SUMMARY
Protein ubiquitination is a dynamic and reversible process of adding single ubiquitin molecules or various ubiquitin chains to target proteins. Here, using multidimensional omic data of 9,125 tumor samples across 33 cancer types from The Cancer Genome Atlas, we perform comprehensive molecular characterization of 929 ubiquitin-related genes and 95 deubiquitinase genes. Among them, we systematically identify top somatic driver candidates, including mutated FBXW7 with cancer-type-specific patterns and amplified MDM2 showing a mutually exclusive pattern with BRAF mutations. Ubiquitin pathway genes tend to be upregulated in cancer mediated by diverse mechanisms. By integrating pan-cancer multiomic data, we identify a group of tumor samples that exhibit worse prognosis. These samples are consistently associated with the upregulation of cell-cycle and DNA repair pathways, characterized by mutated TP53, MYC/TERT amplification, and APC/PTEN deletion. Our analysis highlights the importance of the ubiquitin pathway in cancer development and lays a foundation for developing relevant therapeutic strategies.
In Brief
Ge et al. analyze a cohort of 9,125 TCGA samples across 33 cancer types to provide a comprehensive characterization of the ubiquitin pathway. They detect somatic driver candidates in the ubiquitin pathway and identify a cluster of patients with poor survival, highlighting the importance of this pathway in cancer development.

INTRODUCTION
The highly conserved ubiquitin pathway serves as a crucial regulator, mediating a myriad of cellular events that underlie the development of an assortment of cancer types (Di Fiore et al., 2003; Hoeller and Dikic, 2009; Nakayama and Nakayama, 2006). The ubiquitin molecule is an 8.5-kDa, 76-amino-acid globular protein with a complex 3-dimensional surface topology that is able to form various types of ubiquitin chains, thereby acting as a robust, post-translational protein modifier (Weissman, 2001). The canonical addition of ubiquitin to a protein substrate involves the consecutive actions of three main families of ubiquitination (UBQ) enzymes through a coordinated enzymatic cascade (Fuchs, 2002; Gao et al., 2013). In the first step, a thiol-ester bond is formed between a ubiquitin-activating enzyme (E1) and the carboxy-terminal glycine of ubiquitin. The next step involves the transfer of the activated ubiquitin from the E1 enzyme to a ubiquitin-conjugating enzyme (E2) via a trans-thiolation reaction. Finally, a ubiquitin ligase (E3), which functions as a scaffold protein binding both the E2 enzyme and the target protein, mediates the transfer of ubiquitin from the E2-ubiquitin conjugate, most commonly onto the ε-amino group of a lysine residue on the protein substrate, thus forming an isopeptide bond (Hoeller and Dikic, 2009; Weissman, 2001). Because UBQ is a dynamic and reversible process, deubiquitinating enzymes (DUBs) fulfill a converse role in the pathway by deconjugating ubiquitin from proteins entirely or trimming poly-ubiquitin chains, thereby enhancing regulation in the ability to abrogate or modify ubiquitin protein modifications (Komander et al., 2009; Weissman, 2001). Recognition of distinct UBQ patterns by downstream effectors elicits various cellular functions: it can mark proteins for degradation via the proteasome, alter their subcellular localization, affect their activity, and promote or prevent protein interactions.
In recent years, the role of the ubiquitin pathway in cancer has gained attention for two main reasons. First, both basic and translational studies have shown extensive evidence connecting the malfunction of the ubiquitin pathway with tumor initiation and progression. For example, UBQs have been found to be tightly associated with many cancer-related pathways, including cell-cycle progress, p53 activation, DNA damage repair, apoptosis, nuclear factor κB (NF-κB), and receptor tyrosine kinase pathways (Hoeller and Dikic, 2009; Li et al., 2003; Lind et al., 2006; Massoumi et al., 2006; Meetei et al., 2003; Weissman, 2001); DUBs are implicated in many of the same cancer pathways as UBQs; in addition, they are associated with chromatin remodeling, WNT signaling, and transforming growth factor β (TGF-β) signaling (Dey et al., 2012; Dupont et al., 2009; Luise et al., 2011; Tauriello et al., 2010; Wicks et al., 2005; Zhao et al., 2009). Second, targeting the ubiquitin pathway for regulating target protein levels, rather than for its biological activity, has emerged as a promising therapeutic strategy for cancer patients. Because many oncoproteins are subject to UBQ-dependent degradation, enhancing UBQ or targeting certain DUBs may lead to destabilization or functional inactivation of key oncoproteins, including some undruggable targets such as MYC and β-catenin (Salami and Crews, 2017; Xiao et al., 2016). A few drugs targeting the ubiquitin pathway have been approved by the U.S. Food and Drug Administration (FDA) (Huang and Dixit, 2016; Swisher et al., 2017).
Given the pervasive impact and clinical utility of the ubiquitin pathway across many cancer types, it is important to curate genomic insights into the role of this pathway in cancer development and treatment through a systematic, pan-cancer analysis. The Cancer Genome Atlas (TCGA) has generated genomic, transcriptomic, proteomic, epigenomic, and clinical data over large patient cohorts, providing an unprecedented opportunity for such an analysis (Cancer Genome Atlas Research Network et al., 2013). We performed a molecular characterization of UBQ and DUB genes across 9,125 patients from 33 cancer types (Table S1). To maximize the chance of making scientific and clinical findings, we compiled a comprehensive list of 929 UBQ-related genes (including both validated and computationally predicted E1 and E2 enzymes, as well as E3 ligases and their associated adaptor genes, termed UBQ genes hereafter for simplicity) and 95 DUB genes (see curation details in STAR Methods, Figure S1, and Table S2). Our analysis will not only further elucidate the role of the ubiquitin pathway in cancer development but also directly inform researchers and clinicians as to possible driver genes and eminently druggable targets for future clinical trials and therapeutics.
RESULTS
Mutation Driver Candidates of UBQ and DUB Genes
Based on TCGA mutation data of whole-exome sequencing, we examined the somatic mutation profiles of UBQ and DUB genes in 33 cancer types. Overall, across 8,811 non-hypermutated cancer samples, the mutation frequency was low for both UBQ and DUB genes, with an average mutation number per patient of 4.5 and 0.5, respectively. To identify potential cancer drivers, we employed two complementary computational approaches. First, we used a ratiometric method for nominating cancer driver genes based on the enrichment of hotspot or loss-of-function (LoF) mutations among all mutations observed in a gene (Figure 1A) (Vogelstein et al., 2013). In this pan-cancer analysis, we identified 19 UBQ/DUB genes with >30% hotspot mutations and 29 genes with >30% LoF mutations (FBXW7 was identified by both criteria). Second, we used MutSigCV (Lawrence et al., 2013) to pinpoint UBQ/DUB gene drivers whose mutation rates were significantly higher than the background expectation within each cancer type. Using a q value cutoff of 0.1, we identified 23 such genes in 23 cancer types (Figure 1B). In total, these two methods identified 55 driver candidates, and their overall mutation frequency ranged from 0.2% to 7.2% (Figure S2). We then mapped these 55 putative cancer drivers to different gene categories in the ubiquitin pathway and found no specific enrichment patterns (Figure 1C): there were no E1 enzyme driver genes of the driver genes detected, two were E2 enzyme drivers, four were DUB drivers, and the rest (49) were E3 ligases and associated adaptors. Among 15 driver genes identified by both methods, SPOP, KEAP1, and CHD4 were enriched with hotspot mutations, while BAP1, CDH1, CUL3, EP300, KDM5C, MAP3K1, NSD1, RNF43, TLE1, VHL, and LZTR1 contained excessive LoF mutations. This analysis provides a systematic view of potential mutation drivers among UBQ and DUB genes.
