SUMMARY
Translation initiation is typically restricted to AUG codons, and scanning eukaryotic ribosomes inefficiently recognize near-cognate codons. We show that queuing of scanning ribosomes behind a paused elongating ribosome promotes initiation at upstream weak start sites. Ribosomal profiling reveals polyamine-dependent pausing of elongating ribosomes on a conserved Pro-Pro-Trp (PPW) motif in an inhibitory non-AUG-initiated upstream-Conserved-Coding region (uCC) of the antizyme inhibitor 1 (AZIN1) mRNA, encoding a regulator of cellular polyamine synthesis. Mutation of the PPW motif impairs initiation at the uCC’s upstream near-cognate AUU start site and derepresses AZIN1 synthesis, while substitution of alternate elongation pause sequences restores uCC translation. Impairing ribosome loading reduces uCC translation and paradoxically derepresses AZIN1 synthesis. Finally, we identify translation factor eIF5A as a sensor and effector for polyamine control of uCC translation. We propose that stalling of elongating ribosomes triggers queuing of scanning ribosomes and promotes initiation by positioning a ribosome near the start codon.
TOC image
Ivanov et al. reveal that ribosome queuing upstream of a paused elongating ribosome promotes translation initiation at an upstream near-cognate start codon on the AZIN1 mRNA. This mechanism is exploited during polyamine autoregulation of antizyme inhibitor synthesis and similar strategies may control translation of other mRNAs with poor start sites.
INTRODUCTION
Based on the scanning model of translation initiation in eukaryotes, a 43S ribosomal preinitiation complex (PIC: 40S subunits with associated translation initiation factors eIFs 1, 1A, 2, 3, and 5, and with Met-tRNAiMet bound in the P site) binds at the 5′ end of an mRNA with the help of the eIF4F complex (consisting of eIFs 4G, 4E, and 4A) and then moves in a 3′ direction inspecting the sequence for potential start sites (Hinnebusch, 2014). While an AUG codon is the most prominent signal for a translation start site, flanking or context nucleotides also contribute to start site selection with an A or G at the −3 position and a G at +4, relative to the A of the AUG codon, enhancing start site selection (Kozak, 2005). Upon encountering a start codon, the PIC stops scanning, and a series of conformational changes result in release of the initiation factors and joining of the large subunit to form the elongating 80S ribosome. The vectorial nature of scanning results in ribosomes typically initiating at the start codon closest to the 5′ end of an mRNA (Hinnebusch, 2011; Kozak, 1991). Accordingly, initiation codons present 5′ of the start codon of the main open reading frame (mORF) can have dramatic effects on mORF translation (Kozak, 1989b, 1991). These 5′ initiation codons, depending on their reading frame, can extend the N-terminus of the mORF product or generate upstream open reading frames (uORFs) that terminate prior to the mORF or overlap the mORF in an alternate reading frame. Varying estimates suggest that uORFs are present on as high as 50% of all eukaryotic mRNAs (Hinnebusch et al., 2016; Johnstone et al., 2016).
Recent appreciation of the prevalence of near-cognate (non-AUG) initiation in the 5′ leaders of eukaryotic mRNAs has suggested that the occurrence of uORFs might be even higher than these estimates (Ingolia et al., 2011; Lee et al., 2012). A subset of uORFs initiated at weak start sites, typically a near-cognate start codon, are defined as upstream conserved coding regions (uCCs) (Ivanov et al., 2008). As the name implies these uORFs show patterns of conservation consistent with coding regions including a preponderance of synonymous versus non-synonymous substitutions; however, the uCCs either lack a conventional AUG start codon or initiate at an AUG codon with poor context nucleotides. It is unclear how scanning ribosomes select these weak start codons for initiation. One of the first identified and studied uCCs is present in eukaryotic mRNAs encoding proteins involved in polyamine biosynthesis: antizyme inhibitor (AZIN) and homologs of ornithine decarboxylase (ODC) (Ivanov et al., 2008).
Polyamines are small organic polycations that are essential for a variety of basic cellular functions (Igarashi and Kashiwagi, 2010; Pegg, 2016). Reflecting the importance of their homeostasis, the key enzymes in the biosynthesis of polyamines are tightly regulated, including at the translational level, in response to perturbations in intracellular polyamine content (Ivanov et al., 2010a; Pegg, 2009). The first step in the biosynthesis of polyamines is the decarboxylation of ornithine to produce putrescine, a reaction catalyzed by the enzyme ODC. ODC is inhibited by a protein called ornithine decarboxylase antizyme (Heller and Canellakis, 1981), or antizyme (OAZ), which binds to ODC and presents it for ubiquitin-independent degradation by the 26S proteasome (Murakami et al., 1992). Antizyme, in turn, is inhibited by AZIN, a homolog of ODC that has retained its affinity for antizyme, but has lost its catalytic activity (Murakami et al., 1996). Mammals express two paralogs of AZIN, AZIN1 and AZIN2, which have evolved independently (Ivanov et al., 2010b). The expression of antizyme and AZIN is inversely regulated at the translational level by polyamines. Expression of antizyme is induced under conditions of elevated polyamine levels via programmed +1 translational frameshifting on the antizyme mRNA (Ivanov and Atkins, 2007; Kahana, 2009; Matsufuji et al., 1995). In contrast, experiments with reporters indicate that elevated polyamines cause repression of AZIN synthesis, dependent on the uCC in the AZIN1 mRNA (Ivanov et al., 2008). Introduction of a cognate AUG codon in place of the near-cognate AUU start codon of the uCC results in constitutive repression of AZIN synthesis (Ivanov et al., 2008), consistent with the notion that translation of the 52-amino acid long uCC prevents efficient reinitiation at downstream start codons. In contrast, substitution of a non-cognate UUU codon in place of the uCC start codon results in constitutive derepression of AZIN synthesis (Ivanov et al., 2008). Thus, AZIN synthesis is inversely correlated with uCC translation such that ribosomes that translate the uCC fail to synthesize AZIN. In this report, we reveal the mechanism underlying polyamine regulation of AZIN synthesis.
RESULTS
Coding sequence of the AZIN1 uCC is required for polyamine stimulation of uCC translation initiation
Along with the uCC, the AZIN1 mRNA contains three AUG-initiated uORFs; however, these conventional uORFs are not necessary for polyamine regulation (Ivanov et al., 2008). To assess uCC translation, the last sense codon of the AUU-initiated uCC of the mouse azin1 mRNA was previously fused in-frame with a firefly luciferase reporter lacking its native AUG start codon to generate the uCC-WT-FLuc chimera that retains the AUG-initiated uORF1 (Ivanov et al., 2008). Whereas uCC-WT-FLuc synthesis was low in cells depleted of polyamines by growing in the presence of α-difluoromethyl ornithine (DFMO), a suicide inhibitor of ODC that catalyzes the first step in polyamine biosynthesis (Pegg, 2009), enhanced uCC-WT-FLuc synthesis was observed following supplementation with spermidine (SPD) to achieve high intracellular polyamine levels (Ivanov et al., 2008). Moreover, as this previous study also showed that a highly-conserved sequence near the C-terminus of the uCC was required for polyamine regulation of the mORF (AZIN1) synthesis (Ivanov et al., 2008), we investigated if the C-terminus of the uCC was also involved in the polyamine-dependent enhanced AUU initiation on the uCC. Coupling a single nucleotide deletion in the codon for Phe43 of the 52-residue uCC with a single nucleotide insertion of T after the last sense codon of the uCC generated the uCC out-of-frame (uCC-OF-FLuc) reporter which shifts the reading frame for the last ten codons of the uCC from encoding FNAEPPWEPS in the WT to encoding STLSHLGNLA in the OF mutant. As shown in Figures 1A and S1, the OF mutation abolished the polyamine-dependent stimulation of uCC synthesis, suggesting that the coding capacity of the uCC paradoxically impacted initiation on the same uORF. In this and subsequent experiments in which two different conditions were tested the changes in reporter activity were not due to changes in mRNA levels, as determined by statistical significance tests on qPCR data (Tables S1–S5).
Figure 1. Sequence element near the C-terminus of the AZIN1 uCC is required for polyamine stimulation of translation initiation at the upstream AUU start codon.
