNA Sequencing and Phylogenetic Analyses
PCR products were separated on a 2% agarose gel and products of expected size (330 bp for GI, 344 bp for GII) were excised and purified by QIAquick gel extraction kit (QIAGEN, Valencia, CA) as per manufacturer’s instructions. Purified products were cycle-sequenced in both directions using Big Dye-based sequencing method on an ABI Prism® 3130xl Genetic Analyzer by Macrogen (Seoul, Korea). Raw sequences were edited and assembled subsequently using Sequencher 4.8 software (Gene Codes, Ann Arbor, MI). The phylogenetic trees were based on partial capsid sequences (region C) having amplicon size of 264 and 252 bp for GI and GII, respectively. The phylogenetic relationship was inferred by the Maximum Likelihood method with Jukes–Cantor nucleotide substitution models, and 100 bootstrap replicates, using MEGA5.05 software [Tamura et al., 2011].
Statistical Analysis
Statistical analysis was performed using SPSS v.19 (SPSS, Chicago, IL). Chi-square (χ2) or Fisher exact test and Kruskal Wallis test were used to determine the association among the four different norovirus positive and negative groups for different variables. A P-value of less than 0.05 was considered statistically significant.
RESULTS
Norovirus Prevalence, Demographics, and Clinical Characteristics
As a first step towards determining the prevalence of norovirus infections in Pakistan, 255 samples were collected from hospitalized children younger than 5 years of age with acute diarrhea in Karachi, Lahore, Peshawar, and Rawalpindi. Overall, 41 (16.1%) of the 255 fecal specimens tested positive for norovirus of which 27 (18.1%) of the 149 samples were detected in the 2006–2007 season and 14 (13.2%) of the 106 samples in the 2007–2008 season. Nine (22%) samples tested GI positive, 30 (73%) GII positive and 2 (5%) samples tested positive for both GI and GII. The prevalence for the hospitals in each city was 17.6% (18/102) for Karachi, 15.7% (8/51) for Lahore, 15.7% (8/51) for Peshawar, and 13.7% (7/51) for Rawalpindi (Table I). The prevalence of norovirus was similar 23/143 (16%) in rotavirus-positive samples compared to rotavirus negative samples 18/112 (16%) (Table II).
TABLE I.
Norovirus Prevalence in Children Younger Than 5 Years Hospitalized With Acute Gastroenteritis From Four Cities of Pakistan
| Norovirus | |||||
|---|---|---|---|---|---|
|
|
|||||
| City | No. of specimens tested |
GI and GII (% positivity) |
GI (% of all norovirus) |
GII (% of all norovirus) |
Mixed GI + GII (%) |
| Karachi | 102 | 18 (17.6) | 4 (22.2) | 14 (77.7) | – |
| Lahore | 51 | 8 (15.7) | 3 (37.5) | 5 (62.5) | – |
| Peshawar | 51 | 8 (15.7) | 1 (12.5) | 5 (62.5) | 2 (25) |
| Rawalpindi | 51 | 7 (13.7) | 1 (12.5) | 6 (85.7) | – |
| All cities | 255 | 41 (16.1) | 9 (3.5) | 30 (11.8) | 2 (0.8) |
TABLE II.