Figure 1. Frequently Mutated UBQ and DUB Genes as Potential Cancer Drivers.
(A) UBQ and DUB genes are plotted as fractions of hotspot versus LoF mutations among all non-silent mutations across cancer types. Genes enriched with hotspot mutations are shown in red, genes enriched with LoF mutations are in blue, and FBXW7 is shown in orange, because it is enriched with both hotspot and LoF mutations. The circles represent UBQs, and the squares represent DUBs.
(B) Significantly mutated genes identified by MutSigCV in each cancer type are shown. The circles represent UBQs, and the squares represent DUBs; the circle or square size is proportional to the significance level. The fraction of patients harboring non-silent mutations in each gene is shown by color scale.
(C) UBQ and DUB genes enriched with hotspot and LoF mutations are mapped to different gene categories in the ubiquitin pathway. See also Figure S2.
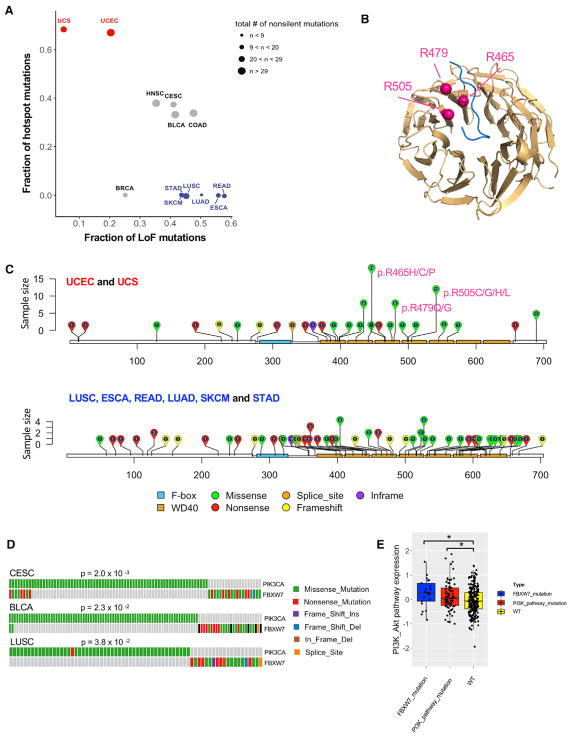
Of particular interest, FBXW7 showed enrichment of both hotspot and LoF mutations (Figure 1A). The FBXW7 protein functions as the substrate recognition component of the SKP1-CUL1-F-box protein (SCF) ubiquitin ligase complex. As an established tumor suppressor gene, it mediates the degradation of cell-cycle promoters or oncoproteins, including cyclin E (Koepp et al., 2001; Siu et al., 2012), c-Myc (Yada et al., 2004), c-Jun (Wei et al., 2005), Notch (Gupta-Rossi et al., 2001), Mcl1 (Ren et al., 2013), and mTOR (Mao et al., 2008). To gain more insight into its mutational profile, we examined the mutation distributions of FBXW7 in different cancer types and found three distinct patterns (Figure 2A): (1) hotspot mutations were enriched in two uterine cancer types, uterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS); (2) LoF mutations were enriched in skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), rectum adenocarcinoma (READ), and esophageal carcinoma (ESCA); and (3) the proportions of both hotspot and LoF mutations were high in head and neck squamous cell carcinoma (HNSC), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), bladder urothelial carcinoma (BLCA), and colon adenocarcinoma (COAD). Consistent with previous studies, FBXW7 contains three notable missense mutation hotspots (R465, R479, and R505) in the second, third, and fourth WD40 domains that recognize the consensus phospho-motif located in its substrate (Figure 2B) (Hao et al., 2007). Figure 2C shows the FBXW7 mutation distributions for the hotspot mutation-enriched cancer types and the LoF mutation-enriched cancer types. The three missense hotspots accounted for 49% (38 of 77) of the FBXW7 mutations observed in UCEC and UCS. The contrasting mutation patterns of FBXW7 mutations may reflect tissue-specific roles of FBXW7 substrates or different FBXW7-mediated oncogenic mechanisms in different tumor contexts. We further assessed the occurrence of FBXW7 mutations with those in clinically actionable cancer genes and revealed that mutations in FBXW7 and PIK3CA showed mutual exclusivity in three cancer types: CESC, BLCA, and LUSC (Figure 2D), suggesting that mutations in these two genes confer similar functional consequences. Patients with FBXW7 or phosphatidylinositol 3-kinase (PI3K) pathway mutations (mutations found in PIK3CA, PTEN, and STK11) had higher PI3K pathway expression activity than patients without such mutations (Figure 2E).
Figure 2. FBXW7 Is Enriched with Both Hotspot and Loss-of-Function Mutations.
(A) Fractions of hotspot mutations versus LoF mutations among all non-silent mutations in FBXW7 are plotted for different cancer types. Cancer types enriched with hotspot mutations are shown in red, those enriched with LoF mutations are in blue, and those enriched with both hotspot and LoF mutations are in gray.
(B) WD40 domain structure of FBXW7 protein in which three arginines (R465, R479, and R505) are mutation hotspots and located at the substrate binding surface.
(C) Distributions of FBXW7 non-silent mutations in cancer types enriched with hotspot mutations (UCEC and UCS) and cancer types enriched with LoF mutations (ESCA, LUAD, LUSC, READ, SKCM, and STAD).
(D) FBXW7 mutations show mutually exclusive patterns with PIK3CA mutations in BLCA, CESC, and LUSC.
(E) Compared to tumors without mutations in FBXW7 or PI3K pathway genes, tumors with either FBXW7 or PI3K pathway mutations show elevated PI3K-Akt pathway activity, with *, p < 0.05.
The bottom and top of the box are the first and third quartiles, and the whiskers extend to 1.5 IQR of the lower quartile and the upper quartile, respectively.
