Abstract
A recombinant vaccine containing Aventis Pasteur’s canarypox vector (ALVAC)–HIV and gp120 alum decreased the risk of HIV acquisition in the RV144 vaccine trial. The substitution of alum with the more immunogenic MF59 adjuvant is under consideration for the next efficacy human trial. We found here that an ALVAC–simian immunodeficiency virus (SIV) and gp120 alum (ALVAC–SIV + gp120) equivalent vaccine, but not an ALVAC–SIV + gp120 MF59 vaccine, was efficacious in delaying the onset of SIVmac251 in rhesus macaques, despite the higher immunogenicity of the latter adjuvant. Vaccine efficacy was associated with alum-induced, but not with MF59-induced, envelope (Env)-dependent mucosal innate lymphoid cells (ILCs) that produce interleukin (IL)-17, as well as with mucosal IgG to the gp120 variable region 2 (V2) and the expression of 12 genes, ten of which are part of the RAS pathway. The association between RAS activation and vaccine efficacy was also observed in an independent efficacious SIV-vaccine approach. Whether RAS activation, mucosal ILCs and antibodies to V2 are also important hallmarks of HIV-vaccine efficacy in humans will require further studies.
The RV144 HIV-vaccine trial, which used ALVAC–HIV and AIDSVAX HIV gp120, clade B and E proteins formulated in alum, resulted in limited but significant (P = 0.04) protection from HIV acquisition1. Serum IgG antibodies against Env variable region 1 (V1) and V2 were inversely correlated with the risk of HIV-1 infection2, and sieve analysis demonstrated genetic markers of immunologic pressure at positions 169 and 181 of V2 (ref. 3). Monomeric serum IgA to HIV-Env was positively correlated with the risk of HIV-1 acquisition and inhibited IgG-mediated, antibody-dependent cell cytotoxicity (ADCC)2, 4. The correlate analyses pointed to the importance of antibodies for protection, which suggests that changing the formulation of the gp120 antigen to include a more immunogenic adjuvant could improve vaccine efficacy. Alum is a T helper (Th) cell 2–inducing adjuvant, whereas the oil-in-water emulsion MF59 adjuvant elicits Th1 and Th2 responses and affects antibody isotypes in an antigen-dependent manner5. Importantly, MF59 has been proposed as the adjuvant of choice for the next set of ALVAC + gp120 vaccine trials in humans, which will be conducted in South Africa (http://vaccineenterprise.org/content/P5Partnership).
Prior macaque studies demonstrated that the administration of ALVAC–SIV, either alone or in combination with gp120, induced protection against SIVmac251 acquisition, depending on the dosage of challenge6–8. Here we tested whether we could recapitulate the protection observed in RV144 by using the ALVAC + gp120 alum–SIV vaccine in macaques, and we took advantage of the similar vaccine efficacy conferred by this model to identify systemic and mucosal correlates of risk of SIVmac251 acquisition. Finally, we tested whether the MF59 adjuvant improves the efficacy of this vaccine regimen. Surprisingly, we observed no vaccine efficacy, despite the ability of MF59 to induce higher immune responses than alum. The reduced risk of virus acquisition in the alum-vaccine group was associated with the induction of mucosal ILCs and mucosal antibodies to V2 that were correlated with the expression of ten of 12 genes that constitute part of the RAS pathway. Further studies will be required to assess whether these results in macaques can be extended to HIV vaccines for humans.
RESULTS
Alum but not MF59 reduced the risk of SIVmac251 acquisition
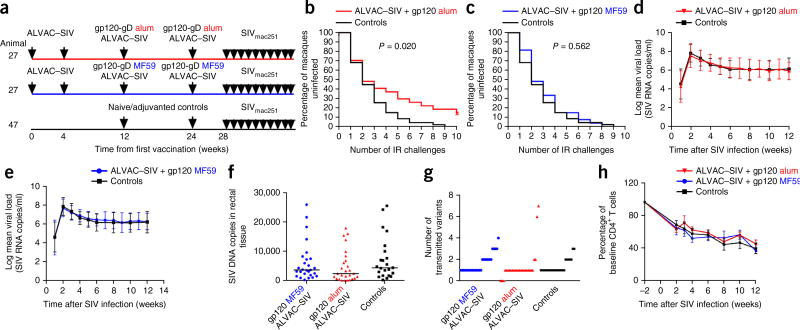
54 rhesus macaques were assigned to two vaccine arms that controlled for the major histocompatibility complex (MHC)-I alleles present, age, weight and gender. All macaques were primed twice, at week 0 and week 4, with ALVAC–SIV, and received two boosters, one each at week 12 and week 24, with ALVAC–SIV together with the gD-g p120 M766 and gD-gp120 CG7V formulated either in alum (n = 27) or MF-59 (n = 27). We used two gp120 proteins that differ in their amino acid sequence to emulate RV144 that used the gp120 clades E and B (ref. 1) (Fig. 1a). Of note, because MF59 is dose sparing, the amount of gp120 in the boosters was halved for the latter group (alum, 200 µg; MF59, 100 µg). An additional 47 unvaccinated macaques—of those, 24 concurrent and 23 historical—were used as controls that matched vaccinated animals (weight, gender and MHC-I alleles; Supplementary Fig. 1a–F). Our study was powered to compare the relative vaccine efficacy in vaccinated macaques with placebo controls, but not to compare vaccine efficacy between the two regimens. We challenged animals intrarectally weekly with ten repeated low doses of SIVmac251, starting 4 weeks after the final immunization. The time of challenge was chosen to model early exposure after vaccination, given that high-risk volunteers in HIV-vaccine trials may be exposed to the virus soon after vaccination. Vaccination with ALVAC–SIV + gp120 alum reduced the risk of SIVmac251 acquisition relative to unvaccinated controls (log–rank test; P = 0.020), showing an estimated vaccine efficacy of 44% at each challenge (Fig. 1b). By contrast, the ALVAC–SIV + gp120 MF59 vaccine regimen did not reduce the risk of infection (log–rank test, P = 0.562; Fig. 1c). Whether the outcome would differ with a later challenge will require further experimentation. Quantitation of mucosal Ki67+CD4+ cells and circulating CD4+ T cells that expressed the activation markers Ki67 and CCR5 was equivalent in the two vaccinated groups at 3 weeks before the first exposure to SIVmac251, and their levels were not correlated with time to SIVmac251 acquisition (Supplementary Fig. 1g–j). By contrast, the frequency of cells expressing the mucosal homing integrin α4β7 within Ki67+CD4+ T cells was indirectly correlated with the number of challenges in the alum group required to infect animals (P = 0.02; Supplementary Fig. 1k). In the MF59 group, we found a correlation between the number of transmitted variants and the frequency of CD38+ cells and of α4β7+Ki67+CD4+ T cells (P = 0.01 and P = 0.04, respectively; Supplementary Fig. 1l,m). We observed no evidence of enhancement or viral suppression in the plasma (Fig. 1d,e) or in the rectal mucosa of the vaccinated macaques that became infected in either arm of the study (Fig. 1f). The numbers of transmitted virus variants did not differ among the three groups, and neither vaccine prevented the loss of CD4+ T cells in macaques that became infected (Fig. 1g,h).
Figure 1.
Study design and vaccine efficacy. Unless stated otherwise, in all the figures, the data for the alum group are in red (n = 27) and those for the MF59 group (n = 27) and the controls (n = 24) are in blue and black, respectively. (a) Study design. Animals were immunized with ALVAC (vCP2432) expressing SIV genes gag-pro and gp120TM, derived from the founder variant of SIVmac251 designated M766r and boosted with the SIVmac251 M766 gp120-gD (referred to in the text as gp120) and SIVsmE660 gp120-gD CG7V (referred to in the text as gp120) proteins formulated in alum (200 mg) or MF59 (100 mg). (b,c) SIVmac251 acquisition. The number of intrarectal (IR) exposures before viral acquisition was assessed in the alum (b) and MF59 (c) groups, relative to controls, using the log–rank test of the discrete-time proportional hazards model. (d,e) SIV RNA levels in the plasma. Error bars show logarithmic mean ± s.d. of animals vaccinated with alum (d) or MF59 (e), and all historical and concurrent control animals (n = 47). (f) SIV DNA copies in rectal mucosa at 2 weeks after infection (horizontal lines, median) in all vaccinated and concurrent controls (n = 23). (g) Number of viral variants in the plasma at 2–4 weeks after infection in all groups. (h) Percentage of CD4+ T cell changes in the blood over time (weeks) (mean ± s.e.m.).
Immunological correlates of risk of SIVmac251 acquisition
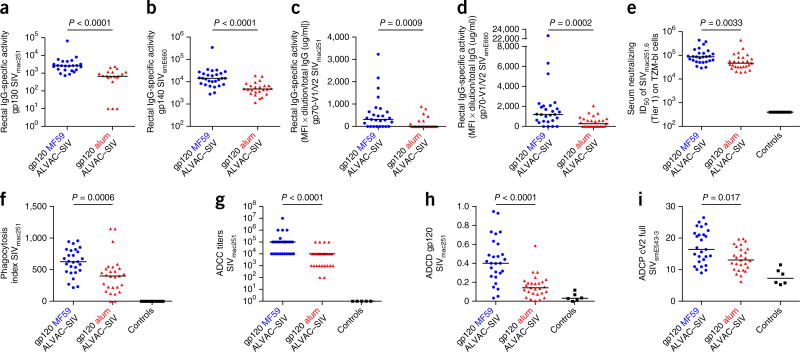
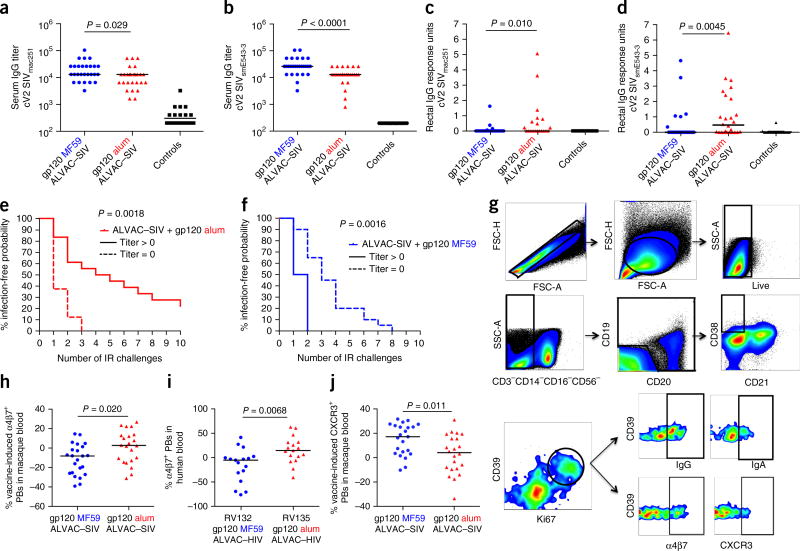
The difference in the protection outcome of the two vaccine regimens provided us with the opportunity to investigate in depth the immunological parameters that are correlated with the risk of SIVmac251 acquisition2, 9. Overall, MF59 induced stronger systemic and mucosal humoral and cellular responses to both Env proteins than did alum, both in magnitude and in function (Fig. 2, Supplementary Fig. 2 and Supplementary Tables 1 and 2). Notably, although the levels of IgG to cyclic V2 (cV2) were higher in the serum of the MF59 group than in that of the alum group (Fig. 3a,b), cV2 IgG was higher in rectal secretions from the alum group than in those from the MF59 group (Fig. 3c,d). Importantly, among the alum-treated macaques, the mucosal cV2 IgG response was associated with a decreased risk of SIV acquisition (log–rank test, P = 0.0018; Fig. 3e). By contrast, rectal IgG antibodies to cV2 were correlated with an increased risk of SIVmac251 acquisition in the MF59 group (log–rank test, P = 0.0016; Fig. 3f). Rectal cV2 IgA levels were low and were not correlated with viral acquisition in either group (Supplementary Fig. 3d,e and data not shown). The discrepancy in the levels of serum and mucosal cV2 IgG in the MF59 and alum arms (Fig. 3a–d) prompted us to investigate whether the two adjuvants affected the surface expression of homing markers on circulating plasmablasts (PBs). The levels of α4β7 and of the chemokine receptor CXCR3 were measured before and after immunization on total PB subsets (Fig. 3g). Alum did not significantly alter the expression of either molecule (Supplementary Fig. 3f,g), as compared to pre-vaccination specimens, whereas MF59 did, resulting in a significantly higher level of α4β7+ PBs in the alum group than in the MF59 group (P = 0.02; Fig. 3h). We observed similar differences, consistent with the macaque results, in α4β7 levels on circulating PBs in humans immunized in the RV135 and RV132 trials, which used an ALVAC (vCP1521)–HIV + rgp120 (MN/A244) that was adjuvanted either with alum10 or MF59, respectively (Fig. 3i). MF59 was also associated with a significant increase (P < 0.001) above the baseline in the number of CXCR3+ PBs in macaques. The recruitment of a significantly higher number of pro-inflammatory neutrophils in lymph nodes of the MF59 group than in the alum recipients (P = 0.023; Supplementary Fig. 3i), as well as the higher level of interferon (IFN)-γ produced by antigen-specific CD4+ T cells (Supplementary Fig. 2a), may have contributed to an increase in the expression of CXCR3 on PBs in the MF59 group (Fig. 3j)11.