Demographic and Clinical Characteristics of Pediatrics Gastroenteritis Patients (Under Five Years) Stratified by Rotavirus and Norovirus Status
| Rotavirus positive (n = 143) | Rotavirus negative (n = 112) | |||
|---|---|---|---|---|
|
|
|
|||
| Norovirus N (%) | Norovirus N (%) | |||
|
|
|
|||
| Characteristics | Positive (n = 23) | Negative (n = 120) | Positive (n = 18) | Negative (n = 94) |
| Age (months)a | ||||
| < = 6 | 7 (30.4) | 28 (23.3) | 3 (16.7) | 30 (31.9) |
| 7–12 | 9 (39.1) | 51 (42.5) | 5 (27.8) | 32 (34.0) |
| 13–24 | 7 (30.4) | 33 (27.5) | 7 (38.9) | 22 (23.4) |
| 25–36 | 0 (0.0) | 3 (2.5) | 3 (16.7) | 7 (7.4) |
| 37–59 | 0 (0.0) | 4 (3.3) | 0 (0.0) | 3 (3.2) |
| Sex (n = 232)a | ||||
| Male | 15 (65.2) | 71 (59.2) | 11 (61.1) | 59 (62.8) |
| Female | 8 (34.8) | 49 (40.8) | 7 (38.9) | 35 (37.2) |
| Vomiting (n = 215)a | ||||
| Yes | 15 (65.2) | 70 (61.9) | 11 (73.3) | 51 (58.6) |
| No | 8 (34.8) | 43 (38.1) | 4 (26.7) | 36 (41.4) |
| Fever (n = 223)a | ||||
| Yes | 8 (34.8) | 66 (57.9) | 8 (47.1) | 54 (58.7) |
| No | 15 (65.2) | 48 (42.1) | 9 (52.9) | 38 (41.3) |
| Dehydration status (n = 221)a | ||||
| Severe | 14 (60.9) | 63 (53.4) | 7 (41.2) | 48 (53.9) |
| Some | 8 (34.8) | 52 (45.2) | 10 (58.8) | 39 (43.8) |
| None | 1 (4.3) | 0 (0.0) | 0 (0.0) | 2 (2.2) |
| Treatment received (n = 225)a | ||||
| IVF | 15 (65.2) | 88 (75.9) | 14 (82.4) | 70 (76.1) |
| ORT | 6 (26.1) | 18 (15.5) | 3 (17.6) | 18 (19.6) |
| Both | 2 (8.7) | 10 (8.0) | 0 (0.0) | 4 (4.3) |
| Median diarrheal episodes/dayb | 8 | 10 | 10 | 10 |
| Median vomiting episodes/dayb | 2 | 2 | 3 | 2 |
| Median duration of diarrheab | 2 | 2 | 2.5 | 2 |
| Median duration of vomitingb | 1 | 1 | 1 | 1 |
N (%): total number of cases (percent).
Chi-square (χ2) or Fisher exact test P>0.05
Kruskal Wallis test P>0.05
Three (16.7%) of 18 norovirus positive rotavirus-negative children were younger than 6 months of age, 12 (66.7%) were between 7 months and 2 years of age, and the remaining 3 (16.7%) children were between 2 and 3 years of age. Norovirus infections were more prevalent in male (63.4%) than in female (36.4%) children (Table II).
Based on results for norovirus and rotavirus, children were separated in four groups. Among patients who were positive for norovirus and rotavirus negative, 73.3% experienced vomiting and 47.1% had fever whereas in patients that were positive for both norovirus and rotavirus, 65.2% had vomiting and 34.8% had fever (Table II). Overall, 82.4% of the children that tested positive for norovirus but negative for rotavirus received intravenous fluid replacement therapy (IVF) and 17.6% received oral rehydration therapy (ORT) while among the norovirus and rotavirus-positive group, 65.2% received IVF, and 26.1% ORT. No statistically significant difference was observed in the vomiting and stool frequency per 24 hr among all the four groups; however, we found minor differences in the median of vomiting and diarrheal episodes in a 24-hr period between norovirus positive rotavirus-negative patients and patients that tested positive for both norovirus and rotavirus (Table II).
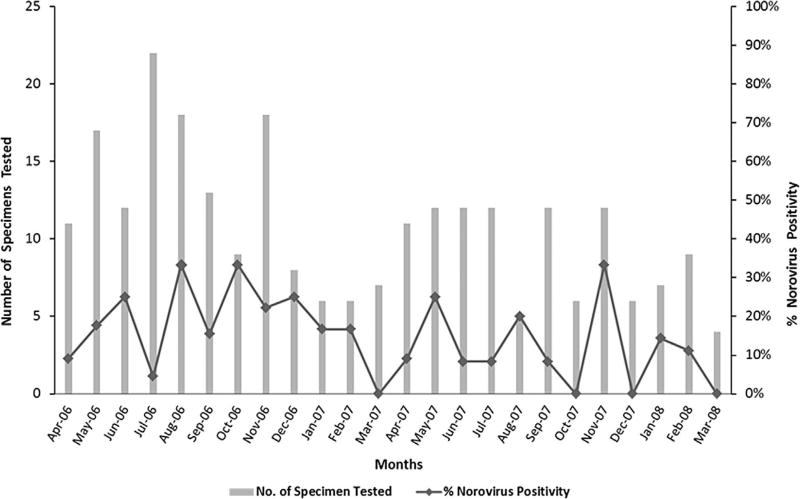
Norovirus was detected throughout the year but fewer infections were observed between December and March, which tend to be the cooler months in Pakistan (Fig. 1).