Somatic Copy-Number Alteration Patterns of UBQ and DUB Genes
To infer somatic copy-number alteration (SCNA) drivers, we used GISTIC2 (Mermel et al., 2011) to identify significant focal deletion and amplification peaks in each of 33 cancer types. UBQ and DUB genes showed similar overall SCNA profiles in terms of the amplification and deletion gene fractions across cancer types (Figures S3A and S3B). To more rigorously assess the SCNA significance of UBQ and DUB genes against the cancer-type-specific background rate, we calculated the enrichment of UBQ and DUB genes that reside in the amplification or deletion peaks identified by GISTIC2 (q < 0.25) using Fisher’s exact test (Figures 3A, S3C, and S3D). Four cancer types (kidney renal clear cell carcinoma [KIRC], SKCM, cholangiocarcinoma [CHOL], and pancreatic adenocarcinoma [PAAD]) showed significant deletion peak enrichments, while no cancer types showed significant amplification peak enrichment (p < 0.01) (Figure 3A). Figure 3B shows the top 30 frequently detected UBQ and DUB genes across different cancer types, including ARNT, MDM2, and FAM63A (amplification) and PARK2, ING5, and ING2 (deletion).
Figure 3. Somatic Copy-Number Alterations of UBQ and DUB Genes.
(A) Fractions of UBQ and DUB genes residing in the amplification or deletion peaks (identified by GISTIC2, q < 0.25) compared to non-UBQ/DUB genes in different cancer types. Significant deletion enrichments are detected with *p < 0.01.
(B) Most frequently amplified or deleted UBQ and DUB genes in multiple cancer types. The circle size is proportional to the significance level of GISTIC2 results.
(C) MDM2 amplification shows a mutually exclusive pattern with BRAF mutations in SKCM. TP53 mutations are shown for comparison. Each bar represents one patient; significance was assessed by Fisher’s exact test.
(D) TP53 protein and mRNA expression of tumor samples with MDM2 amplification versus those with BRAF mutations or wild-type (WT) samples, with *p < 0.05.
The bottom and top of the box are the first and third quartiles, and the whiskers extend to 1.5 IQR of the lower quartile and the upper quartile, respectively.
(E) Graphical model showing the synergistic effect of MDM2 inhibitor and BRAF inhibitor.
See also Figure S3.
Among the top UBQ/DUB genes with frequent SCNAs, MDM2 was significantly amplified in 11 cancer types. The protein product of this gene is a negative regulator of TP53 and a therapeutic target under intensive clinical investigation. We therefore focused on MDM2 to examine its mutually exclusive pattern with (1) amplification of other UBQ/DUB genes and (2) somatic mutations of clinically actionable genes. Among UBQ and DUB genes, we found a mutually exclusive pattern of MDM2 and SKP2 amplifications in LUAD (Figure S3E), suggesting convergence of their functions on the same downstream effectors. In agreement with this notion, previous studies have shown that MDM2 prevents the binding of the E2F1 protein to its E3 ligase SCFskp2, thus inhibiting E2F1 degradation (Zhang et al., 2005). For clinically actionable genes, we found that MDM2 amplifications were mutually exclusive to BRAF and ATM mutations in SKCM and BLCA, respectively (Figures 3C and S3F). BRAF kinase domain mutations, such as V600E, result in a constitutively activated form of the protein in around 50% of SKCM patients (45.1% in this study), which then leads to stimulated mitogen-activated protein kinase (MAPK) signaling and induces tumor cell proliferation. The mechanism through which MDM2 antagonizes p53 functions is acting as the p53-specific E3 ligase and promoting p53 degradation, which then leads to reduced cell apoptosis. We observed MDM2 amplification in 4.1% of the SKCM samples in this study, in which p53 protein levels were significantly lower than in samples with BRAF mutations alone or with neither BRAF mutations nor MDM2 amplifications. This pattern was not observed at the mRNA expression level (Figure 3D). These results confirmed the function of MDM2 acting as an E3 ligase targeting the p53 protein for degradation. Furthermore, the mutually exclusive pattern of MDM2 amplification and BRAF mutation suggests that a reduced p53 pathway or induced MAPK signaling can serve as an impetus for aberrant tumor cell proliferation (Figure 3E). This intriguing pattern implies that restoring p53 function and blocking the MAPK pathway at the same time could be more beneficial to SKCM patients than interfering with either pathway alone. Studies have shown increased apoptosis and inhibition of melanoma growth by combining a BRAF inhibitor and p53 reactivation (Lu et al., 2013; Saiki et al., 2014).
Upregulated mRNA Expression of UBQ/DUB Genes in Cancer
To investigate the patterns of dysregulation of UBQ and DUB genes in cancer, we examined their gene expression using mRNA expression data of paired tumor and normal samples from 16 cancer types, because such paired-sample comparisons help reduce the effects of potential confounding factors. We identified differentially expressed genes (Wilcoxon signed rank test) between tumors and their matched normal samples and performed gene enrichment using gene set enrichment analysis (GSEA) (Subramanian et al., 2005). The combined set of UBQ and DUB genes showed significant enrichment in genes that were upregulated in tumor tissues in 7 of the 16 cancer types examined (CHOL, COAD, liver hepatocellular carcinoma [LIHC], LUAD, LUSC, PRAD, and BLCA), and more genes were upregulated than downregulated in these diseases (q < 0.1) (Figures 4A, 4B, and S4A). In contrast, only thyroid cancer (thyroid carcinoma [THCA]) showed a significant opposite pattern (q < 0.1) (Figures 4A and 4B). We obtained similar results for UBQ and DUB genes separately (Figure 4A).
Figure 4. Multiple Mechanisms Contribute to Upregulation of UBQ and DUB Genes in Cancer.
(A) UBQ and DUB genes showed upregulation in tumor samples in seven cancer types (GSEA, q < 0.1).
(B) Proportions of upregulated, neutral, and downregulated UBQ/DUB genes in the seven cancer types (Wilcoxon signed rank test, q < 0.1).
(C) Top: proportions of copy-number amplification, neutral level, and deletions in upregulated and neutral UBQ/DUB gene groups in each cancer type. Middle: proportions of significantly decreased (paired t test, p < 0.05), decreased, and other expression of miRNA regulators in tumor samples relative to matched normal samples in upregulated and neutral UBQ/DUB gene groups. Bottom: proportions of significantly decreased (paired t test, p < 0.05), decreased, and otherwise DNA methylation level in tumor samples relative to matched normal samples in upregulated and neutral UBQ/DUB gene groups. The asterisks indicate the significant proportion difference between the two groups (chi-square test, *q < 0.01). Right: Venn diagram showing the proportions of upregulated UBQ/DUB genes affected by different regulatory mechanisms.
See also Figures S4 and S5.
