Summary
Growth factor binding to EGFR drives conformational changes that promote homodimerization and transphosphorylation, followed by adaptor recruitment, oligomerization, and signaling through Ras. Whether specific receptor conformations and oligomerization states are necessary for efficient activation of Ras is unclear. We therefore evaluated the sufficiency of a phosphorylated EGFR dimer to activate Ras without growth factor by developing a chemical-genetic strategy to crosslink and “trap” full-length EGFR homodimers on cells. Trapped dimers become phosphorylated and recruit adaptor proteins at stoichiometry equivalent to that of EGF-stimulated receptors. Surprisingly, these phosphorylated dimers do not activate Ras, Erk, or Akt. In the absence of EGF, phosphorylated dimers do not further oligomerize or reorganize on cell membranes. These results suggest that a phosphorylated EGFR dimer loaded with core signaling adapters is not sufficient to activate Ras and that EGFR ligands contribute to conformational changes or receptor dynamics necessary for oligomerization and efficient signal propagation through the SOS-Ras-MAPK pathway.
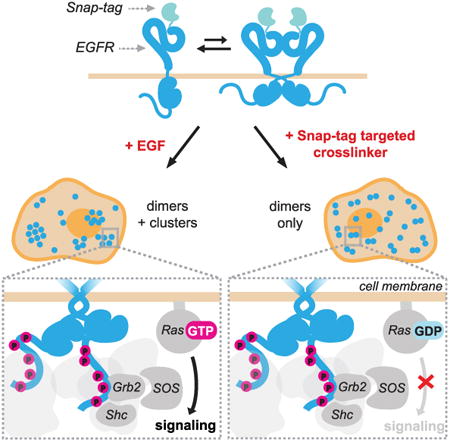
Graphical abstract
Liang et al. demonstrate that the recruitment of key signaling adapters to stable phosphorylated EGFR dimers is not sufficient for the activation of Ras and its downstream pathways. Binding of EGFR ligands induces conformational changes and receptor dynamics necessary for oligomerization and efficient signal propagation through the SOS-Ras-MAPK pathway.
Introduction
Epidermal growth factor receptor (EGFR) is a broadly expressed receptor tyrosine kinase frequently mutated or overexpressed in cancer. The steps of EGFR activation by ligands such as EGF have been extensively studied. Biochemical, imaging, and structural evidence support a model wherein monomers of EGFR are inactive and in equilibrium with a population of inactive dimers (Chung et al., 2010; Jura et al., 2009). Binding of EGF stabilizes receptor conformations that expose an extracellular dimerization interface, triggering accumulation of active EGFR dimers (Ferguson et al., 2003; Ogiso et al., 2002). One intracellular kinase then allosterically activates the other, resulting in phosphorylation of C-terminal tyrosines (Zhang et al., 2006) (Figure 1A). Phosphorylated tyrosines recruit signaling adapters such as Shc, Grb2, and SOS, which stimulate a variety of downstream pathways (Margolis et al., 1989). Among these, the Ras-MAPK (mitogen-activated protein kinase) pathway is a particularly important regulator of cell behaviors such as proliferation and migration.
Figure 1. Decoupling EGFR Dimerization and Transphosphorylation from Other EGFInduced Conformational and Spatial Changes.
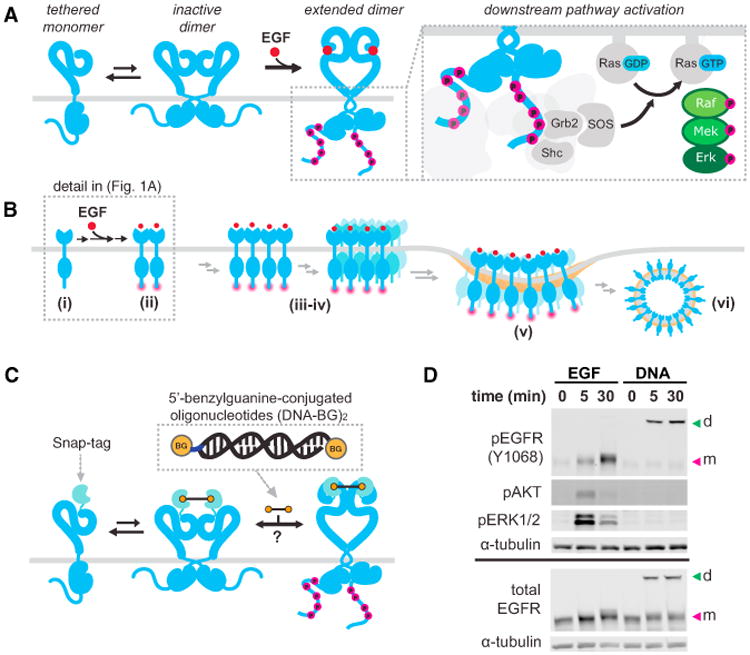
(A) EGFR exists in a tethered monomer or an inactive dimer formation. Upon EGF binding, it adopts an extended dimer conformation and undergoes auto-transphosphorylation. Phosphorylated dimers recruit adaptor proteins to EGFR, resulting in activation of the Ras-MAPK pathway.