Figure 2.
Env-specific IgG responses in mucosa. Unless differently stated, data throughout are in red for the alum group (n = 27), blue for the MF59 group (n = 27) and black for the controls (n = 24). (a,b) Specific activity of rectal IgG-binding antibodies to the gp130 of SIVmac251 (a) and the gp140 of SIVsmE660 (b). (c,d) Specific activity of IgG-binding antibodies to the gp70-V1/V2 scaffold of SIVmac251 (c) or SIVsmE660 (d) in rectal secretion. (e–g) Neutralizing antibodies to Tier 1 SIVmac251.6 (e), phagocytosis index (f) and ADCC (g). (h,i) Gp120-specific complement activation (h) and V2-specific phagocytosis (i). Each symbol represents one animal, and horizontal lines represent the medians. The Mann–Whitney–Wilcoxon test was used to compare continuous factors between two groups.
Figure 3.
Serum and rectal IgG to cyclic V2, PB homing markers and SIVmac251 acquisition. (a–d) IgG to cV2 of SIVmac251 (MF59, n = 26; alum, n = 20; controls, n = 12) (a,c) and SIVsme543.3 (MF59 and alum, n = 26; controls n = 18) (b,d) in sera (a,b) and rectal mucosa (c,d). Titers or response units are shown. Horizontal lines are medians. (e,f) SIVmac251 acquisition in vaccinated animals that had postvaccination rectal IgG response units to cV2 of SIVsmE543 >0 (solid lines) or 0 (dotted lines) in the alum group (e) and in the MF59 group (f). Although IgG responses to cV2 peptides in the mucosa were generally low, and high background was noted in some animals in the pre-vaccination samples, postvaccination data were supported by an independent multiplex assay of mucosal V2 responses (Supplementary Fig. 3). (g) Phenotypic characterization of PBs. Plots represent the gating strategy, where gating is represented by squares or circles and the arrows represent the parent-to-daughter population flow for each line. (h,j) Frequency of vaccine-induced α4β7+ PBs (h) or vaccine-induced CXCR3+ (j) in macaques at day 7 after the final immunization (alum, n = 22; MF59, n = 23. (i) Frequency of vaccine-induced α4β7+ PBs in the blood of 17 humans enrolled in the RV132 and RV135 HIV-vaccine trials. Blood was collected at 14 d from vaccination (Wilcoxon signed–rank test).
The ability of cV2 antibodies to affect the risk of SIV acquisition differently in the two vaccinated groups suggested that there are differences in antibody effector functions. Because glycan influences the functions of antibodies, we analyzed glycoforms in serum antibodies at 1 week after administering the final immunization (week 25). We observed that alum vaccination induced elevations in the fractions of galactose–fucose (G1F) structures that were linked to enhanced ADCC and pro-inflammatory G0 glycans, which are associated with high complement activation and are also enriched in HIV controllers12, 13. MF59 vaccination, by contrast, resulted in the induction of sialylated SIV Env-specific antibodies (Supplementary Fig. 4). Siaylation is associated with a dampening of inflammation14. These data are indicative of functional differences in the gp120-specific antibodies that are elicited by the two adjuvants in serum. A similar analysis on mucosal cV2 IgG was hampered by their low titers.
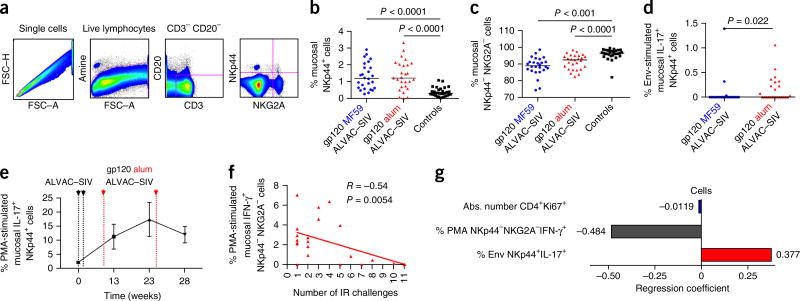
Next, we studied mucosal ILCs comprising natural killer (NK) cells that express NKG2A and NKp44 receptors15, 16. Within the mucosal ILC population, NKG2A+ cells mediate cytotoxicity, whereas NKp44+ cells produce IL-17, IL-22 and B cell–activating factor (BAFF), thereby promoting epithelial integrity and mucosal homeostasis16, 17. NKp44−NKG2A− is a less-defined ILC population that produces cytokines, including IFN-γ. Vaccination with both adjuvants increased the frequency of mucosal NKp44+ cells, and decreased that of NKp44−NKG2A− cells, as compared to the unvaccinated controls (Fig. 4a,b). The stimulation of mucosal mononuclear cells with overlapping peptides that encompass the entire SIVmac251 gp120 proteome revealed a higher proportion of NKp44+IL-17-producing cells in the alum group relative to MF59 recipients (Fig. 4d). In an independent experiment, we assessed whether IL-17+NKp44+ cells are recruited and maintained at mucosal sites after vaccination, by immunizing six naive macaques identically with ALVAC–SIV + gp120 in alum. We found that the frequency of these cells was increased during immunization, and that their levels were maintained up to the time of challenge exposure (Fig. 4e).
Figure 4.
Vaccine-induced changes in the frequency and function of mucosal ILC subsets. (a) Representative flow cytometric plots defining ILCs in the rectal mucosa of rhesus macaques. ILCs were identified using a side-scatter versus forward-scatter gate and phenotypically defined as CD3−CD20− and NKG2A+ or NKp44+ cells, or as NKG2A−NKp44−cells. Purple line defines the gates used to calculate CD20−CD3− cells and NKp44+NKG2A+ cells. (b,c) Relative frequency of NKp44+ (b) and NKG2A−NKp44− (c) cells at week 25 in the MF59-vaccinated (n = 26) and alum-vaccinated animals (n = 27) and unimmunized controls (n = 30). Horizontal lines represent the medians. (d) Frequency of NKp44+ cells producing IL-17 after Env-stimulation (alum, n = 24; MF59, n = 22). (e) Kinetic of the recruitment of NKp44+ ILCs that produce IL-17 after in vitro stimulation in six immunized animals (mean values ± s.d.). (f) Correlation between the percentage of mucosal IFN-γ+NKG2A−NKp44− cells and time of acquisition in the alum group. (g) The ability of individual sets of aggregate data to predict protection in the MF59 and alum groups was compared among animals that were infected in four or fewer challenges (≤4) or after five or more challenges (≥5), using logistic regression and leave-one-out cross-validation (Supplementary Fig. 5). The best model predicting alum-conferred protection from SIV acquisition was obtained when features identified as “cells” were used (accuracy, >81%). The features selected in the “cells” regression model are presented as a bar plot (y axis). The length and direction of each bar is proportional to the relative predictive contribution of each feature in the model (x axis, regression coefficient). Features associated with a high risk of SIV acquisition are in blue; those associated with low risk are in red.
We then conducted logistic-regression analysis, comparing the humoral profiles of macaques that were infected after four or fewer or five or more challenges (Supplementary Tables 3 and 4). We observed that specific glycosylation patterns in antibodies to the SIV envelope were associated with a risk of viral acquisition in the MF59 group (Supplementary Fig. 4 and 5). Notably, functional ILCs and mucosal humoral markers were the features that were significantly associated with protection in the alum group (accuracies of 81.5% and 66.7%, respectively; Supplementary Fig. 5). Among the features associated with acquisition in this group (Supplementary Table 4), NKp44−NKG2A− cells that produced IFN-γ (percentage of phorbol 12 myristate 13 acetate-(PMA)–NKp44−NKG2A− IFN-γ+) were significantly associated with an increased risk of acquisition (R = −0.549; P = 0.0054; Fig. 4f), whereas IL-17+ NK cells (% Env NKp44+IL-17+) were associated with a reduced risk of acquisition in the multivariate analysis (R = 0.377; Fig. 4g). Collectively, these data show that the two adjuvants induced humoral and cellular responses that differ in their location, frequency and function.
Figure 5.
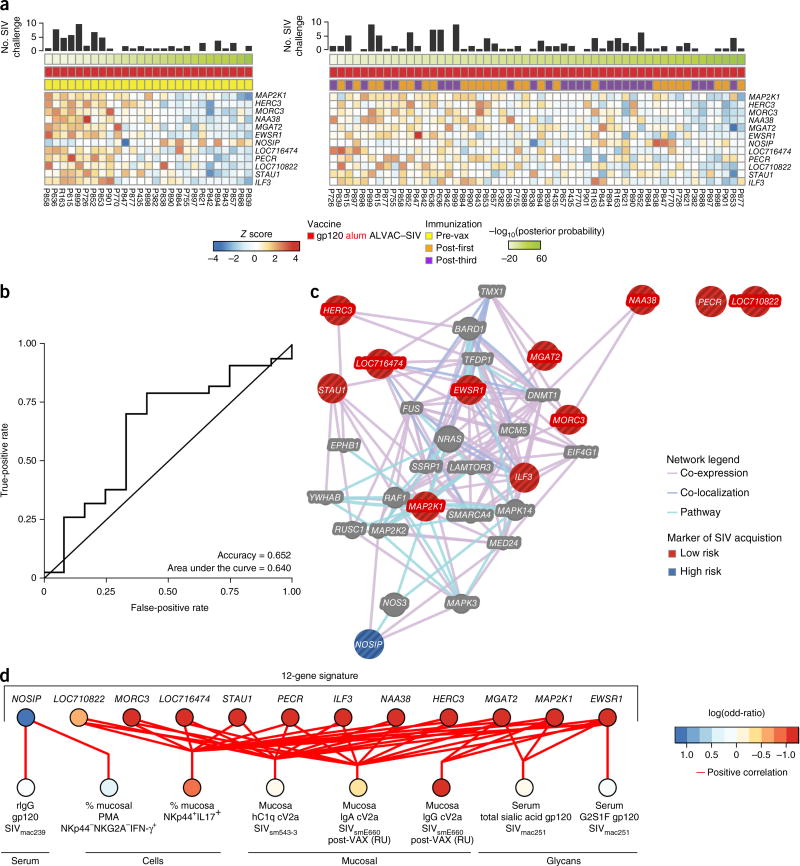
Transcription and immune profiles associated with the risk of SIVmac251 acquisition. (a) Heat map (left) showing the expression of a 12-gene signature predicting protection by the alum group. The 12 genes that compose this signature were identified using the prevax samples and tested on the postvax samples (post-first and post-third) (right). (b) Receiver-operating characteristic curve presenting the accuracies of the 12-gene classifier on the postvax samples. (c) Network inference based on the 12 genes included in the predictive signature. The list of 12 genes was uploaded in GeneMania. Edges are based on co-localization, co-expression and canonical interaction (pathways). Genes associated with a high risk of SIV acquisition in the alum-treated macaques are presented as blue nodes, and genes associated with a low risk of SIV acquisition are presented as red nodes. Gray nodes represent genes associated with the RAS pathway that did not significantly correlate with SIV acquisition. (d) Integrative analysis between the transcriptomic data, the humoral markers and the glycan modifications associated with acquisition. Least-square regression using, as independent variables, the pre-vaccination expression of the 12-gene signature identified to be predictive of protection by the alum-adjuvanted vaccine (c), and as dependent variables, the humoral markers of protection (Supplementary Table 3) was performed using the function spls of the R package mixOmics. The network presents all the pairs of features that are significantly positively correlated with each other (absolute Pearson correlation, R > 0.223; P < 0.05).
