Abstract
The genetic architecture of amyotrophic lateral sclerosis (ALS) is being increasingly understood. In this far-reaching review, we examine what is currently known about ALS genetics and how these genes were initially identified. We also discuss the various types of mutations that might underlie this fatal neurodegenerative condition and outline some of the strategies that might be useful in untangling them. These include expansions of short repeat sequences, common and low-frequency genetic variations, de novo mutations, epigenetic changes, somatic mutations, epistasis, oligogenic and polygenic hypotheses.
Keywords: Amyotrophic lateral sclerosis, Gene discovery, Genetic heterogeneity, GWAS, NGS, Somatic mosaicism
1. INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease affecting upper and lower motor neurons leading to rapidly progressive paralysis and eventually death from respiratory failure. Although this core definition is remarkably straightforward, it is becoming increasingly apparent that ALS is not a monolithic entity, but rather represents a heterogeneous group of diseases that share clinical features.
Examples of the heterogeneity associated with ALS are easy to find: the majority of cases die within three to four years of disease onset, but up to 10% of ALS patients survive for more than 10 years (Chiò et al., 2009a); there is wide variability in disease from population to population and across geographical region (Cronin et al., 2007); age at onset ranges from early twenties to the ninth decade of life; the clinical manifestation of disease differs from patient to patient in terms of clinical onset (bulbar-onset versus spinal-onset disease, proximal versus distal weakness, upper limb versus lower limb predominant), course (upper motor neuron versus lower motor neuron predominant), and frontotemporal lobe involvement (normal cognition versus mild cognitive impairment and/or dementia), to name but a few. Variability in neuropathology has also been observed with TDP-43 positive inclusions dominating most cases, but other cases lacking these inclusions.
Genetics offers a means to dissect out this heterogeneity and understand the cellular mechanisms leading to motor neuron degeneration. Paradoxically, however, it is this very heterogeneity associated with ALS that is the biggest obstacle to unraveling the genetics. (Singleton et al., 2010)
1.1. What portion of ALS is genetic?
For decades after its initial description in the half of 19th century (Aran, 1848; Cruveilhier 1852; Charcot and Joffroy, 1869), ALS was thought to be a non-hereditary disease. It was not until Kurland and Mulder reported on familial aggregation in the 1950s that heritable factors were considered important in ALS etiology (Kurland and Mulder, 1955). Today, a family history of disease is recognized in 10% of cases, whereas the remaining 90% of cases are labeled as sporadic as they appear to occur randomly in the community. Even here, however, the sands are shifting, with an increasing portion of cases recognized as having a family history of related neurodegenerative diseases such as frontotemporal dementia. Autosomal dominant inheritance is by far the most common, but incomplete penetrance appears to be the rule.
To date, the genetic etiology of approximately two thirds of familial ALS and about 10% of sporadic disease has been identified (Renton et al., 2014). Genetic mutations are clearly responsible for the remaining one third of familial disease, but it is not known how much of the remaining sporadic disease is genetic and how much is due to other factors such as environmental exposures, aging or lifestyle choices? Genome-wide data suggest that genetic factors contribute to at least 23% of sporadic ALS (Keller et al., 2014). Even this high value, however, is likely to be an underestimate as the calculation was based on common variants in the human genome and would not capture the portion of disease arising from rare variants.
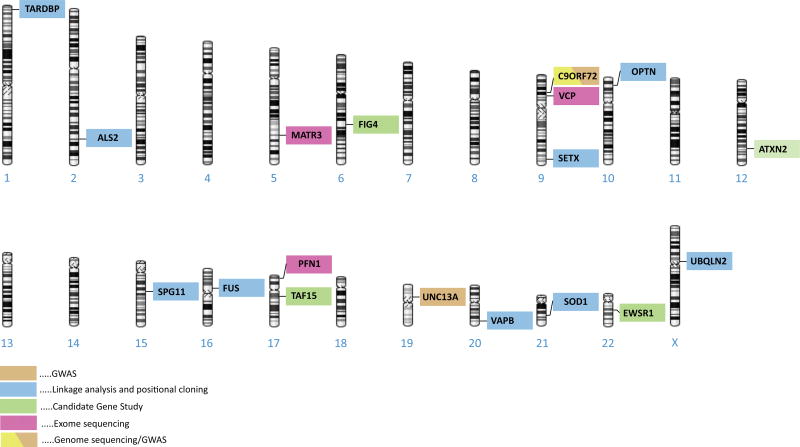
To date, more than twenty-five genes linked to ALS have been identified (table 1, figure 1). We present these genes in two categories, namely (a) genes identified using linkage analysis and positional cloning, and (b) genes identified through the application of advanced genome-wide technologies. Though not every gene fits neatly into this framework, describing the genetic discoveries in ALS in this way provides a historical context and highlights how advances in genomic technologies are revolutionizing the way we think about this fatal neurodegenerative disease.
Table 1.
list of genes claimed to be involved in ALS etiology and pathogenesis
| Gene | Location | FALS locus |
Strategy | Suggested role in ALS* |
Associated phenotypes | OMIM nr |
Refs |
|---|---|---|---|---|---|---|---|
| SOD1 | 21q22.11 | ALS 1 | Linkage analysis | AD, AR, main gene | - | 147450 | Rosen et al., 1993 |
| ALS2 | 2q33.2 | ALS 2 | Linkage analysis | AR, main gene? | Hereditary spastic paraplegia | 606352 | Hadano et al., 2001 |
| Unknown | 18q21 | ALS 3 | Linkage analysis | AD? | ? | (606640) | Hand et al., 2002 |
| SETX | 9q34.13 | ALS 4 | Linkage analysis | AR, main gene? | Spinocerebellar ataxia | 608465 | Chen et al., 2004 |
| SPG11 | 15q21.1 | ALS 5 | Linkage analysis, family study, candidate gene, WES | AR, main gene? | Hereditary spastic paraplegia | 610844 | Orlacchio et al., 2010; Daoud et al., 2012 |
| FUS | 16p11.2 | ALS 6 | Linkage analysis, candidate gene | AD, AR, main gene | - | 137070 | Vance et al., 2009; Kwiatkowski et al., 2009 |
| Unknown | 20p13 | ALS 7 | Linkage analysis | AD? | ? | (608031) | Sapp et al., 2003 |
| VAPB | 20q13.33 | ALS 8 | Linkage analysis | AD, main gene? | Spinal muscular atrophy, late-onset | 605704 | Nishimura et al., 2004 |
| ANG | 14q11.1 | ALS 9 | Candidate gene, association study | AD?, main gene? modifier gene? | - | 105850 | Greenway et al., 2006 |
| TARDBP | 1p36.22 | ALS 10 | Linkage analysis, candidate gene | AD, main gene | FTD | 605078 | Gitcho et al., 2008; Kabashi et al., 2008; Sreedharan et al., 2008 |
| FIG4 | 6q21 | ALS 11 | Candidate gene | AD, AR, main gene? | Charcot-Marie-Tooth disease, type 4J; Yunis-Varon syndrome | 609390 | Chow et al., 2009 |
| OPTN | 10p13 | ALS 12 | Family study: homozygosity mapping | AD, AR, main gene | Open angle glaucoma | 602432 | Maruyama et al., 2010 |
| ATXN2 | 12q24.12 | ALS 13 | Candidate gene | Susceptibility gene | Spinocerebellar ataxia | 601517 | Elden et al., 2010; Daoud et al., 2011 |
| VCP | 9p13.3 | ALS 14 | Family study: WES | AD, main gene | FTD, inclusion body myopathy, Paget’s disease | 601023 | Johnson et al., 2010 |
| UBQLN2 | Xp11.21 | ALS 15 | Linkage analysis | XL, main gene | FTD | 300264 | Deng et al., 2011 |
| SIGMAR1 | 9p13.3 | ALS 16 | Family study: homozygosity mapping | AR, main gene | - | 601978 | Al-Saif et al., 2011 |
| CHMP2B | 3p11.2 | ALS 17 (ALS-FTD3) | Linkage analysis, candidate gene | AD, main gene | FTD | 609512 | Parkinson et al., 2006 |
| PFN1 | 17p13.3 | ALS 18 | Family study: WES | AD, main gene | - | 176610 | Wu et al., 2012 |
| Unknown | 9q21-q22 | ALS-FTD 1 | Linkage analysis | AD? | FTD | Hosler et al.,, 2000 | |
| C9orf72 | 9p21.2 | ASL-FTD 2 | GWAS, linkage analysis | AD, main gene | FTD | 614260 | Renton et al., 2011; DeJesus-Henrnandez et al., 2011 |
| MATR3 | 5q31.2 | Family study: WES | AD, main gene | - | 164015 | Johnson et al., 2014 | |
| CHCHD10 | 22q11.23 | Family study: WES | AD, main gene | FTD, cerebellar ataxia, myopathy | 615903 | Bannwarth et al., 2014; Chaussenot et al., 2014 | |
| SQSTM1 | 5q35.3 | Candidate gene | AD, main gene | FTD, inclusion body myopathy, Paget’s disease | 601530 | Fecto et al., 2011 | |
| TAF15 | 17q12 | Candidate gene | AD, main gene? | - | 601574 | Couthouis et al., 2011 | |
| EWSR1 | 22q12.2 | Candidate gene | AD?, main gene? | - | 133450 | Couthouis et al., 2012 | |
| HNRNPA1 | 12q13.13 | Family study, WES | AD, main gene | FTD, inclusion body myopathy, Paget’s disease | 164017 | Kim et al., 2013 | |
| HNRNPA2B1 | 7p15.2 | Family study, WES | AD, main gene | FTD, inclusion body myopathy, Paget’s disease | 600124 | Kim et al., 2013 | |
| SPAST | 2q22.3 | Candidate gene | AD, main gene? | Hereditary spastic paraplegia | 604277 | Meyer et al., 2005 | |
| VEGF | 6p21.1 | Candidate gene, association study | Susceptibility gene | - | 192240 | Lambrechts et al., 2009 | |
| HFE | 6p22.2 | Candidate gene, association study | Susceptibility gene | Hemochromatosis | 613609 | Goodall et al., 2005 | |
| NEFH | 22q12.2 | Candidate gene, association study | Susceptibility gene? | - | 162230 | Al-Chalabi et al., 1999; Figlewicz et al., 1994 | |
| PRPH | 12q13.12 | Candidate gene | AD?, main gene? | - | 170710 | Leung et al., 2004 | |
| PON1, 2, 3 | 7q21.3 | Candidate gene, association study | Susceptibility gene? | - | 168820 | Wills et al., 2009 | |
| DCTN1 | 2p13.1 | Linkage analysis | AD, main gene? | Perry syndrome | 601143 | Puls et al., 2003 | |
| CHRNA4 | 20q13.33 | Candidate gene, association study | Susceptibility gene | Nocturnal frontal lobe epilepsy | 118504 | Sabatelli et al., 2012 | |
| CHRNA3 CHRNB4 | 15q25.1 | Candidate gene, association study | Susceptibility gene? | - | 118509 118503 | Sabatelli et al., 2009 | |
| ERLIN2 | 8p11.23 | Family study: homozygosity mapping | AR, main gene | Hereditary spastic paraplegia | 611605 | Al-Saif et al., 2012 | |
| UNC13A | 19p13.11 | GWAS | Susceptibility gene, modifier gene | - | 609894 | Chiò et al., 2013a; Diekstra et al., 2012a | |
| DPP6 | 7q36.2 | GWAS | Susceptibility gene? | Ventricular fibrillation | 126141 | Van Es et al., 2008 | |
| ELP3 | 8p21.1 | GWAS | Susceptibility gene? | - | 612722 | Simpson et al., 2009 | |
| ZNF512B | 20q13.33 | GWAS | Susceptibility gene, modifier gene | - | - | Iida et al. 2011; Tetsuka et al., 2013 | |
| ITPR2 | 12p12.1 | GWAS | Susceptibility gene? | - | 600144 | Chiò et al., 2009b | |
| FGGY | 1p32.1 | GWAS | Susceptibility gene? | - | - | Chiò et al., 2009b | |
| CHGB | 20p12.3 | Candidate gene, association study | Susceptibility gene? | - | 118920 | Gros-Louis et al., 2009 | |
| DPYSL3 | 5q32 | Candidate gene, association study | Susceptibility gene? | - | 601168 | Blasco et al., 2013 | |
| GRN | 17q21.31 | Candidate gene, association study | Susceptibility gene, modifier gene | - | 138945 | Sleegers et al., 2008 | |
| KIFAP3 | 1q24.2 | GWAS | Modifier gene | - | 601836 | Landers et al., 2009 | |
| EPHA4 | 2q36.1 | Candidate gene | Modifier gene | - | 602188 | Van Hoecke et al., 2012 | |
| PPARGC1A | 4p15.2 | Candidate gene, association study | Modifier gene | - | 604517 | Eschbach et al., 2013 | |
| APOE | 19q13.32 | Candidate gene, association study | Modifier gene | Alzheimer disease | 107741 | Zetterberg et al., 2008 | |
| MAOB | Xp11.3 | Candidate gene, association study | Modifier gene | - | 309860 | Orrù et al., 1999 | |
| CX3CR1 | 3p22.2 | Candidate gene, association study | Modifier gene | - | 601470 | Lopez-Lopez et al., 2014 | |
| SMN1 (SMN2) | 5q13.2 | Candidate gene, association study | Susceptibility gene | Spinal muscular atrophy | 600354 | Corcia et al., 2002 | |
| EPHA3 | 1q22 | GWAS | Protective factor | - | 601381 | Uyan et al., 2013 | |
| SS18L1 | 20q13.33 | WES (trios study) | AD, main gene | - | 606472 | Chesi et al., 2013 | |
| DAO | 13q33.2 | Linkage analysis | AD, main gene? | - | 124050 | Mitchell et al., 2010 | |
| PNPLA6 | 19p13.2 | Family study: homozygosity mapping | AR, main gene? | Hereditary spastic paraplegia, Boucher-Neuhäuser and Gordon Holmes syndromes | 603197 | Rainier et al., 2008 | |
| MAPT | 17q21.31 | Candidate gene, association study | Susceptibility gene | FTD | 157140 | Fang et al., 2013 | |
| TREM2 | 6p21.1 | Candidate gene, association study | Susceptibility gene | Alzheimer disease | 605086 | Cady et al., 2014 |
GWAS: genome wide association study; WES: whole exome sequencing
AD: autosomal dominant inheritance; AR: autosomal recessive inheritance; X-linked inheritance
OMIM: Online Mendelian Inheritance in Men database (www.omim.org)
Figure 1.
schematic representation of selected ALS genes.