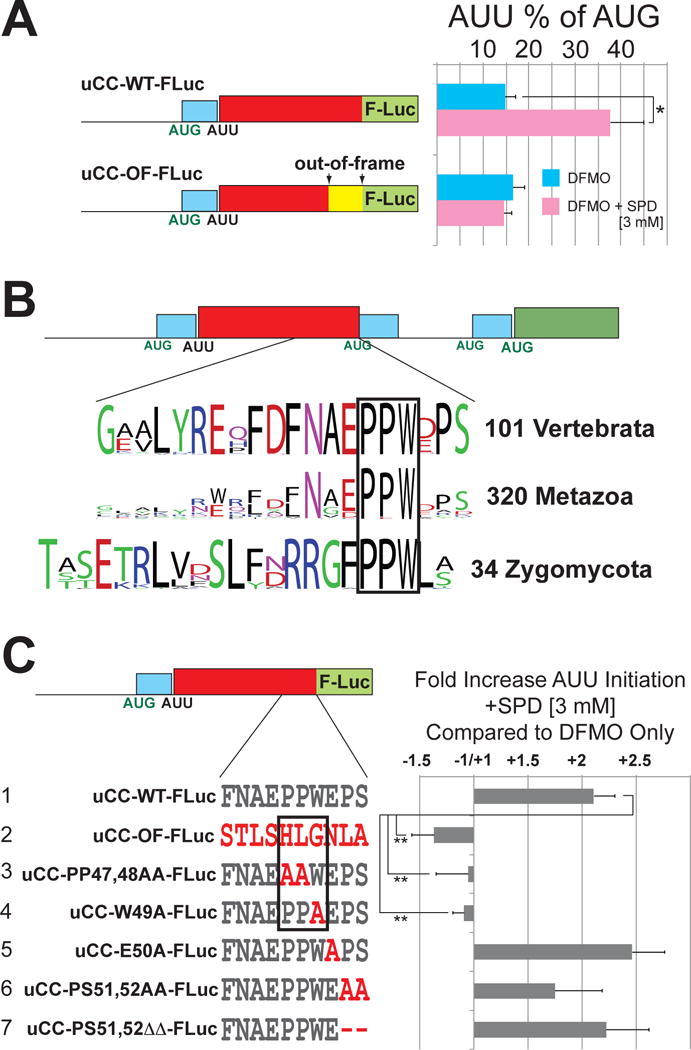
(A) HEK293T cells treated with DFMO (polyamine-depleted) or DFMO+SPD (high polyamines) were transfected with mouse azin1 uCC-luciferase reporters. OF, last 10 codons of the uCC out-of-frame. Percent AUU initiation was calculated relative to corresponding AUG-initiated constructs. Error bars denote standard deviation; *p<0.05 (Student’s two-tailed t-test; n=4, assayed in duplicate). Results from tests of additional SPD concentrations are presented in Figure S1; relative reporter mRNA levels are presented in Table S1. (B) Schematic of the AZIN1 mRNA from mammals: conventional AUG initiated uORFs in cyan, uCC in red, and mORF in green. WebLogo representations of the most conserved C-terminal uCC residues from (top) 101 vertebrate AZIN1 orthologs or (middle) 320 metazoan ODC homologs, including the 101 vertebrate AZIN1 sequences; (bottom) WebLogo of the entire uCC from 34 Zygomycota ODC mRNAs. The highly conserved PPW motif is boxed. (C), azin1 uCC-Luc fusions with the indicated frameshift or point mutations (red) were tested as in panel (A). Mutations that alter the PPW motif are boxed. Firefly reporter values were normalized to a co-transfected Renilla luciferase initiated by AUG in perfect context. Error bars denote standard deviation; **p<0.01 (Student’s two-tailed t-test; n=5, assayed in duplicate). Results in panels (A) and (C) are from independent experiments; relative reporter mRNA levels are presented in Table S1.
To identify uCC residues that are critical for translational control, uCC sequences from diverse organisms were collected and compared. Analysis of the amino acid sequences of the uCCs present in the leaders of 101 AZIN1 orthologs from vertebrates provided limited information, as multiple positions showed high and nearly universal conservation (Figures 1B, S2A). Extending the comparison to include an additional 219 uCC sequences from ODC homologs in invertebrates restricted the conservation to the motif N(G/A)(D/E)PPW in metazoans (Figure 1B, middle WebLogo). Comparison to an independently arising uCC sequence in the ODC mRNA of Zygomycota (fungi) narrowed the conservation to the motif PPW (Figure 1B). Point mutations in this region were examined for their effect on supporting enhanced AUU initiation in the presence of high polyamines (Figure 1C). Mutations altering the PPW motif (PP47,48AA or W49A) abolished the enhanced near-cognate initiation, indicating that these residues are necessary for the effect. By contrast, mutations targeting the PS motif (PS51,52AA or PS51,52ΔΔ) or the conserved Glu residue in the EPPW motif (E50A) did not significantly impact the polyamine induction of uCC reporter synthesis, suggesting that these residues are not necessary, at least in the context of a uCC-Fluc fusion reporter.
If the polyamine-dependent induction of AUU initiation of the uCC contributes to translational regulation of the AZIN1 mORF, then mutations that abolish regulation of the uCC-FLuc fusions would be expected to derepress AZIN1 synthesis. To test this idea, uCC point mutations were introduced in a reporter in which a simplified version of the AZIN1 mRNA leader lacking the three AUG-initiated uORFs (due to point mutations in their AUG start codons) was fused upstream of Renilla luciferase. Notably, the 7.4-fold polyamine repression of AZIN1-Renilla expression observed with the uCC-WT# reporter lacking the conventional AUG-initiated uORFs (Figure S2E, construct 1) was comparable to the 6.5-fold repression previously reported for the uCC-WT reporter containing the conventional uORFs (Ivanov et al., 2008). The mutations PP47,48AA and W49A in the conventional uORF-depleted mRNA leader configuration significantly impaired polyamine-dependent repression of the reporter (Figure S2E, constructs 4–5). While the PP47,48AA mutation blocked the polyamine repression of luciferase synthesis, this double point mutation was significantly less effective than the mutation that shifts the last 10 codons out-of-frame (construct 2). Moreover, while the mutation E46A had no effect (construct 3), the mutations PS51,52AA (construct 6) led to significant, but less drastic, derepression than the mutations PP47,48AA or W49A (constructs 4–5). Combining the PP47,48AA and PS51,52AA in a single compound mutant (construct 7) led to complete loss of repression, which mimicked the results with the out-of-frame mutation. Taken together, these data reveal, surprisingly, that the coding sequence of the uCC is critical for polyamine-regulated initiation at an upstream site. Moreover, the mutational data suggest that PPW and PS-stop motifs act autonomously, and this conclusion is also supported by comparative genomics analysis. The PPW and PS-stop motifs occur invariably together in uCC sequences of vertebrate AZIN1 (Figure S2A) and many other metazoan uCCs (Ivanov et al., 2008). However, in several evolutionary branches one of the motifs is lost. For example, the PS-stop motif is lost in uCC homologs of nematodes and arthropods (Figure S2B,C), while the PPW motif is absent in the uCC elements of ODC mRNAs in cnidarians (Figure S2D).
Elongation pause on the uCC promotes initiation
To assess the translational state of the uCC and the AZIN1 mORF, ribosome profiling (Ingolia et al., 2009) was performed either with cells pretreated with DFMO only (polyamine depletion) or DFMO + 2 mM SPD (high polyamines). Ribosome footprints mapping to the AZIN1 mRNA were quantified to assess the relative translation of the uCC and mORF. As shown in Figure 2, the ribosomal occupancy on the uCC was inversely correlated with translation of the mORF in a manner controlled by the levels of polyamines. Under polyamine depletion conditions few ribosome footprints mapped to the uCC, whereas high ribosome occupancy was observed on the mORF (Figure 2, upper panel; ribo count uCC:mORF = 0.02:1). Consistent with the known polyamine repression of AZIN1 synthesis (Ivanov et al., 2008), the ratio of ribosomal footprints mapping to the uCC versus the mORF increased ~22-fold under high polyamine conditions (Figure 2, lower panel; ribo count uCC:mORF = 0.43:1). The ribosomal footprints near the 3′ end of the uCC in the presence of high polyamines could be separated into three distinct clusters of peaks. The middle cluster of peaks corresponded to ribosomes pausing during elongation with the Trp codon of the PPW motif in the decoding A site of the ribosome and the two proline residues at the end of the nascent peptide and linked directly to the P-site tRNA (Figure 2, inset). The other two clusters of peaks correspond to ribosomes at or 10 codons upstream of the termination codon (Figure 2, inset). This latter peak, which is one ribosome width before the termination codon, could be evidence of 80S ribosomes queuing immediately behind a paused terminating ribosome. These profiling and reporter assay results are consistent with a model in which the PPW and PS-stop motifs confer polyamine-dependent translation elongation and termination pauses, respectively, that impede the flow of scanning ribosomes. Accordingly, queuing of scanning ribosomes upstream of the paused ribosome would promote initiation from the upstream weak (AUU) start site.
Figure 2. Polyamine repression of AZIN1 mORF translation correlates with high ribosome occupancy near the 3′ end of the uCC.
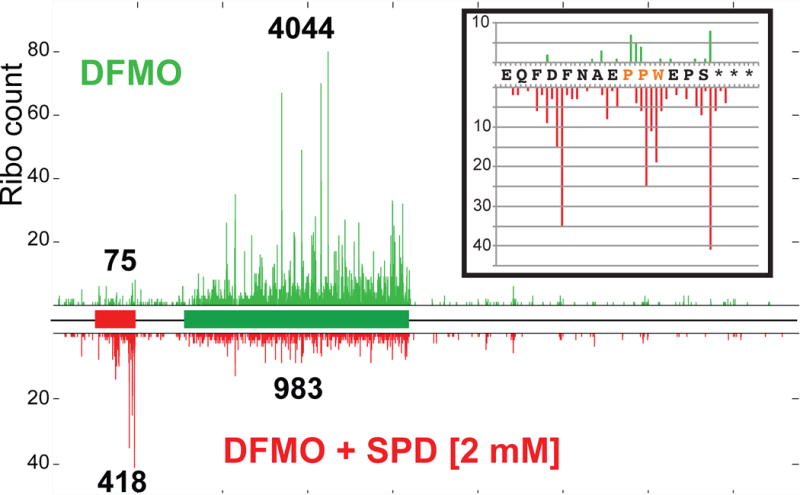
Ribosome protected mRNA fragments from HEK293T cells grown under low (DFMO, upper panel) and high (DFMO + 2 mM SPD, lower panel) polyamine conditions were mapped on the AZIN1 mRNA with the uCC (red) and mORF (green) depicted as rectangles in the schematic. The total (not normalized) ribosome fragment counts were aligned to the AZIN1 mRNA assuming a 15 nt offset from the 5′ end of the protected fragments to the A site. Quantified fragment counts mapping to the uCC and mORF under each condition are indicated above and below the ribosome profiles. (Inset) enlarged view of the fragments mapping within the last 15 codons of the uCC including on the PPW motif (orange).