12 genes are correlated with decreased SIVmac251 acquisition
We performed microarray analysis of the transcriptome in total PBMCs pre-vaccination (pre-vax), 24 h after the first immunization (post-first) and 24 hours after the third immunization (post-third), in all 54 immunized macaques. We selected the 24-h time point because gene-expression analysis in a pilot study (see Methods) revealed that the innate immune response, as monitored by the expression of IFN-regulated genes17, was strongest at 24 h after immunization, when compared to all post-vaccination time points investigated (data not shown). Pathway analysis using gene set enrichment analysis (GSEA) revealed that the MF59 and alum groups could be grouped into six major functional modules that included RAF signaling, JAK–STAT signaling, T cell activation, cell migration, nuclear factor (NF)-κB signaling, and antiviral signaling (Supplementary Fig. 6). Pathways associated with T cell activation, such as the regulation of IL-2 expression in activated and anergic T cells, and CD28, T cell receptor (CD3) and ICos–ICosL (inducible T cell costimulator) signaling, were strongly induced in macaques that received MF59, confirming the FACS and Fluidigm assessments (Supplementary Table 5). Pathways that encompassed cell-migration genes were also differently expressed in the alum and MF59 groups, which parallels our findings of adjuvant-associated changes in the levels of α4β7+ and CXCR3+ on PB (Fig. 3h,j and data not shown).
We assessed whether the changes in transcriptional profiles were associated with the risk of SIVmac251 acquisition. No transcript measured pre-vaccination, and post-third immunization was significantly associated with protection at the univariate level (Supplementary Table 6). At the multivariate level, additional predictors of protection were not identified in samples collected after immunization (Supplementary Fig. 7), whereas pre-vaccination predictors of protection in the alum arm (131 transcripts) maintained a significant predictive power when tested after vaccination (R = 0.321; P = 0.0299). These data suggest that pre-vaccination immune status dictates (at least partially) which animal will be protected by this vaccine strategy (Supplementary Fig. 7). A naive Bayes classifier was built using transcripts that were correlated with protection in the alum arm—identified pre-vaccination—and the classifier was optimized using cross-validation. Only 12 of the initial 131 transcripts were needed to predict the protection status of the macaques (Fig. 5a). This 12-gene signature predicted time to acquisition in macaques treated with alum with 65% accuracy (Fig. 5b), but it was not associated with acquisition in the MF59 group.
Network analysis revealed that the RAS pathway was directly or indirectly linked to the members of the 12-gene signature (Fig. 5c). Importantly, the 12 genes predictive of decreased viral-acquisition risk were also significantly correlated with humoral and cellular markers associated with protection (P ≤ 0.049; Figs. 2–4, 5d). For example, all of the genes associated with delayed SIV acquisition (all except nitric oxide synthase interacting protein (NOSIP)) were correlated with the frequency of mucosal NKp44+IL-17+ cells (P ≤ 0.049). This association may be related to the fact that MAP2K1 (ref. 18) and ILF3 (refs. 19, 20) are involved in the induction of ILC cytotoxic function, whereas HECT and RLD domain-containing E3 ubiquitin protein ligase 3 (HERC3) and MAP2K1 (ref. 21) are involved in enhancing the function of ILCs (i.e., classical and nonclassical MHC class I). Conversely, NOSIP, a marker of acquisition in the alum group, was correlated with IFN-γ-producing NK cells and thus may be a regulator of Innate responses through its action on nitric oxide22. NOSIP may contribute to the augmentation of IFN-γ-producing ILCs that are negatively associated with protective immunity22, 23. In addition, MGAT2, MAP2K1 and EWSR1, all of which were associated with the delayed SIV acquisition, were correlated with the level of rectal IgG against cV2a of the SIVsmE543-3 divergent strain. Notably, MGAT2, which encodes a glycosyltransferase involved in protein glycosylation branching, that affects antibody function, was correlated with the total levels of sialic acid incorporated into SIV-specific antibody glycosylation, which in turn was positively correlated with alum-conferred delayed SIV acquisition. These findings suggest that there are direct links between the 12 genes upstream of RAS and the mucosal antibodies and cellular markers that are associated with alum-conferred delayed SIV acquisition, and they point to RAS signatures as crucial predictors of protective vaccine profiles in macaques.
RAS in protection by an independent SIV-vaccine modality
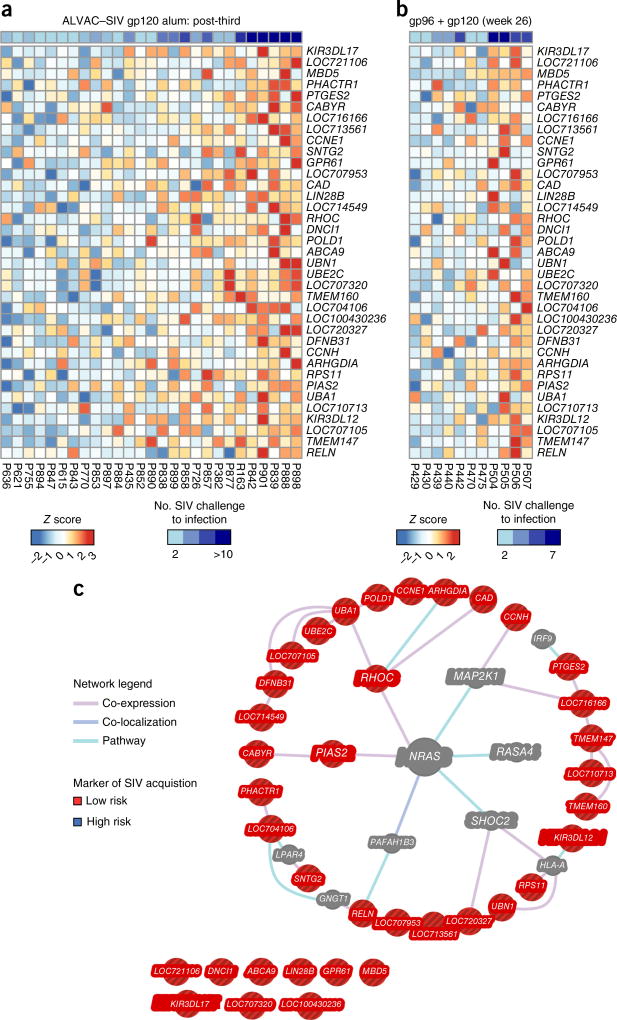
The association of RAS with vaccine-conferred protection from SIV acquisition was observed in an independent data set24, in which we use a nonvectored vaccine composed of intraperitoneal inoculation of HEK293 cells that co-expressed the heat shock protein gp96-Ig chaperoning SIV gag and Env, together with intramuscular boosts with gp120 + gp96-Ig25. This vaccine significantly delayed SIVmac251 acquisition (vaccine efficacy = 68%; P = 0.01) in rhesus macaques25 (Supplementary Table 7). Genes that were positively correlated with delayed SIV acquisition 24 h after the first immunization with ALVAC–SIV (Fig. 6a) were also significantly associated with delayed acquisition induced by the SIV-gp96 + gp120 vaccine24 (GSEA: FDR q value = 0.0435; Fig. 6b). Network analysis (Fig. 6c) revealed that, among the 37 genes associated with vaccine efficacy conferred by both the ALVAC–SIV + gp120 alum and SIV–gp96 + gp120 vaccines, 28 genes were directly or indirectly associated with the RAS pathway. A permutation test was performed to estimate the probability of selecting randomly from all the genes in the macaque genome (14,987 genes) a list of 37 genes in which 28 or more form a part of the RAS pathway. This resulted in a P value of 0.001 (see ‘Vaccine efficacy correlates’ in Methods), which suggests that the association of RAS signaling with vaccine-conferred protection is unlikely to be reproduced by chance alone. Importantly, several components of the RAS pathway, including the MAP2K1 kinase downstream of RAS and the SHOC2 phosphatase that regulates RAS function, are included in the gene-expression signature that is associated with a lowered risk of acquisition (Fig. 6c). Similarly, PIAS-2, an ubiquitin ligase that inhibits JAK–STAT signaling, and RELN (also known as Reelin), a transcription factor that controls plasma cell differentiation, are both linked to RAS signaling26. Finally, the identification of KIR3DL12 and KIR3DL17, which are expressed on NK cells, in this gene signature supports the involvement of ILCs in determining the time of SIV acquisition, as observed in the ALVAC–SIV + gp120 alum arm (Fig. 4e,g). All together, these data demonstrate a complex interplay between a gene signature related to the RAS pathway and innate and adaptive responses that, in turn, are associated with a decreased risk of SIV acquisition.
Figure 6.
RAS-related genes associated with protection conferred by the SIV gp96 + gp120 alum vaccine. Heat-map representation of the expression of the 37 genes associated with protection by ALVAC–SIV + gp120 alum (24-h post-third vaccination) and SIV-gp96 + gp120 (1 week post-third vaccination). Genes that were positively correlated with the number of SIV challenges to infection after ALVAC–SIV + gp120 alum vaccination significantly overlapped with genes associated with the number of SIV challenges to infection after vaccination with SIV-gp96 + gp120 (Supplementary Table 7). Leading-edge analysis revealed that 37 genes were contributing to the enrichment. (a) Heat map of the expression of the 37 genes after vaccination with ALVAC–SIV + gp120 alum. The expression intensities are represented using a blue-white-red color scale. Rows correspond to transcripts, and columns correspond to profiled samples. Samples were ordered by increasing expression of the 37 genes. (b) Heat map of the expression of the 37 genes after SIV-gp96 + gp120 vaccination. (c) Network inference based on the 37 genes associated with protection conferred by the ALVAC–SIV + gp120 alum and SIV-gp96 + gp120 vaccines. List of 37 genes was uploaded in GeneMANIA to test their interaction with NRAS. The principal hub of network, NRAS, was placed in the center of the network, whereas genes directly associated with the RAS pathway were placed in the inner circle, and genes indirectly associated with RAS were placed in the outer circle. Edges correspond to interaction identify by GeneMANIA colored by the type of interaction between genes (co-expression, co-localization or canonical pathway interaction). Nodes represent genes associated with a low risk of SIVmac251 acquisition (red) and intermediary genes identified by GeneMANIA (gray).
DISCUSSION
In this macaque study, we identify aggregate features of innate and adaptive mucosal immune responses as correlates of protection from rectal SIVmac251 challenge by using a vaccine regimen that aims to recapitulate the RV144 trial. We show the importance of the alum and MF59 adjuvants in shaping immune responses, and we demonstrated that the higher vaccine immunogenicity of MF59 does not directly translate to higher vaccine efficacy against SIV acquisition. Whether a delayed exposure to challenge at the end of the immunization might have resulted in a different outcome remains to be ascertained. Similarly, the relative contribution of the adjuvants alone to the transcription profile associated with each vaccine will need further evaluation. We observed that the two regimens studied elicited qualitatively different immune responses, functionally distinct Env-specific systemic CD4+ T cells, ILCs at mucosal sites and Fc-effector profiles. Correlative analysis in the alum group demonstrated associations between the risk of SIVmac251 acquisition and the frequency of mucosal NKp44+IL-17+ ILCs that regulate mucosal integrity and that are functionally impaired in HIV infection27, as well as with the level of mucosal IgG to cyclic V2. Strikingly, within the MF59 group, the mucosal level of IgG to V2 was correlated with an increased risk of virus acquisition, which suggests that antibodies with seemingly similar specificities carry functional differences. Additionally, the two adjuvants differentially affected the fraction of PBs that expressed α4β7. Mechanistically, these differences may relate to the diverse inflammatory profiles of alum and MF59, as demonstrated by a more durable recruitment of neutrophils in the lymph nodes during immunization; the increased production of IFN-γ by SIV-specific CD4+ T cells; and the upregulation of CXCR3 on PBs, all supported by transcriptomic data. The lack of protection conferred by the MF59 regimen was not associated with an increase in the number of mucosal CCR5+ or Ki67+CD4+ T cells elicited by vaccination. Rather, we found that the levels of CD38+ and α4β7+ on vaccine-induced Ki67+CD4+ T cells were correlated with an increased number of virus variants in the MF59 arm (Supplementary Fig. 1l). Thus, it is possible that the qualitative differences in innate and adaptive responses between the two adjuvants, supported also by systems-biology profiling, in combination with the vaccine-induced activation of CD4+ T cell subsets, may account for differences in vaccine efficacy. Ten of the 12 genes that were correlated with vaccine-conferred protection from SIVmac251 acquisition in the alum group were associated with the RAS pathway, which is essential for NK and T cell function28, 29. The RAS pathway acts directly on ILC activity and enhances the expression of NK cell target genes30, 31. Importantly, we confirmed the potential importance of the RAS pathway by demonstrating its association with vaccine efficacy conferred by a mucosal SIV-vaccine regimen based on the combination of heat shock protein gp96-IgG with SIVgp120 (refs. 24, 25). The full understanding of the mechanisms that elicit protective response at mucosal sites, as induced by the SIV-vaccine modalities studied here, will potentially help to guide the design of future vaccines and the choice of adjuvant that is best able to potentiate efficacy and end the HIV epidemic. All our results pertain to this experimental system, and their relevance to humans will require evaluation in human trials.