2. GENES IDENTIFIED USING LINKAGE ANALYSIS AND POSITIONAL CLONING
2.1. SOD1
In 1993, an international consortium identified SOD1 as a gene responsible for autosomal dominant FALS cases, by means of linkage analysis in 18 ALS pedigrees (Rosen et al., 1993). Through this method is possible to map the location of disease-causing loci by testing the co-segregation of genetic markers with the phenotype of interest. Multiple markers across the whole genome are usually screened in large families and a statistical test is performed to determine which markers are inherited by affected subjects more often than would be expected by chance. Candidate regions are eventually studied to identify the causative gene and mutations (posistional cloning). To date, over 150 different mutations have been reported in this gene, consisting primarily of missense mutations, with a smaller number of nonsense and deletion/insertion mutations. Synonymous, intronic and upstream variants are reported, though they are less likely to be pathogenic. SOD1 mutations can be identified in 12 to 20% of FALS cases and in about 1–2% of apparently sporadic cases (Chiò et al., 2008, Millecamps et al., 2010; Rosen et al., 1993). However, significant differences are observed across ethnic groups (Alavi et al., 2013; Brown et al., 2012; Kwon et al., 2012; Tsai et al., 2013).
It is not clear whether all of the reported variants in SOD1 are pathogenic, or instead represent incidental findings in affected subjects. Reliable genetic evidence in the form of segregation in large families or their observations in multiple affected subjects, and not in normal individuals matched for ethnicity, exists only for a portion of them (Andersen et al., 2006).
Most SOD1 mutations show an autosomal dominant, high-penetrance pattern of inheritance. In contrast, recessive inheritance is instead typical for D90A mutation in Scandinavian population (Andersen et al., 1996). Clear genotype-phenotype correlations and prognostic trends can be drawn only for few mutations. A4V, the most common variant in North America, is responsible for an aggressive form of ALS, in which death occurs usually within a year after symptoms onset (Cudkowicz et al., 1997). Other variants consistently associated with a poor survival are G41S, G93A, and R115G. D90A homozygosity is associated with prolonged survival of over 10 years and may present with sensory involvement.
2.2. TARDBP (TDP-43), FUS and the other RNA-binding genes
Following the discovery of TAR DNA-binding protein 43 (TDP-43) as a major component of ubiquitin-positive cytoplasmic inclusions that are the neuropathological hallmark of the disease (Neumann et al., 2006), mutations in the TARDBP gene (encoding the TDP-43 protein) were identified in both sporadic and familial ALS cases (Gitcho et al., 2008; Kabashi et al., 2008; Sreedharan et al., 2008). A total of 47 different missense and one truncating mutations have now been reported (Lattante et al., 2013) and overall TARDBP mutations are found in about 4% of FALS cases (Chiò et al., 2008; Millecamps et al., 2010) and about 1% of sporadic cases (Chiò et al., 2008; Brown et al., 2012, Kwon et al., 2012; Mentula et al. 2012; Tsai et al., 2013). A founder effect may explain the relatively high frequency of the A382T variant in Sardinian population (Chiò et al., 2011a).
In agreement with the fact that TDP-43 inclusions are also typical of FTD, clinical phenotypes associated with TARDBP mutations include ALS with cognitive impairment and FTD (Chiò et al. 2010), FTD without ALS (go et al., 2009), and clinically definite Parkinson disease (Quadri et al., 2011; Rayaprolu et al., 2013).
Shortly after the discovery of TARDBP as a cause of familial ALS, mutations were identified in FUS within a linkage region on chromosome 16 (Vance et al., 2009; Kwiatkowski et al., 2009). FUS protein shares functional homology with TDP-43, and nearly all the reported mutations in both genes affect the protein C-terminus containing ribonucleoprotein binding domain. Mutations in this gene account for ~5% of FALS and about 1% of SALS, with higher rates observed in oriental populations (Brown et al., 2012; Chiò et al., 2008; Lattante et al. 2012; Millecamps et al., 2010; Tsai et al., 2013; Yan et al., 2010). The associated phenotype is typical ALS, often juvenile onset, though some individuals present with FTD. Most ALS cases carrying a mutation in FUS appears to have a peculiar neuropathological signature, in that FUS-immunoreactive cytoplasmic inclusions can be found on autopsy, rather than phosphorylated TDP-43 (Vance et al., 2009; Kwiatkowski et al., 2009).
Both TDP-43 and FUS proteins contain a prion-like domain, a feature that may promote aggregation by acting as a template to induce the misfolding of native proteins and their entrapment into aggregates. Couthouis et al. (2011) performed a systematic survey of human proteins harboring RNA recognition motifs and prion-like domains to find additional candidates similar to TDP-43 and FUS. Among the candidate proteins identified, they performed mutational analysis of TAF15-encoding gene, leading to the identification of missense variants in patients with ALS (Couthouis et al., 2011). Sequencing of EWSR1, another gene belonging to the same group, yielded similar results (Couthouis et al., 2012). Definitive confirmation that TAF15 and EWSR1 are ALS genes based on segregation within a family is eagerly anticipated.
2.3. Other FALS genes identified through linkage analysis and cloning
For some of the identified genes, observed phenotypes can be more consistently related to different neurological disorders: recessive ALS2 (Eymard-Pierre et al., 2002; Hadano et al., 2001) and SPG11 (Daoud et al., 2012) mutations cause hereditary spastic paraplegia, overlapping with juvenile primary lateral sclerosis or juvenile ALS. SETX mutations are more typically associated with ataxia (Chen et al., 2004; Duquette et al., 2005). FIG4 mutations cause autosomal recessive Charcot-Marie-Tooth disease, type 4J, a hereditary motor sensory neuropathy (Campeau et al., 2013; Chow et al., 2009; de Leeuw, 2008).
Optineurin (OPTN) mutations have long been described as a cause of primary open angle glaucoma (Reazaie et al., 2002). Recently, both heterozygous and homozygous mutations have been reported in FALS cases (Maruyama et al., 2010), with either dominant or recessive pattern of inheritance. Nonetheless, mutations in this gene appear to be a rare cause of ALS (Sugihara et al., 2011).
A single missense VAPB mutation (P56S) was initially identified in several Brazilian families presenting with different phenotypes: atypical ALS, late-onset spinal muscular atrophy, and typical severe ALS with rapid progression (Nishimura et al., 2004). To date, only one other VAPB mutation has been described to date in association with ALS (T46I) (Chen et al., 2010).
Following the detection of an association of ALS with the rs11701 SNP in the Irish population, ANG was identified as a candidate gene and several mutations have been reported in both FALS and SALS cases (Greenway et al., 2004; Greenway et al., 2006). However, the causal role of ANG has not yet been defined and remains ambiguous, since some variants have been repeatedly found in healthy controls, and some ANG mutated subjects have been reported bearing mutations in different ALS-causing genes (Luigetti et al., 2011; Lattante et al., 2012).
UBQLN2 is the only ALS gene mapping on chromosome X identified to date (Deng et al., 2011). Even though UBQLN2 mutations are a rare cause of ALS, its role in the pathogenesis of ALS is supported by the observation that ubiquilin-2 is a component of skein-like inclusions that are considered a hallmark of ALS pathology.
The p62/sequestosome 1 protein is another component of pathological inclusions in neurodegentative diseases, including ALS. Based on these findings, sequencing analysis of the p62-encoding gene, SQSTM1, allowed for the detection of mutations in ALS patients, but also in cases with ALS-FTD and isolated FTD. According to various reports, SQSTM1 variants may account for 1–2% of FALS and up to 4% of SALS (Chen et al., 2014; Fecto et al., 2011; Kwok et al., 2014; Rubino et al., 2012; Teyssou et al., 2013).
CHMP2B is another gene whose involvement in ALS was found through a candidate gene approach, supported by the initial discovery of a mutation in an FTD family (Parkinson et al., 2006). CHMP2B mutations have been found in very few ALS cases, but they seem to be more specifically associated with the lower motor neuron predominant variant of ALS (with a detection rate of about 10%) (Cox et al., 2010).
Other genes claimed to be involved in ALS are the following: HFE (Goodall et al., 2005; Li et al., 2014); VEGF (Lambrechts et al., 2009); NEFH (Al-Chalabi et al., 1999, Figlewicz et al., 1994) and PRPH (Corrado et al., 2011; Leung et al., 2004; Gros-Louis et al., 2004), both coding for intermediate filament proteins; the paraoxonase genes PON1, PON2 and PON3 (Wills et al., 2009); DCTN1 (Puls et al., 2003; Münch et al., 2004), whose mutations are now more commonly linked to Perry syndrome (Farrer et al. 2009); SIGMAR1 and genes coding for the subunits of acetylcholine receptors, mainly CHRNA4 (Sabatelli et al., 2009; Sabatelli et al., 2012a); SIGMAR1 (Al-Saif et al., 2011) and ERLIN2 (Al-Saif et al., 2012), are repored to be responsible for juvenile ALS and juvenile primary lateral sclerosis.
2.4. Validation of ALS causative variants
Several genes have been claimed to be someway related to ALS in scientific literature (see table 1). Even if there is consensus about the causal role of a subset of major genes like C9orf72, SOD1, TARDBP, and FUS, for other genes further evidences are required.
It is worth noting that concerns have been raised even regarding the actual pathogenicity of a small number of SOD1 mutations (Felbecker et al. 2010).
It is actually a tough task to provide definitive proof of pathogenicity for ALS-associated variants. The co-segregation of a specific variant with the disease in large families and the presence of the same variant in multiple unrelated patients and not in control subjects are usually considered as self-conclusive evidence, but they can be applied only to a limited number of cases. Functional studies provide insight into pathophysiological mechanisms, but extreme caution should be applied in using functional biological data to support weak genetic data.
Locus and allelic heterogeneity of ALS are the main factors complicating the discovery and validation of ALS genetics, since a single gene may be involved in a very limited number of cases. Furthermore, the relative impact of various genes may be significantly diverse among different populations.
A subset of genes reported to be mutated in ALS patients are primarily known to be responsible for different neurological conditions (e.g.: ALS2, VAPB, SPG11, FIG4, ATXN2, SPAST, DCTN1, SMN1).
Some variants initially detected in ALS patients were subsequently found to be present in healthy controls (e.g.: ANG gene). This highlights the importance of keeping an open mind with respect to reported mutations and a willingness to revise previous closely held opinions of pathogenicity.
Finally, some variants might not be responsible for ALS by themselves, but they could act as predisposing or disease-modifying factors.
3. GENES IDENTIFIED THROUGH THE APPLICATION OF ADVANCED GENOME-WIDE TECHNOLOGIES
3.1. Genome-wide association studies of ALS
Association studies involve comparison of the frequencies of genetic variants between groups of unrelated affected individuals and control subjects. Initial association studies were based on a candidate gene approach, but, with advances in genomic assay technology, several million markers across the genome can now be interrogated in a single experiment. This is known as genome-wide association study (GWAS).
Several GWAS have been published in ALS (Schymick et al., 2007; van Es et al., 2007; Cronin et al., 2008a; van Es et al., 2008; Simpson et al., 2009; Chiò et al., 2009b; Landers et al., 2009; van Es et al., 2009; Laaksovirta et al. 2010; Shatunov et al., 2010; Iida et al., 2011; Kwee et al., 2012; ALSGEN Consortium et al., 2013; Deng et al., 2013; Fogh et al. 2014). In addition, there have been studies of copy number variation (Blauw et al., 2008; Cronin et al., 2008; Wain et al., 2009; Blauw et al., 2010; Uyan et al. 2013), a study specifically focusing on homozygosity segments (Mok et al. 2013), and a pooling GWAS combined with pathway analysis (Xie et al., 2014).
A role as a risk factor has been invoked for several loci, including FGGY (Dunkley et al., 2007) , DPP6 (Van Es et al., 2008), ELP3 (Simpson et al., 2009), UNC13A (van Es et al., 2009; Shatunov et al., 2010), ZNF512B (Iida et al. 2011), ITPR2 and SUNC1 (Chiò et al., 2009b). For most of them, significant associations were not confirmed in subsequent replication studies.
Several factors may be invoked to explain why many of the GWAS hits have not replicated. Most ALS causative variants are limited to very small number of patients. Consequently, there are population stratification issues that are not easily eliminated: GWAS signals may be driven up by very few samples that cannot be significantly represented in different study cohorts. Furthermore, false positive results should always be considered even in presence of statistical significance. These effects are even more likely for small case-control cohorts, as they were in older GWAS studies.
For these reasons, the main goal remains the identification of the actual pathogenic variant: in fact, the most important result obtained by GWAS remains the definition of the C9orf72 locus (see section 3.4).
Genome-wide association studies can be adapted to look for genetic variants that modify phenotype, for example age at symptom onset or prognosis. Among others, PGRN (Sleegers et al., 2008), KIFAP3 (Landers et al., 2009), EPHA4 (Van Hoecke et al., 2012), UNC13A (Chiò et al., 2013; Diekstra et al., 2012a), ZNF512B (Tetsuka et al., 2013), PPARGC1A (Eschbach et al., 2013) have been reported to influence the survival in ALS, while PGRN, PPARGC1A, APOE (Zetterberg et al., 2008) MAO-B (Orrù et al., 1999) have been proposed as modifier of age of onset. These associations should be intrepted cautiously, as they still need confirmation and attempts to replicate the observed effects in some cases led to conflicting results (Traynor et al., 2010).
Genetic and allelic heterogeneity are the most important confounding factors for GWAS, since the presence of multiple risk haplotypes reduces the intensity and significance of detected signals. Clearly, this is the case for ALS that is increasingly recognized to represent a collection of similar neurological diseases rather than a single nosological entity. Future GWAS will need to be designed to maximize the statistical power and minimize the false discovery rate: this will involve larger case-control cohorts, as well as stratification of GWAS data based on different populations and well-defined clinical categories. Furthermore, newer generations of GWAS platforms assay rare variants with potentially larger effects on phenotype.