(B) EGF binding to EGFR also results in rapid changes in spatial organization from monomers (i) to dimers (ii); to higher order multimers and nanoscale clusters (iii-iv); to micron scale clusters in clathrin-coated pits (v); and, finally, to endosomes (vi).
(C) A chemical genetic system utilizing a SNAP-tag on the N terminus of full-length EGFR and BG-modified DNA dimers as crosslinkers.
(D) Representative western blot of lysates from cells treated with 8 nM EGF or 2 μM (DNA-BG)2. To maintain DNA hybridization, SDS-PAGE samples were not boiled. EGFR dimers (d) and monomers (m) are indicated with arrows.
The formation of phosphorylated EGFR dimers is generally considered sufficient to initiate Ras signaling because the dimers recruit the Ras-GEF (guanine nucleotide exchange factor) SOS to the membrane, and membrane-localized SOS is sufficient to activate Ras under a variety of conditions (Aronheim et al., 1994; Christensen et al., 2016; Toettcher et al., 2013). However, conflicting observations raise questions regarding whether a phosphorylated dimer is a competent signaling unit, sufficient to activate Ras, in the absence of EGF. For example, dimerization of a chimeric receptor's intracellular domains with rapamycin derivatives was sufficient to induce EGFR phosphorylation and downstream Erk phosphorylation (Muthuswamy et al., 1999). In contrast, dimerization of EGFR on cancer cell lines with therapeutic antibodies resulted in phosphorylated EGFR but no Erk phosphorylation (Yoshida et al., 2008). While these examples varied greatly in experimental design—for instance, the antibodies specifically targeted EGFR's extracellular domain and locked EGFR dimers in an inactive conformation (Li et al., 2005), whereas the chimeric EGFR had its extracellular domain replaced with the transmembrane and extracellular domain of p75 Neurotrophin receptor—it remains difficult to rationalize how the phosphorylated intracellular domains could be signaling competent in one study but not in another.
A possible resolution of this conundrum is the requirement for a specific tertiary or quaternary structure beyond the dimer, promoted by EGF binding, to efficiently activate Ras. Upon EGF binding, dimers undergo rapid spatial rearrangement into oligomers and nanoscale clusters (Figure 1B) (Ariotti et al., 2010; Clayton et al., 2008; Ichinose et al., 2004; Saffarian et al., 2007; van Lengerich et al., 2017), and these oligomers may promote downstream signaling (Huang et al., 2016; Kozer et al., 2013; Needham et al., 2016). However, because oligomerization and signaling changes occur on a similar timescale, it remains unclear whether specific spatial intermediates are a cause or consequence of downstream signaling.
We sought to determine whether a phosphorylated EGFR dimer is sufficient to activate Ras signaling without EGF. This question is challenging to answer, because receptor overexpression (Avraham and Yarden, 2011;Pedersen et al., 2005), mutations and truncations (Arkhipov et al., 2013; Bessman et al., 2014), and antibodies (Li et al., 2005; Schmiedel et al., 2008) can perturb the conformations adopted by EGFR and have unpredictable consequences on signaling. We addressed these challenges by developing a chemical genetic strategy based on targeted chemical crosslinking that allows for the preparation of a clean population of full-length receptor dimers, expressed at near-WT (wildtype) levels, and dimerized using long and flexible crosslinkers that do not significantly restrict receptor conformations. This strategy effectively decouples EGFR dimerization from other EGF-induced conformation changes and dynamics, allowing us to conclude that the critical function of EGF in Ras signal transduction is not limited to promoting the formation of a phosphorylated EGFR dimer, but also promoting receptor dynamics, conformations, or oligomeric states necessary for downstream signaling.
Results
A Chemical Genetic System for Preparing Full-Length EGFR Dimers without Ligand
To decouple EGF-induced receptor dimerization from other EGF-induced conformation changes, we sought to exploit the equilibrium between monomers and inactive dimers on resting cells. We hypothesized that selectively reducing the off rate of EGFR dimers would stimulate autophosphorylation rates sufficient to overcome high endogenous levels of background phosphatase activity (Kleiman et al., 2011), thereby generating phosphorylated receptor dimers. First, we modified the N terminus of full-length EGFR with a flexible linker and a SNAP-tag, which rapidly forms a covalent bond with benzyl guanine (BG), as the chemical dimerization domain. When this construct was stably expressed in HEK293 cells at physiological levels, we found that it was efficiently activated by the addition of nanomolar concentrations of EGF (Figure 1D). For the chemical dimerizer, we incorporated BG at the 5′-hydroxyl of double-stranded DNA molecules (DNA-BG)2, (20-mer; approximate length, 6.8 nm; Figure 1C). Addition of (DNA-BG)2 to live cells for 5 or 30 min resulted in a higher molecular weight band by western blot, consistent with a trapped dimeric species (Figures 1D and S1). Blotting for phosphorylation of tyrosine 1068 confirmed that the kinase domains of trapped dimers were active. Strikingly, we also observed pronounced differences in phosphorylated Erk between EGF-stimulated and trapped dimer receptors (Figure 1D). Consistently, and in multiple cell lines, we observed strong Erk and Akt signaling from EGF-stimulated SNAP-EGFR, and no signaling above background in the presence of (DNA-BG)2 (Figure S1). These results suggest that selective stabilization of an EGFR dimer is sufficient to stimulate kinase activity independent of additional conformational changes associated with EGF binding. However, receptor phosphorylation alone did not generate Ras-MAPK signaling.