ONLINE METHODS
Vaccination and viral challenge
All animals used in this study were colony-bred Rhesus macaques (Macaca mulatta), obtained from Covance Research Products (Alice, TX). The animals were housed and handled in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care International. Both care and use of the animals were in compliance with all relevant institutional (US National Institutes of Health) guidelines. The protocol (AUP 491) was approved by the Advanced BioScience Laboratories’s Institutional Animal Care and Use Committee.
A total of 54 juvenile macaques were randomized into two groups of 27 macaques, according to their major histocompatibility status, gender and weight (average, 3 kg). Statistical analysis suggested an 80% probability of detecting significant vaccine efficacy in a range of 30–40% (likelihood ratio test)32. Although the investigators were unblinded, the animal handlers were blinded to the vaccine groups. All 54 macaques were immunized at weeks 0, 4, 12 and 24 with intramuscular inoculations of 108 plaque-forming units (PFU) of ALVAC (vCP2432) expressing SIV genes gag-pro and gp120TM (Sanofi Pasteur). The sequence of the SIV genes was obtained from a mucosally transmitted founder variant of SIVmac251 designated M766r (ref. 33). The gag-pro gene was codon-optimized and constructed using human-codon bias, leaving the stem loop and slippery sequences intact to allow for the one frame-shift required for pol translation. The gag-pro encodes the entire gag, followed by pol, through the end of the protease. The gp120TM gene was codon-optimized and constructed using human-codon bias. The native signal peptide was replaced with a synthetic signal based on the human tissue plasminogen activator (tPa) signal peptide (MDAMKRGLCCVLLLCGAVFVTTTEA). The SIVM766 gp120 from I20 through R527, and the 22-residue gp41 transmembrane (TM) domain from Y695 to L716 follows (numbering is based on SIVmac251). Six residues of the HIV-1LAI cytoplasmic tail (N706 to G711) follow the TM domain (numbering based on HIV-1HXB2 numbering convention).
At weeks 12 and 24, 27 animals consisting of 12 females and 15 males, 5 MamuA*01+ and 4 MamuB*17+ (average weight, 5.4 g), were given a bivalent monomeric-gp120 protein boost in the opposite thigh of the vector immunization, containing 200 µg each of the mucosally transmitted founder variants, SIVmac251-M766 (ref. 33) gp120-gD (referred to in the text as gp120) and SIVsmE660 gp120-gD CG7V (ref. 34) (referred to in the text as gp120), both formulated in alum. The diluted proteins were mixed 1:1 with Alhydrogel 2% (Invivogen, Cat# vac-alu-50) by rocking at room temperature for 10 min. An additional group of 27 animals (13 females and 14 males, six MamuA*01+ and four MamuB*17+; average weight, 6.7 g) were given only 100 µg (as per Novartis’s recommendation) of both gp120-gD proteins in MF59.
This study was planned with the provision of using 24 simultaneous controls that received alum (n = 6), MF59 (n = 12) or nothing (n = 6), as well as 23 historical controls that received alum (n = 11)6 or gp96-Ig (n = 12)25. All controls were housed in the same facility and were challenged by the same operators with the same challenge-virus stock. The control group had ten MamuA*01+ and six MamuB*17+ animals and an equivalent number of females and males. Animals were not screened for TRIM-5α because we previously demonstrated a lack of sensitivity to TRIM-5α restriction of this SIVmac251 stock used at the same dose (120 TCID50)35. 4 weeks after the final immunization, all animals were challenged with 120 TCID50 SIVmac251 via the rectal route. Animals testing negative for SIV RNA in plasma were re-exposed, and a maximum of ten weekly challenges were used. SIVmac251 acquisition did not differ in the simultaneous and historical controls (Supplementary Fig. 1a). A second study was performed to define the kinetic of NKp44+ cells and to perform transcriptome analyses in PBMCs. Six animals were vaccinated with the identical ALVAC–SIV vaccine and boosted with the SIVmac251-M766 gp120-gD and SIVsmE660 gp120-gD CG7V (referred to in the text as gp120). These animals were not exposed to SIVmac251 and were euthanized for tissue collection at 4 weeks after the last immunization.
Single-genome amplification (SGA)
Transmitted founder viruses and their progeny were identified by single-genome amplification (SGA) of SIV RNA from plasma at 2–4 weeks from the infection. SIV RNA was extracted, and limiting-dilution PCR of newly synthesized cDNA was performed33, 34. Transmitted founder-virus lineages were identified by low-diversity sequence and by single sequences with unique mutations. Phylogenetic trees were generated using ClustalW.
Measurement of viral RNA, DNA and CD4+ T cells
SIVmac251 RNA in plasma was quantified by nucleic acid sequence-based amplification, as previously described36. SIV DNA was quantified in mucosal tissues 3 weeks after infection by a real-time qPCR assay with sensitivity up to ten copies × 106 cells, as previously described37. CD4+ T cell counts were periodically determined from whole blood by flow cytometry, as previously described38.
Intracellular cytokine staining (ICS)
Cryopreserved peripheral blood mononuclear cells (PBMCs) were thawed in a 37 °C water bath. The cells were then transferred to pre-warmed R10m (RPMI 1640 by BioWhittaker, Walkersville, MD, 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin G, 100 µg/ml streptomycin with 50 U/ml Benzonase by Novagen, Madison, WI) and washed. Cells were then resuspended at 1–2 million cells per ml in R10 and rested overnight in a 37 °C and 5% CO2 incubator. Cells were stimulated at 1–3 million cells per well in a 96-well v-bottom plate with peptide pools (final concentration of 2 µg/ml) in the presence of GolgiPlug, at a final concentration of 10 µg/ml; (BD Biosciences, San Jose, California) for 6 h. Negative controls received an equal concentration of DMSO instead of peptides. At the end of the incubation, the plate was transferred to 4 °C overnight.
ICS was performed as outlined39. The following monoclonal antibodies were used: BV421 anti-CD4 (OKT4; cat. #317433, 2.5/100 µl), BV570 anti-CD8 (RPA-T8; cat. #301037, 0.62 µl/100 µl) from BioLegend; ECD anti-CD69 (TP1.55.3; cat. #6607110, 2.5 µl/100 µl) from Beckman Coulter; APCCy7 anti-CD3 (SP34.2; cat. #557757, 0.31/100 µl), APC anti-IFN-γ(B27; cat. #554702, 0.62 µl/100µl), PE anti-IL-2 (MQ1–17H12; cat. #554566, 0.31 µl/100 µl), FITC anti-TNF-α (Mab 11; cat. #554566, 0.62 µl/100 µl) from BD Biosciences. Aqua LIVE/DEAD kit (Invitrogen, Carlsbad, CA) was used to exclude dead cells. All antibodies were previously titrated to determine the optimal concentration. Samples were acquired on an LSR-II flow cytometer and analyzed using FlowJo version 9.6.3 (Treestar, Inc., Ashland, OR).
Fluidigm
PBMCs from vaccinated NHP (n = 9 per group) were stimulated with 2µg/ml Env m766 overlapping peptide pool or DMSO for 6 h before surface staining. Flow cytometric cell sorting was performed on a 20-parameter FACSAria (BD), running FACSDiVa software (version 6.1.3, BD). T cells were discriminated by staining PBMCs with the following fluorescently labeled, anti-human monoclonal antibodies: BV421-CCR7 (G043H7; cat. #353208, 10 µl), BV510-CD19 (HIB19; cat. #302242, 8 µl), BV510-CD8 (RPA-T8; cat. #301048, 0.3 µl), BV510-CD14 (M5E2: cat. #301842, 2 µl), BV605-PD1 (EH12.2H7; cat. #329924, 3 µl), PE-CCR6 (G034E3; cat. #353410, 0.5 µl) and PECy7-ICOS (C398.4A; cat. #313520, 0.5 µl) from BioLegend; APC-CD95 (DX2; cat. #558814, 0.5 µl), Ax700-CXCR3 (1C6; cat. #561320, 5 µl), APCCy7-CD3 (SP34-2; cat. #557757, 4 µl), PECy5-CD154 (TRAP1; cat. #555701, 10 µl) from BD Bioscience; PECy5.5-CD4 (S3.5; cat. #MHCD0418, 2 µl) from ThermoFisher Scientific; ECD-CD28 (CD28.2; cat. #6607111, 1 µl) from Beckman Coulter, Ax488-CXCR5 (710D82.1; NIH NHP Reagent Program, 5 µl) and Aqua Blue LIVE/DEAD Fixable Dead Cell Stain (Invitrogen). CD154+CD4+ T cells were sorted at 25 cells each well for pooled cell (“nanoarray”) dynamic RT-qPCR array (Fluidigm) and gene-expression profiles were quantified using a panel of 96 gene-specific primers, as previously described40. The results were analyzed using JMP version 10.
IgG- and IgA-binding antibody assay
The SIV Env-specific IgG and IgA antibodies in serum and rectal mucosa were determined by a custom SIV bAb multiplex assay (SIV-BAMA), as previously described3, 41. For mucosal samples, specific activity was calculated by the ratio of MFI (linear range of standard curve)/µg/ml total macaque IgG, which was measured by macaque IgG or IgA ELISA to normalize the data to recovery of antibody. Rectal swab samples in solution were spun, filtered and concentrated down to approximately half of the starting volume. The samples were examined for blood contamination and measured for semiquantitative evaluation of hemoglobin. Purified IgG (DBM5) from a SIV-infected macaque (provided by M. Roederer, VRC, NIH) was used as the positive control to calculate SIV antibody concentration. Positive controls for each antigen were tracked via a Levy Jennings Plot. Specific activity was calculated from the total macaque IgG levels and the SIV-specific concentrations. Antibodies against native V1/V2 epitopes were quantitated by binding assays against native SIV V1/V2 antigens expressed as gp70-fusion proteins related to the CaseA2 antigen used in the RV144 correlate study (provided by A. Pinter). These proteins contained the glycosylated, disulfide-bonded V1/V2 regions of SIVmac239, SIVmac251 and SIVsmE660 (corresponding to AA 120–204 of HXB2 Env), fused to residue 263 of the SU (gp70) protein of Fr-MuLV.
Reagents and surface plasmon resonance
SIV-V2 peptide was synthesized by JPT Peptide Technologies GmbH, Berlin, Germany. The peptide was allowed to fold and cyclize under thermodynamic control at high dilution, and the purity was determined to be greater than 90% by high-performance liquid chromatography and mass spectrometry. The amino acid sequence of the SIV V2 peptide is based on the SIVsmE543-3 V2 domain from GenBank accession number U72748. The SIV-V2 peptide sequence contains an N-terminal biotin tag and the sequence is as follows:
GF SIVsmE543 CIKNNSCAGLEQEPMIGCKFNMTGLKRDKKIEYNETWY SRDLICEQPANGSESKCY. GF SIVmac251 full CIAQNNCTGLEQEQMISCKFN MTGLKRDKTKEYNETWYSTDLVCEQGNSTDNESRCY.
CM5 chips and the Biacore amine coupling kit were purchased from GE Healthcare, Piscataway, NJ, USA. Streptavidin was purchased from Invitrogen, Grand Island, NY. Affinity purified goat anti-monkey IgG and IgA (gamma-chain- or alpha-chain-specific) antibody was purchased from Rockland Immunochemicals, Gilbertsville, PA.