3.2. Copy number variants
Copy number variants constitute an important source of human genetic variability. They mainly consist of the loss (deletion) or gain (duplication) of stretches of DNA sequence, typically 1 kb to several Mbs in size. Similar to single nucleotide polymorphisms (SNPs), copy number variants are detected in healthy people. Several surveys have been performed in attempt to evaluate the involvement of copy number variants in ALS (Blauw et al., 2008; Cronin et al., 2008; Wain et al., 2009; Soichet et al., 2009; Blauw et al., 2010; Uyan et al. 2013), while others investigated specific categories of copy number variants including de novo copy number variants (Pamphlett and Morahan, 2011a), somatic copy number variants (Pamphlett and Morahan, 2011b; Pamphlett and Morahan, 2011c), and copy number variants in ALS-discordant monozygotic twin pairs (Pamphlett and Morahan, 2011c). Although many copy number variants that were specific to patients with ALS were detected, common copy number variants were not significantly associated to ALS in these studies.
Homozygous deletions of SMN1 underlie the most common cause of spinal muscular atrophy and several studies have suggested that duplication of SMN1 gene may contribute to ALS susceptibility (Wang et al., 2014, Kuzma-Kozakiewicz et al., 2013; Blauw et al., 2012; Corcia et al., 2002; Corcia et al.; 2006). Conflicting results have been obtained for SMN2, a homologe of SMN1 (Lee et al., 2012a; Corcia et al., 2012). Finally, a recent study pointed out that deletion of EPHA3 might be a protective factor (Uyan et al., 2013). Taken together, these data indicate that the role of copy number variants in ALS has not been fully resolved and is worth exploring further, possibly with the help of newer genome-wide technologies.
3.3. Next generation sequencing
The recent introduction of high-throughput massive parallel sequencing methods has revolutionized gene-hunting strategies. Whole-exome sequencing allows the identification of coding variants across (nearly) all genes (the so-called “exome”). In so doing, this type of genetic analysis facilitates the rapid identification of pathogenic mutations and is ideally suited to the study of families for which a limited number of samples are available.
By applying WES to a four-generation family with four members presenting with autosomal dominantly inherited ALS, a missense mutation in VCP gene was found segregating with the disease (Johnson et al., 2010). This finding was supported by the identification of other VCP mutations in FALS and SALS cases (Koppers et al., 2012; Abramzon et al., 2012). The causal role of VCP was further strengthened by the fact that it was already known to underlie a syndromic condition characterized by FTD, inclusion body myopathy and Paget’s disease of the bone (IBMPFD). The same approach more recently led to the discovery of mutations in HNRNPA1 and HNRNPA2B1 (Kim et al., 2013).
These recent genetic findings arising from exome sequencing have expanded our understanding of the pleiotropic effects and phenotypic spectrum associated with specific genes. Several kindreds have now been described in which apparently unrelated manifestations involving different organs and systems are observed: Paget’s disease of bone, inclusion body myopathy, and lastly ALS and FTD. The term “multisystem proteinopathy” has been proposed to describe these diverse, but somehow interconnected conditions, based on their shared pathologic features of protein aggregation in affected tissues (Benatar et al., 2013). Four genes have been identified whose mutations are likely responsible for that multifaceted syndrome, VCP, SQSTM1, HNRNPA2B1 and HNRNPA1, and even though their relative involvement in ALS appears to be limited, their discovery sheds new light on pathogenic mechanisms underlying ALS.
Another gene whose mutations were identified by means of WES in large ALS kindreds is PFN1 (Wu et al., 2012). More recently, mutations in another RNA/DNA binding protein, MATR3, have been reported as a cause of familial ALS in several large kindreds. MATR3 binds directly to TDP-43 and at least some of the identified mutations alter this binding in a selective and RNA-dependent manner (Johnson et al., 2014). Overall, this provides further support for the notion that disruption of RNA metabolism is central to motor neuron degeneration.
The last identified gene is CHCHD10, whose mutations may be responsible for ALS alone (Müller et al. 2014) or in association with frontotemporal dementia, cerebellar ataxia and myopathy (Bannwarth et al., 2014; Chaussenot et al., 2014; Johnson et al., 2014). It appears to be involved in mitochondrial stability and its discovery breathes new life into long-standing theories suggesting an important role played by mitochondrial dysfunction in ALS pathogenesis (Cozzolino et al., 2013). Other CHCHD10 mutations were reported to be responsible for autosomal dominant mitochondrial myopathy (Ajroud-Driss et al., 2014).
3.4. C9orf72 repeat expansion
The identification of the GGGGCC-repeat expansion in the first intron of C9orf72 as the cause of 9p21-linked ALS and FTD (Renton et al., 2011; DeJesus-Henrnandez et al., 2011) was the result of the application of both linkage analysis (Pearson et al., 2011; Morita et al., 2006; Le Ber et al., 2009), GWAS (van Es et al., 2009; Shatunov et al., 2010; Laaksovirta et al., 2010) and next generation sequencing. The C9orf72 repeat expansion constitutes the most frequent genetic cause of both FALS (about 40 %) (Majounie et al., 2012; Chiò et al., 2012a) and SALS cases (about 7 %) (Sabatelli et al., 2012b), providing the definitive evidence that the same etiopathogenic mechanisms underlie both SALS and FALS. The expansion also accounts for a remarkable percentage of familial FTD cases (about 25 %) (Majounie et al., 2012), thus consolidating the concept that ALS and FTD are different manifestations of a common neurodegenerative pathway. It is worth noting that the reported rates of C9orf72 expansion are referred to populations of European descent and lower rates have been observed in different ethnic groups (Alavi et al., 2014; Ogaki et al., 2012; Zou et al., 2013a).
The understanding of the molecular mechanisms underlying the pathogenesis of C9orf72-related ALS is important not only for its prognostic and therapeutic implications, but also because it can help the further discovery of other ALS causative factors. For example, urged by these findings, scientist have been searching for other repeat-expansion in ALS, ATXN2 CAG-repeat expansions are associated with increased risk of developing ALS (Liu et al., 2013; Daoud et al., 2011; Van Damme et al., 2011; Elden et al., 2010). Conversely, the analysis of the nucleotide repeat lengths of genes associated with other neurologic or neuromuscular disorders revealed no association with ALS (Groen et al., 2012; Figley et al., 2014).
3.5. De novo mutations
De novo mutations have been reported to play an important role in the pathogenesis of many disorders, such as autism and schizophrenia (Epi4K Consortium et al., 2013; Gratten et al., 2013; Neale et al., 2012; Sanders et al., 2012; Vissers et al., 2013; Xu et al. 2011) and also have been identified in known ALS genes (Alexander et al., 2002; Zou et al., 2013b; Calvo et al., 2014; Conte et al., 2012; Chiò et al. 2011b; DeJesus-Hernandez et al., 2010). The power of applying exome-sequencing to parents-case offspring trios is that it allow the investigator to look for de novo mutations across the genome and not just confined to known ALS genes. To this end, an exome sequencing study involving 47 ALS patient-parents trios identified CREST as a new gene possibly involved in ALS (Chesi et al., 2013). However, each individual in the general population carries up to four de novo coding mutations, meaning that identification of a de novo variant in a gene does not constitute proof of its pathogenicity. Additional genetic data are required to validate that mutations in the nominated gene are truly causative, something that is lacking in current publications.
4. FURTHER GENETIC MECHANISMS/ANALYSES YET TO BE FULLY EXPLORED
4.1 Epigenetics
Epigenetic modifications influence the expression pattern of the genome, Alteration of epigenetic processes, including DNA methylation, have been long known as causes of human diseases (e.g. imprinting syndromes, conditions caused by mutation in genes regulating epigenetic modifications, and cancer). Epigenetic mechanisms also appear to be involved in motor neuron cell death (Chestnut et al. 2011). Furthermore, epigenetics changes are the consequence of a dynamic process influenced by the interaction between genes and environment, and a fraction of those changes might even be transmitted to the offspring (Gu et al., 2012).
Genome-wide analyses have been performed to date in ALS investigating epigenetic changes or differences in epigenetic signature (i.e.: DNA-methylation pattern) between patients and controls (Morahan et al., 2009; Figueroa-Romero et al., 2012). Despite statistically significant results, suggesting epigenetically altered genes in ALS, confirmatory evidences are still pending.
4.2 Oligogenic and polygenic models of ALS
Interaction of multiple risk variants at different loci in single individuals is an important aspect to consider when dissecting the genetics bases of a complex disease such as ALS. The fact that mutations in ALS causative genes display a classic Mendelian pattern of inheritance may lead to the belief that ALS is a monogenic disorder. Nevertheless, a significant part of ALS heritability cannot be easily explained by only considering a simple monogenic model (Singleton et al., 2010). Identification of epistatic interactions among multiple ALS-related genes might explain the substantial phenotypic differences observed among subjects carrying identical mutations and the incomplete penetrance observed in some families.
Supporting an oligogenic basis of ALS, there are reports of the co-occurrence of mutations in two ALS-related genes (i.e.: C9orf72, SOD1, FUS, TARDBP, and ANG) in both isolated patients and multiple individuals from ALS families (Luigetti et al., 2011; van Blitterswijk et al., 2012; Chiò et al., 2012b). Compound inheritance of two variants with different effect sizes and frequencies (i.e.: a high risk rare mutation and a more frequent allele acting as a modifier) in the same ALS gene might account for incomplete penetrance observed in ALS families, as already demonstrated in a different genetic condition (Albers et al., 2011).
4.3 Somatic mutations
Somatic mutations are a well-known cause of human disease, with cancer being the most striking example. Advances in next generation sequencing has allowed the discovery of mutations involving a small fraction of brain cells as a cause of severe neurological disorders (Rivière et al, 2012; Lee et al., 2012; Poduri et al., 2012). Several studies also show that mosaicism is a feature of “normal” brain (McConnell et al., 2012; Bushman et al., 2013; Baillie et al., 2011). These observations support the hypothesis that brain mosaicism may be a cause of some complex neurological and psychiatric diseases. Due to cortical architecture and developmental processes, it is also possible that neurons carrying a mosaic mutation are not clustered together, but are interspersed with “normal” neurons (Poduri et al., 2013). For example, a mosaic mutation may involve pyramidal neurons throughout the cortex, sparing all the other types of cortical neurons and glial cells. It is worth citing a reported case of sporadic, early onset Alzheimer’s disease attributed to a somatic presenilin-1 mutation in brain cells (Beck et al., 2004) and a case of Creutzfeld-Jacob disease caused by an early embryonic somatic mutation in PRNP gene (Alzualde et al., 2010). To our knowledge, no such an example has yet been described in ALS, but it remains an interesting hypothesis to be tested in the next future. Furthermore, prion-like mechanisms have been proposed in ALS pathogenesis (Grad et al., 2011; Munch et al., 2011; Furukawa et al., 2011; Nonaka et al., 2013) and, at least conceptually, mosaic mutations would fit with that notion.
7. UNRAVELING THE GENETICS OF ALS: THE WAY FORWARD
Recent years has seen a boom in the identification of new ALS genes. The discovery of the C9orf72 repeat expansion had the biggest impact, explaining a significant proportion of both FALS and SALS cases and of the observed overlap between ALS and FTD. It has been argued that are unlikely to be other genetic discoveries with as high a frequency as the C9orf72 mutation and that, given the high cost of genetic studies, the ALS research community should focus their efforts elsewhere. Perhaps not unsurprisingly, we disagree with that sentiment and counter that a more complete knowledge of the underlying genetic defects is essential in studying and understanding pathogenic mechanisms leading to ALS. Furthermore, frequency should not necessarily be taken as a barometer of importance. For example, mutations in the TARDBP gene account for barely 4% of familial ALS cases, and yet the presence of TDP-43 pathology has come to define the disease.
This leaves us with the question as to how to move forward in the increasingly complicated ALS genetic space. Key to this will be larger cohorts of cases and controls and this will necessitate national and international collaboration to collect these samples. In that regard, making raw genomic data publicly available is an important component, as it allows researchers around the world to access the data and combine it with their own results, thereby increasing the power of their dataset for free. The infrastructure for this socially conscious data sharing has already been established in the form of the dbGaP repository (www. http://www.ncbi.nlm.nih.gov).
The discovery of new genes is in some ways a self-sustaining process: it facilitates further discoveries through many procedures. The characterization of the genetic defects underlying ALS will help to define nosologically the multifaceted nature of ALS as a spectrum of disease, allowing for a classification of cases into clearer phenotypically categories, in which a common genetic cause is more likely to be identified: the “overlapping phenotype” strategy has already demonstrated to be effective in studies relying on WES (Gilissen et al., 2012). A more precise stratification of ALS patients based on the presence of causative genetic mutations will help the identification of further genetic variants acting as modifiers of the phenotype. Similarly, studies aimed to define environmental risk factor will largely benefit form a better characterization of the genetic background.
Finally, but most importantly, the identification of the genetic etiology of ALS allows for the development of targeted therapeutic interventions. Gene therapy in the form of personalized medicine holds great promise and already early stage clinical trials involving antisense oligonucleotides against mutated SOD1 have been completed (Miller et al., 2013). Similar therapy has been proposed also for the C9orf72 repeat expansion (Fernandes et al., 2013).
Highlights.