Fig. 1.
Seasonality of norovirus. Seasonal distribution of norovirus infections in hospitalized children with acute gastroenteritis, Pakistan from April 2006 to March 2008. Bars represent number of specimens tested each month and the line diagram shows number of norovirus positive specimens.
Norovirus Genotypes
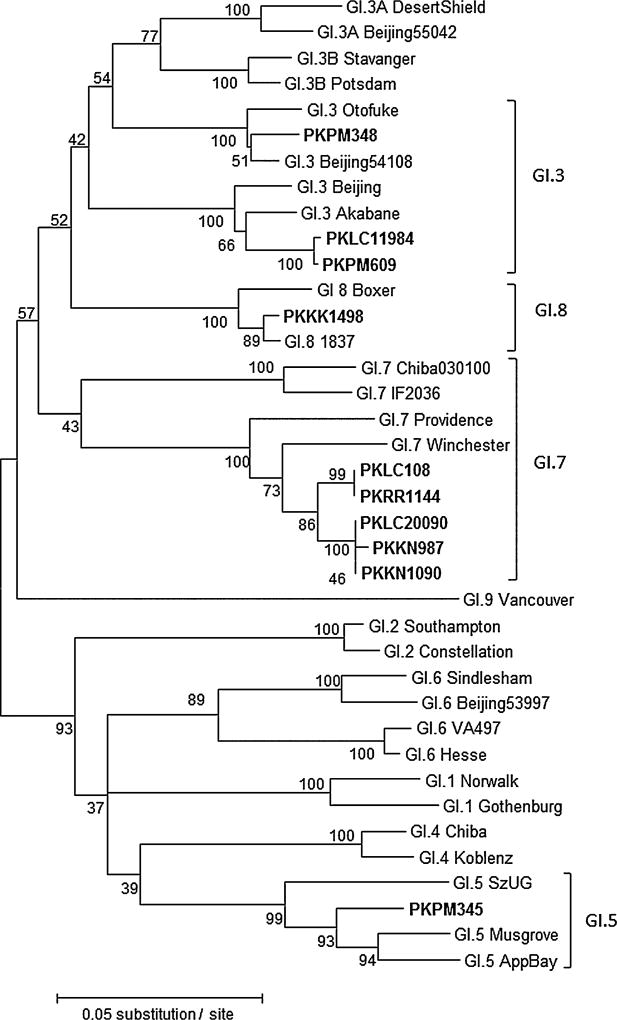
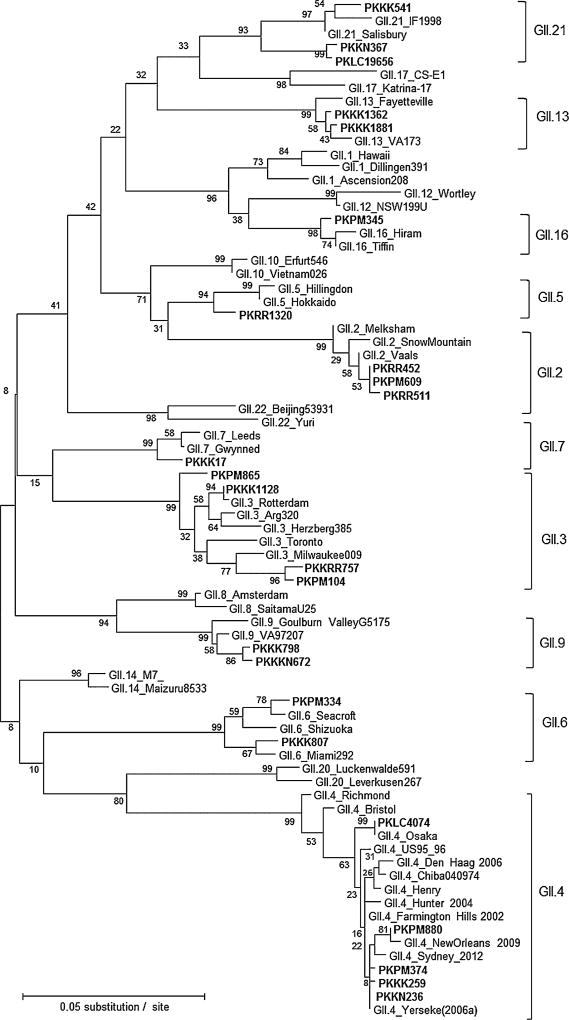
Of the 41 norovirus real-time RT-PCR positive stool samples, 35 (85%) could be genotyped including two samples having mix infection. A total of 14 different norovirus genotypes were detected (Figs. 2 and 3). More genotypes were found in the 2006–2007 season (n = 12) compared to the 2007–2008 season (n = 8) (Table III). Within GI, GI.7 viruses were most prevalent (50%), followed by GI.3 (30%), GI.5 (10%), and GI.8 (10%). Within GII, GII.4 viruses were the most frequently identified genotype (20%) including three different GII variant strains (GII.4 Yerseke 2006a, GII.4 Osaka, and GII.4 New Orleans), followed by GII.3 (16%) and GII.2 (16%). Several rare genotypes such as GII.21 (12%), GII.9 (8%), and GII.5 (4%) were also detected. The two samples containing mixed GI and GII strains (GI.5/GII.16, GI.3/GII.2) were from Peshawar city in 2006–2007 season.
Fig. 2.