If the polyamine-induced elongation pause on the PPW motif contributes to enhanced initiation at the upstream AUU codon, an elongation pause generated by other means could be expected to have a similar effect. To test this hypothesis the last 10 codons of the AZIN1 uCC were replaced with a sequence encoding the regulatory portion of the Arginine Attenuator Peptide (AAP) (Wang et al., 1998) from the arg-2 mRNA of N. crassa (Figure 3A). Ribosomes translating the AAP both in fungi (where it is endogenous) and in vertebrates (where it is not present naturally) pause in the presence of excess arginine (Fang et al., 2004; Wang and Sachs, 1997). As predicted, adding arginine to the media significantly enhanced AUU initiation on the uCC-FLuc fusion containing the AAP sequence (Figure 3B, construct 1, uCC-AAP-FLuc), but not on the WT (construct 3), out-of-frame (construct 4), or polyamine-regulated OAZ1 frameshift (construct 5) reporters. Introducing the point mutation D12N, which is known to alleviate the arginine-induced elongation pause on the AAP (Wang and Sachs, 1997), abolished the arginine-specific enhancement of AUU initiation by the uCC-AAP fusion (Figure 3B, construct 2). As expected, the uCC-AAP-FLuc and the uCC-AAP-D12N-Fluc fusions, like the uCC-OF-FLuc construct, failed to respond to polyamines (Figure 3B), consistent with the removal of the PPW motif from these reporters. Taken together, these data support the notion that an elongation pause can create an impediment to scanning ribosomes leading to enhanced translation initiation from an upstream weak start site, and has certain similarities with previous reports showing that introduction of a downstream stem-loop structure (Kozak, 1989a, 1990) or RNA-binding protein (Iwasaki et al., 2016; Medenbach et al., 2011) to impede scanning can enhance recognition of a weak start site.
Figure 3. Translation elongation pause enhances initiation at upstream AUU codon.
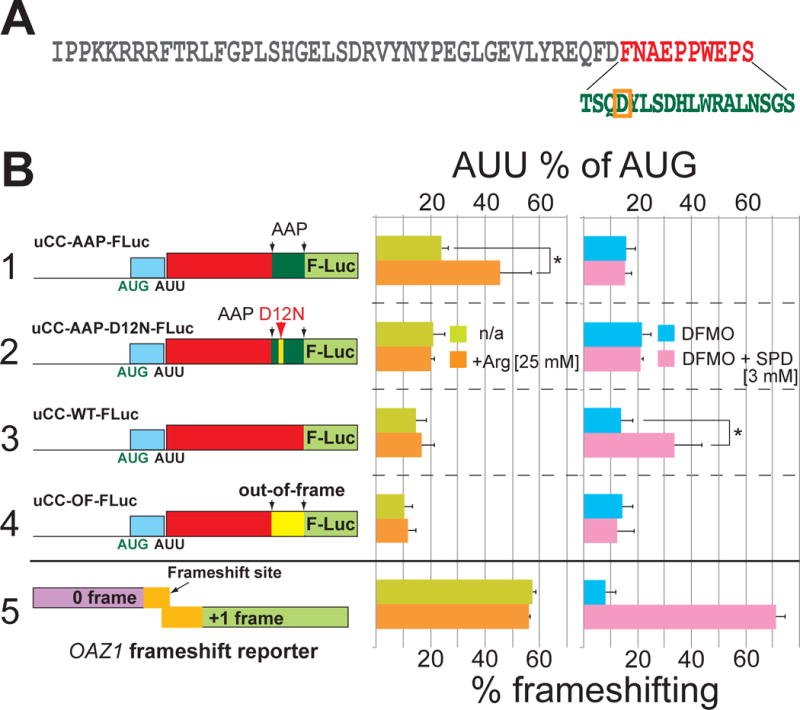
(A) Amino acid sequence of the mouse azin1 uCC. The last 10 residues (red) were replaced by (bottom) the C-terminal two-thirds of the N. crassa AAP sequence (green) in uCC-AAP hybrid constructs; critical APP residue D12 is boxed in orange. (B) The indicated uCC-Luc reporters or a dual-luciferase human OAZ1 frameshift reporter (to monitor polyamine levels) were transfected either in HEK293T cells (right panel) pretreated with DFMO or DFMO + 3 mM SPD or in U2OS cells (middle panel) cultured in DMEM (400 μM Arg) media +/- 25 mM Arg. Percent AUU initiation calculation and firefly normalization were performed as in Figure 1. Error bars denote standard deviation; *p<0.05 (Student’s two-tailed t-test; n=4, assayed in duplicate); relative reporter mRNA levels are presented in Table S2.
Polyamines target eIF5A function to stimulate uCC translation initiation
The comparative genomic and mutagenic analyses identified the PPW motif as crucial for the polyamine regulation of uCC and mORF translation, and the profiling revealed polyamine-dependent ribosomal pausing on this same motif. Interestingly, PPW motifs were the most prominent pause sites detected in bacteria lacking EF-P (Woolstenhulme et al., 2015), a translation elongation factor known to be important for polyproline synthesis (Doerfel et al., 2013; Ude et al., 2013; Woolstenhulme et al., 2015). As the homologous translation factor in eukaryotes, eIF5A (Dever et al., 2014; Saini et al., 2009), has also been shown to promote polyproline synthesis (Gutierrez et al., 2013), we hypothesized that eIF5A is required for elongation at the PPW motif in the uCC and that polyamines exert their action on this motif by inhibiting eIF5A activity. This hypothesis was tested using reconstituted yeast in vitro translation assays, and peptide synthesis was monitored by electrophoretic thin-layer chromatography (TLC). As shown in Figure 4A, synthesis of the peptide MEPPWK was at background levels in the absence of eIF5A and was stimulated in a dose-dependent manner by addition of eIF5A. When peptide synthesis was performed in the presence of 0.1 μM eIF5A, near the K1/2 for MEPPWK synthesis (Figure 4B), increasing the concentration of SPD from 1 mM to 4 mM completely inhibited MEPPWK production (Figure 4C). The inhibition of MEPPWK synthesis by added SPD was not due to a general inhibition of translation because synthesis of the peptide MEFFWK, which is synthesized efficiently in the absence of eIF5A, was generally resistant to added SPD (Figure 4D). We hypothesized that polyamines are interfering with eIF5A function on the ribosome. In support of this idea, it is noteworthy that the polyamine spermine was found to crosslink to helices H74 and H93 of the 23S rRNA near the peptidyl transferase center (PTC) of bacterial ribosomes (Xaplanteri et al., 2005) and that eIF5A interacts with the same region of eukaryotic ribosomes (Gutierrez et al., 2013; Melnikov et al., 2016; Schmidt et al., 2016). Consistent with a model in which eIF5A and polyamines compete for a common site on the ribosome, MEPPWK synthesis, like MEFFWK synthesis, was maintained at high levels even under conditions of high polyamines when the eIF5A concentration was increased 100-fold (Figure 4E,F).
Figure 4. High polyamine levels block eIF5A stimulation of PPW peptide synthesis.

(A) Fractions of MEPPWK synthesis in yeast in vitro reconstituted elongation assays performed in the presence of the indicated concentration of eIF5A were quantified and fit to a single exponential equation. (B) Maximum fraction of peptide synthesis (Ymax) values from panel (A) were fit to the Michaelis-Menten equation to calculate the k1/2 value for eIF5A (~0.1 μM). (C-F) Fractions of MEPPWK (C,E) or MEFFWK (D,F) synthesis in elongation assays performed in the presence of the indicated concentration of SPD, and either 0.1 μM (C,D) or 100 μM (E,F) eIF5A were plotted and fit to a single exponential equation or a simple two-step process for a sigmoidal curve (Lorsch and Herschlag, 1999). Biochemical data in panels (A-F) are representative of results obtained from three independent experiments. (G) HEK293T cells pretreated in 1 mM DFMO for 4 days were co-transfected with the indicated uCC-Luc reporters and either control or DHPS-targeted shRNA. After 48 hr, cells were supplemented with 1 μM SPD and then harvested after 24 hr. Percent AUU initiation calculation was performed as in Figure 1. Error bars denote standard deviation; ***p<0.001 (Student’s two-tailed t-test; n=4, assayed in duplicate).