Surface plasmon resonance (SPR) measurements were conducted with a Biacore T200 using the CM5 chip, as described previously6. Streptavidin was immobilized onto the chip using the amine coupling kit, as directed by the immobilization wizard packaged within the T200 control software. 6,700 reported response units (RU) of 1 µM streptavidin in 20 mM sodium formate, pH 4.2 (10 min contact time, 10 µl/min flow rate) was immobilized. The biotinylated peptide was prepared at a concentration of 1 µM in 20 mM TRIS, pH 7.4 and allowed to flow (at 10 µl/min) over the streptavidin-coated surface of flow cell 4, until 3,500 RU of SIV V2 peptide were captured.
The mucosal swabs were thawed on ice and centrifuged at 16,100 rcf, 4 °C, for 5 min. The supernatant was diluted tenfold in TBS, pH 7.4, and then analyzed on the Biacore. The diluted mucosal samples were passed over the chip surface at a flow rate of 30 µl/min for 3 min, followed by a 5-min dissociation period. A 20 µg/ml solution of affinity-purified, gamma-chain-specific goat anti-monkey IgG or IgA antibody was passed over the peptide-coated, Ig-bound surface for 2 min at a flow rate of 10 µl/min. After a 70-s dissociation period, the chip surface was regenerated and data were analyzed, as previously described, using the BIAevaluation 4.1 software6. The RU for the IgG-specific or IgA-specific values are the difference between the average value of a 5-s window taken 60 s after the end of the anti-IgG or anti-IgA injection and the average value of a 5-s window taken 10 s before the beginning of the anti-IgG or anti-IgA injection. The data (RU) are presented as dot plots for individual mucosal samples.
To determine total IgG antibodies in the mucosal samples, anti-IgG immobilization on a CM5 chip was performed using 100 nM of unconjugated gamma-chain-specific goat anti-monkey IgG (Rockland, Inc., Gilbertsville, PA) in 20 mM sodium acetate, pH 4.2, with a 5-min contact time and 10 µl/min flow rate, resulting in the immobilization of 9,100 RU. Centrifuged mucosal samples (diluted 1:10) were passed over the chip surface at a flow rate of 30 µl/min for 3 min, followed by a 5-min dissociation period. The relative amount of monkey IgG was determined using the same secondary injection and analysis strategy described above.
Chemstrips were used to determine the blood contamination in mucosal samples. 10 µl of the mucosal supernatant sample was spotted onto a Chemstrip 5 OB Urine Test Strip (Roche, cat. #11893467-160). After 60 s, any change in color was recorded for comparison to the manufacturer’s color chart.
Neutralizing antibodies
Neutralization was measured as a reduction in luciferase-reporter gene expression after a single round of infection in TZM-bl cells, as described previously42, 43. TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu. 200 TCID50 of virus were incubated with serial threefold dilutions of test sample in duplicate, in a total volume of 150 µl for 1 h out of 18 h at 37 °C, as indicated, in 96-well flat-bottom culture plates. Freshly trypsinized cells (10,000 cells in 100 µl of growth medium containing 75 µg/ml DEAE-dextran) were added to each well. One set of control wells received cells and virus (virus control), and another set received cells only (background control). After 48 h, 100 µl of cells was transferred to 96-well, black solid plates (Costar) for measurements of luminescence using the Britelite luminescence reporter-gene assay system (PerkinElmer Life Sciences). Neutralization titers are the dilution at which relative luminescence units (RLU) were reduced by 50%, as compared to those in virus control wells, after the subtraction of background RLUs.
Assay stocks of molecularly cloned Env-pseudotyped viruses (SIVmac251 CS.41, SIVmac239 0.23, SIVmac251 WY:30) were prepared by transfection in HEK293 T cells and were titrated in TZM-bl cells, as previously described44. Assay stocks of uncloned TCLA-SIVmac251 and SIVmac251 CS/2002 letvin were produced in H9 and human PBMCs, respectively, and were titrated in TZM-bl cells.
Antibody-dependent, cell-mediated cytotoxic activity
ADCC activity in serum samples collected at week 27 was measured using the RFADCC assay, as previously described45. Briefly, SIVmac251 gp120 protein (Advanced Bioscience Laboratories, Inc., Rockville, MD) was used to coat CEM.NKr target cells, which were then co-cultured with human PBMC effectors at an E:T ratio of 50:1. CEM. NKr cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: CEM.NKR Cells. Serial dilutions of macaque sera were tested for ADCC activity in a 4-h assay, and the ADCC titer was defined as the highest tenfold serum dilution that generated ADCC activity above the background cutoff (mean ADCC activity over a dilution series of a pool of macaque-negative sera plus 3 s.d.). ADCC-mediated maximum percent killing of target cells (% ADCC max killing) was defined for each positive sample as the highest percent killing mediated at any of the dilutions tested. Sera with percent killing below the cutoff value were scored negatively.
Phagocytosis
SIVmac251 virus stock was concentrated with Lenti-X Concentrator (Clontech, Mountain View, CA), according to the manufacturer’s protocol, and resuspended in complete medium (RPMI-1640 supplemented with 10% FBS and Pen-Strep l-glutamine. Fluorescein isothiocyanate (FITC)-labeled virus was prepared by following the protocol of Hartshorn et al.46. Briefly, concentrated virus stock was incubated with FITC at a 10:1 (virus: FITC) ratio by volume for 1 h and dialyzed overnight against PBS. FITC-labeled virus was stored in single-use aliquots in liquid nitrogen until use.
FITC-labeled virus (10 µl, based on previous titration; data not shown) was incubated in screw-cap Eppendorf tubes with heat-inactivated serum from pre-immunized and week 26 samples (1:100 final) in complete medium for 1 h at 37 °C in 250 µl total volume. Tubes were spun at 21,920g at 4 °C for 1 h. Without touching the bottom of the tube, the top 200 µl of supernatant from each tube was removed and discarded. 200 µl of fresh complete medium was added to each tube and vigorously vortexed. 100 µl of from each tube was dispensed into duplicate wells of a 96-well round-bottom microtiter plate. 100 µl of complete medium was added to two separate wells to serve as the cell-only control. 100 µl of THP-1 cells (20,000 cells) were added to each well, and the plate was incubated for 1 h on an orbital shaker at 37 °C. THP-1 cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID and were routinely tested for mycoplasma contamination with a Sigma Mycoplasm PCR kit. After 1 h, the cells were washed three times with PBS, trypsinized to remove bound but not internalized FITC-labeled virus, and resuspended in 150 µl of 4% paraformaldehyde. The microtiter plate was stored at 4 °C in the dark overnight. The next day, the entire volume of each well was transferred to snap-cap Eppendorf tubes and read on an Accuri C6 flow cytometer using CFlow Plus software. Data were collected and analyzed by setting the FL1 gate at less than 1% for the cell-only control and multiplying the percent positive cells by the median MFI to obtain the phagocytic index. Net phagocytic index is reported and was obtained by subtracting the mean of the indices from a pool of 16 pre-immunized serum samples.
Phenotypic analyses of plasmablasts
The frequency of PBs was measured in the blood of 23 macaques vaccinated with ALVAC–SIV + gp120 MF59 and 22 macaques vaccinated with the ALVAC–SIV + gp120 alum regimen, before vaccination and at 7 d after the last immunization. Not less than 10 million cells at a time were stained with Ax700 anti-CD3 (SP34-2; cat. #557917, 1:80 dilution), anti-CD14 (M5E2; cat. #557923, 1:50 dilution), anti-CD16 (3G8; cat. #557920, 1:85 dilution), anti-CD56 (B159, cat. #557919, 1:50 dilution), PECy7 anti-CD21 (B-ly4; cat. #561374, 1:85 dilution), PE anti-Ki67 (B56, cat. #556027, 1:25 dilution), PerCPCy5.5 anti-IgM (G20–127; cat. #561285, 1:50 dilution); Qdot605 anti-IgG (G18–145; cat. #562025, 1:50 dilution) and PECF594 anti-CXCR3 (1C6; cat. #562451, 1:50 dilution) from BD Biosciences; PECy5-CD19 (J3–119; cat. #IM2643U, 1:30 dilution, Beckman Coulter); 650-CD20 (2H7; cat. #95–0209, 1:50 dilution, eBiosciences); FITC-CD38 (AT-1; cat. #10415, 1:250 dilution, StemCell); BV421-CD39 (MOCP-21; cat. #328214, 1:50 dilution, BioLegend); IgA-Texas Red (polyclonal; cat. #2050-07, 1:50 dilution, SouthernBiotech) and APC-α4β7 (cat. #11718, 1:50 dilution, NIH AIDS Reagent Program, Division of AIDS, NIAID). Cells were permeabilized with Cytofix/Cytoperm (BD Biosciences). Acquisition was performed on LSRII (BD Biosciences), and data were analyzed by FlowJo software (TreeStar). PBs were gated as lineage-negative (CD3-CD14-CD16-CD56-) CD20+CD21-Ki67pos/high (CD38pos/high CD39+)47. Cells in this gate were positive for IgG, IgA and IgM, thus validating this panel to detect both PBs and plasma cells in macaques. The frequency of PBs expressing CXCR3 or α4β7 was calculated.
The frequency of PBs expressing CXCR3 or α4β7 was similarly assessed in PBMCs collected at 2 weeks after the final immunization from 17 volunteers vaccinated with ALVAC-HIV + gp 120 alum (RV135) and 17 vaccinated with ALVAC-HIV+ gp120 MF59 regimen (RV132). Both RV135 and RV132 studies had a double-blind placebo-controlled phase 2, and were approved by the appropriate Institutional Review Boards, specifically by four ethical-review committees in Thailand and the United States ((i) Ministry of Public Health Ethical Review Committee of Research in Human Subjects (MoPH EC)—FWA00001953; (ii) Institutional Review Board, Royal Thai Army Medical Department (IRBRTA)—FWA00001813; (iii) Mahidol University Central Institutional Review Board (MU-CIRB)—FWA 00000926; (iv) Office of Research Protections, Human Research Protection Office (ORP HRPO)) and by the National AIDS Commission of Thailand. Volunteers were healthy, HIV-negative Thai adults, and written informed consent was obtained. The vaccines used in those trials were ALVAC-HIV (vCP1521): Recombinant canary-pox vector expressing gag and protease subtype B (LAI) and Env-gp120 CRF01_AE (92TH023) linked to the transmembrane-anchoring portion of subtype B gp41 (LAI) genes (Sanofi Pasteur), bivalent HIV-1 gp120 SF2 50 µg (subtype B) and gp120 CM235 100µg (CRF01_AE) adjuvanted with MF59 (Novartis) and AIDSVAX B/E: bivalent gp120 MN 300 µg (subtype B) and gp120 A244 300 µg (CRF01_AE) adjuvanted with alum (Global Solutions for Infectious Diseases, GSID, South San Francisco, CA)10, 48.
In the human samples, B cells were defined as CD20+CD19+ CD27+. PBMCs were collected from vaccines before and 2 weeks after the last immunization.
Phenotypic and functional analysis of mucosal ILCs
Rectal biopsies were obtained 1 week after the last immunization (week 25). Mononuclear cells were isolated from rectal biopsies. For the phenotypic characterization, 3 × 106 cells were used, and flow cytometry staining was carried out for cell-surface and intracellular molecules using standard protocols. After this, anti-human fluorochrome-conjugated mAbs known to cross-react with rhesus macaques were used for the staining: AlexaFluor 700 anti-CD3 (SP34-2, cat. #557917, 0.2 mg/ml), APCCy7 anti-CD8 (SK1, cat. #561945, 5 µl) PerCPCy5.5 anti-CD4 (L200; cat. #552838, 5 µl) from BD Biosciences (San Jose, CA); V450 anti-TNF-α (Mab11; cat. #502920, 5 µl), NKp44 (P44-8; cat. #325110, 5 µl), PECy5 anti-CD40L (24–31; cat. #310808, 5 µl) 605NC anti-IL-2, (17H12; cat. #500331, 50 µg/ml), 605NC anti-CD20 (2H7, cat. #302334, 50 µg/ml) from BioLegend (San Diego, CA); PE-CY7 anti-IL-17 (eBio64DEC17; cat. #25-7179-42, 0.125 µg), FITC anti-CD107a (eBioH4A3; cat. #11-1079-42, 5 µl, 0.5 µg) from eBioscience (San Diego, CA); PE anti-NKG2A (Z199; cat. #PN A60797, 5 µl) from Beckman Coulter (Fullerton, CA). The yellow and aqua LIVE/DEAD viability dyes (Invitrogen, cat. #L34959, 1 µl) were used to exclude dead cells. At least 50,000 CD3− singlet events (were acquired on a LSR II (BD) and analyzed using FlowJo Software (TreeStar). Mucosal NK cell were gated on live lymphocytes, which were negative for CD20 and CD3 and further classified on the basis of the expression of NKG2A and NKp44 surface markers15, 16.