Genotyping of norovirus GI strains. Phylogenetic analysis of GI norovirus strains detected in Pakistan from April 2006 to March 2008. The phylogenetic tree was constructed by the maximum likelihood method with 100 bootstrap replicates using MEGA5.05. Sequences of the following GI norovirus reference strains were included in the tree: GI.1_Gothenburg_SWE04 (EU085529), GI.1_Norwalk_USA68 (M87661), GI.2_Constellation59_USA99 (AF435807), GI.2_Southampton_GBR91 (L07418), GI.3A_Beijing55042_CHN07 (GQ856473), GI.3A_DesertShield_USA90 (U04469), GI.3B_Potsdam_DEU00 (AF439267), GI.3B_Stavanger_NOR95 (AF145709), GI.3_Beijing 54108_CHN07 (GQ856470), GI.3_Otofuke_JPN79 (AB187514), GI.3_Akabane991130_JPN99 (EF547396), GI.3_Beijing54114_C HN07 (GQ856471), GI.4_Chiba407_JPN87 (AB042808), GI.4_-Koblenz433_DEU00 (AF394960), GI.5_AppalachicolaBay318_USA95 (AF414406), GI.5_Musgrove_GRB89 (AJ277614), GI.5_SzU G1_JPN99 (AB039774), GI.6_Beijing53997_CHN07 (GQ856463), GI.6_Sindlesham_GBR95 (AJ277615), GI.6_Hesse_DEU97 (AF0937970), GI.6_VA497_USA99 (AF538678), GI.7_Winchester_GBR94 (AJ277609), GI.7_Providence191_USA1 (JN899243), GI.7_Chiba030100_JPN03 (AJ844469), GI.7_IF2036_IRQ03 (AY675555), GI.8_1837_USA08 (GU299761), GI.8_Boxer_USA01 (AF538679), and GI.9_Vancouver730_CAN04 (HQ637267). Norovirus strains (in bold) detected in this study are indicated with unique ID’s representing the city and hospital origin (PKPM, Mercy Hospital Peshwar; PKLC, Children’s Hospital Lahore; PKRR, Rawalpindi General Hospital; PKKK, Kharadar General Hospital; and PKKN, National Institute of Child Health Karachi).