To examine the molecular mechanisms underlying the UBQ/DUB mRNA upregulation in the seven cancer types, we further integrated SCNA, DNA methylation, and microRNA (miRNA) expression data and compared the patterns of upregulated UBQ/DUB genes to those of neutral ones (i.e., genes showing no significant differential mRNA expression). First, in 6 of the 7 cancer types, upregulated genes showed a significantly higher proportion of copy-number amplifications than did neutral genes (chi-square test, q < 0.01) (Figure 4C, top), highlighting the significant role of somatic copy-number gain in increasing UBQ/DUB gene expression in tumor samples. Second, for four cancer types with miRNA expression data and sufficient matched tumor and normal pairs (n > 20), compared to neutral genes, upregulated genes showed a significantly higher proportion of their reduced miRNA regulators in tumor samples of LIHC and LUAD (chi-square test, q < 0.1) (Figure 4C, middle), suggesting that miRNA-mediated gene repression contributes to the increased UBQ/DUB mRNA expression. Third, for six cancer types with DNA methylation data and sufficient matched tumor and normal pairs (n > 20), compared to neutral genes, upregulated genes showed a significantly higher proportion of reduced methylation levels in tumor samples of COAD and LUAD (chi-square test, q < 0.01) (Figure 4C, bottom), suggesting notable contributions of methylation-mediated gene silencing in these two cancer types. Finally, across cancer types, ~71% of upregulated UBQ/DUB genes were affected by these mechanisms and ~10% of them were mediated by more than one mechanism (Figures 4C, right, and S4B). These results provide a quantitative view of how different mechanisms contribute to the dysregulation of UBQ and DUB genes in tumor samples.
In addition, we performed an analysis to identify key miRNA regulators of UBQ and DUB genes. We inferred coding gene targets of 1,855 miRNAs by integrating both sequence information of target genes and the co-expression of the corresponding mRNA-miRNA pairs (STAR Methods). Several master miRNA regulators for UBQ and DUB genes emerged from this analysis (Figure S5), including the mir-200 family (mir-200a, mir-200b, mir-200c, mir-141, and mir-429), the mir-17/92 cluster (mir-17, mir-18a, mir-19a, mir-20a, and mir-19b-1), and mir-7-1.
Integrated Genomic Analysis of UBQ and DUB Genes
In addition to the preceding single-platform-oriented analysis, we integrated the data from mRNA expression, SCNA profiles, and DNA methylation to gain a more comprehensive picture of UBQ/DUB molecular patterns. We first normalized mRNA expression and DNA methylation data by Z scores within each cancer type to minimize tissue effects and then clustered the samples based on three data types separately. For each data type, all samples were appropriately clustered into four groups (Figure 5A–5C). Using the cluster-of-cluster assignment (COCA) strategy (Hoadley et al., 2014), we then represented each platform-specific cluster as binary vectors and re-clustered all samples across the three data types, which revealed three robust clusters (COCA1, COCA2, and COCA3) (Figures 5D and S6A). The samples were relatively evenly distributed among different cancers, and overall, 55%, 25%, and 20% of the samples belonged to COCA1, COCA2, and COCA3, respectively.
Figure 5. Integrative Genomic Clustering and Patient Survival Analysis.
(A–C) Heatmaps of consensus clustering for three platforms: RNA sequencing (RNA-seq)-based mRNA expression (A), somatic copy-number alterations (B), and DNA methylation (C).
(D) Consensus matrix of integrative clustering showing three robust clusters (COCA1, COCA2, and COCA3).
(E) COCA clusters correlate with patient overall survival and disease-specific survival times in 10 and 9 cancer types, respectively.
(F) Kaplan-Meier plots of nine cancer types showing overall survival curves for three clusters of patients with log-rank p values.
See also Figure S6.
We next assessed the clinical relevance of these three clusters. Because patient survival time generally reflects the progression of the disease and represents a key clinical variable, we performed survival analysis within each cancer type (log-rank test). The COCA clusters showed significant association with overall survival for 10 of 30 cancer types with a sufficient sample size and follow-up time (Figure 5E). In addition, the COCA clusters showed similar significant associations with the disease-specific survival times for nine cancer types (Figure 5E). Strikingly, in all these cancer types, COCA2 was always associated with worse prognosis (Figures 5F, S6B, and S6C). In addition, we examined the correlations of COCA clusters with established tumor subtypes and found significant correlations in multiple cancer types (Figure S6D). Altogether, these results highlight the potential clinical utility of this UBQ/DUB-driven subtyping.
Biological Pathways and Molecular Drivers Associated with COCA2
To gain biological insights into the intriguing subtype COCA2, we first identified the most associated hallmark pathways by GSEA based on mRNA expression data (q < 0.1). Across the 33 cancer types, upregulated genes in COCA2 (relative to COCA1 and COCA3) showed consistently significant enrichment in the G2M checkpoint and DNA repair pathways; COCA2 correlated with other hallmark pathways but did so less consistently (Figure 6A). To confirm the preceding strong pathway associations, we further analyzed the pathway scores of the cell-cycle and DNA damage pathways derived from TCGA protein expression data for 19 major cancer types with >100 samples (Akbani et al., 2014). COCA2 showed significantly higher cell-cycle and DNA damage response pathway scores in 12 and 9 cancer types, respectively (p < 0.05) (Figure 6B).
Figure 6. Biological Pathways and Somatic Drivers Associated with the Poor Prognostic Tumor Subtypes (COCA2).
(A) Association of COCA2 with GSEA hallmark gene sets. Significant positive associations are shown in red, significant negative associations are shown in blue, and non-significant ones are shown in gray.
(B) Reverse-phase protein array (RPPA)-based pathway scores of cell-cycle and DNA repair between COCA2 samples (red box) and other samples (blue box).
The bottom and top of the box are the first and third quartiles, and the whiskers extend to 1.5 IQR of the lower quartile and the upper quartile, respectively.
(C) Significantly mutated genes identified by MutSigCV, in which the mutations are significantly enriched (red) or depleted (blue) in COCA2 compared to COCA1 and COCA3 in different cancer types (q < 0.1).
(D) SCNA drivers identified by GISTIC2, in which amplifications (for oncogenes) and deletions (for tumor suppressors) are significantly enriched in COCA2 compared to COCA1 and COCA3 in different cancer types (q < 0.001).
We next sought to identify somatic alternations that potentially drive the COCA2 subtypes. We first focused on significantly mutated genes (identified by MutSigCV) in each cancer type and assessed whether their mutation rates were different between the COCA2 samples and the remaining samples (Figure 6C). TP53 was differentially mutated in 13 cancer types (q < 0.1), including UCEC, LUAD, lower grade glioma (LGG), HNSC, COAD, and LIHC, in which significant survival patterns were observed (Figure 5F). Then we examined SCNA drivers by focusing on known oncogenes and tumor suppressors residing in amplification or deletion peaks (identified by GISTIC2) in each cancer type (Mermel et al., 2011; Zack et al., 2013). We found that COCA2 was associated with the amplifications of MYC and TERT and the deletions of PTEN and APC in multiple cancer types (q < 0.001) (Figure 6D). These potential SCNA drivers also showed consistent gene expression patterns across cancer types (Figure S7).