Amyotrophic lateral sclerosis constitutes a heterogeneous neurodegenerative disorder
Genetics factors play a significant role in ALS etiology and pathogenesis
Genetic causes of ~65% of familial and ~10% of sporadic ALS have been identified
Different genetic mechanisms underlie ALS etiology
NGS techniques may favor the discovery of new genes
List of abbreviations
- ALS
Amyotrophic lateral sclerosis
- FALS
Familial amyotrophic lateral sclerosis
- SALS
Sporadic amyotrophic lateral sclerosis
- FTD
Frontotemporal dementia
- GWAS
Genome-wide association study
- SNP
Single-nucleotide polymorphism
- NGS
Next generation sequencing
- WES
Whole exome sequencing
- dbGaP
database of Genotypes and Phenotypes
- OMIM
Online Mendelian Inheritance in Man
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramzon Y, Johnson JO, Scholz SW, Taylor JP, Brunetti M, Calvo A, Mandrioli J, Benatar M, Mora G, Restagno G, Chiò A, Traynor BJ. Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2012;33(9):2231.e1–2231.e6. doi: 10.1016/j.neurobiolaging.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajroud-Driss S, Fecto F, Ajroud K, Lalani I, Calvo SE, Mootha VK, Deng HX, Siddique N, Tahmoush AJ, Heiman-Patterson TD, Siddique T. Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics. 2014 doi: 10.1007/s10048-014-0421-1. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavi A, Nafissi S, Rohani M, Zamani B, Sedighi B, Shamshiri H, Fan JB, Ronaghi M, Elahi E. Genetic analysis and SOD1 mutation screening in Iranian amyotrophic lateral sclerosis patients. Neurobiol. Aging. 2013;34(5) doi: 10.1016/j.neurobiolaging.2012.09.006. 1516.e1-8. [DOI] [PubMed] [Google Scholar]
- Alavi A, Nafissi S, Rohani M, Shahidi G, Zamani B, Shamshiri H, Safari I, Elahi E. Repeat expansion in C9ORF72 is not a major cause of amyotrophic lateral sclerosis among Iranian patients. Neurobiol. Aging. 2014;35(1) doi: 10.1016/j.neurobiolaging.2013.07.016. 267.e1-7. [DOI] [PubMed] [Google Scholar]
- Albers CA, Paul DS, Schulze H, Freson K, Stephens JC, Smethurst PA, Jolley JD, Cvejic A, Kostadima M, Bertone P, Breuning MH, Debili N, Deloukas P, Favier R, Fiedler J, Hobbs CM, Huang N, Hurles ME, Kiddle G, Krapels I, Nurden P, Ruivenkamp CA, Sambrook JG, Smith K, Stemple DL, Strauss G, Thys C, van Geet C, Newbury-Ecob R, Ouwehand WH, Ghevaert C. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat. Genet. 2012;44(4):435–439. doi: 10.1038/ng.1083. S1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, Andersen PM, Nilsson P, Chioza B, Andersson JL, Russ C, Shaw CE, Powell JF, Leigh PN. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum. Mol. Genet. 1999;8(2):157–164. doi: 10.1093/hmg/8.2.157. [DOI] [PubMed] [Google Scholar]
- Alexander MD, Traynor BJ, Miller N, Corr B, Frost E, McQuaid S, Brett FM, Green A, Hardiman O. “True” sporadic ALS associated with a novel SOD-1 mutation. Ann. Neurol. 2002;52(5):680–683. doi: 10.1002/ana.10369. [DOI] [PubMed] [Google Scholar]
- Al-Saif A, Al-Mohanna F, Bohlega S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann. Neurol. 2011;70(6):913–919. doi: 10.1002/ana.22534. [DOI] [PubMed] [Google Scholar]
- Al-Saif A, Bohlega S, Al-Mohanna F. Loss of ERLIN2 function leads to juvenile primary lateral sclerosis. Ann. Neurol. 2012;72(4):510–516. doi: 10.1002/ana.23641. [DOI] [PubMed] [Google Scholar]
- ALSGEN Consortium. Ahmeti KB, Ajroud-Driss S, Al-Chalabi A, Andersen PM, Armstrong J, Birve A, Blauw HM, Brown RH, Bruijn L, Chen W, Chiò A, Comeau MC, Cronin S, Diekstra FP, Soraya Gkazi A, Glass JD, Grab JD, Groen EJ, Haines JL, Hardiman O, Heller S, Huang J, Hung WY, ITALSGEN consortium. Jaworski JM, Jones A, Khan H, Landers JE, Langefeld CD, Leigh PN, Marion MC, McLaughlin RL, Meininger V, Melki J, Miller JW, Mora G, Pericak-Vance MA, Rampersaud E, Robberecht W, Russell LP, Salachas F, Saris CG, Shatunov A, Shaw CE, Siddique N, Siddique T, Smith BN, Sufit R, Topp S, Traynor BJ, Vance C, van Damme P, van den Berg LH, van Es MA, van Vught PW, Veldink JH, Yang Y, Zheng JG. Age of onset of amyotrophic lateral sclerosis is modulated by a locus on 1p34.1. Neurobiol. Aging. 2013;34(1) doi: 10.1016/j.neurobiolaging.2012.07.017. 357.e7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzualde A, Moreno F, Martínez-Lage P, Ferrer I, Gorostidi A, Otaegui D, Blázquez L, Atares B, Cardoso S, Martínez de Pancorbo M, Juste R, Rodríguez-Martínez AB, Indakoetxea B, López de Munain A. Somatic mosaicism in a case of apparently sporadic Creutzfeldt-Jakob disease carrying a de novo D178N mutation in the PRNP gene. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B(7):1283–1291. doi: 10.1002/ajmg.b.31099. [DOI] [PubMed] [Google Scholar]
- Andersen PM. Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr. Neurol. Neurosci. Rep. 2006;6(1):37–46. doi: 10.1007/s11910-996-0008-9. [DOI] [PubMed] [Google Scholar]
- Andersen PM, Forsgren L, Binzer M, Nilsson P, Ala-Hurula V, Keränen ML, Bergmark L, Saarinen A, Haltia T, Tarvainen I, Kinnunen E, Udd B, Marklund SL. Autosomal recessive adult-onset amyotrophic lateral sclerosis associated with homozygosity for Asp90Ala CuZn-superoxide dismutase mutation. A clinical and genealogical study of 36 patients. Brain. 1996;119(Pt 4):1153–1172. doi: 10.1093/brain/119.4.1153. [DOI] [PubMed] [Google Scholar]
- Aran FA. Research on an as yet undescribed disease of the muscular system (progressive muscular atrophy) Arch. Gen. Med. 1848;24:15–35. [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, Talbot RT, Gustincich S, Freeman TC, Mattick JS, Hume DA, Heutink P, Carninci P, Jeddeloh JA, Faulkner GJ. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479(7374):534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K, Berg-Alonso L, Kageyama Y, Serre V, Moore DG, Verschueren A, Rouzier C, Le Ber I, Augé G, Cochaud C, Lespinasse F, N’Guyen K, de Septenville A, Brice A, Yu-Wai-Man P, Sesaki H, Pouget J, Paquis-Flucklinger V. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137(Pt 8):2329–2345. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JA, Poulter M, Campbell TA, Uphill JB, Adamson G, Geddes JF, Revesz T, Davis MB, Wood NW, Collinge J, Tabrizi SJ. Somatic and germline mosaicism in sporadic early-onset Alzheimer’s disease. Hum. Mol. Genet. 2004;13(12):1219–1224. doi: 10.1093/hmg/ddh134. [DOI] [PubMed] [Google Scholar]
- Benatar M, Wuu J, Fernandez C, Weihl CC, Katzen H, Steele J, Oskarsson B, Taylor JP. Motor neuron involvement in multisystem proteinopathy: implications for ALS. Neurology. 2013;80(20):1874–1880. doi: 10.1212/WNL.0b013e3182929fc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco H, Bernard-Marissal N, Vourc’h P, Guettard YO, Sunyach C, Augereau O, Khederchah J, Mouzat K, Antar C, Gordon PH, Veyrat-Durebex C, Besson G, Andersen PM, Salachas F, Meininger V, Camu W, Pettmann B, Andres CR, Corcia P, French ALS Study Group A rare motor neuron deleterious missense mutation in the DPYSL3 (CRMP4) gene is associated with ALS. Hum. Mutat. 2013;34(7):953–960. doi: 10.1002/humu.22329. [DOI] [PubMed] [Google Scholar]
- Blauw HM, Veldink JH, van Es MA, van Vught PW, Saris CG, van der Zwaag B, Franke L, Burbach JP, Wokke JH, Ophoff RA, van den Berg LH. Copy-number variation in sporadic amyotrophic lateral sclerosis: a genome-wide screen. Lancet Neurol. 2008;7(4):319–326. doi: 10.1016/S1474-4422(08)70048-6. [DOI] [PubMed] [Google Scholar]
- Blauw HM, Al-Chalabi A, Andersen PM, van Vught PW, Diekstra FP, van Es MA, Saris CG, Groen EJ, van Rheenen W, Koppers M, Van’t Slot R, Strengman E, Estrada K, Rivadeneira F, Hofman A, Uitterlinden AG, Kiemeney LA, Vermeulen SH, Birve A, Waibel S, Meyer T, Cronin S, McLaughlin RL, Hardiman O, Sapp PC, Tobin MD, Wain LV, Tomik B, Slowik A, Lemmens R, Rujescu D, Schulte C, Gasser T, Brown RH, Jr, Landers JE, Robberecht W, Ludolph AC, Ophoff RA, Veldink JH, van den Berg LH. A large genome scan for rare CNVs in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2010;19(20):4091–4099. doi: 10.1093/hmg/ddq323. [DOI] [PubMed] [Google Scholar]
- Blauw HM, Barnes CP, van Vught PW, van Rheenen W, Verheul M, Cuppen E, Veldink JH, van den Berg LH. SMN1 gene duplications are associated with sporadic ALS. Neurology. 2012;78(11):776–780. doi: 10.1212/WNL.0b013e318249f697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroni B, Bonvicini C, Alberici A, Buratti E, Agosti C, Archetti S, Papetti A, Stuani C, Di Luca M, Gennarelli M, Padovani A. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum. Mutat. 2009;30(11):E974–983. doi: 10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- Brown JA, Min J, Staropoli JF, Collin E, Bi S, Feng X, Barone R, Cao Y, O’Malley L, Xin W, Mullen TE, Sims KB. SOD1, ANG, TARDBP and FUS mutations in amyotrophic lateral sclerosis: a United States clinical testing lab experience. Amyotroph. Lateral Scler. 2012;13(2):217–222. doi: 10.3109/17482968.2011.643899. [DOI] [PubMed] [Google Scholar]
- Bushman DM, Chun J. The genomically mosaic brain: aneuploidy and more in neural diversity and disease. Semin. Cell. Dev. Biol. 2013;24(4):357–369. doi: 10.1016/j.semcdb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady J, Koval ED, Benitez BA, Zaidman C, Jockel-Balsarotti J, Allred P, Baloh RH, Ravits J, Simpson E, Appel SH, Pestronk A, Goate AM, Miller TM, Cruchaga C, Harms MB. TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. J.A.M.A. Neurol. 2014;71(4):449–453. doi: 10.1001/jamaneurol.2013.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo A, Moglia C, Canosa A, Brunetti M, Barberis M, Traynor BJ, Carrara G, Valentini C, Restagno G, Chiò A. A de novo nonsense mutation of the FUS gene in an apparently familial amyotrophic lateral sclerosis case. Neurobiol. Aging. 2014;35(6):1513.e7–1513.e11. doi: 10.1016/j.neurobiolaging.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau PM, Lenk GM, Lu JT, Bae Y, Burrage L, Turnpenny P, Román Corona-Rivera J, Morandi L, Mora M, Reutter H, Vulto-van Silfhout AT, Faivre L, Haan E, Gibbs RA, Meisler MH, Lee BH. Yunis-Varón syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am. J. Hum. Genet. 2013;92(5):781–791. doi: 10.1016/j.ajhg.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charcot JM, Joffroy A. Deux cas d’atrophie musculaire progressive avec lesions de la substance grise et des faisceaux antero-lateraux de la moelle epiniere [French] Arch. Physiol. Neurol. Pathol. 1869;2:744. [Google Scholar]
- Chaussenot A, Le Ber I, Ait-El-Mkadem S, Camuzat A, de Septenville A, Bannwarth S, Genin EC, Serre V, Augé G, The French research network on FTD and FTD-ALS. Brice A, Pouget J, Paquis-Flucklinger V. Screening of CHCHD10 in a French cohort confirms the involvement of this gene in frontotemporal dementia with amyotrophic lateral sclerosis patients. Neurobiol. Aging. 2014 doi: 10.1016/j.neurobiolaging.2014.07.022. pii:S0197-4580(14)00491-6. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, Nicholson GA, Auer-Grumbach M, Wagner K, De Jonghe P, Griffin JW, Fischbeck KH, Timmerman V, Cornblath DR, Chance PF. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am. J. Hum. Genet. 2004;74(6):1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HJ, Anagnostou G, Chai A, Withers J, Morris A, Adhikaree J, Pennetta G, de Belleroche JS. Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. J. Biol. Chem. 2010;285(51):40266–40281. doi: 10.1074/jbc.M110.161398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zheng ZZ, Chen X, Huang R, Yang Y, Yuan L, Pan L, Hadano S, Shang HF. SQSTM1 mutations in Han Chinese populations with sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2014;35(3) doi: 10.1016/j.neurobiolaging.2013.09.008. 726.e7-9. [DOI] [PubMed] [Google Scholar]
- Chesi A, Staahl BT, Jovičić A, Couthouis J, Fasolino M, Raphael AR, Yamazaki T, Elias L, Polak M, Kelly C, Williams KL, Fifita JA, Maragakis NJ, Nicholson GA, King OD, Reed R, Crabtree GR, Blair IP, Glass JD, Gitler AD. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 2013;16(7):851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 2011;31(46):16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Traynor BJ, Lombardo F, Fimognari M, Calvo A, Ghiglione P, Mutani R, Restagno G. Prevalence of SOD1 mutations in the Italian ALS population. Neurology. 2008;70(7):533–537. doi: 10.1212/01.wnl.0000299187.90432.3f. [DOI] [PubMed] [Google Scholar]