Fig. 3.
Genotyping of norovirus GII strains. Phylogenetic analysis of GII norovirus strains detected in Pakistan from April 2006 to March 2008. Sequences of the following GII norovirus reference strains were included in the tree: GII.1_Hawaii_USA71 (U07611), GII.1_Dillingen391_DEU01 (AF425767), GII.1_Ascension208_USA10 (JN797508), GII.2_Melksham_GBR94 (X81879), GII.2_SnowMountain_USA76 (AY134748), GII.2_Vaals_NLD05 (AB281090), GII.3_Toronto_CAN91 (U02030), GII.3_Milwaukee009_USA10 (JN565063), GII.3_Arg320_ARG99 (AF190817), GII.3_Herzberg385_DEU01 (AF539439), GII.3_Rotterdam_NLD06 (AB385626), GII.4_Bristol_GBR93 (X76716), GII.4_Chiba040974_JPN04 (AB294782), GII.4_Farmington Hills_USA02 (AY502023), GII.4_Henry_USA00 (FJ411170), GII.4_Hunter_AUS04 (DQ078794), GII.4_DenHaag_NL06 [2006b] (EF126965), GII.4_NewOrleans [NO1805a_USA09] (GU445325), GII.4_Osaka_JPN07 [Riviera] (AB434770), GII.4_Richmond_USA94 (EU078406), GII.4_Yerseke_NLD06 [2006a] (EF126963), GII.4 Sydney_AUS12 (JX459908), GII.5_Hillingdon_GBR90 (AJ277607), GII.5_Hokkaido133_JPN03 (AB212306), GII.6_Shizuoka_JPN08 (HM633213), GII.6_Miami292_USA94 (AF414410), GII.6_Seacroft_GBR90 (AJ277620) GII.7_Gwynedd_USA94 (AF414409), GII.7_Leeds_GBR90 (AJ277608), GII.8_Amsterdam_NLD98 (AF195848), GII.8_SaitamaU25_JPN00 (AB039780), GII.9_Goulburn ValleyG5175_AUS83 (DQ379715), GII.9_VA97207_USA97 (AY038599), GII.10_Erfurt546_DEU00 (AF427118), GII.10_Vietnam026_VNM02 (AF504671), GII.12_NSW199U_AUS08 (GQ845370), GII.12_Wortley_GBR90 (AJ277618), GII.13_Fayetteville_USA98 (AY113106), GII.13_VA173_USA10 (JN899242), GII.14_M7_USA99 (AY130761), GII.14_Maizuru8533_JPN08 (GU017903), GII.16_Hiram_USA00 (AY502006), GII.16_Tiffin_USA99 (AY502010), GII.17_CSE1_USA02 (AY502009), GII.17_Katrina-17_USA05 (DQ438972), GII.20_Leverkusen267_DEU05 (EU424333), GII.20_Luckenwalde591_DEU02 (EU373815), GII.21_IF1998_IRQ03 (AY675554) and GII.21_Salisbury150_USA11(JN899245), GII.22_Yuri_JPN02 (AB083780), GII.22_Beijing53931_CN07 (GQ856469). The phylogenetic tree was constructed by maximum likelihood method with 100 bootstrap replicates using MEGA5.05.norovirus. The key of the Pakistani strains is the same as in Figure 2.
TABLE III.