A unique feature of eIF5A is the post-translationally modified residue hypusine (Nε-(4-amino-2-hydroxybutyl)lysine) (Wolff et al., 2007). The hypusine residue, which is essential for eIF5A stimulation of polyproline synthesis (Gutierrez et al., 2013), as well as for MEPPWK synthesis (Figure S3A–B), is synthesized in two steps by the enzymes deoxyhypusine synthase (DHPS) and deoxyhypusine hydroxylase (DOHH) (Dever et al., 2014; Wolff et al., 2007). To test the role of eIF5A inactivation to facilitate uCC translation in vivo, DHPS was targeted for partial inactivation by shRNA knockdown. To potentiate the effects of DHPS knockdown, cells were treated with low levels of spermidine (1 μM) that were, on their own, insufficient to confer enhanced AUU initiation with the uCC-WT-FLuc construct (Figure S1). The reduction in eIF5A hypusination upon DHPS shRNA knockdown (Figure S3C–D) mimicked the effects of high polyamines and lead to significantly enhanced AUU initiation on the WT uCC reporter, but not on the uCC-OF-FLuc construct that lacks the PPW motif (Figure 4G).
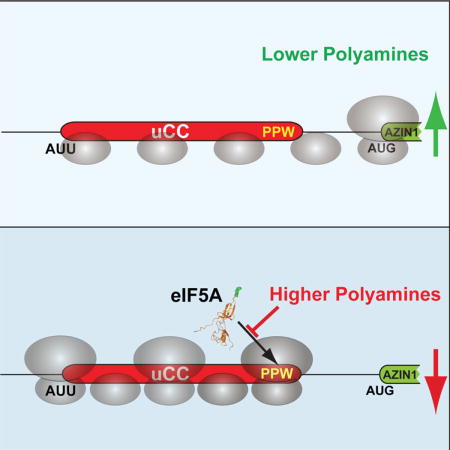
Ribosome queuing model for regulation of translation start site selection
The experimental results and comparative genomics analysis presented above suggest the following model for homeostatic control of AZIN1 mRNA translation when polyamine levels fluctuate to lower or higher levels (Figure 5). Under lower polyamine conditions, most ribosomes scan across the AUU codon of the uCC without initiating. These scanning ribosomes initiate at the AUG codon of the mORF and synthesize AZIN1. The occasional ribosome that initiates at the AUU codon translates the uCC and then is efficiently released upon reaching the stop codon. As eIF5A activity is high under lower polyamine conditions (Figure 4C), ribosomes translating the uCC do not pause while either decoding the PPW motif or terminating at the PS-stop motif. Moreover, consistent with the notion that translation of a long uORF is incompatible with reinitiation at downstream start sites (Jackson et al., 2012), previous studies showed that forced translation of the uCC, by mutating the near cognate AUU initiation codon to a cognate AUG codon, precluded AZIN1 synthesis under polyamine permissive and repressive conditions (Ivanov et al., 2008). Thus, we propose that the subset of scanning ribosomes that translate the uCC likely disengage from the mRNA at the uCC stop codon and do not reinitiate at the mORF to synthesize AZIN1. In contrast, under higher polyamine conditions we propose that interference of eIF5A function causes elongating ribosomes on the uCC to pause at two discrete sites: first, while decoding the PPW motif; and second, while decoding PS-stop during termination. We propose that these pauses, in turn, serve two purposes. First, they create a roadblock that obstructs scanning ribosomes from reaching the start codon of the mORF. Second, the paused ribosomes cause queuing of upstream scanning ribosomes leading to prolonged positioning of a scanning ribosome in the vicinity of the AUU codon, which enables the ribosome to initiate at this otherwise inefficient near-cognate start site.
Figure 5. Schematic model of polyamine-induced and eIF5A-mediated ribosomal queuing leading to enhanced initiation at the AUU start codon of the uCC on the AZIN1 mRNA.
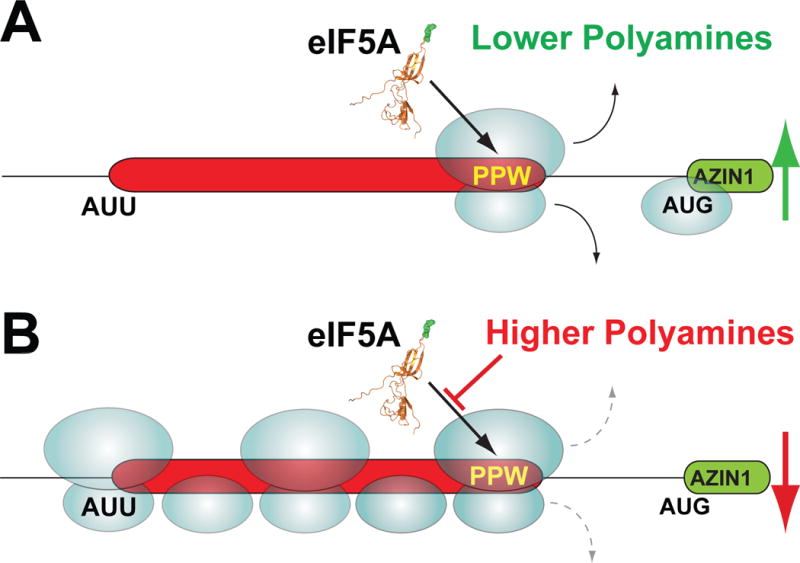
(A) Under conditions of lower polyamines most scanning 40S ribosomes skip over the uCC (red) start codon (AUU) without initiating and then initiate downstream on the AZIN1 mORF (green). The occasional ribosome that initiates on the uCC AUU start codon synthesizes the uCC peptide and then disengages from the mRNA. (B) Under conditions of higher polyamines any ribosome that initiates on the uCC start codon elongates down the uORF and encounters the PPW sequence. The high polyamines interfere with eIF5A function and cause the ribosome to stall. Subsequent scanning ribosomes, which mostly skip over the AUU start codon, and the occasional elongating ribosome form a queue behind the stalled ribosome. A queued subunit spends an extended time traversing and in the vicinity of the near cognate start codon allowing greater opportunities for initiation. The enhanced rate of initiation on the uCC reinforces the elongation stall, prevents ribosomes from scanning to the AZIN1 mORF, and effectively suppresses AZIN1 synthesis.
According to our model for uCC translation, ribosome queuing is critical to position a ribosome in the vicinity of the uCC start codon. Two experimental strategies have been used to test the importance of queuing for translational control of AZIN1 synthesis. First, we reasoned that reducing the rate of ribosome loading on mRNA would impair queue formation and subvert the effects of higher polyamines, leading to reduced uCC translation and derepressed mORF translation on the AZIN1 mRNA. To test this hypothesis, a simplified version of the uCC-WT-FLuc reporter lacking the AUG-initiated conventional uORF1 was expressed in cells in the presence of 1 mM SPD. As expected, this high level of SPD stimulated uCC translation dependent on the PPW motif (Figure 6A, purple bars) and promoted high levels of frameshifting on the human antizyme OAZ1 mRNA (Figure 6C), a sensitive in vivo reporter of polyamine levels. To interfere with ribosome binding to mRNA, we modulated the activity of two translation factors. In an early step of translation initiation, an eIF2–GTP–Met-tRNAiMet ternary complex binds to the 40S subunit, a necessary prerequisite for subsequent binding of the ribosome to mRNA in a reaction mediated by the cap-binding complex eIF4F. The drug 4EGI-1 blocks ribosome binding to mRNAs by disrupting the interaction between the cap-binding factor eIF4E and the scaffolding protein eIF4G within the eIF4F complex (Moerke et al., 2007). As shown in Figure 6A, addition of 4EGI-1 inhibited uCC expression. Notably, this impact of 4EGI-1 was not simply due to a general effect on protein synthesis or AUU initiation. At the concentrations used in these experiments, 4EGI-1 inhibited general translation by 80–95% (Figure S4A); however, the uCC reporter activity was normalized to an AUG control (which was also inhibited), so we conclude that AUU initiation on the uCC was hypersensitive to impaired ribosome loading. Moreover, 4EGI-1 had no effect on the uCC-OF-FLuc reporter that lacks the PPW motif (Figure 6A, bottom construct), further demonstrating the unique sensitivity of uCC translation to reduced ribosome loading. In addition to repressing uCC translation, treatment with 4EGI-1 derepressed mORF (Renilla luciferase) expression on an mRNA containing the 5′ leader from AZIN1 (Figure 6B). Though 4EGI-1 was identified as an eIF4F inhibitor, immunoblot analyses revealed a modest increase in eIF2α phosphorylation in cells treated with 4EGI-1 (Figure S4B–C), thus in these experiments 4EGI-1 may be impairing ribosome loading by both impairing eIF4F function and by inducing eIF2α phosphorylation.
Figure 6. Ribosome queuing potentiates translation of the uCC to regulate AZIN1 mRNA translation.