Trapped EGFR Dimers Are Phosphorylated to a Similar Extent as EGF-Activated EGFR
Differences in EGFR phosphorylation levels between (DNA-BG)2 and EGF stimuli, as well as the pattern of phosphorylation (Ronan et al., 2016), could explain differences in downstream Erk activation. This hypothesis could be tested by quantitative western blotting, but quantitative comparison can be challenging between monomeric and crosslinked species, because large differences in molecular weight impact the transfer efficiency of proteins (Towbin et al., 1979). We therefore selectively melted (DNA-BG)2 crosslinks by boiling samples after cell lysis but prior to SDS-PAGE (Figure 2A).
Figure 2. Quantitative Comparison of Tyrosine Phosphorylation after Dimerization by EGF or (DNA-BG)2.
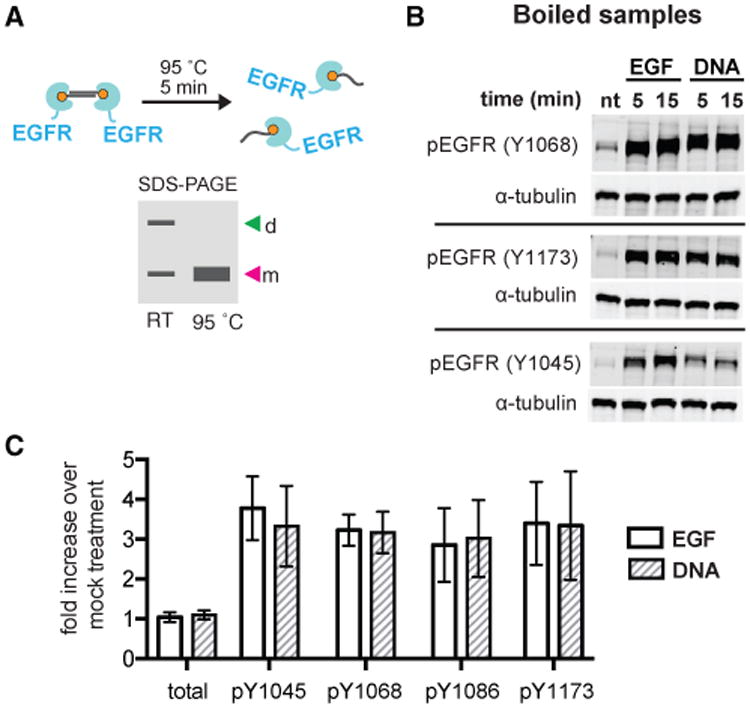
(A) DNA-dimerized receptors can be revealed by PAGE without boiling or can be boiled to reveal a monomer for direct comparison to EGFR monomers. d, dimer; m, monomer; RT, room temperature.
(B) Representative western blot of boiled lysates from cells treated with serum-free media (nt, no treatment), 8 nM EGF or 2 μM (DNA-BG)2 at various tyrosines.
(C) Mean fold increase of total EGFR and phosphotyrosines upon EGF or (DNA-BG)2 treatment compared to no treatment control (n = 3; error bars indicate SD).
Using the concentration of EGF and (DNA-BG)2 in Figure 1D, which gave EGFR phosphorylation in both conditions but Erk phosphorylation only with EGF, we used quantitative western blotting to compare the phosphorylation levels of a suite of tyrosines: Y1045, Y1068, Y1086, and Y1173. Notably, we observed phosphorylation to a similar extent for both conditions (Figures 2B and 2C) at a time point and EGF concentration sufficient for propagation of downstream signals. Increasing the concentration of (DNA-BG)2 gives a similar result, illustrating the crosslinker was working near saturating conditions (Figure S2).