Distribution of Norovirus Genotypes Identified in Hospitalized Children in Pakistan
| Norovirus genogroup |
Genotype | No. of samples |
April 2006–March 2007 (%) |
April 2007–March 2008 (%) |
% of all genotypes |
|---|---|---|---|---|---|
| GI | GI.3 | 2 | 1 (3.7) | 1 (7.1) | 4.9 |
| GI.7 | 5 | 3 (11.1) | 2 (14.3) | 12.2 | |
| GI.8 | 1 | – | 1 (7.1) | 2.4 | |
| GII | GII.2 | 3 | 2 (7.4) | 1 (7.1) | 7.3 |
| GII.3 | 4 | 2 (7.4) | 2 (14.3) | 9.8 | |
| GII.4 (Osaka, New Orleans, Yerseke) | 5 (1, 1, 3) | 4 (14.8) | 1 (7.1) | 12.2 | |
| GII.5 | 1 | 1 (3.7) | – | 2.4 | |
| GII.6 | 2 | 2 (7.4) | – | 4.9 | |
| GII.7 | 1 | 1 (3.7) | – | 2.4 | |
| GII.9 | 2 | 2 (7.4) | – | 4.9 | |
| GII.13 | 2 | – | 2 (14.3) | 4.9 | |
| GII.21 | 3 | 2 (7.4) | 1 (7.1) | 7.3 | |
| GI + GII | GI.5 + GII.16 | 1 | 1 (3.7) | – | 2.4 |
| GI.3 + GII.2 | 1 | – | 1 (7.1) | 2.4 | |
| Could not be typeda | 8 | 6 (22.2) | 2 (14.3) | 19.5 | |
| Total | 41 | 27 (100) | 14 (100) | 100.0 |
No PCR products obtained or unreadable sequence data.
DISCUSSION
The main objective of this study was to assess the norovirus disease burden in hospitalized children younger than 5 years of age with acute gastroenteritis and to determine the genetic diversity of norovirus strains circulating in Pakistan. Overall, 16.1% of all samples tested positive for norovirus with 73% positive for GII and 22% for GI whereas 5% of the samples were positive for both GI and GII. This is the first nationwide report on norovirus from Pakistan. This study reported substantially higher disease burden compared to a previous study conducted on rotavirus negative samples collected from a hospital in Karachi from 1990 to 1994 [Phan et al., 2004]. Although it may be possible that the prevalence of norovirus infections has increased over the last 12 years in Karachi, a more plausible explanation is that conventional PCR that was used in the previous study was not as sensitive as the real-time PCR used in our study. Prevalence data reported here falls in the range (8–30%) that has been reported in other low resource countries [Rahman et al., 2010; Yang et al., 2010; Kirby et al., 2011; Zeng et al., 2012; Nahar et al., 2013].
In agreement with previous reports, most infections in this study were caused by GII viruses [Chhabra et al., 2009; Rahman et al., 2010; Yang et al., 2010; Zeng et al., 2012]. Diverse GI and GII genotypes of norovirus were found in this study, including GI.5, GI.8, GI.3, GII.5, GII.7, GII.9, GII.13, GII.16, and GII.21. Interestingly, GI.7 viruses were the most frequently GI viruses, a finding concordant with a previous report from Pakistan [Phan et al., 2004]. A total of 12.2% of the norovirus infections were caused by GII.4 viruses, which is significantly lower than what has typically been reported [Kirby et al., 2011; Zeng et al., 2012; Estévez et al., 2013; Payne et al., 2013]. This may be partly caused by the fact that our study does not include samples from nosocomial outbreaks, which typically are caused by GII.4 viruses.
GII.4 New Orleans viruses were detected in samples collected in November 2007, which is almost 2 years prior to the emergence of this GII.4 variant globally. For example, in United States, GII.4 New Orleans emerged in October 2009 and accounted for 75% of all norovirus outbreaks from October 2009 to January 2011 [Vega et al., 2011]. Young children in developing countries may therefore perhaps serve as a reservoir from which new norovirus strains emerge and at times cause epidemic spreads between continents possibly due to frequent traveling. In contrast to countries with a temperate climate, norovirus infections were found throughout the year with no distinct increase during the winter season [Ahmed et al., 2013]. Most clinical characteristics of norovirus infection reported in this report were consistent with published data from other developing countries [Rahman et al., 2010; Nataraju et al., 2011]. Of note, 56% (23/41) of norovirus infected children were co-infected with rotavirus (Table II), a finding that is contradictory to what has been reported previously where co-infections ranged from 1.5% to 18% [Nataraju et al., 2011; Zeng et al., 2012; Estévez et al., 2013; Trainor et al., 2013].