Low expression or mutated TP53 and MYC amplification are well-established drivers of cell-cycle and DNA damage repair response dysregulation (Campaner and Amati, 2012; Dang, 2012; Nakayama and Nakayama, 2006; Williams and Schumacher, 2016). Deletion or low expression of the tumor suppressor PTEN has been shown to drive cell-cycle progression, proliferation, and cell survival (Chalhoub and Baker, 2009; Minami et al., 2014; Ming and He, 2012). High expression of TERT, the catalytic subunit of telomerase, immortalizes cancer cells by promoting cell-cycle progression and increased survival. Furthermore, low expression of anaphase-promoting complex (APC) drives uncontrolled cell-cycle progression and proliferation. Therefore, we put forward a model in which mutated TP53 and amplified MYC are closely associated with primarily an upregulation of key ubiquitin-related enzymes, leading to an uncontrolled cell cycle, elevated DNA damage response, and ultimately poor survival for COCA2 patients (Figure 7). For the cell-cycle pathway, in addition to the upregulated core backbone components of SCF and the anaphase-promoting complex (APC/C), substrate recognition components such as CDH1, CDC20, and SKP2 showed upregulation, while FBXW7 and BTRC were downregulated. This result is consistent with the nature of the complexes’ substrates. For example, substrates of FBXW7 are oncoproteins, such as cyclin E, c-Myc, and Notch, while substrates of SKP2 are tumor suppressors, such as p21, p27, and p57. For the DNA damage pathway, there was increased mRNA expression of RNF8, RNF168, RAD18, BRCA1, and UBE2N; the exception was HERC2. Increased DNA damage response intimately integrates with the dysregulation of cell-cycle progression and checkpoint control. This can potentially create a deleterious feedback loop, in which dysregulation of cell-cycle checkpoints, coupled with elevated DNA damage repair, leads to cells with unrepaired DNA damage entering replication, thereby amplifying the subsequent DNA damage response.
Figure 7. Mechanistic Model Describing the Biological Process Underlying COCA2 Subtypes.
Somatic drivers identified for COCA2 subtypes (top) cause the expression-level changes of key UBQ and DUB genes in SCF complex, APC/C complex, and DNA damage response that underlie the aberrant activities of cell-cycle and DNA damage pathways (middle), thereby leading to poor patient survival of COCA2 subtypes (bottom). See also Figure S7.
DISCUSSION
Using the latest TCGA multidimensional molecular profiling data, we performed comprehensive molecular characterization of the ubiquitin pathway of 1,024 genes across 9,125 samples of 33 cancer types. There are three key findings in our study. First, we systematically cataloged driver candidates with significant mutation and SCNA patterns. Compared with top SCNA drivers, the profiles of mutation drivers are diverse across different cancer types. For example, BAP1 and VHL are frequently mutated in mesothelioma (MESO) and KIRC, respectively, whereas FBXW7 is enriched with hotspot mutations in UCEC and UCS but enriched with LoF mutations in ESCA, LUAD, LUSC, READ, SKCM, and STAD. These results suggest context-dependent oncogenic mechanisms of UBQ/DUB mutation drivers, which have been less appreciated in the field. Second, we show that compared to matched normal tissues, genes in the ubiquitin pathway tend to be overexpressed in a range of cancer types, and collectively, 71% of the upregulated genes are contributed by one of three mechanisms: somatic copy-number gain, reduced methylation-mediated gene silencing, and reduced miRNA-mediated gene regulation in tumors. Finally, the cross-platform integrative analysis reveals a group of patients that is consistently correlated with worse prognosis across nine cancer types. These tumor samples are associated with differential UBQ/DUB expression underlying the perturbation of many fundamental signaling pathways, notably cell-cycle progression and DNA damage repair, likely resulting from key molecular drivers such as TP53, MYC, TERT, PTEN, and APC. These striking and robust patterns highlight a unique value of the ubiquitin pathway in patient classification, conferring potential prognostic utility.
One major motivation for studying UBQs and DUBs is the potential to develop drugs that target the ubiquitin system (Salami and Crews, 2017). Thus far, the progress has been limited. This could be largely due to the lack of systemic characterization of significant driver mutations, SCNA patterns, and dysregulated expression profiles in the ubiquitin pathway across cancer types through an integrated genomic analysis that would provide clinically relevant drug candidates to the pharmaceutical industry. Another challenge is that unlike kinases, many components of the ubiquitin pathway lack a well-defined catalytic pocket, which makes them difficult to target by small molecules, although this obstacle might be overcome by the development of inhibitors that block specific protein-protein interaction. However, given its widespread impact, the potential for targeting some key components in the ubiquitin pathway for drug development through the controlled proteostasis mechanism is immense. Studies have provided a proof of principle that certain E3 ligases and DUBs are potential therapeutic targets that are amenable to inhibition by small molecules. For instance, MDM2 and SKP2, two oncogenic E3 ligases overexpressed in multiple cancer types, can be inhibited by Nutlins and Compound 25, respectively; these compounds have shown promising anti-tumor effects in xenograft tumor models (Chan et al., 2013; Vassilev et al., 2004). Moreover, the deubiquitinase USP7 has been shown to deubiquitinate several key cancer proteins, and P5091, a highly specific inhibitor of USP7, induced apoptosis in multiple myeloma cells (Chauhan et al., 2012). Our study suggests that targeting the ubiquitin pathway components involved in cell-cycle progression and DNA damage response pathways may offer promising opportunities for drug interventions, because these two pathways tightly correlate with the prognostically relevant tumor subtypes. In addition, mutually exclusive patterns between ubiquitin pathway genes and known actionable cancer genes suggest potential combination therapeutic strategies. This focused, systematic analysis of UBQ and DUB genes will lay a critical foundation for understanding the dysregulation of UBQ in cancer and provide unique insights into the development of related therapeutic approaches.
STAR★METHODS
KEY RESOURCES TABLE.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Han Liang (hliang1@mdanderson.org).
METHOD DETAILS
Curation of UBQ and DUB gene sets
Given the diverse and integrative nature of the ubiquitin pathway, it was difficult to properly curate a definitive UBQ gene list. After an initial rigorous literature search, no established consensus was observed in the field beyond E1 and E2 enzymes, and the discovery of important E3 enzymes, adapters, and other E3-associated genes was both escalating and in high debate. We therefore started our curation with a centralized, thorough database of UBQ and UBQ-associated genes, the Ubiquitin and Ubiquitin-like Conjugation Database (UUCD, http://uucd.biocuckoo.org/), which is continually updated and optimized as well as the most detailed and comprehensive available (Gao et al., 2013).