Phosphorylated EGFR Dimers Are Not Sufficient to Stimulate Ras Activation
Mechanistically, several steps occur between the formation of a phosphorylated EGFR dimer and Erk activation. We therefore sought to identify the specific step at which the signaling capacity of EGF- and (DNA-BG)2-stimulated dimers diverged. Because Erk activation requires Ras-GTP formation, we first investigated whether signaling breakdown occurred at or before the level of Ras activation. To evaluate the activation status of Ras, we used the Ras-binding domain of Raf, which selectively binds Ras-GTP, to pull down GTP-bound activated Ras from wholecell lysates. We used the same concentrations of EGF and (DNA-BG)2 as in our earlier assays and confirmed that, while EGF and (DNA-BG)2 stimulated similar levels of Y1068 phosphorylation after 5 min, only EGF-activated EGFR was capable of activating Erk signaling. Analyzing the same lysates for Ras activation, we observed efficient pulldown of Ras-GTP in EGF-treated cells, while little to no Ras-GTP was detected in cells treated with (DNA-BG)2 (Figures 3A and 3B). This was particularly surprising, given that Y1068 is widely considered the primary site responsible for recruiting the Grb2/Sos complex that activates Ras (Yamauchi et al., 1997). Thus, phosphorylated EGFR dimers are not sufficient to activate Ras.
Figure 3. Trapped EGFR Dimers Recruit Adaptors with Similar Stoichiometry to EGF-Stimulated Cells but Do Not Activate Ras.
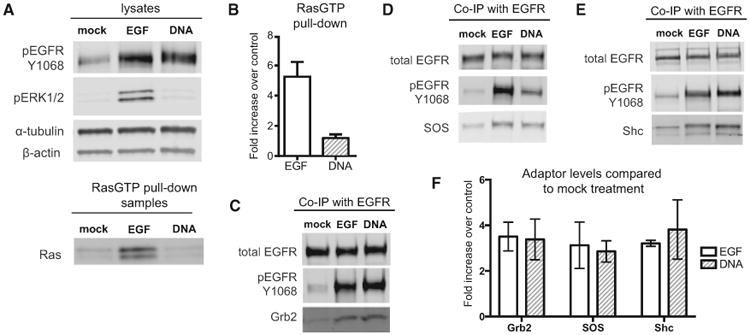
(A) Representative western blot showing lysates from cells treated with either 8 nM EGF, 2 μM (DNA-BG)2, or serum-free media (mock) for 5 min. The same lysates were used in a RasGTP pull-down, and samples were blotted for total Ras.
(B) Mean RasGTP levels in each treatment compared to negative control (n = 3; error bars indicate SD).
(C) Representative blot of Grb2 co-immunoprecipitation (coIP) with EGFR on lysates from treated cells.
(D) Representative blot of SOS coIP with EGFR.
(E) Representative blot of Shc coIP with EGFR.
(F) Quantification of adaptor coIP in treated cells compared to negative controls. Signals for each adaptor were normalized to total EGFR levels in the pull-down sample and plotted as mean fold increase over mock treatment (n = 3; error bars indicate SD).
Trapped EGFR Dimers Recruit Key Adaptor Proteins for Ras Signaling
An inability to activate Ras could be explained by an inability of phosphorylated EGFR to recruit core signaling adaptors such as SOS, Shc, and Grb2. We therefore investigated adaptor recruitment to EGF-stimulated and trapped dimer receptors using co-immunoprecipitation. We treated SNAP-EGFR-expressing cells with EGF or (DNA-BG)2 to generate similar levels of phosphorylated receptor, immunoprecipitated the total EGFR, and then compared the quantity of adaptor proteins that coprecipitated after 5 min. Surprisingly, we did not observe a difference in the quantity of precipitated Grb2, SOS, and Shc between receptors stimulated with EGF or (DNA-BG)2, despite striking changes in the level of Ras-GTP observed under the same conditions (Figures 3C–3F). These results show that differential recruitment of core adaptor proteins to EGFR cannot explain the differences in Ras signaling between our two conditions.
The Structure and Charge of the Crosslinker Do Not Significantly Impact EGFR Transphosphorylation or Signal Propagation
Given these surprising findings, we next investigated whether (DNA-BG)2 was contributing to the lack of Ras signaling in trapped EGFR dimers. Adding the reagents sequentially, with (DNA-BG)2 followed by EGF, resulted in Erk and Akt activation (Figure S2), suggesting that (DNA-BG)2 was not broadly inactivating EGFR. Next, we removed both charge and rigidity from the dimerizer by substituting the nucleic acid portion of (DNA-BG)2 with a highly flexible and uncharged polyethylene glycol (PEG) molecule. Treatment of SNAP-EGFR-expressing cells with PEG26-BG2 triggered efficient formation of phosphorylated dimers but no Erk phosphorylation to levels above control (Figure S2). Similar results were observed for shorter PEG crosslinkers, including PEG9 and PEG5. Moreover, crosslinking a mutant EGFR (V924R), which is unable to form active asymmetric kinase dimers, did not result in receptor phosphorylation (Figure S2). These findings demonstrate that crosslinkers activate the receptor by promoting canonical interactions between the kinase domains but that they are deficient in their ability to promote specific EGF-dependent active conformations necessary for Ras activation.