This report confirms the significant role that norovirus infection plays as the cause of acute pediatric gastroenteritis in Pakistan. However, several limitations can be noted including the small sample size and selected study patients (hospitalized children under 5 years of age) Low resource country-specific features such as poor hygiene, lack of proper healthcare facilities, and compromised immunity especially among children do not necessarily result in higher prevalence rates compared to high-resource countries. Norovirus positive samples were genotyped by sequencing a small region of the capsid gene but since recombination often occurs among noroviruses, additional polymerase-based typing would provide more information if strains from the same genotype are similar or are potential recombinant viruses [Lu et al., 2015].
This work sets the stage for a large-scale community and hospital-based epidemiological study spanning several years, which could provide much-needed insight into the prevalence of norovirus infections in the pediatric population, as well as changes in strain distribution over time in Pakistan. Results from such an evidence-based study are required to be able to influence policy makers to consider appropriate measures that will reduce the number of diarrhea-related deaths in the Pakistani pediatric population.
Acknowledgments
Grant sponsor: National Institute of Health’s Fogarty International Center; Grant number: 1 D43 TW007585-01
We are grateful to Drs. Syed Ali, Syed Hani Abidi (Department of Biological and Biomedical Sciences), and Najeeha Talat (Department of Pediatrics and Child Health) for critical reviewing of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Centers for Disease Control and Prevention.
Footnotes
Disclaimer: The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- Ahmed SM, Lopman BA, Levy K. A systematic review and meta-analysis of the global seasonality of norovirus. PLoS ONE. 2013;8:e75922. doi: 10.1371/journal.pone.0075922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, Jha P, Campbell H, Walker CF, Cibulskis R, Eisele T, Liu L, Mathers C. Global, regional, and national causes of child mortality in 2008: A systematic analysis. Lancet. 2010;375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- Chhabra P, Dhongade RK, Kalrao VR, Bavdekar AR, Chitambar SD. Epidemiological, clinical, and molecular features of norovirus infections in Western India. J Med Virol. 2009;81:922–932. doi: 10.1002/jmv.21458. [DOI] [PubMed] [Google Scholar]
- Estévez A, Arvelo W, Hall AJ, Lopez MaR, Lopez B, Reyes L, Moir JC, Gregoricus N, Vinjé J, Parashar UD. Prevalence and genetic diversity of norovirus among patients with acute diarrhea in Guatemala. J Med Virol. 2013;85:1293–1298. doi: 10.1002/jmv.23578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry J, Vinje J, Lipp EK. A rapid and efficient method for quantitation of genogroups I and II norovirus from oysters and application in other complex environmental samples. J Virol Methods. 2009;156:59–65. doi: 10.1016/j.jviromet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Green KY. Caliciviridae: The noroviruses. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B, editors. Fields’ virology. 6. Philadelphia, PA: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2013. pp. 583–609. [Google Scholar]
- Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. Broadly reactive and highly sensitive assay for norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan NM, Kirby A, Abd-Eldayem SA, Dove W, Nakagomi T, Nakagomi O, Cunliffe NA. Detection and molecular characterisation of rotavirus and norovirus infections in Jordanian children with acute gastroenteritis. Arch Virol. 2011;156:1477–1480. doi: 10.1007/s00705-011-0996-x. [DOI] [PubMed] [Google Scholar]
- Kazi AM, Warraich GJ, Qureshi S, Qureshi H, Khan MM, Zaidi AK. Sentinel hospital-based surveillance for assessment of burden of rotavirus gastroenteritis in children in Pakistan. PLoS ONE. 2014;9:e108221. doi: 10.1371/journal.pone.0108221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A, Al-Eryani A, Al-Sonboli N, Hafiz T, Beyer M, Al-Aghbari N, Al-Moheri N, Dove W, Cunliffe NA, Cuevas LE. Rotavirus and norovirus infections in children in Sana’a, Yemen. Trop Med Int Health. 2011;16:680–684. doi: 10.1111/j.1365-3156.2011.02756.x. [DOI] [PubMed] [Google Scholar]
- Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–114. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- Lu QB, Huang DD, Zhao J, Wang HY, Zhang XA, Xu HM, Qu F, Liu W, Cao WC. An increasing prevalence of recombinant GII norovirus in pediatric patients with diarrhea during 2010–2013 in China. Infect Genet Evol. 2015;31C:48–52. doi: 10.1016/j.meegid.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Menon VK, Sarkar R, Moses PD, Agarwal I, Simon A, Kang G. Norovirus genogroup II gastroenteritis in hospitalized children in South India. Am J Trop Med Hyg. 2013;89:1019–1022. doi: 10.4269/ajtmh.13-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar S, Afrad MH, Begum N, Al-Mamun F, Sarker AK, Das SK, Faruque ASG, Pourkarim MR, Choudhuri MSK, Azim T. High prevalence of noroviruses among hospitalized diarrheal patients in Bangladesh, 2011. J Infect Dev Countries. 2013;7:892–896. doi: 10.3855/jidc.2944. [DOI] [PubMed] [Google Scholar]
- Nataraju SM, Pativada M, Chatterjee D, Nayak MK, Ganesh B, Bhattacharya MK, Ramamurthy T, Ganguly S, Saha DR, Rajendran K, Ghosh M, Kobayashi N, Krishnan T. Molecular epidemiology of norovirus infections in children and adults: Sequence analysis of Region C indicates genetic diversity of NVGII strains in Kolkata, India. Epidemiol Infect. 2011;139:910–918. doi: 10.1017/S0950268810001731. [DOI] [PubMed] [Google Scholar]
- Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne DC, Vinjé J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT. Norovirus and medically attended gastroenteritis in US children. N Engl J Med. 2013;368:1121–1130. doi: 10.1056/NEJMsa1206589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Okame M, Nguyen TA, Maneekarn N, Nishio O, Okitsu S, Ushijima H. Human astrovirus, norovirus (GI, GII), and sapovirus infections in Pakistani children with diarrhea. J Med Virol. 2004;73:256–261. doi: 10.1002/jmv.20084. [DOI] [PubMed] [Google Scholar]
- Rahman M, Hassan Z, Nahar Z, Faruque AS, Van Ranst M, Rahman SR, Azim T. Molecular detection of noroviruses in hospitalized patients in Bangladesh. Eur J Clin Microbiol Infect Dis. 2010;29:937–945. doi: 10.1007/s10096-010-0948-5. [DOI] [PubMed] [Google Scholar]
- Romani S, Mohebbi SR, Hosseini SM, Azimzadeh P, Vahedi M, Derakhshan F, Zali MR. Prevalence of norovirus infection in children and adults with acute gastroenteritis, Tehran, Iran, 2008–2009. Food Environ Virol. 2012;4:1–5. doi: 10.1007/s12560-011-9071-8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor E, Lopman B, Iturriza-Gomara M, Dove W, Ngwira B, Nakagomi O, Nakagomi T, Parashar U, Cunliffe N. Detection and molecular characterisation of noroviruses in hospitalised children in Malawi, 1997–2007. J Med Virol. 2013;85:1299–1306. doi: 10.1002/jmv.23589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega E, Barclay L, Gregoricus N, Williams K, Lee D, Vinje J. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerg Infect Dis. 2011;17:1389–1395. doi: 10.3201/eid1708.101837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol. 2015;53:373–381. doi: 10.1128/JCM.01535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Health Statistics 2012. Geneva: WHO Press; 2012. URL: http://www.who.int/gho/publications/world_health_statistics/EN_WHS2012_Full.pdf. [Google Scholar]
- Yang SY, Hwang KP, Wu FT, Wu HS, Hsiung CA, Chang WC, Lin JS, Yang SC, Huang SL, Huang YC. Epidemiology and clinical peculiarities of norovirus and rotavirus infection in hospitalized young children with acute diarrhea in Taiwan, 2009. J Microbiol Immunol Infect. 2010;43:506–514. doi: 10.1016/S1684-1182(10)60078-3. [DOI] [PubMed] [Google Scholar]
- Zeng M, Xu X, Zhu C, Chen J, Zhu Q, Lin S, Jie Y, Shu X. Clinical and molecular epidemiology of norovirus infection in childhood diarrhea in China. J Med Virol. 2012;84:145–151. doi: 10.1002/jmv.22248. [DOI] [PubMed] [Google Scholar]