- Chiò A, Logroscino G, Hardiman O, Swingler R, Mitchell D, Beghi E, Traynor BG, Eurals Consortium Prognostic factors in ALS: A critical review. Amyotroph. Lateral Scler. 2009a;10(5–6):310–323. doi: 10.3109/17482960802566824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Schymick JC, Restagno G, Scholz SW, Lombardo F, Lai SL, Mora G, Fung HC, Britton A, Arepalli S, Gibbs JR, Nalls M, Berger S, Kwee LC, Oddone EZ, Ding J, Crews C, Rafferty I, Washecka N, Hernandez D, Ferrucci L, Bandinelli S, Guralnik J, Macciardi F, Torri F, Lupoli S, Chanock SJ, Thomas G, Hunter DJ, Gieger C, Wichmann HE, Calvo A, Mutani R, Battistini S, Giannini F, Caponnetto C, Mancardi GL, La Bella V, Valentino F, Monsurrò MR, Tedeschi G, Marinou K, Sabatelli M, Conte A, Mandrioli J, Sola P, Salvi F, Bartolomei I, Siciliano G, Carlesi C, Orrell RW, Talbot K, Simmons Z, Connor J, Pioro EP, Dunkley T, Stephan DA, Kasperaviciute D, Fisher EM, Jabonka S, Sendtner M, Beck M, Bruijn L, Rothstein J, Schmidt S, Singleton A, Hardy J, Traynor BJ. A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum. Mol. Genet. 2009b;18(8):1524–1532. doi: 10.1093/hmg/ddp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Calvo A, Moglia C, Restagno G, Ossola I, Brunetti M, Montuschi A, Cistaro A, Ticca A, Traynor BJ, Schymick JC, Mutani R, Marrosu MG, Murru MR, Borghero G. Amyotrophic lateral sclerosis-frontotemporal lobar dementia in 3 families with p.Ala382Thr TARDBP mutations. Arch. Neurol. 2010;67(8):1002–1009. doi: 10.1001/archneurol.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Borghero G, Pugliatti M, Ticca A, Calvo A, Moglia C, Mutani R, Brunetti M, Ossola I, Marrosu MG, Murru MR, Floris G, Cannas A, Parish LD, Cossu P, Abramzon Y, Johnson JO, Nalls MA, Arepalli S, Chong S, Hernandez DG, Traynor BJ, Restagno G, Italian Amyotrophic Lateral Sclerosis Genetic (ITALSGEN) Consortium Large proportion of amyotrophic lateral sclerosis cases in Sardinia due to a single founder mutation of the TARDBP gene. Arch. Neurol. 2011a;68(5):594–598. doi: 10.1001/archneurol.2010.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Calvo A, Moglia C, Ossola I, Brunetti M, Sbaiz L, Lai SL, Abramzon Y, Traynor BJ, Restagno G. A de novo missense mutation of the FUS gene in a “true” sporadic ALS case. Neurobiol. Aging. 2011b;32(3) doi: 10.1016/j.neurobiolaging.2010.05.016. 553.e23-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Borghero G, Restagno G, Mora G, Drepper C, Traynor BJ, Sendtner M, Brunetti M, Ossola I, Calvo A, Pugliatti M, Sotgiu MA, Murru MR, Marrosu MG, Marrosu F, Marinou K, Mandrioli J, Sola P, Caponnetto C, Mancardi G, Mandich P, La Bella V, Spataro R, Conte A, Monsurrò MR, Tedeschi G, Pisano F, Bartolomei I, Salvi F, Lauria Pinter G, Simone I, Logroscino G, Gambardella A, Quattrone A, Lunetta C, Volanti P, Zollino M, Penco S, Battistini S, ITALSGEN consortium. Renton AE, Majounie E, Abramzon Y, Conforti FL, Giannini F, Corbo M, Sabatelli M. Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9ORF72. Brain. 2012a;135(Pt 3):784–793. doi: 10.1093/brain/awr366. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Restagno G, Brunetti M, Ossola I, Calvo A, Canosa A, Moglia C, Floris G, Tacconi P, Marrosu F, Marrosu MG, Murru MR, Majounie E, Renton AE, Abramzon Y, Pugliatti M, Sotgiu MA, Traynor BJ, Borghero G, SARDINIALS Consortium ALS/FTD phenotype in two Sardinian families carrying both C9ORF72 and TARDBP mutations. J. Neurol. Neurosurg. Psychiatry. 2012b;83(7):730–733. doi: 10.1136/jnnp-2012-302219. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Mora G, Restagno G, Brunetti M, Ossola I, Barberis M, Ferrucci L, Canosa A, Manera U, Moglia C, Fuda G, Traynor BJ, Calvo A. UNC13A influences survival in Italian amyotrophic lateral sclerosis patients: a population-based study. Neurobiol. Aging. 2013a;34(1) doi: 10.1016/j.neurobiolaging.2012.07.016. 357.e1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, Figlewicz D, Brown RH, Meisler MH. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am. J. Hum. Genet. 2009;84(1):85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte A, Lattante S, Zollino M, Marangi G, Luigetti M, Del Grande A, Servidei S, Trombetta F, Sabatelli M. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul. Disord. 2012;22(1):73–75. doi: 10.1016/j.nmd.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Corcia P, Mayeux-Portas V, Khoris J, de Toffol B, Autret A, Müh JP, Camu W, Andres C, French ALS Research Group Amyotrophic Lateral Sclerosis. Abnormal SMN1 gene copy number is a susceptibility factor for amyotrophic lateral sclerosis. Ann. Neurol. 2002;51(2):243–246. doi: 10.1002/ana.10104. [DOI] [PubMed] [Google Scholar]
- Corcia P, Camu W, Halimi JM, Vourc’h P, Antar C, Vedrine S, Giraudeau B, de Toffol B, Andres CR, French ALS Study Group SMN1 gene, but not SMN2, is a risk factor for sporadic ALS. Neurology. 2006;67(7):1147–1150. doi: 10.1212/01.wnl.0000233830.85206.1e. [DOI] [PubMed] [Google Scholar]
- Corcia P, Ingre C, Blasco H, Press R, Praline J, Antar C, Veyrat-Durebex C, Guettard YO, Camu W, Andersen PM, Vourc’h P, Andres CR. Homozygous SMN2 deletion is a protective factor in the Swedish ALS population. Eur. J. Hum. Genet. 2012;20(5):588–591. doi: 10.1038/ejhg.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrado L, Carlomagno Y, Falasco L, Mellone S, Godi M, Cova E, Cereda C, Testa L, Mazzini L, D’Alfonso S. A novel peripherin gene (PRPH) mutation identified in one sporadic amyotrophic lateral sclerosis patient. Neurobiol. Aging. 2011;32(3) doi: 10.1016/j.neurobiolaging.2010.02.011. 552.e1-6. [DOI] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, Epstein J, Chiang A, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Solski JA, Williams KL, Mojsilovic-Petrovic J, Ingre C, Boylan K, Graff-Radford NR, Dickson DW, Clay-Falcone D, Elman L, McCluskey L, Greene R, Kalb RG, Lee VM, Trojanowski JQ, Ludolph A, Robberecht W, Andersen PM, Nicholson GA, Blair IP, King OD, Bonini NM, Van Deerlin V, Rademakers R, Mourelatos Z, Gitler AD. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. U.S.A. 2011;108(52):20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, Kim CE, Frackelton EC, Solski JA, Williams KL, Clay-Falcone D, Elman L, McCluskey L, Greene R, Hakonarson H, Kalb RG, Lee VM, Trojanowski JQ, Nicholson GA, Blair IP, Bonini NM, Van Deerlin VM, Mourelatos Z, Shorter J, Gitler AD. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012;21(13):2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LE, Ferraiuolo L, Goodall EF, Heath PR, Higginbottom A, Mortiboys H, Hollinger HC, Hartley JA, Brockington A, Burness CE, Morrison KE, Wharton SB, Grierson AJ, Ince PG, Kirby J, Shaw PJ. Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS) P.L.o.S. One. 2010;5(3):e9872. doi: 10.1371/journal.pone.0009872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino M, Ferri A, Valle C, Carrì MT. Mitochondria and ALS: implications from novel genes and pathways. Mol. Cell. Neurosci. 2013;55:44–49. doi: 10.1016/j.mcn.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Cronin S, Hardiman O, Traynor BJ. Ethnic variation in the incidence of ALS: a systematic review. Neurology. 2007;68(13):1002–1007. doi: 10.1212/01.wnl.0000258551.96893.6f. [DOI] [PubMed] [Google Scholar]
- Cronin S, Berger S, Ding J, Schymick JC, Washecka N, Hernandez DG, Greenway MJ, Bradley DG, Traynor BJ, Hardiman O. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum. Mol. Genet. 2008a;17(5):768–774. doi: 10.1093/hmg/ddm361. [DOI] [PubMed] [Google Scholar]
- Cronin S, Blauw HM, Veldink JH, van Es MA, Ophoff RA, Bradley DG, van den Berg LH, Hardiman O. Analysis of genome-wide copy number variation in Irish and Dutch ALS populations. Hum. Mol. Genet. 2008b;17(21):3392–3398. doi: 10.1093/hmg/ddn233. [DOI] [PubMed] [Google Scholar]
- Cruveilhier J. Sur la paralysie musculaire, progressive, atrophique [French] Bull. Acad. Med. (Paris) 1852;18:490–502. 546–583. [Google Scholar]
- Cudkowicz ME, Warren L, Francis JW, Lloyd KJ, Friedlander RM, Borges LF, Kassem N, Munsat TL, Brown RH., Jr Intrathecal administration of recombinant human superoxide dismutase 1 in amyotrophic lateral sclerosis: a preliminary safety and pharmacokinetic study. Neurology. 1997;49(1):213–222. doi: 10.1212/wnl.49.1.213. [DOI] [PubMed] [Google Scholar]
- Daoud H, Belzil V, Martins S, Sabbagh M, Provencher P, Lacomblez L, Meininger V, Camu W, Dupré N, Dion PA, Rouleau GA. Association of long ATXN2 CAG repeat sizes with increased risk of amyotrophic lateral sclerosis. Arch. Neurol. 2011;68(6):739–742. doi: 10.1001/archneurol.2011.111. [DOI] [PubMed] [Google Scholar]
- Daoud H, Zhou S, Noreau A, Sabbagh M, Belzil V, Dionne-Laporte A, Tranchant C, Dion P, Rouleau GA. Exome sequencing reveals SPG11 mutations causing juvenile ALS. Neurobiol. Aging. 2012;33(4) doi: 10.1016/j.neurobiolaging.2011.11.012. 839.e5-9. [DOI] [PubMed] [Google Scholar]
- de Leeuw CN. CMT4J: Charcot-Marie-Tooth disorder caused by mutations in FIG4. Clin. Genet. 2008;73(4):318–319. doi: 10.1111/j.1399-0004.2008.00962_3.x. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Kocerha J, Finch N, Crook R, Baker M, Desaro P, Johnston A, Rutherford N, Wojtas A, Kennelly K, Wszolek ZK, Graff-Radford N, Boylan K, Rademakers R. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum. Mutat. 2010;31(5):E1377–1389. doi: 10.1002/humu.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p–linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477(7363):211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Wei L, Zuo X, Tian Y, Xie F, Hu P, Zhu C, Yu F, Meng Y, Wang H, Zhang F, Ma H, Ye R, Cheng H, Du J, Dong W, Zhou S, Wang C, Wang Y, Wang J, Chen X, Sun Z, Zhou N, Jiang Y, Liu X, Li X, Zhang N, Liu N, Guan Y, Han Y, Han Y, Lv X, Fu Y, Yu H, Xi C, Xie D, Zhao Q, Xie P, Wang X, Zhang Z, Shen L, Cui Y, Yin X, Cheng H, Liang B, Zheng X, Lee TM, Chen G, Zhou F, Veldink JH, Robberecht W, Landers JE, Andersen PM, Al-Chalabi A, Shaw C, Liu C, Tang B, Xiao S, Robertson J, Zhang F, van den Berg LH, Sun L, Liu J, Yang S, Ju X, Wang K, Zhang X. Genome-wide association analyses in Han Chinese identify two new susceptibility loci for amyotrophic lateral sclerosis. Nat. Genet. 2013;45(6):697–700. doi: 10.1038/ng.2627. [DOI] [PubMed] [Google Scholar]
- Diekstra FP, van Vught PW, van Rheenen W, Koppers M, Pasterkamp RJ, van Es MA, Schelhaas HJ, de Visser M, Robberecht W, Van Damme P, Andersen PM, van den Berg LH, Veldink JH. UNC13A is a modifier of survival in amyotrophic lateral sclerosis. Neurobiol. Aging. 2012a;33(3) doi: 10.1016/j.neurobiolaging.2011.10.029. 630.e3-8. [DOI] [PubMed] [Google Scholar]
- Dunckley T, Huentelman MJ, Craig DW, Pearson JV, Szelinger S, Joshipura K, Halperin RF, Stamper C, Jensen KR, Letizia D, Hesterlee SE, Pestronk A, Levine T, Bertorini T, Graves MC, Mozaffar T, Jackson CE, Bosch P, McVey A, Dick A, Barohn R, Lomen-Hoerth C, Rosenfeld J, O’connor DT, Zhang K, Crook R, Ryberg H, Hutton M, Katz J, Simpson EP, Mitsumoto H, Bowser R, Miller RG, Appel SH, Stephan DA. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N. Engl. J. Med. 2007;357(8):775–788. doi: 10.1056/NEJMoa070174. [DOI] [PubMed] [Google Scholar]
- Duquette A, Roddier K, McNabb-Baltar J, Gosselin I, St-Denis A, Dicaire MJ, Loisel L, Labuda D, Marchand L, Mathieu J, Bouchard JP, Brais B. Mutations in senataxin responsible for Quebec cluster of ataxia with neuropathy. Ann. Neurol. 2005;57(3):408–414. doi: 10.1002/ana.20408. [DOI] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rüb U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, Gitler AD. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466(7310):1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi4K Consortium; Epilepsy Phenome/Genome Project. Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE, Epstein MP, Glauser T, Goldstein DB, Han Y, Heinzen EL, Hitomi Y, Howell KB, Johnson MR, Kuzniecky R, Lowenstein DH, Lu YF, Madou MR, Marson AG, Mefford HC, Esmaeeli Nieh S, O’Brien TJ, Ottman R, Petrovski S, Poduri A, Ruzzo EK, Scheffer IE, Sherr EH, Yuskaitis CJ, Abou-Khalil B, Alldredge BK, Bautista JF, Berkovic SF, Boro A, Cascino GD, Consalvo D, Crumrine P, Devinsky O, Dlugos D, Epstein MP, Fiol M, Fountain NB, French J, Friedman D, Geller EB, Glauser T, Glynn S, Haut SR, Hayward J, Helmers SL, Joshi S, Kanner A, Kirsch HE, Knowlton RC, Kossoff EH, Kuperman R, Kuzniecky R, Lowenstein DH, McGuire SM, Motika PV, Novotny EJ, Ottman R, Paolicchi JM, Parent JM, Park K, Poduri A, Scheffer IE, Shellhaas RA, Sherr EH, Shih JJ, Singh R, Sirven J, Smith MC, Sullivan J, Lin Thio L, Venkat A, Vining EP, Von Allmen GK, Weisenberg JL, Widdess-Walsh P, Winawer MR. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschbach J, Schwalenstöcker B, Soyal SM, Bayer H, Wiesner D, Akimoto C, Nilsson AC, Birve A, Meyer T, Dupuis L, Danzer KM, Andersen PM, Witting A, Ludolph AC, Patsch W, Weydt P. PGC-1α is a male-specific disease modifier of human and experimental amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013;22(17):3477–3484. doi: 10.1093/hmg/ddt202. [DOI] [PubMed] [Google Scholar]