Phenotypic analysis of activated CD4+ T cells in mucosa and blood
Mononuclear cells isolated from blood at week 13 from nine animals per group were stained with PerCPCy5.5 anti-CD4, Ax700 anti-CD3, FITC anti-Ki67 and BV650 anti-CCR5 (3A9, 5 µl) all from BD Bioscience and APC anti-α4β7 and PE-anti-CD38, provided by the NIH Nonhuman Primate Reagent Resource (R24 OD010976, and NIAID contract HHSN 272201300031C) to assess CD4+ T cell activation. The same antibody concentrations and clones, staining methods, live/dead discriminant, instruments and analysis software described in the PB section above were used here. Gating was done on live CD3+CD4+ cells, and the frequency of Ki67+ and CCR5+ cells was calculated on CD4+ T cells. The frequency of α4β7 and CD38 was calculated on Ki67+CD4+ T cells.
Cells isolated from rectal mucosa of six animals per group were stained to measure the expression of CCR5+ on CD4+ T cells. Cells were stained with CD3, CD8 and CCR5 (CTC5; 1:20 dilution, R&D), and the frequency of live CD20-CD3+CD8-CCR5+ cells was calculated.
Immunohistochemistry on rectal mucosa and lymph nodes
Rectal mucosa tissue was collected from all macaques at 1 week after the last immunization and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) and embedded in paraffin.
TBS with 0.05% Tween 20 was used for all washes. Antibody diluent (Dako, Inc., Carpinteria CA) was used for all antibody dilutions. The primary antibodies used included monoclonal antibody (mAb) anti-CD4 mouse IgG1 (IF6; Leica Microsystems, Bannockburn, IL) dilution 1:20, and polyclonal anti-Ki67 rabbit serum (Neomarker, Fremont, CA) dilution 1:300. For all primary antibodies, slides were subjected to an antigen-retrieval step consisting of incubation in AR10 (Biogenex Inc, San Ramon, CA) for 2 min at 125 °C in the Digital Decloaking Chamber (Biocare Medical, Concord, CA), followed by cooling for 10 min to 90 °C before rinsing in water and a final buffer rinse. Primary antibodies were replaced by normal rabbit IgG or mouse IgG (Invitrogen, Grand Island, NY) and included with each staining series as the negative control. Nonspecific binding sites were blocked with 10% goat serum and 5% bovine serum albumin (BSA, Jackson ImmunoResearch, West Grove, PA). Binding of the primary antibodies was detected simultaneously using 1:500 dilution Alexa Fluor 488-labeled polyclonal goat anti-rabbit IgG and 1:500 dilution Alexa Fluor 568-labeled polyclonal goat anti-mouse IgG (Molecular probes, Grand Island, NY), for 1 h. All slides were coverslipped using ProLong Gold with 4′, 6-diamidino-2-phenylindole dihydrochloride hydrate (DAPI, Molecular probes, Grand Island, NY) to stain nuclei. All the control experiments gave appropriate results with minimal nonspecific staining (data not shown).
Slides were visualized with epifluorescent illumination using a Zeiss Axioplan 2 microscope (Carl Zeiss, Inc., Thornwood, NY) and appropriate filters. Digital images were captured and analyzed by using Openlab software (Inprovision, Waltham, MA).
Five high-power (40×) microscope field lamina propria from rectal sections were randomly chosen and captured digitally with the system described above. Each captured field includes an area of approximately 0.04 mm2. Only clearly positive cells with distinctly labeled nuclei (DAPI) and bright staining were considered to be positive. Individual positive cells in the five captured high-power microscope fields of the immunohistochemical stained tissue sections were counted manually by a single observer. The numbers of positive cells are presented as cells per square millimeter.
For the detection of neutrophils in lymph nodes, sections from biopsies (8 µm) were deparaffinized and rehydrated in xylene and ethanol gradient and incubated in acetone before staining, as described49. The sections were incubated with neutrophil elastase (Dako followed by the anti-mouse (Dako) or anti-rabbit (Vector Laboratories)) biotinylated secondary antibodies. The peroxidase system Vectastain Elite ABC kit together with substrate diaminobenzidine tetrahydrochloride (Vector Laboratories) was used for detection. Cell nuclei were counterstained with Mayer’s Haematoxylin. Quantification was performed blinded using a Leica DMR-X microscope (Leica Microsystems GmbH, Wetzlar, Germany).
The number of positive mAb-labeled cells/mm2 of tissue sample was quantified using a Zeiss AxioCam HRc digital camera mounted on a Zeiss microscope (Zeiss, Jena, Germany) fitted with a 40× plan neofluar objective and a polarizing filter cube with appropriate filters (Omega Optical, Brattleboro, VT). Digital images were captured with Openlab software (Improvision, Lexington, MA). One section per tissue with representative histomorphological components (cortex, paracortex, follicles, medulla for LN; and epithelium, lamina propria, and muscularis mucosa for rectum) was analyzed. Five high-power (×40) microscope fields of the T cell-rich zone (paracortex) per LN section and five high-power fields of lamina propria from rectal sections were randomly chosen and captured digitally with the system described above. Each captured field includes an area of approximately 0.04 mm2. Only clearly positive cells with distinctly labeled nuclei (4′, 6-diamidino-2-phenylindole dihydrochloride hydrate) and bright staining were considered to be positive. Individual positive cells in the five captured high-power microscope fields of the immunohistochemical stained tissue sections were counted manually by a single observer. The numbers of positive cells are presented as cells per square millimeter.
Antibody-dependent cellular phagocytosis (ADCP)
SIV-specific phagocytic activity was assessed using a flow cytometry-based phagocytic assay as described previously50. Briefly fluorescent streptavidin microspheres were coated with biotinylated rgp120SIVmac251 protein (Immune Technology), SIVsmE543-3 cV2a or SIVsmE543-3 cV2c cyclic peptide, and the ability of purified IgG antibodies to drive uptake by the monocytic THP-1 cells was assessed.
Antibody-dependent complement deposition (ADCD)
Antibody-dependent complement deposition was assessed by measurement of complement component C3b on the surface of target cells. CD4-expressing target cells were pulsed with rgp 120 SIVmac251 (60 mg/ml) and incubated with antibodies. Purified IgG was combined with pulsed or unpulsed target cells and freshly harvested HIV-negative donor plasma diluted with veronal buffer 0.1% gelatin (1:10 dilution) to allow for complement deposition. Replicates using heat-inactivated donor plasma were used as negative controls. Cells were incubated for 20 min at 37 °C and washed with 15 mM EDTA in PBS. Complement deposition was detected by staining for C3b-FITC (Cedarlane). Cells were fixed and the proportion of target cells with C3b-FITC deposition was analyzed by flow cytometry.
Antibody-dependent NK cell activation
An assay to determine the expression of surface CD 107a and intracellular production of IFN-γ and MIP-1β was performed by pulsing the CEM-NKr CCR5+ T lymphoblast cell line with rgp120SIVmac251 (60 mg/ml), as previously described51. NK cells were isolated from whole blood from healthy donors using negative selection with RosetteSep (STEMCELL Technologies), as recommended by the manufacturer. The CEM-NKr cells and isolated primary NK cells were mixed at a ratio of 1:5, and purified IgG, anti- CD107a-phycoerythrin (PE)-Cy5 (BD), brefeldin A (10 mg/ml) (Sigma-Aldrich), and GolgiStop (BD) were added for 5 h at 37 °C. The cells were then first stained for surface markers using anti-CD 16-allophycocyanin (APC)-Cy7 (BD), anti-CD56-PE-Cy7 (BD) and anti-CD3-Alexa Fluor 700 (BD) and then stained intracellularly with anti-IFN-γ-APC (BD) and anti-MIP-1β-PE (BD) using Fix and Perm A and B solutions (Invitrogen). The cells were then fixed in 4% paraformaldehyde and analyzed using flow cytometry. NK cells were defined as CD3− and CD16− and/or CD56+.
SIV-specific antibody-effector array
A customized multivariate Luminex assay was developed using a panel of SIV antigens coupled to carboxylated fluorescent beads (Luminex Corp.), as described previously52. Fc-gamma receptors (human FcgRIIa, FcgRIIb, FcgRIIIa, FcgRIIIb and Rhesus R2A-1 and 2A-2) were produced in HEK293 cells, and purified via Ni2+ ion-affinity chromatography, as described previously53. Human C1q was purchased from Sigma-Aldrich (C1740-.5MG). FcRs and C1q were biotinylated using EZ-Link Sulfo-NHS-SS-Biotin (Pierce 21331) at a molar ratio of 5 M biotin per M of protein. Biotinylation was carried out for 2 h at RT in Tris pH 8.2, with a protein concentration of 0.2 mg/ml. Afterwards, the excess biotin was removed via dialysis. Immediately before use, the biotinylated FcR or C1q was mixed with a 1:4 molar ratio of Streptavadin-PE (Prozyme cat. #PJ31S), and diluted to a final concentration of 1.0 µg/ml FcR/C1q in Luminex Assay Buffer (PBS + 0.1% BSA + 0.05% Tween).
Coupled microspheres were premixed in Assay Buffer, creating a working mixture of 12.5 microspheres per bead type, per µl. Using a black, clear bottom 384-well plate (Greiner Bio One, 781906) 40 µl of the working microsphere mixture (500 beads of each type/well) were added to 10 µl of either 100× diluted serum or rectal mucosal supernatant (all dilutions in PBS). The plate was covered and incubated for 2 h at room temperature on an XYZ plate shaker (IKA). The plate was washed five times with 65 µl of Assay Wash (PBS-1X, 0.1% BSA, 0.5% Triton-100X) using a plate-washing system (BioTek 405). Antigen-specific antibody binding to FcgRs or lectins was detected using the tetrameric PE-conjugated detection reagents described above, at 1.0 µg/ml with 50 µl/well. After 1 h of incubation at room temperature on a shaker, the plate was washed five times with 65 µl of Luminex sheath fluid, and microspheres were resuspended in 35 µl of sheath fluid.
A Bio-plex array reader (FlexMap 3D, Bio-Plex Manager 5.0, Bio-Rad) detected the microspheres, and binding of PE detector antibody was measured to calculate a median fluorescence intensity (MFI). Background signal, defined as the average MFI observed for each microsphere set when incubated with the PE-conjugated detection reagent in the absence of clinical antibody sample, was subtracted from the MFI for each sample.
Glycan analysis
20 µg of purified IgG were denatured and treated with PNGase enzyme (NEB) to release N-linked glycans. Proteins were precipitated in ice-cold ethanol, and the glycan-containing supernatants were dried in a Centrivap. Dried glycans were fluorescently labeled with a 1:1 ratio of 50 mM APTS (8-aminoinopyrene-1,3,6-trisulfonic acid; Life Technologies) in 1.2 M citric acid and 1 M sodium cyanoborohydride in tetrahydrofuran (Sigma-Aldrich) at 55 °C for 2 h. Labeled glycans were dissolved in ultrapure water, and excess unbound APTS was removed using Bio-Gel P-2 (Bio-Rad) size-exclusion columns. Samples were run with a LIZ 600 DNA ladder in Hi-Di formamide (Life Technologies) on an ABI 3130Xl DNA sequencer. Data was analyzed using GeneMapper software, and peaks were assigned on the basis of the migration of known standards and glycan digests. Peak area was calculated and used to determine the relative percentage of each glycan structure.
Integrated humoral profiles and correlates of protection analyses
All vaccine data was centered and scaled using the R statistical language (Version 3.0.2) (Team, R.C. A language and environment for statistical computing, 2013). Luminex assay data was preprocessed, as previously described54, and then organized and labeled to quantify antibody-binding signals between antibody-detection reagents (for example, IgG, C1q, rhFc-γR2a, etc.) and specific antigens (for example, gp120, gp140, PR55 gag, etc.) tested.