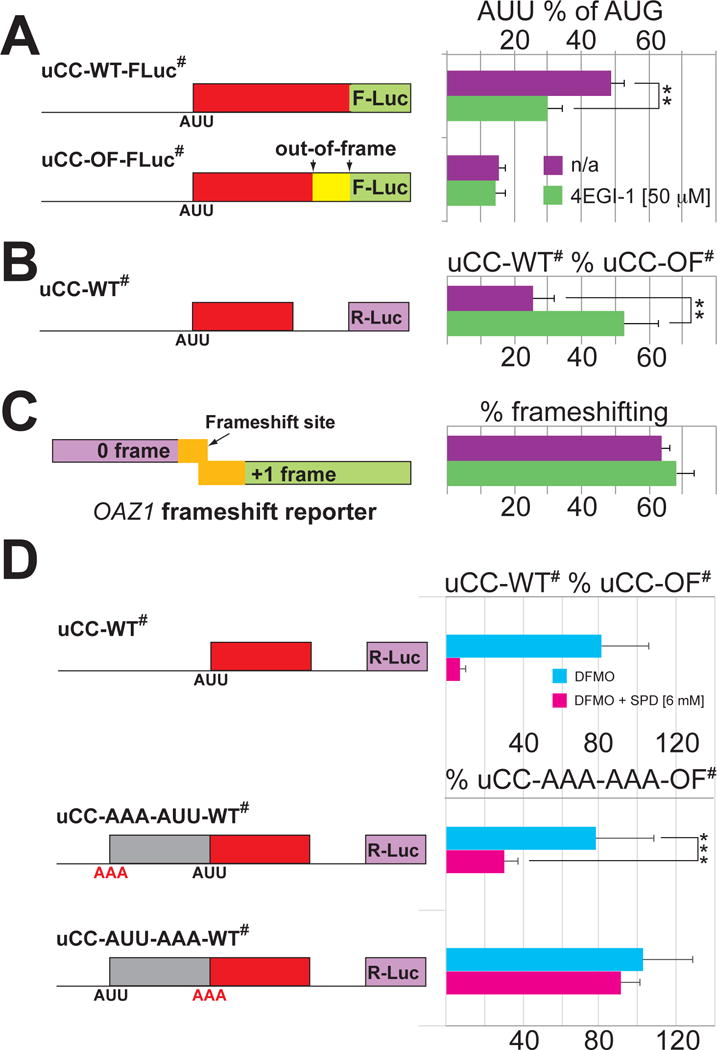
(A–C) Impairing 40S ribosome loading mitigates the effects of high polyamines on AZIN1 regulation. HEK293T cells were transfected with the indicated uCC-Luc, azin1 leader-Renilla or dual-luciferase OAZ1 reporter and incubated for 24 hr in DMEM supplemented with 1 mM aminoguanidine and 1 mM SPD in the absence or presence of 4EGI-1. Percent AUU initiation (A) was calculated as described in Figure 1A; relative reporter mRNA levels are presented in Table S3. To monitor AZIN1 synthesis (B), the full 5′ leader of mouse azin1 mRNA was fused upstream of Renilla luciferase. Wild type reporter activity was calculated as a percent of a reporter containing the out-of-frame uCC mutation and normalized to a co-transfected firefly luciferase reporter as in Figure 1C; relative reporter mRNA levels are presented in Table S3. To monitor levels of free polyamines, percent OAZ1 +1 frameshifting (C) was calculated relative to an in-frame reporter. Error bars denote standard deviation; **p<0.01 (Student’s two-tailed t-test; n=5, assayed in duplicate); # symbol in reporter names indicates that the AUG start codon of conventional uORFs were mutated to AAA. (D) Extending the length of the AZIN1 uCC impairs polyamine regulation. HEK293T cells were transfected with the indicated uCC-Luc reporters and grown as described in Figure 1A, except that cells were supplemented with 6 mM SPD. The grey box in the bottom two constructs indicates a 186 nt insertion (Table S6). In the middle construct the 62-codon extension starts with a non-initiating AAA codon and the uCC retains its normal AUU start codon; in the bottom construct the extension starts with an AUU codon and uCC start codon is changed to AAA. Luciferase activity of each reporter is compared to a reporter with AAA codons at both positions and the last 10 codons of the uCC out-of-frame. Error bars denote standard deviation; ***p<0.001 (Student’s two-tailed t-test; n=10, assayed in duplicate); relative reporter mRNA levels are presented in Table S3.
In an alternate approach to reduce ribosome queuing, we examined the impact of impairing ribosome loading by directly targeting eIF2 activity. Arsenite causes oxidative stress leading to phosphorylation of eIF2α (Figure S4B–C), reduced ternary complex formation, and fewer ribosomes binding to mRNA. Thus, arsenite is expected to mimic 4EGI-1 by reducing ribosome queuing and altering AZIN1 mRNA translation. As observed with 4EGI-1, arsenite treatment inhibited general translation by 96–97% (Figure S4A). However, within the context of this general inhibition, uCC translation was hypersensitive to arsenite treatment (Figure S5A), which also led to derepression of mORF translation on the reporter containing the AZIN1 5′ leader (Figure S5A). As arsenite did not affect frameshifting on an antizyme reporter (Figure S5A), these impacts of arsenite on AZIN1 translation are not due to alterations in polyamine levels. In support of these results using AZIN1 reporters, published ribosome profiling studies (Andreev et al., 2015) revealed arsenite induction of AZIN1 mRNA translation. In cells containing normal (repressive) levels of polyamines, treatment with arsenite induced a 2.4-fold increase in the translation of the AZIN1 mORF relative to the uCC (Andreev et al., 2015) (Figure S5B). Taken together, these studies employing 4EGI-1 or arsenite to inhibit ribosome loading on an mRNA support the notion that a ribosome queue is critical for uCC translation. We propose that impaired ribosome loading diminishes the queuing of scanning ribosomes upstream of the ribosome paused during translation of the PPW motif in the uCC. The absence of a ribosome queued near the AUU start codon of the uCC results in diminished uCC translation and enables more ribosomes to scan past the uCC and initiate translation at the downstream mORF (Figure 5).
In a second strategy to test the importance of ribosome queuing for translational control of AZIN1 synthesis, we examined the impact of lengthening the uCC. We reasoned that the length of the uCC has been optimized during evolution to enable precise control of uCC translation in response to varying polyamine levels. Accordingly, increasing the distance between the ribosome pause at the PPW motif and suboptimal AUU start codon of the uCC is expected to disrupt translation control. To test this prediction, reporters were constructed by inserting 186 nucleotides (62 codon) at the 5′ end of the uCC. This extension more than doubles the length of the uCC and makes it substantially longer than any naturally occurring uCC in AZIN1 or ODC mRNAs, which in vertebrates range in length from 46 to 54 codons. The sequence used for the extension was obtained from the mouse Trip12 mRNA, which shows signatures of leaky scanning including an AUG start codon in poor context followed by more than 1000 nucleotides without an AUG codon in all three reading frames. We reasoned that sequences downstream of the poor context start in the Trip12 mRNA would be conducive to scanning, and we selected a 186-nt sequence to insert in the uCC-WT# reporter. As the uCC initiates at a near-cognate start codon, three in-frame near-cognate start codons within the inserted sequence were changed to codons that do not support initiation. Three reporters were constructed to assess the impact of the inserted sequence on AZIN1 mORF (Renilla luciferase) synthesis (Figure 6D). In one construct the extension starts with a non-initiating AAA codon and the natural AUU start codon of the UCC is left intact (uCC-AAA-AUU-WT#). In a second construct the extension initiates with the near cognate AUU codon and the natural uCC start codon was changed to AAA (uCC-AUU-AAA-WT#). Finally, a control construct was generated in which both the extension and the natural start codons were replaced by AAA and the out-of-frame mutation was inserted in the uCC to enable maximal unregulated reporter expression (uCC-AAA-AAA-OF#). In all three cases, the 4 nucleotides upstream and 3 nucleotides downstream of both the extension and the natural start codons matched the nucleotides flanking the natural AUU start codon of the uCC. As shown in Figure 6D, polyamine regulation of reporter expression was maintained when the 186 nts were inserted upstream of the intact uCC (uCC-AAA-AUU-WT#). Thus, extending the length of the 5′ leader upstream of the uCC did not impact polyamine control of uCC or mORF translation. In contrast, moving the uCC start codon 62 codons further upstream impaired polyamine repression of reporter expression (uCC-AUU-AAA-WT#), even though the stalling potential of the PPW sequence within the uCC was unaltered (Figure 6D). We conclude that lengthening the uCC impairs polyamine control, presumably because the ribosome queue is of insufficient length to position a ribosome in the vicinity of the repositioned uCC start site.
DISCUSSION
The 5′ leader is a common target for controlling mRNA translation (Hinnebusch et al., 2016). In addition to mRNA secondary structure, upstream AUG codons and uORFs impact the efficiency of translation. Moreover, selection of the translation start site by scanning ribosomes is sensitive to the context nucleotides flanking the start codon as well as to the use of near-cognate start codons. In this report, we revealed a mechanism of translational control employing a uORF initiated with a near-cognate start codon. Whereas scanning ribosomes typically fail to recognize near-cognate codons, as well as codons with poor context nucleotides, we showed that elongation pausing during translation of the uORF and subsequent queuing of ribosomes upstream of the paused ribosomes can enhance selection of the weak start codon.
The paradoxical stimulation of mORF translation by inhibiting ribosome loading via 4EGI-1 or arsenite is reminiscent of the stimulation of GCN4 (Hinnebusch, 2005) and ATF4 (Lu et al., 2004; Vattem and Wek, 2004) mRNA translation by phosphorylation of eIF2α. Whereas translation of the GCN4 and ATF4 mRNAs is controlled by multiple uORFs and by regulated reinitiation on inhibitory uORFs upstream of the mORF (Hinnebusch et al., 2016), the AZIN1 uCC functions as a solo element in the 5′ leader of an mRNA. Our model for AZIN1 translational control could explain how translation of mRNAs bearing an inhibitory (elongation pause) uORF with a suboptimal start codon like CHOP and GADD34 mRNAs in mammals (Palam et al., 2011; Young et al., 2016; Young et al., 2015) is resistant to, or induced by, eIF2α phosphorylation. Thus, the uCC model can explain regulation by elongation-pausing inhibitory uORFs that are initiated not only by near-cognate start codons but also by AUG codons with suboptimal context. In fact, the uCCs in ODC homologs from several metazoa are initiated by AUG codons in poor context (Ivanov et al., 2008). In addition, the AAP uORF in fungi, which is initiated at an AUG codon in poor context (Wang and Sachs, 1997), could be considered a uCC, thus expanding the uCC model beyond eIF5A and polyamines to regulation by other small molecules. Likewise, the recently described translational regulation of the GGP gene in plants by ascorbate also appears to fit the uCC paradigm (Laing et al., 2015). Moreover, in addition to functioning in regulatory uORFs, we can imagine that similar elongation-pausing sequences in mORFs or in alternate reading frames could lead to utilization of alternate (weak) start codons and thus expand the coding potential of mRNAs.