We also considered that the irreversible nature of BG-based crosslinker dimerization versus reversible EGF-induced dimerization could be a factor in the difference in downstream signaling. To address this, we made versions of the 20-bp (DNA-BG)2 with only 6, 8, or 10 contiguous complementary bases to increase the off rate of the duplex. If the irreversibility of crosslinks was the explanation of the observed defect in signaling, we would expect an increase in Erk phosphorylation per unit of receptor phosphorylation as the duplex melting temperature approached 37°C. However, we did not observe an increase with any of the mismatched duplexes (Figure S2).
Phosphorylation of EGFR Dimers Is Not Sufficient for Nanoscale Oligomer Formation
Our findings demonstrate that, when EGFR dimers are trapped with linkers that do not significantly constrain receptor conformation, they can autophosphorylate and recruit key signaling adapters such as SOS but, surprisingly, do not stimulate Ras. We therefore sought to better understand how trapped receptor dimers might differ from EGF-induced complexes in events downstream of receptor phosphorylation, such as oligomerization and trafficking. We investigated this question by imaging cells treated with (DNA-BG)2 or EGF using stochastic optical reconstruction microscopy (STORM). We used an EGFR construct with a photoswitchable fluorescent protein (mEos 3.2) fused to the C terminus to resolve oligomers that might only be visible by imaging below the diffraction limit and expressed this construct at levels similar to those of receptors in previous experiments (Figure S1). We observed that, upon the addition of (DNA-BG)2 to SNAP-EGFR-mEos-expressing cells, the spatial arrangement of receptors was similar to that of unstimulated controls, whereas receptors stimulated with EGF led to rapid accumulation of bright foci after 10 min (Figures 4A and S3). We quantified images of cells treated with media alone, EGF, or (DNA-BG)2 and constructed pairwise distance histograms for each condition. A peak in the histogram indicates an increase in receptor local density compared to random at a given length scale. Compared to untreated cells, we found an increase in the histogram height (indicative of increased dimers and small oligomers) as well as width (indicative of the formation of larger oligomers) in cells treated with EGF. In contrast, the analysis of receptor distribution in the (DNA-BG)2-treated cells showed only a modest increase in histogram height, consistent with an increased dimer fraction, but no increase in histogram width, indicative of no change in the size of clusters when compared to untreated cells (Figure 4B).
Figure 4. Trapped EGFR Dimers Do Not Form Nanoscale Spatial Intermediates or Traffic to Clathrin-Coated Pits.
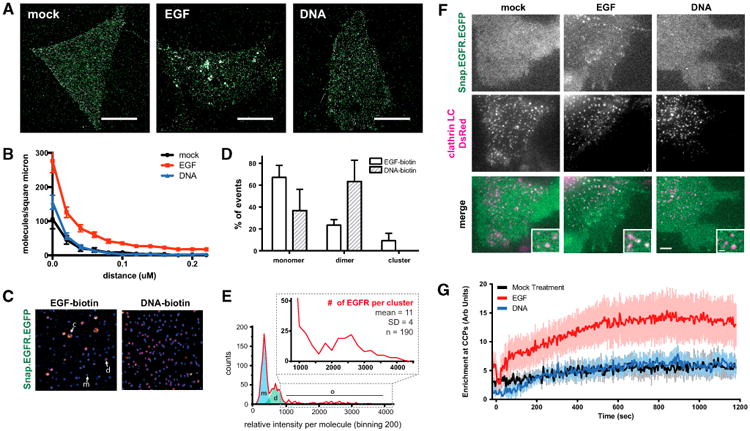
(A) Representative images of HEK293-SNAP-EGFR-mEos cells incubated with serum-free media (mock), 8 nM EGF or 2 μM (DNA-BG)2 for 10 min and then imaged by STORM. Scale bars, 10 μm.
(B) Pairwise correlation analysis of STORM images graphed as median and standard error (n = 10 cells per condition).
(C) Representative images of single-molecule IP of SNAP-EGFR-EGFP cells treated with biotin (bt)-EGF or bt-(DNA-BG)2 for 5 min. Ligand-bound receptors from lysates were immobilized on neutravidin-coated slides and imaged. EGFR monomers (m) appear as blue spots, dimers (d) appear as pink spots, and clusters (c) appear as larger yellow spots.
(D) Mean monomer, dimer, and cluster populations of EGFR graphed as a percentage of the sample (n = 3 independent experiments; error bars indicate SD).
(E) Representative EGF-biotin-treated sample with counts of relative intensity per molecule. The blue shaded region represents the monomer portion, the green shaded region represents the dimer portion, and zoom represents the cluster portion. The average number of EGFR molecules per cluster was estimated by dividing the average intensity of the clusters by the intensity of a monomer.
(F) TIRF images of HEK293 cells co-transfected with SNAP-EGFR-EGFP and clathrin-light chain-dsRed after treatment with 8 nM EGF, 2 μM DNA, or serum-free media at 15 min. Scale bar, 1 μm.
(G) Enrichment of SNAP-EGFR-EGFP at clathrin-coated pits over time after treatment with 8 nM EGF, 2 μM (DNA-BG)2, or serum-free media graphed as mean and SD (n = 10 cells per condition).