Unsupervised analysis of antibody and functional-assay correlations with conferred protection, as determined by the number of challenges to infection, was performed in R, using the stats package (Version 3.0.2) cor function. Pearson-correlation coefficients were calculated using pairwise complete observations and visualized in correlation plots generated with the corrplot package (Version 0.71).
Hierarchical-clustering analysis of antibody-binding measurements observed in plasma and mucosal samples for alum and MF59 was performed in R. Scaled and normalized measurements for antibody features were truncated at ± 6 s.d. (SD), and Pearson-correlation coefficients were calculated for all plasma and mucosal antibody features measured. Antibody features were clustered using the Ward Linkage. Correlations and clustering were visualized using the gplots (Version 2.11.3) heatmap.2 function.
Classification analyses was performed both by Partial Least-squares Discriminant Analysis (PLS-DA) using the Mathworks Statistics Toolbox for MATLAB (version R2014a), and by Regularized Logistic Regression, using the R glmnet package (Version 2.02).
PLS-DA is an asymmetric ‘discriminative’ form of supervised principal-component analysis in which the response variable indicates the class or category of the samples, in this case, alum or MF59. PLS-DA was performed using all measured parameters, as well as subsets of parameters, to explore how well subject class could be predicted using Fc Function data, plasma and mucosal Luminex data, combined V2-specific plasma Function and Luminex data, as well as plasma gp120-specific glycan profiles. PLS-DA models were illustrated using MATLAB.
Logistic regression is a generalized linear model for binary classification. Here it was used to identify which measured parameters (Fc Function, Luminex antibody-binding affinities observed in plasma and mucosal samples, glycan profiles, V2-specific Fc Function and Luminex, etc.) best predict protection in alum or MF59 animals that became infected after ≤4 challenges (nonprotective) versus ≥5 challenges (protective). Logistic regression models were constructed using a Lasso-style regularization penalty to promote sparse models (relying on a few dominant discriminative features and mitigating the risk of over fitting); the penalty was chosen via cross-validation, identifying the largest penalty, such that classification error is within 1 standard error of the minimum (i.e., yielding a sparse model with low error). Subjects were weighted to account for class imbalance.
Note that “full models” using all samples may be overly optimistic in characterizing the ability to differentiate classes. To assess generalizability of the differentiation, within the limits of the available data, a leave-one-out cross-validation (LOOCV) was performed with the same regularized logistic regression approach, fixing the penalty to that used for the full models. In LOOCV, a model is trained on all but one sample and the predictive accuracy is assessed for that left-out sample, repeating to leave out each sample and average the accuracies.
Logistic-regression predictions were illustrated in terms of log-odds ratios between the two classes, with negative numbers for one class and positive numbers for the other and relative magnitudes indicative of confidence. The results were visualized using GraphPad Prism (Version 6.0) and the R package ggplot2 (version 1.0.1).
Gene-expression analysis
A preliminary study including six rhesus macaques immunized at week 0 and week 4 with ALVAC-SIV and boosted at week 12 and week 24 with ALVAC + alum was conducted to identify the best time point to investigate immune response after immunization. Samples were taken pre-immunization, 16, 24, 48 and 72 h after each of the four immunizations. Gene-expression analysis revealed that regulation of IFN-regulated genes17, a mark of innate immune response, was the strongest at the 24 h time point, as compared to pre-immunization, versus the other three time points investigated (data not shown). Thus, the 24-h post-immunization was used for the full transcriptomic study. Microarray analysis was conducted using biotinylated cRNA hybridized to Human HT-12 version 4 BeadChips (Illumina). The arrays were scanned using Illumina’s iSCAN and quantified using Genome Studio (Illumina). Analysis of the Genome Studio output data was conducted using R/Bioconductor software packages55. Briefly, missing bead intensities (<2% of the expression data) were imputed using the nearest-neighbor averaging method56. Quantile normalization was applied to the raw intensities, followed by a log2 transformation. Principal-component analysis was used to reduce the dimensionality of the expression data and to generate plots for evaluation of similarities between sample sets. The LIMMA package was used to fit a linear model to each probe and to perform moderated t tests and F tests on the compared groups (Smyth, G.K. “Bioinformatics and computational biology solutions using R and Bioconductor”, 2005). To control the expected proportions of false positives, each nominal P value was adjusted using the Benjamini–Hochberg method51. Transcripts were deemed to be significantly associated with a treatment if the correlation between their expression and the class distinction (for example, postvax versus prevax) resulted in an adjusted P value below 0.05. GSEA was used to annotate genes and rank canonical pathways57. In brief, genes were ranked by the absolute correlation (depicted by the LIMMA absolute t statistic) between their expression and the class distinction. Given a defined set of genes (for example, genes members of a signaling pathway), the goal was to determine whether the members of that gene set are found at the top of the list, implying they are not randomly distributed across the ranked list. An Enrichment Score was calculated to quantify the degree to which the gene set is over-represented at the top of the entire ranked list. A gene-based permutation test procedure was used to estimate a false-discovery rate for a given enrichment score. The collection of gene sets used in this analysis consisted of Ingenuity canonical pathways. Significantly enriched pathways were organized in networks using the enrichment maps strategy58. This was accomplished by linking gene sets by the number of genes differently expressed overlapping between gene sets. Overlap between significant gene sets is computed according to the Jaccard index. A Jaccard index of 0.5 was used to generate the enrichment maps presented in Supplementary Figure 6.
The microarray data presented in this article have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GEO: GSE72624.
Vaccine-efficacy correlates
Gene-expression signatures of protection were built for each combination of vaccination and immunization step and tested on the remaining samples. As a first step, genes significantly associated with the number of challenges to infection (P ≤ 0.01), in a given combination, were identified. Transcripts not annotated to any genes were omitted from further analysis. As a second step, this set of genes was used to order the samples from other combinations on the basis of their mean–rank expression of those genes (i.e., samples ordered from the one expressing the fewest genes associated with protection to the one expressing the most genes associated with protection). Pearson’s t test was used to assess the significance of the association between the mean–rank ordering of the tested samples and protection. To build a classifier for protection by ALVAC +alum the number of challenges to protection had to be discretized. To do so, samples taken before vaccination with ALVAC alum (ALVAC-alum prevax) were clustered according to the expression of the genes identified as being significantly associated with the number of challenges to infection (i.e., hierarchical clustering using Euclidean distance and complete linkage). The two main clusters identified were used to separate macaques in a “nonprotected” and “protected” group (containing respectively three-quarters and one-quarter of the macaques). As a third step, a naive Bayes classifier, able to classify samples into the “nonprotected” and “protected” groups, was built. ‘Leave-one-out’ cross-validation was used to identify the number of features maximizing the accuracy of the naive Bayes classifier. The accuracy was calculated as the mean of the sensitivity and specificity, and undersampling was used to correct for class imbalance between the “nonprotected” and “protected” group of macaques. As a fourth step, ROC analysis was used to assess the accuracy of the resulting build classifier on the postvax samples. GeneMANIA version 3.3.1 was used to identify relations (co-localization, co-expression and pathways) between the selected features of the naive Bayes classifier. To that end, the human orthologs and homologs of the macaques genes included in the classifier were obtained from the NCBI gene and homologene portal. The human homologs were then imported into GeneMANIA, and a network was generated with default parameters (equal weight of network, 20 added nodes to consolidate the network). The Selinger et al.24 raw gene-expression data, samples annotations and features annotation were downloaded from the GEO (GSE59068). The raw probe intensities were normalized, as described in the Selinger et al.24 original publication. LIMMA was used to fit linear models with gene expression as the dependent variable, and the number of SIV challenges to infection was used as the independent variable, separately for each time point after immunization (prevaccination, post-week 1, week 26 and week 32. Time points after challenge were excluded from further analysis. GSEA was used to test for the enrichment of genes correlated with the number of SIV challenges identified in the study. Briefly, the top-200 genes positively and negatively correlated with the number of SIV challenges (ordered by LIMMA t statistic) separately for each immunization time point were identified (prevaccination, 24 h post-first immunization and 24 h post-third immunization). Enrichment of those lists of genes among genes correlated with the number of SIV challenges to infection after SIV-gp96 + gp120 immunization was evaluated by GSEA. To evaluate the statistical significance of the enrichment, a gene-wise permutation was performed, and the calculated false-discovery rate (FDR q-value) is reported. For significant enrichment corresponding to an FDR q value below 5%, the genes contributing to the enrichment (i.e., leading-edge genes) were identified, and their expression was used to visually inspect their association with number of SIV challenges to infection: heat maps, with samples ordered by the mean–rank of the leading-edge genes, are presented in Figure 6a,b. GeneMANIA version 3.3.1 was used with default parameters to identify relations (co-localization, co-expression and pathways) between the leading-edge genes and NRAS. To test for the enrichment of proteins known to interact with NRAS (co-localization, co-expression and pathways) among the leading-edge genes, a 1,000-fold permutation test was performed. During each iteration of the permutation test, a set of 37 genes was randomly selected among all the genes represented on the bead array. That list of randomly selected genes was supplemented with the NRAS gene and imported to GeneMANIA. A network was generated with default parameters. The number of genes linked directly or indirectly to NRAS was extracted. The observed number of genes linked to NRAS was then compared to the simulated number of genes linked to NRAS obtained from the permutations test, and a P value was derived and given in the main text of the paper.
Integrative analysis of gene expression and humoral data sets
A partial least-square regression as implemented by the function spls of the R package mixOmics was used to project the omics data in the same space54. Gene expression, being the data set containing the most features, was used as the independent variable, and the humoral data set was used as dependent variable. Once the two omics are projected in the same space, a Pearson correlation between features of the first and second omics are calculated. All absolute Pearson correlations R ≥ 0.223 corresponding to a P value below 0.05 were used to generate a correlation network between omics. The R package igraph (version 1.0.1) was used to generate a network representation with the significant correlation between gene-expression prevaccination and humoral markers associated with protection in the alum arm.
Statistical analysis
The Mann–Whitney–Wilcoxon test was used to compare continuous factors between two groups. Correlation analyses were performed using the Spearman rank correlation method with exact permutation P values calculated. The number of challenges before viral acquisition was assessed using the log–rank test of the discrete–time proportional hazards model. The Wilcoxon signed–rank test was applied to changes in PB levels from prevaccination to postvaccination. Mean neutrophil counts from multiple views were tested using repeated measures analysis of variance.
Code availability
Source code to reproduce the Figures 5 and 6 can be accessed at https://sites.google.com/a/case.edu/fouslim/publication/p162.
Supplementary Material
Acknowledgments
We would like to thank T. Nolan and D. Abram for their editorial assistance, N. Miller and A. Shultz for their help in the study design, and J. Warren for support with systems biology and several antibody assays under Simian Vaccine Evaluation Unit (SVEU) contract number HHSN266200600005C, awarded to Advanced BioScience Laboratories, Inc. (ABL). This work was supported by the intramural US National Cancer Institute (NCI) program and the extramural US National Institute of Allergy and Infectious Diseases (NIAID) program, together with the US Army Medical Research and Material Command (USAMRMC) (W81XWH-07-2-0067), the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., the US Department of Defense, the Collaboration for AIDS Vaccine Discovery (CAVD) grants OPP1032325 and OPP1032817 from the Bill and Melinda Gates Foundation, and in part, with federal funds from the NCI, US National Institutes of Health (NIH), under contract no. HHSN261200800001E (to the whole team). We would like to acknowledge the following institutions for the grants supporting the authors: NIH primate grant HHSN27201100016C (D.M.); Center for AIDS Research grant AI064518 (G.F.); Bill and Melinda Gates’ Foundation grant OPP111572 (M.E.A., C.B.K., E.F.B., K.G.D.); NIH/NIAID grant 5R01AI102691 (M.E.A, E.F.B.); Intramural Research Program of the Vaccine Research Center, NIAID, NIH, and CAVD from the Bill and Melinda Gates Foundation grants OPP1032325 (R.A.K.), R01 AI118581 and R01 AI102715 (D.F.). The views expressed are those of the authors and should not be construed to represent the positions of the US Army, the Department of Defense or the Department of Health and Human Services. Mention of trade names, commercial products, or organizations do not imply endorsement by the US Government. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: CEM.NKR CCR5+ by P. Creswell. The anti-α4β7 in APC was kindly provided by A. A. Ansari (cat. #11718, 1:50 dilution) through the NIH AIDS Reagent Program, Division of AIDS, NIAID.