An intriguing feature of the queuing model for uCC translation is the importance of the distance between the elongation pause and the inefficient start codon. When the distance between the PPW or PS-stop motif and the initiation site is short, as in the ODC uCC in Zygomycota, the distance appears invariable over large evolutionary time-scales. Moreover, the length of these short uCCs is divisible by the length of mRNA protected by an elongating 80S ribosome (approximately 30 nts or 10 codons (Wolin and Walter, 1988)). In the case of Zygomycota the uCC is 19 codons (Figure 1B), which would enable near perfect positioning of the queued ribosome over the start codon. In contrast, for longer uCCs, as in the uCCs of vertebrate AZIN1 mRNAs (~50 codons), the distance between the pause motif and start codon is variable and not always divisible by 10 codons (Figure S6). Notably, the queued ribosomes on the uCC could consist of both 80S elongating complexes and 40S scanning complexes, which protect variable lengths from ~19 to ~75 nucleotides (Archer et al., 2016). Thus, stimulation of translation initiation by queued ribosomes may not require precise positioning of a ribosome on the start codon and instead rely on the initiating ribosome spending a longer time in the vicinity of the start codon, perhaps migrating back-and-forth, or oscillating (Matsuda and Dreher, 2006), across the site as it bumps into the downstream queue. Finally, we note that while the length of the uCCs varies, there appears to be an upper limit on the length. The longest naturally occurring uCCs in vertebrate ODC or AZIN1 mRNAs are around 54 codons, and we showed that increasing the length of the uCC to 114 codons abolishes regulation (Figure 6D). We propose that the uCC length and the duration of the polyamine-induced pause on the PPW motif have been exquisitely matched to enable scanning ribosomes to efficiently skip over the uCC and translate the AZIN1 mORF under low polyamine conditions, while enabling queuing and enhanced uCC translation when polyamine levels increase.
STAR ٭ METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Please direct any requests for further information or reagents to the lead contact, Tom Dever (thomas.dever@nih.gov).
METHOD DETAILS
Sequence compilation and analysis
The uCC sequences were obtained from GenBank using the BLAST algorithm as described previously (Ivanov et al., 2008) except that the Transcriptome Shotgun Assembly (TSA) was also queried. After the sequences were compiled the conceptually translated uCCs were aligned using ClustalX (Larkin et al., 2007). In the Arthropod alignment, minor manual adjustments were performed upon visual inspection. The alignments were then used to generate WebLogo logograms (Crooks et al., 2004). Files with the compiled and aligned sequences are available upon request.
Cell Culture and Transfections
HEK293T cells were obtained from American Type Culture Collection and maintained in DMEM (Corning) supplemented with 1 mM L-glutamine, 10% FBS (Gibco), and penicillin/streptomycin (Quality Biological). For polyamine depletion experiments, cells were split and grown overnight to ~70% confluence, split again, and then 4×106 cells were placed in a 10-cm plate in medium supplemented with 2.5 mM α-difluoromethylornithine (DFMO; a kind gift from P. Woster via Dr. Michael Howard, University of Utah). Cells were incubated for 3 days in DFMO supplemented media at 37°C in 5% CO2 and then transfected using Lipofectamine 2000 reagent (Invitrogen) and the one-day protocol in which suspension cells are added directly to the DNA complexes in white 96-well half-area plates (Costar). Briefly, for the uCC-firefly luciferase fusion experiments 0.2 μl Lipofectamine, 5 ng firefly tester reporter plasmid and 1 ng normalizing Renilla reporter (or 5 ng dual luciferase reporter) were mixed in 25 μl Opti-MEM (Gibco) and dispensed to each well along with 2×104 cells suspended in 25 μl DMEM supplemented with 5 mM DFMO. After another 24 hr of incubation 50 μl of media supplemented to final concentrations of 2.5 mM DFMO, 1 mM aminoguanidine hydrochloride (Sigma), or the same plus variable concentrations of spermidine (Sigma) were added to each well and the cells were returned to the incubator. Luciferase activity was measured after 18 hr of spermidine treatment. For the uCC-Renilla luciferase experiments an identical protocol was followed except that 1 ng of tester reporter and 5 ng of normalizing firefly plasmid DNA were used per well.
For the experiments with arginine treated cells, U2OS cells (kind gift from Dr. Nancy Kedersha, Harvard University) were used. The cells were maintained the same as described above for HEK293T cells. Prior to the day of transfection, cells were split and then grown to ~70% confluence. Transfections were performed as described above except 10 ng firefly reporter and 2.5 ng normalizing Renilla luciferase reporters were used per well. For the dual luciferase reporter 10 ng per well was used. 1×104 cells were added per well and either standard DMEM media was added (already containing 400 μM arginine) or the standard media was supplemented with L-arginine monohydrochloride (Sigma) to a final concentration of 25 mM. The cells were then incubated for 20 hr and the luciferase activities were measured.
For the shRNA knockdown experiments, two shRNAs, TRCN0000330796 and TRCN0000330717, targeting human DHPS and a control shRNA, shc002, without known targets (all purchased from Sigma), were used. Cells, pretreated in DFMO for 4 days, were transfected with 1 ng reporter, 2.5 ng normalizing Renilla expressing plasmids, and 5 ng of each DHPS shRNA plasmid or 10 ng control shRNA and then maintained in DFMO-treated media for another 48 hr. The cells were then either incubated in medium containing DFMO and 1 mM aminoguanidine or the DFMO media was supplemented with 1 μM spermidine and 1 mM aminoguanidine. The cells were lysed and luciferase activity read after another 24 hr of incubation.
For the experiments with 4EGI-1, 20,000 HEK293T cells were transfected with 5 ng firefly reporter and 5 ng normalizing Renilla plasmid, or with 1 ng Renilla reporter and 10 ng normalizing firefly plasmid, or with 5 ng dual luciferase reporter plasmid per well. At the time of transfection 50 μM 4EGI-1 (Santa Cruz Biotechnology) was added to half of the wells. All transfected cells were grown in DMEM media supplemented with 1 mM spermidine and 1 mM aminoguanidine. The cells were lysed and luciferase activity measured after 24 hr of incubation.
For the experiments with arsenite, cells were transfected with 10 ng firefly reporter, 1 ng Renilla reporter or 10 ng of dual luciferase reporter plasmid per well. A normalizing co-transfected reporter was used only with the with dual luciferase reporter. At the time of transfection 25 μM arsenite (Sigma-Aldrich) was added to half of the wells. All transfected cells were grown in DMEM media supplemented with 1 mM spermidine and 1 mM aminoguanidine. The cells were lysed and luciferase activity measured after 18 hr of incubation.
Plasmid Construction
The plasmids listed in Table S5 were in most cases constructed by cloning PCR products generated using the indicated primer (Table S4) and template combinations into the vector p2luc (Grentzmann et al., 1998) using HindIII and BamHI restriction enzymes. Templates phRL-WT, phRL-M4, phRL-M1M4, phRL-M5 and phRL-M4M5 were described previously(Ivanov et al., 2008). The plasmids uCC-AAP-D12N-FLuc (AUU) and uCC-AAP-D12N-FLuc (AUG) were generated using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) using the primers and template combinations indicated in Table S5.
The plasmids listed in Table S5 were constructed by cloning PCR products generated using the indicated primer (Table S4) and template combinations into phRL-WT using HindIII and AvaI restriction enzymes. Templates phRL-M1M2M3M6, phRL-M1M2M5, and phRL-M1M2 were described previously (Ivanov et al., 2008). Plasmids hAZ1-WT and hAZ1-IF for assaying human antizyme frameshifting were constructed by cloning PCR products generated using the indicated primers (Table S4) and templates AZ-1wt and AZ-1if (Ivanov et al., 1998), respectively, between the XhoI and BglII sites of pDluc(Fixsen and Howard, 2010). Sequences of the extended uCC reporters containing Trip12 sequences are presented in Table S6.
Dual Luciferase Assay
Luciferase activities were determined by the Dual Luciferase Stop and Glo Reporter assay system using reagents as described by Dyer et al. (Dyer et al., 2000), with a slight modification (the homemade Stop and Glo buffer was supplemented with 5% commercial Stop and Glo buffer purchased from Promega). Relative light units were measured on a CentroXS3 LB960 microplate luminometer fitted with two injectors (Berthold Technologies). After removing the media, transfected cells were lysed in 25 μl 1× passive lysis buffer (PLB; Promega), and light emission was measured after injection of 50 μl firefly luciferase substrate followed by the same volume of Renilla luciferase substrate. For phRL mutant series, the Renilla luciferase activity was normalized relative to the activity of an internal control p2luc-based plasmid expressing firefly luciferase from an AUG codon in perfect context (Ivanov et al., 2010c). For p2luc mutant series, the firefly luciferase activity was normalized relative to the activity of a plasmid, pSV40-Renilla, expressing Renilla luciferase (Loughran et al., 2012). For the antizyme dual luciferase reporters, firefly was normalized to Renilla luciferase made upstream from the same cistron and the values of “wild type” were compared to “in-frame” control to calculate percent frameshifting.