To further investigate the degree of oligomerization, we performed single-molecule precipitation of SNAP-EGFR-EGFP receptors treated with biotinylated EGF or biotinylated (DNA-BG)2 after 5 min. We precipitated Triton X-100-disrupted cells onto neutravidin-coated slides as previously described (Lee et al., 2013) to immobilize ligand-bound receptor complexes, and we imaged the intensity of individual fluorescent spots as a proxy for the number of EGFR molecules in each complex (Figure 4C). We observed an increased ratio of dimers to monomers in both conditions compared to controls but uniquely observed the formation of multiple brighter spots only when cells were treated with biotinylated-EGF (Figure 4D). Comparing the fluorescence intensity of these spots to those of the putative monomer and dimer peaks suggested an average cluster size of approximately 7–15 molecules for EGF-stimulated receptors (Figure 4E) after 5 min.
In addition to studying receptor arrangements at fixed time points, we also imaged the dynamic reorganization of EGFR on live cells. To do so, we used total internal reflection fluorescence (TIRF) microscopy to follow the EGFR trafficking in real time, using clathrin-coated pits as a frame of reference. Although non-clathrin mediated endocytosis may contribute to EGFR dynamics under certain conditions, we found that a significant fraction of EGF-stimulated receptors were recruited to clathrin-coated pits at the EGF concentration used in our studies, whereas the localization of (DNA-BG)2-treated receptors was largely unchanged compared to controls over the course of 20 min (Figures 4F, 4G, and S4). Therefore, EGF triggers conformational changes in the EGFR that are necessary for the oligomerization of phosphorylated receptors and their reorganization on live-cell membranes.
Discussion
If EGFR oligomerization and other cell-surface dynamics are necessary for efficient MAPK signal transduction, how might they be coupled to Ras activity? EGFR oligomerization could stimulate Ras activity by concentrating Ras and SOS in common signaling complexes, thereby increasing their effective molarity relative to broadly distributed GAPs, and cooperatively stimulating the formation of Ras-GTP. Consistent with this model, Ras dimerization and nanoclustering have been shown to affect downstream Erk signaling (Nan et al., 2015; Tian et al., 2007; Zhou et al., 2014). Alternatively, EGF binding may cause changes in EGFR transmembrane conformation associated with clustering and the formation of lipid microdomains required for signaling. For example, EGFR clusters have been shown to colocalize with membrane regions enriched for PIP2(4,5) (Laketa et al., 2014), and the GEF activity of SOS can be modulated by charged lipids, including PIP2(4,5) (Gureasko et al., 2008; Zhao et al., 2007).
Independent of the detailed mechanism, our findings have important implications for understanding the regulation of Ras—and, possibly, other signaling molecules—by EGFR. We can conclude that conformational changes and/or other processes associated with EGF binding are necessary for oligomer formation and that these higher order EGFR oligomers may be more potent activators of Ras, on a molecule-to-molecule basis, when compared to phosphorylated dimers. Such EGF-dependent formation of EGFR nanoclusters may add an additional layer of spatial regulation to growth factor signaling, which aligns with an emerging view of how Ras regulates downstream pathways, through the formation of similar higher order species. Our findings also emphasize that not all EGFR dimers (or oligomers) are the same, and, depending on the initiating signal, receptor activation may evolve very differently. For example, a recent elegant dissection of structural and functional properties of EGFR dimers induced by different ligands suggests that more stable receptor dimers induce more transient profiles of receptor phosphorylation and downstream pathway activation, presumably by being long lived enough to recruit negative-feedback regulators (Freed et al., 2017). Our findings argue that, in addition to the kinetics of receptor activation, the spatial distribution of receptors following their activation is a critical determinant of downstream signal propagation. Long-lived trapped dimers are signaling deficient not because they fail to accumulate substantial phosphorylation and recruit adapters but, perhaps, because tertiary or quaternary interaction are structurally incompatible with subsequent organization into effective signaling platforms. Finally, our results demonstrate that receptor activation and signal transduction can be mechanistically decoupled. This finding has important implications for the development of future therapeutics, which could specifically target receptor organization rather than activation to modulate signal transduction through specific pathways.
Experimental Procedures
Details are provided in the Supplemental Information.
Cell Signaling Assays
Cells stably expressing SNAP-EGFR were grown to 70%–80% confluency and then serum starved for 6–8 hr prior to stimulus with EGF or (DNA-BG)2 at 37°C, lysed, and prepared for western blots. For quantitative western blotting, secondary antibodies conjugated to either Alexa Fluor 680 or DyLight 800 were used, and blots were imaged on a LI-COR Biosciences imaging system. Scans were quantified and analyzed by densitometry. Measurements were normalized to loading controls and shown as the mean and SD of 3 independent experiments.