Footnotes
Accession codes. Gene Expression Omnibus: the microarray data presented in this article have been deposited under accession code GSE72624.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
G. Franchini, M.V. and S.N.G. designed and coordinated the study, analyzed the data, prepared the figures and wrote the manuscript. R.-P.S. and S.F. analyzed microarray data and correlates of protection, prepared figures and wrote the manuscript. L.S. performed PB analysis, and N.P.M.L. analyzed ILCs. M.C. performed microarray analysis and analyzed data; B.F.K. did the DNA sequencing and analyzed the transmitted founder variants; D.V. and D.M.S. provided statistical support for study design and data analysis. X.S., G.D.T., E.B., M. Rao, A.W.C., K.G.D., C.B.-K., E.P.B., M.E.A., G.A., D.C.M., T.B.P., G. Ferrari, D.N.F., D.A.V.-I. and M.R.-G. performed binding and functional antibody assays on sera and mucosal secretions. S.W. expressed, scaled up and purified the gp120 proteins. K.F., D.S.Q., M.D., M. Roederer, R.A.K., and A.M. performed T cell assays (ICS), Fluidigm. F.L. and K.L. performed neutrophil staining. Z.-M.M. and C.J.M. provided immunohistochemistry data in mucosa, and M.N.D, N.B, P.P., M. Bissa., F.C. and M. Blackburn helped with cell preparations from blood and tissues. V.K., M.G.F. and D.T. performed envelope-specific ELISA on sera and quantitative assays on viral RNA in plasma and viral DNA tissues. S.R.-K., J.H.K. and N.L.M. provided human samples. S.P., S.W.B. and J.T. provided MF59 and generated a new recombinant ALVAC–SIV.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolland M, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomaras GD, et al. Initial B cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox CB, Haensler J. An update on safety and immunogenicity of vaccines containing emulsion-based adjuvants. Expert Rev. Vaccines. 2013;12:747–758. doi: 10.1586/14760584.2013.811188. [DOI] [PubMed] [Google Scholar]
- 6.Pegu P, et al. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIVmac251 acquisition in an immunization regimen that mimics the RV-144 Thai trial. J. Virol. 2013;87:1708–1719. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franchini G, Gurunathan S, Baglyos L, Plotkin S, Tartaglia J. Poxvirus-based vaccine candidates for HIV: two decades of experience with special emphasis on canarypox vectors. Expert Rev. Vaccines. 2004;3(suppl. 4):S75–S88. doi: 10.1586/14760584.3.4.s75. [DOI] [PubMed] [Google Scholar]
- 8.Vaccari M, et al. Protection afforded by an HIV vaccine candidate in macaques depends on the dose of SIVmac251 at challenge exposure. J. Virol. 2013;87:3538–3548. doi: 10.1128/JVI.02863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottardo R, et al. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV-144 vaccine efficacy trial. PLoS One. 2013;8:e75665. doi: 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nitayaphan S, et al. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J. Infect. Dis. 2004;190:702–706. doi: 10.1086/422258. [DOI] [PubMed] [Google Scholar]
- 11.Nakajima C, et al. Induction of the chemokine receptor CXCR3 on TCR-stimulated T cells: dependence on the release from persistent TCR triggering and requirement for IFN-γ stimulation. Eur. J. Immunol. 2002;32:1792–1801. doi: 10.1002/1521-4141(200206)32:6<1792::AID-IMMU1792>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Chung AW, et al. Identification of antibody glycosylation structures that predict monoclonal antibody Fc-effector function. AIDS. 2014;28:2523–2530. doi: 10.1097/QAD.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ackerman ME, et al. Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J. Clin. Invest. 2013;123:2183–2192. doi: 10.1172/JCI65708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 15.Liyanage NP, et al. Antiretroviral therapy partly reverses the systemic and mucosal distribution of NK cell subsets that is altered by SIVmac251 infection of macaques. Virology. 2014;450–451:359–368. doi: 10.1016/j.virol.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves RK, et al. Gut inflammation and indoleamine deoxygenase inhibit IL-17 production and promote cytotoxic potential in NKp44+ mucosal NK cells during SIV infection. Blood. 2011;118:3321–3330. doi: 10.1182/blood-2011-04-347260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waddell SJ, et al. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS One. 2010;5:e9753. doi: 10.1371/journal.pone.0009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Šedý JR, et al. CD160 activation by herpesvirus entry mediator augments inflammatory cytokine production and cytolytic function by NK cells. J. Immunol. 2013;191:828–836. doi: 10.4049/jimmunol.1300894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fric J, et al. NFAT control of innate immunity. Blood. 2012;120:1380–1389. doi: 10.1182/blood-2012-02-404475. [DOI] [PubMed] [Google Scholar]
- 20.Bambard ND, Mathew SO, Mathew PA. LLT1-mediated activation of IFN-γ production in human natural killer cells involves ERK signaling pathway. Scand. J. Immunol. 2010;71:210–219. doi: 10.1111/j.1365-3083.2009.02367.x. [DOI] [PubMed] [Google Scholar]
- 21.Awad A, et al. Natural killer cells induce eosinophil activation and apoptosis. PLoS One. 2014;9:e94492. doi: 10.1371/journal.pone.0094492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dedio J, et al. NOSIP, a novel modulator of endothelial nitric oxide synthase activity. FASEB J. 2001;15:79–89. doi: 10.1096/fj.00-0078com. [DOI] [PubMed] [Google Scholar]
- 23.Bogdan C. Regulation of lymphocytes by nitric oxide. Methods Mol. Biol. 2011;677:375–393. doi: 10.1007/978-1-60761-869-0_24. [DOI] [PubMed] [Google Scholar]
- 24.Selinger C, et al. Multiple low-dose challenges in a rhesus macaque AIDS vaccine trial result in an evolving host response that affects protective outcome. Clin. Vaccine Immunol. 2014;21:1650–1660. doi: 10.1128/CVI.00455-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strbo N, et al. Cutting edge: novel vaccination modality provides significant protection against mucosal infection by highly pathogenic simian immunodeficiency virus. J. Immunol. 2013;190:2495–2499. doi: 10.4049/jimmunol.1202655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Underhill GH, George D, Bremer EG, Kansas GS. Gene expression profiling reveals a highly specialized genetic program of plasma cells. Blood. 2003;101:4013–4021. doi: 10.1182/blood-2002-08-2673. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, et al. Plasmacytoid dendritic cells promote HIV-1-induced group 3 innate lymphoid cell depletion. J. Clin. Invest. 2015;125:3692–3703. doi: 10.1172/JCI82124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragab A, et al. Drosophila Ras-MAPK signaling regulates innate immune responses in immune and intestinal stem cells. EMBO J. 2011;30:1123–1136. doi: 10.1038/emboj.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iborra S, et al. Hras and Nras are dispensable for T cell development and activation but critical for protective TH1 immunity. Blood. 2011;117:5102–5111. doi: 10.1182/blood-2010-10-315770. [DOI] [PubMed] [Google Scholar]
- 30.Liu XV, Ho SS, Tan JJ, Kamran N, Gasser S. Ras activation induces expression of Raet1 family NK receptor ligands. J. Immunol. 2012;189:1826–1834. doi: 10.4049/jimmunol.1200965. [DOI] [PubMed] [Google Scholar]
- 31.Sánchez-Ruiz J, Mejías R, García-Belando M, Barber DF, González-García A. Ral GTPases regulate cell-mediated cytotoxicity in NK cells. J. Immunol. 2011;187:2433–2441. doi: 10.4049/jimmunol.1003089. [DOI] [PubMed] [Google Scholar]
- 32.Hudgens MG, et al. Power to detect the effects of HIV vaccination in repeated low-dose challenge experiments. J. Infect. Dis. 2009;200:609–613. doi: 10.1086/600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keele BF, et al. Identifcation and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keele BF, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 2009;206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fenizia C, et al. TRIM5-α does not affect simian immunodefciency virus SIVmac251 replication in vaccinated or unvaccinated Indian rhesus macaques following intrarectal challenge exposure. J. Virol. 2011;85:12399–12409. doi: 10.1128/JVI.05707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romano JW, Williams KG, Shurtliff RN, Ginocchio C, Kaplan M. NASBA technology: isothermal RNA amplifcation in qualitative and quantitative diagnostics. Immunol. Invest. 1997;26:15–28. doi: 10.3109/08820139709048912. [DOI] [PubMed] [Google Scholar]
- 37.Lee EM, et al. Molecular methods for evaluation of virological status of nonhuman primates challenged with simian immunodefciency or simian-human immunodefciency viruses. J. Virol. Methods. 2010;163:287–294. doi: 10.1016/j.jviromet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 38.Vaccari M, Trindade CJ, Venzon D, Zanetti M, Franchini G. Vaccine-induced CD8+ central memory T cells in protection from simian AIDS. J. Immunol. 2005;175:3502–3507. doi: 10.4049/jimmunol.175.6.3502. [DOI] [PubMed] [Google Scholar]
- 39.Foulds KE, Donaldson M, Roederer M. OMIP-005: quality and phenotype of antigen-responsive rhesus macaque T cells. Cytometry A. 2012;81:360–361. doi: 10.1002/cyto.a.22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez MH, et al. Highly multiplexed quantitation of gene expression on single cells. J. Immunol. Methods. 2013;391:133–145. doi: 10.1016/j.jim.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomaras GD, et al. Vaccine-induced plasma IgA specifc for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc. Natl. Acad. Sci. USA. 2013;110:9019–9024. doi: 10.1073/pnas.1301456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montefori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. 2005 doi: 10.1002/0471142735.im1211s64. Ch, 12. [DOI] [PubMed] [Google Scholar]
- 43.Schiffner T, et al. Immune focusing and enhanced neutralization induced by HIV-1 gp140 chemical cross-linking. J. Virol. 2013;87:10163–10172. doi: 10.1128/JVI.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M, et al. Human immunodefciency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gómez-Román VR, et al. A simplifed method for the rapid fuorometric assessment of antibody-dependent cell-mediated cytotoxicity. J. Immunol. Methods. 2006;308:53–67. doi: 10.1016/j.jim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 46.Hartshorn KL, et al. Neutrophil deactivation by infuenza A viruses: mechanisms of protection after viral opsonization with collectins and hemagglutination-inhibiting antibodies. Blood. 1996;87:3450–3461. [PubMed] [Google Scholar]
- 47.De Vos J, et al. Microarray-based understanding of normal and malignant plasma cells. Immunol. Rev. 2006;210:86–104. doi: 10.1111/j.0105-2896.2006.00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thongcharoen P, et al. A phase 1–2 comparative vaccine trial of the safety and immunogenicity of a CRF01_AE (subtype E) candidate vaccine: ALVAC-HIV (vCP1521) prime with oligomeric gp160 (92TH023/LAI-DID) or bivalent gp120 (CM235/SF2) boost. J. Acquir. Immune Defc. Syndr. 2007;46:48–55. doi: 10.1097/QAI.0b013e3181354bd7. [DOI] [PubMed] [Google Scholar]
- 49.Liang F, et al. Dendritic cell recruitment in response to skin antigen tests in HIV-1-infected individuals correlates with the level of T cell infltration. AIDS. 2013;27:1071–1080. doi: 10.1097/QAD.0b013e32835ecaca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ackerman ME, et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods. 2011;366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjamini Y, Drai D, Elmer G, Kafkaf N, Golani I. Controlling the false-discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 52.Brown EP, et al. High-throughput, multiplexed IgG subclassing of antigen-specifc antibodies from clinical samples. J. Immunol. Methods. 2012;386:117–123. doi: 10.1016/j.jim.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boesch AW, et al. Highly parallel characterization of IgG Fc binding interactions. MAbs. 2014;6:915–927. doi: 10.4161/mabs.28808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liquet B, LêCao KA, Hocini H, Thiébaut R. A novel approach for biomarker selection and the integration of repeated measures experiments from two assays. BMC Bioinformatics. 2012;13:325. doi: 10.1186/1471-2105-13-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Troyanskaya O, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 57.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.