The UUCD database uses a multi-fold process to curate its UBQ genes relying on a combination of manual and computational methods. The first step in the process is a manual literature search using key words related to UBQ and UBQ-associated genes that can covalently recognize and modify other molecules, such as “ubiquitin,” “ubiquitination,” etc. This substantial, amassed list of genes was then distinguished into E1 gene (ThiF/MoeB), E2 gene (UBC, UEV), and E3 gene categories based on the classification of their functional domains in the literature, thereby establishing a dual publication (PMID - Table S2) and UBQ domain correlation criterion for curation (Gao et al., 2013). Since a significant number of integral proteins participate in the function of E3 ligase complexes as adapters/receptors, UUCD further categorized the E3-associated genes into two classifications based on domain sequence and associated function: E3 activity and E3 adaptor, where E3 activity refers to a functioning E3 enzyme in contrast to E3 adapters (E3-complex adaptor, substrate receptor, etc.) integrally participating in E3 enzymatic function (Gao et al., 2013). After properly categorizing the manually curated UBQ genes from the literature, UUCD applied a computational approach using UBQ protein sequences and hidden Markov models (HMM) to predict other possible UBQ enzymes and adapters yet to be discovered (HMM –Table S2). After aligning the protein sequences by MUSCLE, HMMER 3.0 was utilized to create hidden Markov model profiles of 1, 1, and 15 (http://uucd.biocuckoo.org/download/HMM) for E1, E2, and E3 UBQ genes, respectively. Moreover, these HMM models were further utilized along with hmmsearch to search all protein sequences to identify unknown UBQ genes and adapters, thus providing a much more expanded gene list of possible UBQ functionality (Eddy, 1998; Edgar, 2004; Gao et al., 2013). In this study, we chose to cast a wide net for potential E3 adapters/receptors in order for the most comprehensive analysis available.
Our final UBQ gene list was based on extracting the UBQ genes from the UUCD database (downloaded in March 2017) and then adding a filtering step to remove ubiquitin-like genes (enzymes and adapters) not directly associated with UBQ pathway function, such as sumoylation and ISGylation associated genes. As a result, our UBQ gene set included 929 genes in total, including pre-established UUCD gene categories of E1 (8 genes, 1 predicted), E2 (39 genes, 2 predicted), and E3 (882 genes, 368 predicted). For E3 UBQ genes, they were further divided into E3 activity (387 genes, 78 predicted) and E3 adaptor (495 genes, 290 predicted) (Figure S1A, Table S2).
In contrast to the complexity of the UBQ gene curation, DUB genes are far fewer in number and much easier to curate. A general consensus has been reached on their classification, thus a simple mining of the literature detailing an inventory of DUB genes leads to comprehensive gene coverage. In this study, we intersected 3 major DUB review articles and found a substantial overlap of DUB genes, where 80 of 91 total DUB genes were found in all 3 articles (Fraile et al., 2012; Komander et al., 2009; Nijman et al., 2005) (Figure S1B, Table S2). Notably, 4 genes from the MCPIP family, originally reported as DUB genes in Fraile et al., 2012, were later shown to exhibit no DUB activity and therefore filtered first before generating the final union of 91 DUB genes (Niu et al., 2013). In addition, we included four DUB genes recently discovered (Abdul Rehman et al., 2016). As a result, we created a final list of 95 DUB genes comprised of six major classes, including ubiquitin C-terminal hydrolase (UCH), ubiquitin-specific protease (USP), Machado-Joseph domain (MJD), ovarian tumor (OTU), JAB1/MPN/Mov34 metalloprotease (JAMM), and motif interacting with Ub-containing DUB family (MINDY) (Table S2).
Somatic mutation analysis
We obtained TCGA pan-cancer somatic mutation data from Genomic Data Commons. Further filtering steps were used to eliminate artifacts and reduce false-positive calls. A) Only mutations with “PASS” in the “FILTER” column were retained for all cancer types except for ovarian serous cystadenocarcinoma (OV) and acute myeloid leukemia (LAML), for which we allowed “wga.” B) Hypermutated samples with > 1,000 somatic mutations were removed, resulting in somatic mutation data for 8,811 samples for further analyses. Only non-silent somatic mutations were used to calculate mutation frequency. For each cancer type, MutSigCV (v1.4) was used to identify significantly mutated genes in each cancer cohort at a q value of 0.1. Across the pan-cancer cohort, hotspot mutations were defined as missense or in-frame mutations at the same protein amino acid in > 2 patient samples. The fraction of hotspot mutations per gene was calculated as the total number of hotspot mutations over the total number of non-silent mutations found in that gene. The fraction of LoF mutations (defined as Frame_Shift_Ins, Frame_Shift_Del, Nonsense_Mutation, Nonstop_Mutation, Splice_Site, and Tanslation_Start_Site) per gene was calculated as the total number of LoF mutations over the total number of non-silent mutations in that gene. Genes with > 30% hotspot mutations, < 20% LoF mutations, and ≥ 5 unique hotspot mutation positions were identified as enriched with hotspot mutations, while genes containing > 30% LoF mutations, < 30% hotspot mutations, and ≥ 10 LoF mutations were identified as enriched with LoF mutations. Mutual exclusivity for FBXW7 mutations and mutations in clinically actionable genes (annotated as in OncoKB, http://oncokb.org) was performed with the R package “cometExactTest.” To study the effects of FBXW7 mutations, PI3K pathway expression was calculated from protein levels of the PI3K/Akt pathway components as measured by RPPA with the following formula where E means expression:
Somatic copy-number alteration analysis
We obtained SCNA data of 9,125 patient samples from Genomic Data Commons and applied GISTIC2. For each cancer type, genes were considered to be amplified or deleted if they were located in the amplification peak or deletion peak at a q value of 0.25. The SCNA mutual exclusivity test was performed by employing the R package cometExactTest using the integer copy number data. Mutual exclusivity for MDM2 amplification and non-silent mutations in clinically actionable genes was carried out using the R package “cometExactTest.” For the pan-cancer SCNA clustering analysis, the integer copy numbers of UBQ and DUB genes were used with Ward’s method for consensus clustering.
RNA-seq analysis
We obtained normalized gene expression data from Genomic Data Commons. For the tumor-normal comparison, we performed a differential expression analysis between tumor and their matched normal samples for each of 16 cancer types using the Wilcoxon signed rank test and built the pre-ranked gene lists based on signed −log10 p values. For GSEA, the pre-ranked gene lists were then run against the UBQ, DUB and UBQ/DUB gene sets using GSEA Java GUI (version 2.3.3), respectively. For the pan-cancer expression-level clustering analysis, the normalized values from the root squared error method were log2-transformed and Z-normalized within each cancer type. Then, Pearson’s correlation and hierarchical average linkage clustering were applied to the top 800 most variable UBQ/DUB genes for consensus clustering.
MiRNA expression analysis
We obtained normalized miRNA expression data from Genomic Data Commons. To study the mechanisms underlying dysregulated UBQ/DUB genes in cancer, based on the miRNA expression data of paired tumor–normal samples from 562 patients, a paired t test was performed for each gene within each cancer type (5 cancer types with a sample size of > 20 pairs), where genes with significantly high or low expression in tumor samples were determined using a p-value cutoff of 0.05. We identified master miRNA regulators for UBQ and DUB genes based on two criteria. First, a miRNA has at least one seed region (2–8-mer) matched to the 3′UTR of any UBQ/DUB gene. Second, the Spearmen correlation of miRNA with the expression of the target gene was statistically significant (q < 10−5 and rho < −0.5). Cytoscape was used to visualize miRNA and the UBQ/DUB gene network.