Storm
HEK293-SNAP-EGFR-mEos cells were serum starved for 6 hr and then incubated with the indicated stimuli at 37°C for 10 min. Cells were fixed and imaged with an inverted microscope using TIRF illumination, 1003 magnification, and a 561-nm laser at 60 Hz. Once every 10 frames, mEos was converted from green to red state with 405-nm illumination. Images from 10 cells per condition were corrected for blinking as previously described (Puchner et al., 2013), and the molecular positions were then used to calculate all the pairwise distances as previously described (van Lengerich et al., 2017).
Single-Molecule IP
HEK293-SNAP-EGFR-EGFP cells were treated with 8 nm of EGF-biotin or 2 μM of (BG-DNA)2-biotin for 5 min at 37°C, and then lysed with 1% Triton X-100 buffer. Lysates were incubated on neutravidin-coated PEG slides and imaged by TIRF microscopy. Over 3 independent experiments, monomer, dimer, and cluster populations were identified by bleaching steps and analysis of pixel intensity histograms.
Clathrin Colocalization
HEK293 cells were transfected with SNAP-EGFR-EGFP and clathrin light chain-dsRed using Lipofectamine 2000. Cells were imaged 48 hr later, live at 37°C with various stimuli. SNAP-EGFR-EGFP enrichment at clathrin structures was calculated as the difference between the average fluorescence inside and outside regions enriched for dsRed. Each condition represents 10 cells pooled across 7 independent experiments.
Supplementary Material
Highlights.
Chemical-genetic approach to form stable phosphorylated EGFR dimers on cells
Chemically induced dimers recruit adaptors to a similar extent as EGF-activated EGFR
SOS, Grb2, and Shc recruitment to EGFR dimers is not sufficient for Ras activation
EGF induces conformational changes necessary for higher order oligomer formation
Acknowledgments
We thank the Gartner lab members for insightful discussions. We thank B. Huang, K. Shokat, J.Taunton, P. England, and D. Fujimori for sharing instruments and facilities. This work is supported by an Achievement Rewards for College Scientists fellowship and a Genentech Foundation fellowship to S.I.L., an American Cancer Society postdoctoral fellowship (124801-PF-13-365-01-TBE) to B.v.L., a grant from the National Institute of General Medical Sciences (R01 GM109176) to N.J., a UCSF CTSI-SOS pilot grant (1 UL1 RR024131-01), an NIGMS Systems Biology Center grant (P50 GM081879), NSF grant MCB-1330864, and the NSF Center for Cellular Construction (DBI-1548297). Z.J.G. is a Chan Zuckerberg Biohub Investigator. M.C. and T.-Y.Y. were supported by the Institute for Basic Science (IBS; IBS-R0216-D1).
Footnotes
Supplemental Information: Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.02.031.
Author Contributions: S.I.L. and Z.J.G. conceived the study. S.I.L, Z.J.G., and N.J. supervised the study and drafted the manuscript. B.v.L. and S.I.L. cloned plasmids and made cell lines. S.I.L. and D.M.P. made the chemical dimerizers. S.I.L. performed cell-signaling assays. B.v.L. performed STORM. K.E. performed live TIRF. M.C. performed single-molecule IP. T.-Y.Y. and M.v.Z. provided key insight and additional supervision for experiments. All authors contributed to writing and editing the final manuscript.
Declaration of Interests: The authors declare no competing interests.
References
- Ariotti N, Liang H, Xu Y, Zhang Y, Yonekubo Y, Inder K, Du G, Parton RG, Hancock JF, Plowman SJ. Epidermal growth factor receptor activation remodels the plasma membrane lipid environment to induce nanocluster formation. Mol Cell Biol. 2010;30:3795–3804. doi: 10.1128/MCB.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim A, Engelberg D, Li N, al-Alawi N, Schlessinger J, Karin M. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- Bessman NJ, Freed DM, Lemmon MA. Putting together structures of epidermal growth factor receptors. Curr Opin Struct Biol. 2014;29:95–101. doi: 10.1016/j.sbi.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SM, Tu HL, Jun JE, Alvarez S, Triplet MG, Iwig JS, Yadav KK, Bar-Sagi D, Roose JP, Groves JT. One-way membrane trafficking of SOS in receptor-triggered Ras activation. Nat Struct Mol Biol. 2016;23:838–846. doi: 10.1038/nsmb.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- Clayton AHA, Orchard SG, Nice EC, Posner RG, Burgess AW. Predominance of activated EGFR higher-order oligomers on the cell surface. Growth Factors. 2008;26:316–324. doi: 10.1080/08977190802442187. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, Valley CC, Ferguson KM, Leahy DJ, Lidke DS, Lemmon MA. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell. 2017;171:683–695.e18. doi: 10.1016/j.cell.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gureasko J, Galush WJ, Boykevisch S, Sondermann H, Bar-Sagi D, Groves JT, Kuriyan J. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat Struct Mol Biol. 2008;15:452–461. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Bharill S, Karandur D, Peterson SM, Marita M, Shi X, Kaliszewski MJ, Smith AW, Isacoff EY, Kuriyan J. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. eLife. 2016;5:e14107. doi: 10.7554/eLife.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose J, Murata M, Yanagida T, Sako Y. EGF signalling amplification induced by dynamic clustering of EGFR. Biochem Biophys Res Commun. 2004;324:1143–1149. doi: 10.1016/j.bbrc.2004.09.173. [DOI] [PubMed] [Google Scholar]
- Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman LB, Maiwald T, Conzelmann H, Lauffenburger DA, Sorger PK. Rapid phospho-turnover by receptor tyrosine kinases impacts downstream signaling and drug binding. Mol Cell. 2011;43:723–737. doi: 10.1016/j.molcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozer N, Barua D, Orchard S, Nice EC, Burgess AW, Hlavacek WS, Clayton AHA. Exploring higher-order EGFR oligomerisation and phosphorylation–a combined experimental and theoretical approach. Mol Biosyst. 2013;9:1849–1863. doi: 10.1039/c3mb70073a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laketa V, Zarbakhsh S, Traynor-Kaplan A, Macnamara A, Subramanian D, Putyrski M, Mueller R, Nadler A, Mentel M, Saez-Rodriguez J, et al. PIP3 induces the recycling of receptor tyrosine kinases. Sci Signal. 2014;7:ra5. doi: 10.1126/scisignal.2004532. [DOI] [PubMed] [Google Scholar]
- Lee HW, Kyung T, Yoo J, Kim T, Chung C, Ryu JY, Lee H, Park K, Lee S, Jones WD, et al. Real-time single-molecule co-immunoprecipitation analyses reveal cancer-specific Ras signalling dynamics. Nat Commun. 2013;4:1505. doi: 10.1038/ncomms2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Schmitz KR, Jeffrey PD, Wiltzius JJW, Kussie P, Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–311. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Margolis BL, Lax I, Kris R, Dombalagian M, Honegger AM, Howk R, Givol D, Ullrich A, Schlessinger J. All autophosphorylation sites of epidermal growth factor (EGF) receptor and HER2/neu are located in their carboxyl-terminal tails. Identification of a novel site in EGF receptor. J Biol Chem. 1989;264:10667–10671. [PubMed] [Google Scholar]
- Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homoand heterodimers. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Tamguüney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, McCormick F, Chu S. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc Natl Acad Sci USA. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham SR, Roberts SK, Arkhipov A, Mysore VP, Tynan CJ, Zanetti-Domingues LC, Kim ET, Losasso V, Korovesis D, Hirsch M, et al. EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat Commun. 2016;7:13307. doi: 10.1038/ncomms13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- Pedersen MW, Pedersen N, Damstrup L, Villingshøj M, Sønder SU, Rieneck K, Bovin LF, Spang-Thomsen M, Poulsen HS. Analysis of the epidermal growth factor receptor specific transcriptome: effect of receptor expression level and an activating mutation. J Cell Biochem. 2005;96:412–427. doi: 10.1002/jcb.20554. [DOI] [PubMed] [Google Scholar]
- Puchner EM, Walter JM, Kasper R, Huang B, Lim WA. Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory. Proc Natl Acad Sci USA. 2013;110:16015–16020. doi: 10.1073/pnas.1309676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan T, Macdonald-Obermann JL, Huelsmann L, Bessman NJ, Naegle KM, Pike LJ. Different epidermal growth factor receptor (EGFR) agonists produce unique signatures for the recruitment of downstream signaling proteins. J Biol Chem. 2016;291:5528–5540. doi: 10.1074/jbc.M115.710087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarian S, Li Y, Elson EL, Pike LJ. Oligomerization of the EGF receptor investigated by live cell fluorescence intensity distribution analysis. Biophys J. 2007;93:1021–1031. doi: 10.1529/biophysj.107.105494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel J, Blaukat A, Li S, Knöchel T, Ferguson KM. Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization. Cancer Cell. 2008;13:365–373. doi: 10.1016/j.ccr.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lengerich B, Agnew C, Puchner EM, Huang B, Jura N. EGF and NRG induce phosphorylation of HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. Proc Natl Acad Sci USA. 2017;114:E2836–E2845. doi: 10.1073/pnas.1617994114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Ueki K, Tobe K, Tamemoto H, Sekine N, Wada M, Honjo M, Takahashi M, Takahashi T, Hirai H, et al. Tyrosine phosphorylation of the EGF receptor by the kinase Jak2 is induced by growth hormone. Nature. 1997;390:91–96. doi: 10.1038/36369. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Okamoto I, Okabe T, Iwasa T, Satoh T, Nishio K, Fukuoka M, Nakagawa K. Matuzumab and cetuximab activate the epidermal growth factor receptor but fail to trigger downstream signaling by Akt or Erk. Int J Cancer. 2008;122:1530–1538. doi: 10.1002/ijc.23253. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat Cell Biol. 2007;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Liang H, Rodkey T, Ariotti N, Parton RG, Hancock JF. Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol Cell Biol. 2014;34:862–876. doi: 10.1128/MCB.01227-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