- Eymard-Pierre E, Lesca G, Dollet S, Santorelli FM, di Capua M, Bertini E, Boespflug-Tanguy O. Infantile-onset ascending hereditary spastic paralysis is associated with mutations in the alsin gene. Am. J. Hum. Genet. 2002;71(3):518–527. doi: 10.1086/342359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Xu W, Wu C, Zhu M, Li X, Hong D. MAPT as a predisposing gene for sporadic amyotrophic lateral sclerosis in the Chinese Han population. Neural Regen. Res. 2013;8(33):3116–3123. doi: 10.3969/j.issn.1673-5374.2013.33.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer MJ, Hulihan MM, Kachergus JM, Dächsel JC, Stoessl AJ, Grantier LL, Calne S, Calne DB, Lechevalier B, Chapon F, Tsuboi Y, Yamada T, Gutmann L, Elibol B, Bhatia KP, Wider C, Vilariño-Güell C, Ross OA, Brown LA, Castanedes-Casey M, Dickson DW, Wszolek ZK. DCTN1 mutations in Perry syndrome. Nat. Genet. 2009;41(2):163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, Zheng JG, Shi Y, Siddique N, Arrat H, Donkervoort S, Ajroud-Driss S, Sufit RL, Heller SL, Deng HX, Siddique T. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 2011;68(11):1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- Felbecker A, Camu W, Valdmanis PN, Sperfeld AD, Waibel S, Steinbach P, Rouleau GA, Ludolph AC, Andersen PM. Four familial ALS pedigrees discordant for two SOD1 mutations: are all SOD1 mutations pathogenic? J. Neurol. Neurosurg. Psychiatry. 2010;81(5):572–577. doi: 10.1136/jnnp.2009.192310. [DOI] [PubMed] [Google Scholar]
- Figlewicz DA, Krizus A, Martinoli MG, Meininger V, Dib M, Rouleau GA, Julien JP. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum. Mol. Genet. 1994;3(10):1757–1761. doi: 10.1093/hmg/3.10.1757. [DOI] [PubMed] [Google Scholar]
- Figley MD, Thomas A, Gitler AD. Evaluating noncoding nucleotide repeat expansions in amyotrophic lateral sclerosis. Neurobiol. Aging. 2014;35(4) doi: 10.1016/j.neurobiolaging.2013.09.024. 936.e1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Romero C, Hur J, Bender DE, Delaney CE, Cataldo MD, Smith AL, Yung R, Ruden DM, Callaghan BC, Feldman EL. Identification of epigenetically altered genes in sporadic amyotrophic lateral sclerosis. P.L.o.S. One. 2012;7(12):e52672. doi: 10.1371/journal.pone.0052672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogh I, Ratti A, Gellera C, Lin K, Tiloca C, Moskvina V, Corrado L, Sorarù G, Cereda C, Corti S, Gentilini D, Calini D, Castellotti B, Mazzini L, Querin G, Gagliardi S, Del Bo R, Conforti FL, Siciliano G, Inghilleri M, Saccà F, Bongioanni P, Penco S, Corbo M, Sorbi S, Filosto M, Ferlini A, Di Blasio AM, Signorini S, Shatunov A, Jones A, Shaw PJ, Morrison KE, Farmer AE, Van Damme P, Robberecht W, Chiò A, Traynor BJ, Sendtner M, Melki J, Meininger V, Hardiman O, Andersen PM, Leigh NP, Glass JD, Overste D, Diekstra FP, Veldink JH, van Es MA, Shaw CE, Weale ME, Lewis CM, Williams J, Brown RH, Landers JE, Ticozzi N, Ceroni M, Pegoraro E, Comi GP, D’Alfonso S, van den Berg LH, Taroni F, Al-Chalabi A, Powell J, Silani V, the SLAGEN Consortium and Collaborators A genome-wide association meta-analysis identifies a novel locus at 17q11.2 associated with sporadic amyotrophic lateral sclerosis. Hum. Mol. Genet. 2014;23(8):2220–2231. doi: 10.1093/hmg/ddt587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J. Biol. Chem. 2011;286:18664–18672. doi: 10.1074/jbc.M111.231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Disease gene identification strategies for exome sequencing. Eur. J. Hum. Genet. 2012;20(5):490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, 3rd, Bigio EH, Caselli R, Baker M, Al-Lozi MT, Morris JC, Pestronk A, Rademakers R, Goate AM, Cairns NJ. TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 2008;63(4):535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall EF, Greenway MJ, van Marion I, Carroll CB, Hardiman O, Morrison KE. Association of the H63D polymorphism in the hemochromatosis gene with sporadic ALS. Neurology. 2005;65(6):934–937. doi: 10.1212/01.wnl.0000176032.94434.d4. [DOI] [PubMed] [Google Scholar]
- Grad LI, Guest WC, Yanai A, Pokrishevsky E, O’Neill MA, Gibbs E, Semenchenko V, Yousefi M, Wishart DS, Plotkin SS, Cashman NR. Intermolecular transmission of superoxide dismutase 1 misfolding in living cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16398–16403. doi: 10.1073/pnas.1102645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Visscher PM, Mowry BJ, Wray NR. Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nat. Genet. 2013;45(3):234–238. doi: 10.1038/ng.2555. [DOI] [PubMed] [Google Scholar]
- Greenway MJ, Alexander MD, Ennis S, Traynor BJ, Corr B, Frost E, Green A, Hardiman O. A novel candidate region for ALS on chromosome 14q11.2. Neurology. 2004;(6310):1936–1938. doi: 10.1212/01.wnl.0000144344.39103.f6. [DOI] [PubMed] [Google Scholar]
- Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, Morrison KE, Green A, Acharya KR, Brown RH, Jr, Hardiman O. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat. Genet. 2006;38(4):411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- Groen EJ, van Rheenen W, Koppers M, van Doormaal PT, Vlam L, Diekstra FP, Dooijes D, Pasterkamp RJ, van den Berg LH, Veldink JH. CGG-repeat expansion in FMR1 is not associated with amyotrophic lateral sclerosis. Neurobiol. Aging. 2012;33(8) doi: 10.1016/j.neurobiolaging.2012.03.007. 1852.e1-3. [DOI] [PubMed] [Google Scholar]
- Gros-Louis F, Larivière R, Gowing G, Laurent S, Camu W, Bouchard JP, Meininger V, Rouleau GA, Julien JP. A frameshift deletion in peripherin gene associated with amyotrophic lateral sclerosis. J. Biol. Chem. 2004;279(44):45951–45956. doi: 10.1074/jbc.M408139200. [DOI] [PubMed] [Google Scholar]
- Gros-Louis F, Andersen PM, Dupre N, Urushitani M, Dion P, Souchon F, D’Amour M, Camu W, Meininger V, Bouchard JP, Rouleau GA, Julien JP. Chromogranin B P413L variant as risk factor and modifier of disease onset for amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 2009;106(51):21777–21782. doi: 10.1073/pnas.0902174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat. Genet. 2012;44(2):157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH, Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat. Genet. 2001;29(2):166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- Hand CK, Khoris J, Salachas F, Gros-Louis F, Lopes AA, Mayeux-Portas V, Brewer CG, Brown RH, Jr, Meininger V, Camu W, Rouleau GA. A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am. J. Hum. Genet. 2002;70(1):251–256. doi: 10.1086/337945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosler BA, Siddique T, Sapp PC, Sailor W, Huang MC, Hossain A, Daube JR, Nance M, Fan C, Kaplan J, Hung WY, McKenna-Yasek D, Haines JL, Pericak-Vance MA, Horvitz HR, Brown RH., Jr Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. J.A.M.A. 2000;284(13):1664–1669. doi: 10.1001/jama.284.13.1664. [DOI] [PubMed] [Google Scholar]
- Iida A, Takahashi A, Kubo M, Saito S, Hosono N, Ohnishi Y, Kiyotani K, Mushiroda T, Nakajima M, Ozaki K, Tanaka T, Tsunoda T, Oshima S, Sano M, Kamei T, Tokuda T, Aoki M, Hasegawa K, Mizoguchi K, Morita M, Takahashi Y, Katsuno M, Atsuta N, Watanabe H, Tanaka F, Kaji R, Nakano I, Kamatani N, Tsuji S, Sobue G, Nakamura Y, Ikegawa S. A functional variant in ZNF512B is associated with susceptibility to amyotrophic lateral sclerosis in Japanese. Hum. Mol. Genet. 2011;20(18):3684–3692. doi: 10.1093/hmg/ddr268. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Glynn SM, Gibbs JR, Nalls MA, Sabatelli M, Restagno G, Drory VE, Chio A, Rogaeva E, Traynor BJ. Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain. 2014 doi: 10.1093/brain/awu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurrò MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, ITALSGEN Consortium. Galassi G, Scholz SW, Taylor JP, Restagno G, Chiò A, Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68(5):857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, Pliner HA, Abramzon Y, Marangi G, Winborn BJ, Gibbs JR, Nalls MA, Morgan S, Shoai M, Hardy J, Pittman A, Orrell RW, Malaspina A, Sidle KC, Fratta P, Harms MB, Baloh RH, Pestronk A, Weihl CC, Rogaeva E, Zinman L, Drory VE, Borghero G, Mora G, Calvo A, Rothstein JD;ITALSGEN, Drepper C, Sendtner M, Singleton AB, Taylor JP, Cookson MR, Restagno G, Sabatelli M, Bowser R, Chiò A, Traynor BJ. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 2014;17(5):664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40(5):572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Keller MF, Ferrucci L, Singleton AB, Tienari PJ, Laaksovirta H, Restagno G, Chiò A, Traynor BJ, Nall MA. Heritability in ALS: Meta-analysis identifies highly heritable regions associated with risk of ALS. J.A.M.A. Neurology. 2014;71(9):1123–1134. doi: 10.1001/jamaneurol.2014.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers M, van Blitterswijk MM, Vlam L, Rowicka PA, van Vught PW, Groen EJ, Spliet WG, Engelen-Lee J, Schelhaas HJ, de Visser M, van der Kooi AJ, van der Pol WL, Pasterkamp RJ, Veldink JH, van den Berg LH. VCP mutations in familial and sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2012;33(4) doi: 10.1016/j.neurobiolaging.2011.10.006. 837.e7-13. [DOI] [PubMed] [Google Scholar]
- Kurland LT, Mulder DW. Epidemiologic investigations of amyotrophic lateral sclerosis. 2. Familial aggregations indicative of dominant inheritance II. Neurology. 1955;5:249–268. doi: 10.1212/wnl.5.4.249. [DOI] [PubMed] [Google Scholar]
- Kuźma-Kozakiewicz M, Jędrzejowska M, Kaźmierczak B. SMN1 gene duplications are more frequent in patients with progressive muscular atrophy. Amyotroph. Lateral Scler. Frontotemporal Degener. 2013;14(5–6):457–462. doi: 10.3109/21678421.2013.771367. [DOI] [PubMed] [Google Scholar]
- Kwee LC, Liu Y, Haynes C, Gibson JR, Stone A, Schichman SA, Kamel F, Nelson LM, Topol B, Van den Eeden SK, Tanner CM, Cudkowicz ME, Grasso DL, Lawson R, Muralidhar S, Oddone EZ, Schmidt S, Hauser MA. A high-density genome-wide association screen of sporadic ALS in US veterans. P.L.o.S. One. 2012;7(3):e32768. doi: 10.1371/journal.pone.0032768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kwok CT, Morris A, de Belleroche JS. Sequestosome-1 (SQSTM1) sequence variants in ALS cases in the UK: prevalence and coexistence of SQSTM1 mutations in ALS kindred with PDB. Eur. J. Hum. Genet. 2014;22(4):492–496. doi: 10.1038/ejhg.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon MJ, Baek W, Ki CS, Kim HY, Koh SH, Kim JW, Kim SH. Screening of the SOD1, FUS, TARDBP, ANG, and OPTN mutations in Korean patients with familial and sporadic ALS. Neurobiol. Aging. 2012;33(5) doi: 10.1016/j.neurobiolaging.2011.12.003. 1017.e17-23. [DOI] [PubMed] [Google Scholar]
- Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai SL, Myllykangas L, Sulkava R, Jansson L, Hernandez DG, Gibbs JR, Nalls MA, Heckerman D, Tienari PJ, Traynor BJ. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9(10):978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts D, Poesen K, Fernández-Santiago R, Al-Chalabi A, Del Bo R, Van Vught PW, Khan S, Marklund SL, Brockington A, van Marion I, Anneser J, Shaw C, Ludolph AC, Leigh NP, Comi GP, Gasser T, Shaw PJ, Morrison KE, Andersen PM, Van den Berg LH, Thijs V, Siddique T, Robberecht W, Carmeliet P. Meta-analysis of vascular endothelial growth factor variations in amyotrophic lateral sclerosis: increased susceptibility in male carriers of the −2578AA genotype. J. Med. Genet. 2009;46(12):840–846. doi: 10.1136/jmg.2008.058222. [DOI] [PubMed] [Google Scholar]