Analysis of reporter mRNA levels
For all qPCR experiments, cells were grown in 12-well plates. For each well, transfections reagents, including the amount of transfected DNA, were scaled up 40× (i.e. total volume of 500 μl Opti-MEM solution, plus 500 μl DMEM). For HEK293T cell lines, 4×105 cells were used per well. For U2OS cell lines, 2×105 cells were used per well. Otherwise, incubation times and conditions were identical to transfections in half-area plates. RNA was isolated from HEK293T or U2OS cells using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. 2μg of total RNA from each sample was used in single-stranded cDNA synthesis reactions conducted with SuperScript III First-Strand Synthesis SuperMix (Invitrogen) following the manufacturer’s protocol. Following reverse transcription, cDNA reactions were diluted 1:6, and 5μL of dilute cDNA was utilized in each subsequent qPCR reaction. Transcript levels were quantified by qPCR using Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies) on a LightCycler 480 II (Roche). Relative quantification of transcripts was calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001) in which β-actin levels were used for normalization.
Ribosome profiling
Ribosome profiling was carried out according to Ingolia et al. (Ingolia et al., 2012) with some modifications. HEK293T cells stably expressing pcDNA4-TO-Ren-mAZ-FF were maintained in DMEM supplemented with 10% FBS, 1 mM L-glutamine and antibiotics and plated onto 150 mm petri-dishes at ~30% confluency. To deplete endogenous polyamines, cells were maintained for 5 days in the presence of 2.5 mM DFMO and 1mM aminoguanidine, and then the cells were either left untreated or stimulated with 2 mM spermidine for 24 hr. To harvest cells, dishes were chilled on ice and the cells were washed with ice cold PBS plus cycloheximide (100 μg/ml) prior to resuspension in polysome lysis buffer (20 mM Tris–HCl (pH 7.5), 250 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 100 μg/ml cycloheximide, 20 U/ml TURBO DNAse (Ambion, Waltham, MA)). Importantly, to avoid artificial accumulation of initiation complexes at translation initiation starts cells were not pre-treated with cycloheximide (Gerashchenko and Gladyshev, 2014). Cell lysates were incubated on ice for 10 min, and then spun at 16,000 × g for 10 min at 4°C to pellet cell debris. The supernatant was divided for Riboseq and RNAseq library preparation. The lysate for Riboseq was treated with RNAse I (Ambion: 100 U per 3.14 OD260 of lysate) at 23°C for 50 min. Digestion was stopped with SuperasIN (Ambion). Monosomes were isolated using Illustra MicroSpin S-400 HR Columns (GE Healthcare) according to the manufacturer’s instructions.
Total RNA from the monosomes was extracted with phenol/chloroform followed by ethanol precipitation. For the RNAseq control, total RNA was extracted from the second lysate aliquot with Trizol-LS (Life Technologies) and then mRNA was isolated using the Oligotex mRNA kit (Qiagen). Two rounds of polyA(+)-mRNA selection were applied to decrease rRNA contamination to approximately 3%. Purified mRNA was subjected to alkaline hydrolysis as described by Ingolia et al. (Ingolia et al., 2009). Both Riboseq and RNAseq samples were fractionated by 15% denaturing urea PAGE (containing 1 × TBE, 7 M urea, acrylamide (20):bis-acrylamide (1)). Bands corresponding to RNA fragments of 28–34 nts were excised for both Riboseq and RNAseq samples. The RNA was extracted by shaking overnight in a buffer containing 0.3 M NaOAc (pH 5.1), 1 mM EDTA and 0.1% SDS and then precipitated with one volume of isopropanol and 2 μl of GlycoBlue (Life Technologies).
Library preparation was carried out as previously described(Ingolia et al., 2012) with the following modifications. First, the circularization reaction was performed for 2 hr. Second, during PCR library amplification, the temperature ramping speed was set at 2.2°C/s to reduce bias associated with GC content (Aird et al., 2011). Libraries prepared from two independent biological replicates were sequenced on an Illumina HiSeq 4000 system at Genewiz (New Jersey, US).
Processing of the sequence data was performed using the web browser RiboGalaxy tools(Michel et al., 2016). Briefly, after removal of the adapter sequence (CTGTAGGCACCATCAATAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC) reads that could be aligned to rRNA were removed from further consideration. The remaining reads were aligned to the human refGene database using Bowtie. One base from the high-quality 5′ end of each read was removed before alignment, and a maximum of 2 mismatches were permitted in the seed. Reads were aligned to the positive strand, and up to 10 valid alignments were allowed per read. The minimum seed length was set at 25 nts. The reads from each biological replicate were analyzed separately; however, because the reads mapping to AZIN1 under low and high polyamines in the two datasets were very similar, the data from the two replicates were pooled together. CSV files were generated with the reads mapped to each gene, and the files corresponding to NM_015878.5 (AZIN1) were used to generate Figure 2.
Preparation of aminoacyl-tRNA
Yeast initiator tRNAi Met was prepared by T7 in vitro transcription and the tRNAiMet was aminoacylated using purified His-tagged E. coli MetRS as described previously (Murray et al., 2016). Yeast tRNAGlu(UUC) and tRNATrp(CCA) were purified from bulk S. cerevisiae tRNA (Roche) using the biotinylated oligonucleotides 5′- GAAAGCGTGATGTGATAGCCGTTACA-3′-biotin and 5′-TTGGAGTCGAAAGCTCTACCATTGAG-3′-biotin, respectively, as described previously (Gutierrez et al., 2013). Yeast tRNAPro(UGG) was also purified from bulk S. cerevisiae tRNA as previously described (Gutierrez et al., 2013). Yeast tRNAPhe (Sigma) and tRNALys (tRNA Probes, Texas) were obtained from commercial vendors.
To purify yeast glutamyl tRNA synthetase (GUS1) and tryptophanyl tRNA synthetase (WRS1), His-tagged GUS1 and WRS1 were cloned in pQE-80L (Qiagen) to generate pC4857 and pC4853, respectively. To express and purify His-tagged GUS1 or WRS1, E. coli BL21(DE3) transformants carrying pC4857 or pC4853 were grown in 500 ml LB medium containing 100 μg/ml ampicillin at 37 °C to OD600 = 0.5. Following addition of 0.2 mM IPTG, the culture was incubated at 20 °C for 16 hr. Following harvesting, the cell pellet was suspended in 20 ml lysis buffer (50 mM sodium phosphate (pH 8.0), 300 mM sodium chloride and 10 mM imidazole), and cells were broken by sonication using a microtip (5 cycles of 30 s pulse followed by 30 s cooling at 4°C). The cell lysate was cleared by centrifugation at 27,000 × g for 30 min and then mixed gently with 1 ml Ni-NTA resin (Qiagen) at 4 °C for 1 hr. The resin was transferred to a 1 ml disposable column (Qiagen), washed sequentially with 10 ml lysis buffer and 20 ml lysis buffer containing 20 mM imidazole, and then the protein was eluted in 4 ml lysis buffer containing 250 mM imidazole. The elute was dialyzed overnight against 50 mM potassium phosphate (pH 8.0), 5 mM magnesium acetate, 2 mM DTT and 10% glycerol. After dialysis, the protein solutions were concentrated using a Microcon Ultracel YM-50 (EMD Millipore). Prolyl-tRNA synthetase, phenylalanyl-tRNA synthetase and lysyl-tRNA synthetase were prepared as described previously (Gutierrez et al., 2013; Murray et al., 2016).
For aminoacylation of tRNA, 5 μM tRNA was mixed with 2 mM ATP, 0.1 mM amino acid, 10 mM MgCl2 and 1 μM aminoacyl tRNA synthetase in reaction buffer (100 mM HEPES-KOH [pH 7.5], 10 mM KCl and 1 mM DTT), and then incubated at 30 °C for 30 min.
Peptide formation assay
Purified ribosomes and translation factors were prepared and in vitro reconstituted peptide formation assays were performed according to procedures described previously (Gutierrez et al., 2013; Shin et al., 2017). To examine the spermidine effect on peptide synthesis, both initiation and elongation factors were purified in the absence of polyamines. Initiation complexes were prepared in 1× Recon Buffer A (30 mM HEPES-KOH [pH 7.5], 100 mM potassium acetate, 3 mM magnesium acetate, 2 mM DTT) as described previously (Gutierrez et al., 2013). Peptide formation assays contained 4 nM initiation complex, 2 μM eEF1A, 1 μM eEF2, 1 μM eEF3, 1 μM aminoacyl tRNA, 1 mM GTP-Mg2+, 1 mM ATP-Mg2+ and varying amounts of eIF5A and spermidine in 1× Recon Buffer C (30 mM HEPES-KOH [pH 7.5], 100 mM potassium acetate, 1 mM magnesium acetate, 2 mM DTT). The elongation assay components were preincubated for 5 min on ice before adding the initiation complex, and then reactions were incubated at 26 °C. Progress of peptide formation was examined by electrophoretic TLC as described previously (Gutierrez et al., 2013). The fractional yields of peptide products in each reaction at different times were quantified and fit using KaleidaGraph (Synergy Software).