DNA methylation analysis
We obtained DNA methylation 450K data from Genomic Data Commons. For each gene, one DNA methylation probe was selected based on the correlation with its mRNA expression level, and if multiple probes for a gene were available, the probe that had the most negative correlation value was selected. To study the mechanisms underlying dysregulated UBQ/DUB genes in cancer, based on the DNA methylation data of paired tumor–normal samples from 624 patients, a paired t test was performed for each gene within each cancer type (6 cancer types with a sample size of > 20 pairs), where genes significantly hypermethylated or hypomethylated were determined using a p-value cutoff of 0.05. For the pan-cancer methylation-based clustering, consensus clustering was performed for the top 1,000 most variable probes for the UBQ/DUB genes, using the Euclidean distance and partitioning around medoids method.
Integrative clustering and patient survival analysis
We obtained TCGA patient clinical data from Genomic Data Commons. Clusters defined from individual platforms (SCNA, mRNA and DNA methylation) were coded into binary variables for each platform-specific cluster. The matrix of 0 s and 1 s was then used as the input data matrix in the ConsensusClusterPlus R package to identify integrated relationships for the 9,125 patient samples. Pearson’s correlation and hierarchical clustering were used. Overall survival or disease-specific survival curves were compared using log-rank tests in the R package “survival.” To detect biological pathways associated with COCA clusters, for each cancer type, we performed a differential expression analysis between COCA2 and COCA1/COCA3 samples using a t test and built the pre-ranked gene lists based on signed −log10 p values. For GSEA, the pre-ranked gene lists were then run against the seven cancer hallmark gene sets (MSigDB Collections: H) using GSEA Java GUI (version 2.3.3).
RPPA pathway score calculation
We obtained the normalized RPPA data from Genomic Data Commons and Z-normalized the data within each cancer type. Pathway scores for the cell cycle and DNA damage response pathways were calculated for each patient (total of 6,441 patients). To detect biological pathways associated with COCA clusters, RPPA pathway scores were used to test the pathway perturbations between COCA2 and COCA1/COCA3 samples using a t test.
QUANTIFICATION AND STATISTICAL ANALYSES
Somatic mutation, SCNA, and RNA-seq analyses were based on 9,125 tumor samples; and miRNA expression, DNA methylation, and RPPA analyses were respectively based on 7,939, 8,058, and 6,441 tumor samples due to limited data availability. Definitions of significance for various statistical tests are described and referenced in their respective sections in Methods.
DATA AND SOFTWARE AVAILABILITY
The raw data, processed data, and clinical data can be found at the legacy archive of the GDC (https://portal.gdc.cancer.gov/legacy-archive/search/f) and the PancanAtlas publication page (https://gdc.cancer.gov/about-data/publications/pancanatlas). The mutation data can be found here (https://gdc.cancer.gov/about-data/publications/mc3-2017). TCGA data can also be explored through the Broad Institute FireBrowse portal (http://gdac.broadinstitute.org) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org). Details for software availability are in the Key Resources Table.
Supplementary Material
Highlights.
Full molecular characterization of 1,024 ubiquitin pathway genes in 33 cancer types
Systematically identify somatic driver candidates in the ubiquitin pathway
Consistent prognostic patterns of tumor subtypes defined by ubiquitin pathway genes
Propose a ubiquitin pathway mechanistic model underlying poor patient survival
Acknowledgments
This study was supported in part by the NIH (R01CA175486 and U24CA209851 to H.L., R01CA166051 and R01CA181029 to L.M., U54HG003273, U54HG003067, U54HG003079, U24CA143799, U24CA143835, U24CA143840, U24CA143843, U24CA143845, U24CA143848, U24CA143858, U24CA143866, U24CA143867, U24CA143882, U24CA143883, U24CA144025, and P30CA016672), the Cancer Prevention and Research Institute of Texas (RP140462 to H.L. and RP150319 to L.M.), a Stand Up To Cancer Innovative Research Grant (to L.M.), a University of Texas System STARS award (to H.L.), and the US Department of Defense through the Henry M. Jackson Foundation for the Advancement of Military Medicine (W81XWH-12-2-0050, HU0001-16-2-0004). We thank the MD Anderson high-performance computing core facility for computing and LeeAnn Chastain for editorial assistance. The views expressed in this article are those of the authors and do not reflect the official policy of the U.S. Army, Navy, Air Force, Department of Defense, or government.
Footnotes
Supplemental Information includes seven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.047.
AUTHOR CONTRIBUTIONS
H.L. conceived and designed the study; Z.G., J.S.L., Y.W., X.P., Z.C., H.C., Y.S., F.Y., J. Li, H.Z., J. Liu, C.D.S., H.H., H.P.W., L.M., and H.L. performed data analysis; G.Z., L.J., and L.H. wrote the manuscript with input from other co-authors; and L.H. supervised the project.
DECLARATION OF INTERESTS
Michael Seiler, Peter G. Smith, Ping Zhu, Silvia Buonamici, and Lihua Yu are employees of H3 Biomedicine, Inc. Parts of this work are the subject of a patent application: WO2017040526 titled “Splice variants associated with neomorphic sf3b1 mutants.” Shouyoung Peng, Anant A. Agrawal, James Palacino, and Teng Teng are employees of H3 Biomedicine, Inc. Andrew D. Cherniack, Ashton C. Berger, and Galen F. Gao receive research support from Bayer Pharmaceuticals. Gordon B. Mills serves on the External Scientific Review Board of Astrazeneca. Anil Sood is on the Scientific Advisory Board for Kiyatec and is a shareholder in BioPath. Jonathan S. Serody receives funding from Merck, Inc. Kyle R. Covington is an employee of Castle Biosciences, Inc. Preethi H. Gunaratne is founder, CSO, and shareholder of NextmiRNA Therapeutics. Christina Yau is a part-time employee/consultant at NantOmics. Franz X. Schaub is an employee and shareholder of SEngine Precision Medicine, Inc. Carla Grandori is an employee, founder, and shareholder of SEngine Precision Medicine, Inc. Robert N. Eisenman is a member of the Scientific Advisory Boards and shareholder of Shenogen Pharma and Kronos Bio. Daniel J. Weisenberger is a consultant for Zymo Research Corporation. Joshua M. Stuart is the founder of Five3 Genomics and shareholder of NantOmics. Marc T. Goodman receives research support from Merck, Inc. Andrew J. Gentles is a consultant for Cibermed. Charles M. Perou is an equity stock holder, consultant, and Board of Directors member of BioClassifier and GeneCentric Diagnostics and is also listed as an inventor on patent applications on the Breast PAM50 and Lung Cancer Subtyping assays. Matthew Meyerson receives research support from Bayer Pharmaceuticals; is an equity holder in, consultant for, and Scientific Advisory Board chair for OrigiMed; and is an inventor of a patent for EGFR mutation diagnosis in lung cancer, licensed to LabCorp. Eduard Porta-Pardo is an inventor of a patent for domainXplorer. Han Liang is a shareholder and scientific advisor of Precision Scientific and Eagle Nebula. Da Yang is an inventor on a pending patent application describing the use of antisense oligonucleotides against specific lncRNA sequence as diagnostic and therapeutic tools. Yonghong Xiao was an employee and shareholder of TESARO, Inc. Bin Feng is an employee and shareholder of TESARO, Inc. Carter Van Waes received research funding for the study of IAP inhibitor ASTX660 through a Cooperative Agreement between NIDCD, NIH, and Astex Pharmaceuticals. Raunaq Malhotra is an employee and shareholder of Seven Bridges, Inc. Peter W. Laird serves on the Scientific Advisory Board for AnchorDx. Joel Tepper is a consultant at EMD Serono. Kenneth Wang serves on the Advisory Board for Boston Scientific, Microtech, and Olympus. Andrea Califano is a founder, shareholder, and advisory board member of DarwinHealth, Inc. and a shareholder and advisory board member of Tempus, Inc. Toni K. Choueiri serves as needed on advisory boards for Bristol-Myers Squibb, Merck, and Roche. Lawrence Kwong receives research support from Array BioPharma. Sharon E. Plon is a member of the Scientific Advisory Board for Baylor Genetics Laboratory. Beth Y. Karlan serves on the Advisory Board of Invitae.