- Landers JE, Melki J, Meininger V, Glass JD, van den Berg LH, van Es MA, Sapp PC, van Vught PW, McKenna-Yasek DM, Blauw HM, Cho TJ, Polak M, Shi L, Wills AM, Broom WJ, Ticozzi N, Silani V, Ozoguz A, Rodriguez-Leyva I, Veldink JH, Ivinson AJ, Saris CG, Hosler BA, Barnes-Nessa A, Couture N, Wokke JH, Kwiatkowski TJ, Jr, Ophoff RA, Cronin S, Hardiman O, Diekstra FP, Leigh PN, Shaw CE, Simpson CL, Hansen VK, Powell JF, Corcia P, Salachas F, Heath S, Galan P, Georges F, Horvitz HR, Lathrop M, Purcell S, Al-Chalabi A, Brown RH., Jr Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 2009;106(22):9004–9009. doi: 10.1073/pnas.0812937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattante S, Conte A, Zollino M, Luigetti M, Del Grande A, Marangi G, Romano A, Marcaccio A, Meleo E, Bisogni G, Rossini PM, Sabatelli M. Contribution of major amyotrophic lateral sclerosis genes to the etiology of sporadic disease. Neurology. 2012;79(1):66–72. doi: 10.1212/WNL.0b013e31825dceca. [DOI] [PubMed] [Google Scholar]
- Lattante S, Rouleau GA, Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum. Mutat. 2013;34(6):812–826. doi: 10.1002/humu.22319. [DOI] [PubMed] [Google Scholar]
- Le Ber I, Camuzat A, Berger E, Hannequin D, Laquerrière A, Golfier V, Seilhean D, Viennet G, Couratier P, Verpillat P, Heath S, Camu W, Martinaud O, Lacomblez L, Vercelletto M, Salachas F, Sellal F, Didic M, Thomas-Anterion C, Puel M, Michel BF, Besse C, Duyckaerts C, Meininger V, Campion D, Dubois B, Brice A, French Research Network on FTD/FTD-MND Chromosome 9p–linked families with frontotemporal dementia associated with motor neuron disease. Neurology. 2009;72(19):1669–1676. doi: 10.1212/WNL.0b013e3181a55f1c. [DOI] [PubMed] [Google Scholar]
- Lee JB, Lee KA, Hong JM, Suh GI, Choi YC. Homozygous SMN2 deletion is a major risk factor among twenty-five Korean sporadic amyotrophic lateral sclerosis patients. Yonsei. Med. J. 2012a;53(1):53–57. doi: 10.3349/ymj.2012.53.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, Scott E, Bafna V, Hill KJ, Collazo A, Funari V, Russ C, Gabriel SB, Mathern GW, Gleeson JG. De novo somatic mutations in components of the PI3K–AKT3-mTOR pathway cause hemimegalencephaly. Nat. Genet. 2012;44(8):941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CL, He CZ, Kaufmann P, Chin SS, Naini A, Liem RK, Mitsumoto H, Hays AP. A pathogenic peripherin gene mutation in a patient with amyotrophic lateral sclerosis. Brain Pathol. 2004;14(3):290–296. doi: 10.1111/j.1750-3639.2004.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang L, Wang W, Qi XL, Tang ZY. Mutations in the HFE gene and sporadic amyotrophic lateral sclerosis risk: a meta-analysis of observational studies. Braz. J. Med. Biol. Res. 2014;47(3):215–222. doi: 10.1590/1414-431X20133296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lu M, Tang L, Zhang N, Chui D, Fan D. ATXN2 CAG repeat expansions increase the risk for Chinese patients with amyotrophic lateral sclerosis. Neurobiol. Aging. 2013;34(9) doi: 10.1016/j.neurobiolaging.2013.04.009. 2236.e5-8. [DOI] [PubMed] [Google Scholar]
- Luigetti M, Lattante S, Zollino M, Conte A, Marangi G, Del Grande A, Sabatelli M. SOD1 G93D sporadic amyotrophic lateral sclerosis (SALS) patient with rapid progression and concomitant novel ANG variant. Neurobiol. Aging. 2011;32(10) doi: 10.1016/j.neurobiolaging.2011.04.004. 1924.e15-18. [DOI] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chiò A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Chromosome 9-ALS/FTD Consortium; French research network on FTLD/FTLD/ALS; ITALSGEN Consortium. Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465(7295):223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, Gage FH. Mosaic copy number variation in human neurons. Science. 2013;342(6158):632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentula HK, Tuovinen L, Penttilä S, Suominen T, Udd B, Palmio J. TARDBP mutations are not a frequent cause of ALS in Finnish patients. Acta Myol. 2012;31(2):134–138. [PMC free article] [PubMed] [Google Scholar]
- Meyer T, Schwan A, Dullinger JS, Brocke J, Hoffmann KT, Nolte CH, Hopt A, Kopp U, Andersen P, Epplen JT, Linke P. Early-onset ALS with long-term survival associated with spastin gene mutation. Neurology. 2005;65(1):141–143. doi: 10.1212/01.wnl.0000167130.31618.0a. [DOI] [PubMed] [Google Scholar]
- Millecamps S, Salachas F, Cazeneuve C, Gordon P, Bricka B, Camuzat A, Guillot-Noël L, Russaouen O, Bruneteau G, Pradat PF, Le Forestier N, Vandenberghe N, Danel-Brunaud V, Guy N, Thauvin-Robinet C, Lacomblez L, Couratier P, Hannequin D, Seilhean D, Le Ber I, Corcia P, Camu W, Brice A, Rouleau G, LeGuern E, Meininger V. SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: genotype-phenotype correlations. J. Med. Genet. 2010;47(8):554–560. doi: 10.1136/jmg.2010.077180. [DOI] [PubMed] [Google Scholar]
- Mitchell J, Paul P, Chen HJ, Morris A, Payling M, Falchi M, Habgood J, Panoutsou S, Winkler S, Tisato V, Hajitou A, Smith B, Vance C, Shaw C, Mazarakis ND, de Belleroche J. Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase. Proc. Natl. Acad. Sci. U.S.A. 2010;107(16):7556–7561. doi: 10.1073/pnas.0914128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok K, Laaksovirta H, Tienari PJ, Peuralinna T, Myllykangas L, Chiò A, Traynor BJ, Nalls MA, Gurunlian N, Shatunov A, Restagno G, Mora G, Nigel Leigh P, Shaw CE, Morrison KE, Shaw PJ, Al-Chalabi A, Hardy J, Orrell RW. Homozygosity analysis in amyotrophic lateral sclerosis. Eur. J. Hum. Genet. 2013;21(12):1429–1435. doi: 10.1038/ejhg.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan JM, Yu B, Trent RJ, Pamphlett R. A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2009;10(5–6):418–429. doi: 10.3109/17482960802635397. [DOI] [PubMed] [Google Scholar]
- Morita M, Al-Chalabi A, Andersen PM, Hosler B, Sapp P, Englund E, Mitchell JE, Habgood JJ, de Belleroche J, Xi J, Jongjaroenprasert W, Horvitz HR, Gunnarsson LG, Brown RH., Jr A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66(6):839–844. doi: 10.1212/01.wnl.0000200048.53766.b4. [DOI] [PubMed] [Google Scholar]
- Müller K, Andersen PM, Hübers A, Marroquin N, Volk AE, Danzer KM, Meitinger T, Ludolph AC, Strom TM, Weishaupt JH. Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease. Brain. 2014 doi: 10.1093/brain/awu227. pii: awu227. [DOI] [PubMed] [Google Scholar]
- Münch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, Prudlo J, Peraus G, Hanemann CO, Stumm G, Ludolph AC. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 2004;63(4):724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- Munch C, O’Brien J, Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3548–3553. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, Polak P, Yoon S, Maguire J, Crawford EL, Campbell NG, Geller ET, Valladares O, Schafer C, Liu H, Zhao T, Cai G, Lihm J, Dannenfelser R, Jabado O, Peralta Z, Nagaswamy U, Muzny D, Reid JG, Newsham I, Wu Y, Lewis L, Han Y, Voight BF, Lim E, Rossin E, Kirby A, Flannick J, Fromer M, Shakir K, Fennell T, Garimella K, Banks E, Poplin R, Gabriel S, DePristo M, Wimbish JR, Boone BE, Levy SE, Betancur C, Sunyaev S, Boerwinkle E, Buxbaum JD, Cook EH, Jr, Devlin B, Gibbs RA, Roeder K, Schellenberg GD, Sutcliffe JS, Daly MJ. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P, Zatz M. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004;75(5):822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T, Masuda-Suzukake M, Arai T, Hasegawa Y, Akatsu H, Obi T, Yoshida M, Murayama S, Mann DMA, Akiyama H, Hasegawa M. Prion-like properties of of pathological TDP-43 aggregates from diseased brains. Cell Rep. 2013;4:124–134. doi: 10.1016/j.celrep.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Ogaki K, Li Y, Atsuta N, Tomiyama H, Funayama M, Watanabe H, Nakamura R, Yoshino H, Yato S, Tamura A, Naito Y, Taniguchi A, Fujita K, Izumi Y, Kaji R, Hattori N, Sobue G, Japanese Consortium for Amyotrophic Lateral Sclerosis research (JaCALS) Analysis of C9orf72 repeat expansion in 563 Japanese patients with amyotrophic lateral sclerosis. Neurobiol. Aging. 2012;33(10) doi: 10.1016/j.neurobiolaging.2012.05.011. 2527.e11-6. [DOI] [PubMed] [Google Scholar]
- Orlacchio A, Babalini C, Borreca A, Patrono C, Massa R, Basaran S, Munhoz RP, Rogaeva EA, St George-Hyslop PH, Bernardi G, Kawarai T. SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain. 2010;133(Pt 2):591–598. doi: 10.1093/brain/awp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrù S, Mascia V, Casula M, Giuressi E, Loizedda A, Carcassi C, Giagheddu M, Contu L. Association of monoamine oxidase B alleles with age at onset in amyotrophic lateral sclerosis. Neuromuscul. Disord. 1999;9(8):593–597. doi: 10.1016/s0960-8966(99)00052-8. [DOI] [PubMed] [Google Scholar]
- Pamphlett R, Morahan JM, Yu B. Using case-parent trios to look for rare de novo genetic variants in adult-onset neurodegenerative diseases. J. Neurosci. Methods. 2011a;197(2):297–301. doi: 10.1016/j.jneumeth.2011.02.028. [DOI] [PubMed] [Google Scholar]
- Pamphlett R, Morahan JM, Luquin N, Yu B. Looking for differences in copy number between blood and brain in sporadic amyotrophic lateral sclerosis. Muscle Nerve. 2011b;44(4):492–498. doi: 10.1002/mus.22095. [DOI] [PubMed] [Google Scholar]
- Pamphlett R, Morahan JM. Copy number imbalances in blood and hair in monozygotic twins discordant for amyotrophic lateral sclerosis. J. Clin. Neurosci. 2011c;18(9):1231–1234. doi: 10.1016/j.jocn.2010.12.049. [DOI] [PubMed] [Google Scholar]
- Parkinson N, Ince PG, Smith MO, Highley R, Skibinski G, Andersen PM, Morrison KE, Pall HS, Hardiman O, Collinge J, Shaw PJ, Fisher EM, MRC Proteomics in ALS Study; FReJA Consortium ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B) Neurology. 2006;67(6):1074–1077. doi: 10.1212/01.wnl.0000231510.89311.8b. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Williams NM, Majounie E, Waite A, Stott J, Newsway V, Murray A, Hernandez D, Guerreiro R, Singleton AB, Neal J, Morris HR. Familial frontotemporal dementia with amyotrophic lateral sclerosis and a shared haplotype on chromosome 9p. J. Neurol. 2011;258(4):647–655. doi: 10.1007/s00415-010-5815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, Barry BJ, Bourgeois BF, Riviello JJ, Barkovich AJ, Black PM, Ligon KL, Walsh CA. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74(1):41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341(6141):1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH, Jr, Ludlow CL, Fischbeck KH. Mutant dynactin in motor neuron disease. Nat. Genet. 2003;33(4):455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Quadri M, Cossu G, Saddi V, Simons EJ, Murgia D, Melis M, Ticca A, Oostra BA, Bonifati V. Broadening the phenotype of TARDBP mutations: the TARDBP Ala382Thr mutation and Parkinson’s disease in Sardinia. Neurogenetics. 2011;12(3):203–209. doi: 10.1007/s10048-011-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainier S, Bui M, Mark E, Thomas D, Tokarz D, Ming L, Delaney C, Richardson RJ, Albers JW, Matsunami N, Stevens J, Coon H, Leppert M, Fink JK. Neuropathy target esterase gene mutations cause motor neuron disease. Am. J. Hum. Genet. 2008;82(3):780–785. doi: 10.1016/j.ajhg.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayaprolu S, Fujioka S, Traynor S, Soto-Ortolaza AI, Petrucelli L, Dickson DW, Rademakers R, Boylan KB, Graff-Radford NR, Uitti RJ, Wszolek ZK, Ross OA. TARDBP mutations in Parkinson’s disease. Parkinsonism Relat. Disord. 2013;19(3):312–315. doi: 10.1016/j.parkreldis.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, ITALSGEN Consortium. Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, Héon E, Krupin T, Ritch R, Kreutzer D, Crick RP, Sarfarazi M. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295(5557):1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- Rivière JB, Mirzaa GM, O’Roak BJ, Beddaoui M, Alcantara D, Conway RL, St-Onge J, Schwartzentruber JA, Gripp KW, Nikkel SM, Worthylake T, Sullivan CT, Ward TR, Butler HE, Kramer NA, Albrecht B, Armour CM, Armstrong L, Caluseriu O, Cytrynbaum C, Drolet BA, Innes AM, Lauzon JL, Lin AE, Mancini GM, Meschino WS, Reggin JD, Saggar AK, Lerman-Sagie T, Uyanik G, Weksberg R, Zirn B, Beaulieu CL, Finding of Rare Disease Genes (FORGE) Canada Consortium. Majewski J, Bulman DE, O’Driscoll M, Shendure J, Graham JM, Jr, Boycott KM, Dobyns WB. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 2012;44(8):934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung WY, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak-Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH., Jr Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Rubino E, Rainero I, Chiò A, Rogaeva E, Galimberti D, Fenoglio P, Grinberg Y, Isaia G, Calvo A, Gentile S, Bruni AC, St George-Hyslop PH, Scarpini E, Gallone S, Pinessi L, TODEM Study Group SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79(15):1556–1562. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli M, Eusebi F, Al-Chalabi A, Conte A, Madia F, Luigetti M, Mancuso I, Limatola C, Trettel F, Sobrero F, Di Angelantonio S, Grassi F, Di Castro A, Moriconi C, Fucile S, Lattante S, Marangi G, Murdolo M, Orteschi D, Del Grande A, Tonali P, Neri G, Zollino M. Rare missense variants of neuronal nicotinic acetylcholine receptor altering receptor function are associated with sporadic amyotrophic lateral sclerosis. Hum. Mol. Genet. 2009;18(20):3997–4006. doi: 10.1093/hmg/ddp339. [DOI] [PubMed] [Google Scholar]
- Sabatelli M, Lattante S, Conte A, Marangi G, Luigetti M, Del Grande A, Chiò A, Corbo M, Giannini F, Mandrioli J, Mora G, Calvo A, Restagno G, Lunetta C, Penco S, Battistini S, Zeppilli P, Bizzarro A, Capoluongo E, Neri G, Rossini PM, Zollino M. Replication of association of CHRNA4 rare variants with sporadic amyotrophic lateral sclerosis: the Italian multicentre study. Amyotroph. Lateral Scler. 2012a;13(6):580–584. doi: 10.3109/17482968.2012.704926. [DOI] [PubMed] [Google Scholar]
- Sabatelli M, Conforti FL, Zollino M, Mora G, Monsurrò MR, Volanti P, Marinou K, Salvi F, Corbo M, Giannini F, Battistini S, Penco S, Lunetta C, Quattrone A, Gambardella A, Logroscino G, Simone I, Bartolomei I, Pisano F, Tedeschi G, Conte A, Spataro R, La Bella V, Caponnetto C, Mancardi G, Mandich P, Sola P, Mandrioli J, Renton AE, Majounie E, Abramzon Y, Marrosu F, Marrosu MG, Murru MR, Sotgiu MA, Pugliatti M, Rodolico C, ITALSGEN Consortium. Moglia C, Calvo A, Ossola I, Brunetti M, Traynor BJ, Borghero G, Restagno G, Chiò A. C9ORF72 hexanucleotide repeat expansions in the Italian sporadic ALS population. Neurobiol. Aging. 2012a;33(8) doi: 10.1016/j.neurobiolaging.2012.02.011. 1848.e15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, Teran NA, Song Y, El-Fishawy P, Murtha RC, Choi M, Overton JD, Bjornson RD, Carriero NJ, Meyer KA, Bilguvar K, Mane SM, Sestan N, Lifton RP, Günel M, Roeder K, Geschwind DH, Devlin B, State MW. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp PC, Hosler BA, McKenna-Yasek D, Chin W, Gann A, Genise H, Gorenstein J, Huang M, Sailer W, Scheffler M, Valesky M, Haines JL, Pericak-Vance M, Siddique T, Horvitz HR, Brown RH., Jr Identification of two novel loci for dominantly inherited familial amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2003;73(2):397–403. doi: 10.1086/377158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymick JC, Scholz SW, Fung HC, Britton A, Arepalli S, Gibbs JR, Lombardo F, Matarin M, Kasperaviciute D, Hernandez DG, Crews C, Bruijn L, Rothstein J, Mora G, Restagno G, Chiò A, Singleton A, Hardy J, Traynor BJ. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2007;6(4):322–328. doi: 10.1016/S1474-4422(07)70037-6. [DOI] [PubMed] [Google Scholar]
- Shatunov A, Mok K, Newhouse S, Weale ME, Smith B, Vance C, Johnson L, Veldink JH, van Es MA, van den Berg LH, Robberecht W, Van Damme P, Hardiman O, Farmer AE, Lewis CM, Butler AW, Abel O, Andersen PM, Fogh I, Silani V, Chiò A, Traynor BJ, Melki J, Meininger V, Landers JE, McGuffin P, Glass JD, Pall H, Leigh PN, Hardy J, Brown RH, Jr, Powell JF, Orrell RW, Morrison KE, Shaw PJ, Shaw CE, Al-Chalabi A. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9(10):986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet SA, Waibel S, Endruhn S, Sperfeld AD, Vorwerk B, Müller I, Erdogan F, Ludolph AC, Ropers HH, Ullmann R. Identification of candidate genes for sporadic amyotrophic lateral sclerosis by array comparative genomic hybridization. Amyotroph. Lateral Scler. 2009;10(3):162–169. doi: 10.1080/17482960802535001. [DOI] [PubMed] [Google Scholar]
- Simpson CL, Lemmens R, Miskiewicz K, Broom WJ, Hansen VK, van Vught PW, Landers JE, Sapp P, Van Den Bosch L, Knight J, Neale BM, Turner MR, Veldink JH, Ophoff RA, Tripathi VB, Beleza A, Shah MN, Proitsi P, Van Hoecke A, Carmeliet P, Horvitz HR, Leigh PN, Shaw CE, van den Berg LH, Sham PC, Powell JF, Verstreken P, Brown RH, Jr, Robberecht W, Al-Chalabi A. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 2009;18(3):472–481. doi: 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Hardy J, Traynor BJ, Houlden H. Towards a complete resolution of the genetic architecture of disease. Trends in Genetics. 2010;26(10):438–442. doi: 10.1016/j.tig.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K, Brouwers N, Maurer-Stroh S, van Es MA, Van Damme P, van Vught PW, van der Zee J, Serneels S, De Pooter T, Van den Broeck M, Cruts M, Schymkowitz J, De Jonghe P, Rousseau F, van den Berg LH, Robberecht W, Van Broeckhoven C. Progranulin genetic variability contributes to amyotrophic lateral sclerosis. Neurology. 2008;71(4):253–259. doi: 10.1212/01.wnl.0000289191.54852.75. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara K, Maruyama H, Kamada M, Morino H, Kawakami H. Screening for OPTN mutations in amyotrophic lateral sclerosis in a mainly Caucasian population. Neurobiol. Aging. 2011;32(10) doi: 10.1016/j.neurobiolaging.2011.03.024. 1923.e9-10. [DOI] [PubMed] [Google Scholar]
- Tetsuka S, Morita M, Iida A, Uehara R, Ikegawa S, Nakano I. ZNF512B gene is a prognostic factor in patients with amyotrophic lateral sclerosis. J. Neurol. Sci. 2013;324(1–2):163–166. doi: 10.1016/j.jns.2012.10.029. [DOI] [PubMed] [Google Scholar]
- Teyssou E, Takeda T, Lebon V, Boillée S, Doukouré B, Bataillon G, Sazdovitch V, Cazeneuve C, Meininger V, LeGuern E, Salachas F, Seilhean D, Millecamps S. Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: genetics and neuropathology. Acta Neuropathol. 2013;125(4):511–522. doi: 10.1007/s00401-013-1090-0. [DOI] [PubMed] [Google Scholar]
- Traynor BJ, Nalls M, Lai SL, Gibbs RJ, Schymick JC, Arepalli S, Hernandez D, van der Brug MP, Johnson JO, Dillman A, Cookson M, Moglia C, Calvo A, Restagno G, Mora G, Chiò A. Kinesin-associated protein 3 (KIFAP3) has no effect on survival in a population-based cohort of ALS patients. Proc. Natl. Acad. Sci. U.S.A. 2010;107(27):12335–12338. doi: 10.1073/pnas.0914079107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CP, Soong BW, Lin KP, Tu PH, Lin JL, Lee YC. FUS, TARDBP, and SOD1 mutations in a Taiwanese cohort with familial ALS. Neurobiol. Aging. 2011;32(3) doi: 10.1016/j.neurobiolaging.2010.04.009. 553.e13-21. [DOI] [PubMed] [Google Scholar]
- Uyan Ö, Ömür Ö, Ağım ZS, Özoğuz A, Li H, Parman Y, Deymeer F, Oflazer P, Koç F, Tan E, Özçelik H, Başak AN. Genome-wide copy number variation in sporadic amyotrophic lateral sclerosis in the Turkish population: deletion of EPHA3 is a possible protective factor. P.L.o.S. One. 2013;8(8):e72381. doi: 10.1371/journal.pone.0072381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, van Es MA, Hennekam EA, Dooijes D, van Rheenen W, Medic J, Bourque PR, Schelhaas HJ, van der Kooi AJ, de Visser M, de Bakker PI, Veldink JH, van den Berg LH. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012;21(17):3776–3784. doi: 10.1093/hmg/dds199. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Veldink JH, van Blitterswijk M, Corveleyn A, van Vught PW, Thijs V, Dubois B, Matthijs G, van den Berg LH, Robberecht W. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 2011;76(24):2066–2072. doi: 10.1212/WNL.0b013e31821f445b. [DOI] [PubMed] [Google Scholar]
- van Es MA, Van Vught PW, Blauw HM, Franke L, Saris CG, Andersen PM, Van Den Bosch L, de Jong SW, van ‘t Slot R, Birve A, Lemmens R, de Jong V, Baas F, Schelhaas HJ, Sleegers K, Van Broeckhoven C, Wokke JH, Wijmenga C, Robberecht W, Veldink JH, Ophoff RA, van den Berg LH. ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: a genome-wide association study. Lancet Neurol. 2007;6(10):869–877. doi: 10.1016/S1474-4422(07)70222-3. [DOI] [PubMed] [Google Scholar]
- van Es MA, van Vught PW, Blauw HM, Franke L, Saris CG, Van den Bosch L, de Jong SW, de Jong V, Baas F, van’t Slot R, Lemmens R, Schelhaas HJ, Birve A, Sleegers K, Van Broeckhoven C, Schymick JC, Traynor BJ, Wokke JH, Wijmenga C, Robberecht W, Andersen PM, Veldink JH, Ophoff RA, van den Berg LH. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 2008;40(1):29–31. doi: 10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- van Es MA, Veldink JH, Saris CG, Blauw HM, van Vught PW, Birve A, Lemmens R, Schelhaas HJ, Groen EJ, Huisman MH, van der Kooi AJ, de Visser M, Dahlberg C, Estrada K, Rivadeneira F, Hofman A, Zwarts MJ, van Doormaal PT, Rujescu D, Strengman E, Giegling I, Muglia P, Tomik B, Slowik A, Uitterlinden AG, Hendrich C, Waibel S, Meyer T, Ludolph AC, Glass JD, Purcell S, Cichon S, Nöthen MM, Wichmann HE, Schreiber S, Vermeulen SH, Kiemeney LA, Wokke JH, Cronin S, McLaughlin RL, Hardiman O, Fumoto K, Pasterkamp RJ, Meininger V, Melki J, Leigh PN, Shaw CE, Landers JE, Al-Chalabi A, Brown RH, Jr, Robberecht W, Andersen PM, Ophoff RA, van den Berg LH. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 2009;41(10):1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- Van Hoecke A, Schoonaert L, Lemmens R, Timmers M, Staats KA, Laird AS, Peeters E, Philips T, Goris A, Dubois B, Andersen PM, Al-Chalabi A, Thijs V, Turnley AM, van Vught PW, Veldink JH, Hardiman O, Van Den Bosch L, Gonzalez-Perez P, Van Damme P, Brown RH, Jr, van den Berg LH, Robberecht W. EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nat. Med. 2012;18(9):1418–1422. doi: 10.1038/nm.2901. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nat. Genet. 2010;42(12):1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Wain LV, Pedroso I, Landers JE, Breen G, Shaw CE, Leigh PN, Brown RH, Tobin MD, Al-Chalabi A. The role of copy number variation in susceptibility to amyotrophic lateral sclerosis: genome-wide association study and comparison with published loci. P.L.o.S. One. 2009;4(12):e8175. doi: 10.1371/journal.pone.0008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Cui NH, Gao JJ, Qiu XP, Zheng F. SMN1 duplications contribute to sporadic amyotrophic lateral sclerosis susceptibility: Evidence from a meta-analysis. J. Neurol. Sci. 2014;340(1–2):63–68. doi: 10.1016/j.jns.2014.02.026. [DOI] [PubMed] [Google Scholar]
- Wills AM, Cronin S, Slowik A, Kasperaviciute D, Van Es MA, Morahan JM, Valdmanis PN, Meininger V, Melki J, Shaw CE, Rouleau GA, Fisher EM, Shaw PJ, Morrison KE, Pamphlett R, Van den Berg LH, Figlewicz DA, Andersen PM, Al-Chalabi A, Hardiman O, Purcell S, Landers JE, Brown RH., Jr A large-scale international meta-analysis of paraoxonase gene polymorphisms in sporadic ALS. Neurology. 2009;73(1):16–24. doi: 10.1212/WNL.0b013e3181a18674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, Lowe P, Koppers M, McKenna-Yasek D, Baron DM, Kost JE, Gonzalez-Perez P, Fox AD, Adams J, Taroni F, Tiloca C, Leclerc AL, Chafe SC, Mangroo D, Moore MJ, Zitzewitz JA, Xu ZS, van den Berg LH, Glass JD, Siciliano G, Cirulli ET, Goldstein DB, Salachas F, Meininger V, Rossoll W, Ratti A, Gellera C, Bosco DA, Bassell GJ, Silani V, Drory VE, Brown RH, Jr, Landers JE. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488(7412):499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Deng L, Mei P, Zhou Y, Wang B, Zhang J, Lin J, Wei Y, Zhang X, Xu R. A genome-wide association study combining pathway analysis for typical sporadic amyotrophic lateral sclerosis in Chinese Han populations. Neurobiol. Aging. 2014 Jan 17; doi: 10.1016/j.neurobiolaging.2014.01.014. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43(9):864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Deng HX, Siddique N, Fecto F, Chen W, Yang Y, Liu E, Donkervoort S, Zheng JG, Shi Y, Ahmeti KB, Brooks B, Engel WK, Siddique T. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology. 2010;75(9):807–814. doi: 10.1212/WNL.0b013e3181f07e0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg H, Jacobsson J, Rosengren L, Blennow K, Andersen PM. Association of APOE with age at onset of sporadic amyotrophic lateral sclerosis. J. Neurol. Sci. 2008;273(1–2):67–69. doi: 10.1016/j.jns.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Zou ZY, Li XG, Liu MS, Cui LY. Screening for C9orf72 repeat expansions in Chinese amyotrophic lateral sclerosis patients. Neurobiol. Aging. 2013a;34(6) doi: 10.1016/j.neurobiolaging.2012.11.018. 1710.e5-6. [DOI] [PubMed] [Google Scholar]
- Zou ZY, Cui LY, Sun Q, Li XG, Liu MS, Xu Y, Zhou Y, Yang XZ. De novo FUS gene mutations are associated with juvenile-onset sporadic amyotrophic lateral sclerosis in China. Neurobiol. Aging. 2013b;34(4) doi: 10.1016/j.neurobiolaging.2012.09.005. 1312.e1-8. [DOI] [PubMed] [Google Scholar]