References
- Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, Hofmann K, Kulathu Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campaner S, Amati B. Two sides of the Myc-induced DNA damage response: from tumor suppression to tumor maintenance. Cell Div. 2012;7:6. doi: 10.1186/1747-1028-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, McDermott JL, Leach CA, Fulcinniti M, Kodrasov MP, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile JM, Quesada V, Rodriguez D, Freije JM, López-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- Fuchs SY. The role of ubiquitin-proteasome pathway in oncogenic signaling. Cancer Biol Ther. 2002;1:337–341. [PubMed] [Google Scholar]
- Gao T, Liu Z, Wang Y, Cheng H, Yang Q, Guo A, Ren J, Xue Y. UUCD: a family-based database of ubiquitin and ubiquitin-like conjugation. Nucleic Acids Res. 2013;41:D445–D451. doi: 10.1093/nar/gks1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israël A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan MD, Uzunangelov V, et al. Cancer Genome Atlas Research Network. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- Huang X, Dixit VM. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 2016;26:484–498. doi: 10.1038/cr.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Wu HT, Vandin F, Raphael BJ. CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol. 2015;16:160. doi: 10.1186/s13059-015-0700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Monoversus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Lind H, Zienolddiny S, Ekstrøm PO, Skaug V, Haugen A. Association of a functional polymorphism in the promoter of the MDM2 gene with risk of nonsmall cell lung cancer. Int J Cancer. 2006;119:718–721. doi: 10.1002/ijc.21872. [DOI] [PubMed] [Google Scholar]
- Lu M, Breyssens H, Salter V, Zhong S, Hu Y, Baer C, Ratnayaka I, Sullivan A, Brown NR, Endicott J, et al. Restoring p53 function in human melanoma cells by inhibiting MDM2 and cyclin B1/CDK1-phosphorylated nuclear iASPP. Cancer Cell. 2013;23:618–633. doi: 10.1016/j.ccr.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Luise C, Capra M, Donzelli M, Mazzarol G, Jodice MG, Nuciforo P, Viale G, Di Fiore PP, Confalonieri S. An atlas of altered expression of deubiquitinating enzymes in human cancer. PLoS ONE. 2011;6:e15891. doi: 10.1371/journal.pone.0015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, Balmain A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fässler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami A, Nakanishi A, Ogura Y, Kitagishi Y, Matsuda S. Connection between Tumor Suppressor BRCA1 and PTEN in Damaged DNA Repair. Front Oncol. 2014;4:318. doi: 10.3389/fonc.2014.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming M, He YY. PTEN in DNA damage repair. Cancer Lett. 2012;319:125–129. doi: 10.1016/j.canlet.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- Nijman SMB, Luna-Vargas MPA, Velds A, Brummelkamp TR, Dirac AMG, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Niu J, Shi Y, Xue J, Miao R, Huang S, Wang T, Wu J, Fu M, Wu ZH. USP10 inhibits genotoxic NF-κB activation by MCPIP1-facilitated deubiquitination of NEMO. EMBO J. 2013;32:3206–3219. doi: 10.1038/emboj.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Zhao L, Li Y, Yue P, Deng X, Owonikoko TK, Chen M, Khuri FR, Sun SY. The PI3 kinase inhibitor NVP-BKM120 induces GSK3/FBXW7-dependent Mcl-1 degradation, contributing to induction of apoptosis and enhancement of TRAIL-induced apoptosis. Cancer Lett. 2013;338:229–238. doi: 10.1016/j.canlet.2013.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki AY, Caenepeel S, Yu D, Lofgren JA, Osgood T, Robertson R, Canon J, Su C, Jones A, Zhao X, et al. MDM2 antagonists synergize broadly and robustly with compounds targeting fundamental oncogenic signaling pathways. Oncotarget. 2014;5:2030–2043. doi: 10.18632/oncotarget.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salami J, Crews CM. Waste disposal-An attractive strategy for cancer therapy. Science. 2017;355:1163–1167. doi: 10.1126/science.aam7340. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Balinga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KT, Rosner MR, Minella AC. An integrated view of cyclin E function and regulation. Cell Cycle. 2012;11:57–64. doi: 10.4161/cc.11.1.18775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, Konecny GE, Coleman RL, Tinker AV, O’Malley DM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- Tauriello DV, Haegebarth A, Kuper I, Edelmann MJ, Henraat M, Canninga-van Dijk MR, Kessler BM, Clevers H, Maurice MM. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell. 2010;37:607–619. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Dijke PT, Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24:8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- Williams AB, Schumacher B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb Perspect Med. 2016;6:a026070. doi: 10.1101/cshperspect.a026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Zhang P, Ma L. The role of deubiquitinases in breast cancer. Cancer Metastasis Rev. 2016;35:589–600. doi: 10.1007/s10555-016-9640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang H, Li M, Rayburn ER, Agrawal S, Zhang R. Stabilization of E2F1 protein by MDM2 through the E2F1 ubiquitination pathway. Oncogene. 2005;24:7238–7247. doi: 10.1038/sj.onc.1208814. [DOI] [PubMed] [Google Scholar]
- Zhao B, Schlesiger C, Masucci MG, Lindsten K. The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. J Cell Mol Med. 2009;13(8B):1886–1895. doi: 10.1111/j.1582-4934.2008.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.