Western analysis
Transfected HEK293T cells were grown in clear 12-well plates (Costar) with reagent amounts and volumes scaled up 20× per well. Following incubation cells were washed once with 1× PBS and then taken up in 100 μl RIPA lysis buffer supplemented with 1× Complete EDTA-free Protease Inhibitor Cocktail (Roche). Cells were lysed by sonication for 5 seconds at output level 3 using a microtip (Fisher Scientific 550 Sonic Dismembrator), and then lysates were mixed with an equivalent volume 2× Laemmli sample buffer (Bio-Rad) and heated at 80 °C for 5 min. Following separation on a 4–20% SDS-polyacrylamide gel (Bio-Rad), proteins were detected by immunoblotting with mouse monoclonal antibody against human DHPS (Origene, Rockville, MD), rabbit polyclonal antibodies against hypusine (EMD Millipore), purified mouse anti-eIF5A antibodies (BD Biosciences), rabbit monoclonal antibodies against eIF2α (Cell Signaling Technology), or mouse monoclonal anti-eIF2α(Phospho-Ser51) (Abcam). Membranes were blocked with 5% skim milk (Blotting grade blocker, Bio-Rad) in 1× TBST (1× TBS plus 0.1% Tween 20). Horseradish peroxidase conjugated anti-rabbit IgG or anti-mouse IgG (GE Healthcare) secondary antibodies and Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) were used for protein detection.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative data are presented as the mean and standard deviation from biological replicates. To determine statistical significance between groups, comparisons were made using Student’s two-tailed t tests, using the formula imbedded in Microsoft Excel. The number of biological replicates (n) is indicated in the legend of each figure. p values less than 0.05 were considered significant. Sample size estimates were not used. Studies were not conducted blind. For immunoblots, the reported images are representative of at least three independent experiments.
Supplementary Material
Highlights.
Polyamines induce near cognate initiation at a uORF in the AZIN1 mRNA
Elevated polyamines cause ribosomes to pause when translating PPW motif in the uORF
Polyamines competitively inhibit elongation factor eIF5A to block PPW translation
Ribosome queue upstream of the pause promotes initiation at the uORF AUU start codon
Acknowledgments
We thank Patrick B.F. O’Connor for providing the processed mapped reads to AZIN1 from the published arsenite ribosomal profiling study. We thank Alan Hinnebusch, Jon Lorsch, Audrey Michel, Patrick O’Connor, Pavel Baranov, Joo-Ran Kim, and members of the Atkins, Dever, Guydosh and Lorsch labs for helpful discussions. This work was supported in part by the Intramural Research Program of the National Institutes of Health, NICHD and the UCC part was supported by Science Foundation Ireland grants [13/IA/1853;12/IP/1492] and by a Health Research Board Cancer scholarship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
I.P.I. G.L. and J.F.A. conceived the queuing model of uCC initiation, and I.P.I. and T.E.D. conceived the eIF5A regulation. I.P.I., G.L., B.S.S., S.K.Y.B., and I.T. performed the experiments and analyzed the data. C.C. contributed to reporter gene construction. I.P.I. and T.E.D. wrote the manuscript. T.E.D. and J.F.A. supervised the NIH and UCC components, respectively. All authors discussed the results and edited the manuscript.
DECLARATION OF INTERESTS
G.L. is a founder and shareholder of RiboMaps Ltd, a company providing ribosome profiling as a service. As the findings reported here are supported by ribosome profiling, its publication may increase the attractiveness of this technology and indirectly benefit the company.
References
- Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, Gnirke A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RW, Nordin BE, Schimmel P. Activation of microhelix charging by localized helix destabilization. Proc Natl Acad Sci U S A. 1998;95:12214–12219. doi: 10.1073/pnas.95.21.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire MA, Maag D, Savio P, Acker MG, Tarun SZ, Jr, Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, et al. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand M, Balar B, Ulloque R, Gross SR, Kinzy TG. Domain and nucleotide dependence of the interaction between Saccharomyces cerevisiae translation elongation factors 3 and 1A. J Biol Chem. 2006;281:32318–32326. doi: 10.1074/jbc.M601899200. [DOI] [PubMed] [Google Scholar]
- Andreev DE, O’Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, Preiss T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016;535:570–574. doi: 10.1038/nature18647. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Gutierrez E, Shin BS. The hypusine-containing translation factor eIF5A. Crit Rev Biochem Mol Biol. 2014;49:413–425. doi: 10.3109/10409238.2014.939608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- Fang P, Spevak CC, Wu C, Sachs MS. A nascent polypeptide domain that can regulate translation elongation. Proc Natl Acad Sci U S A. 2004;101:4059–4064. doi: 10.1073/pnas.0400554101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixsen SM, Howard MT. Processive selenocysteine incorporation during synthesis of eukaryotic selenoproteins. J Mol Biol. 2010;399:385–396. doi: 10.1016/j.jmb.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014;42:e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L, Beelman CA, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller JS, Canellakis ES. Cellular control of ornithine decarboxylase activity by its antizyme. J Cell Physiol. 1981;107:209–217. doi: 10.1002/jcp.1041070206. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Modulation of cellular function by polyamines. Int J Biochem Cell Biol. 2010;42:39–51. doi: 10.1016/j.biocel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Atkins JF, Michael AJ. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res. 2010a;38:353–359. doi: 10.1093/nar/gkp1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Firth AE, Atkins JF. Recurrent emergence of catalytically inactive ornithine decarboxylase homologous forms that likely have regulatory function. J Mol Evol. 2010b;70:289–302. doi: 10.1007/s00239-010-9331-5. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Gesteland RF, Atkins JF. A second mammalian antizyme: conservation of programmed ribosomal frameshifting. Genomics. 1998;52:119–129. doi: 10.1006/geno.1998.5434. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Atkins JF. UORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc Natl Acad Sci U S A. 2008;105:10079–10084. doi: 10.1073/pnas.0801590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc Natl Acad Sci U S A. 2010c;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, Ingolia NT. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016;534:558–561. doi: 10.1038/nature17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, Giraldez AJ. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 2016;35:706–723. doi: 10.15252/embj.201592759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana C. Regulation of cellular polyamine levels and cellular proliferation by antizyme and antizyme inhibitor. Essays Biochem. 2009;46:47–61. doi: 10.1042/bse0460004. [DOI] [PubMed] [Google Scholar]
- Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989a;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989b;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Kurosinski MA, Luersen K, Ndjonka D, Younis AE, Brattig NW, Liebau E. Filarial parasites possess an antizyme but lack a functional ornithine decarboxylase. Acta Trop. 2013;126:167–176. doi: 10.1016/j.actatropica.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Laing WA, Martinez-Sanchez M, Wright MA, Bulley SM, Brewster D, Dare AP, Rassam M, Wang D, Storey R, Macknight RC, et al. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell. 2015;27:772–786. doi: 10.1105/tpc.114.133777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A. 2012;109:E2424–2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−DDCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D. Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J. 1999;18:6705–6717. doi: 10.1093/emboj/18.23.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 2012;40:2898–2906. doi: 10.1093/nar/gkr1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA. 2006;12:1338–1349. doi: 10.1261/rna.67906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze MW. Translational control via protein-regulated upstream open reading frames. Cell. 2011;145:902–913. doi: 10.1016/j.cell.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Melnikov S, Mailliot J, Shin BS, Rigger L, Yusupova G, Micura R, Dever TE, Yusupov M. Crystal structure of hypusine-containing translation factor eIF5A bound to a rotated eukaryotic ribosome. J Mol Biol. 2016;428:3570–3576. doi: 10.1016/j.jmb.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Mullan JP, Velayudhan V, O’Connor PB, Donohue CA, Baranov PV. RiboGalaxy: A browser based platform for the alignment, analysis and visualization of ribosome profiling data. RNA Biol. 2016;13:316–319. doi: 10.1080/15476286.2016.1141862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Ichiba T, Matsufuji S, Hayashi S. Cloning of antizyme inhibitor, a highly homologous protein to ornithine decarboxylase. J Biol Chem. 1996;271:3340–3342. doi: 10.1074/jbc.271.7.3340. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, Fernandez IS. Structural characterization of ribosome recruitment and translocation by type IV IRES. Elife. 2016;5:e13567. doi: 10.7554/eLife.13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem. 2006;281:32639–32648. doi: 10.1074/jbc.M607076200. [DOI] [PubMed] [Google Scholar]
- Palam LR, Baird TD, Wek RC. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61:880–894. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. Functions of polyamines in mammals. J Biol Chem. 2016;291:14904–14912. doi: 10.1074/jbc.R116.731661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Becker T, Heuer A, Braunger K, Shanmuganathan V, Pech M, Berninghausen O, Wilson DN, Beckmann R. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 2016;44:1944–1951. doi: 10.1093/nar/gkv1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Dever TE. Molecular genetic structure-function analysis of translation initiation factor eIF5B. Methods Enzymol. 2007;429:185–201. doi: 10.1016/S0076-6879(07)29009-3. [DOI] [PubMed] [Google Scholar]
- Shin BS, Katoh T, Gutierrez E, Kim JR, Suga H, Dever TE. Amino acid substrates impose polyamine, eIF5A, or hypusine requirement for peptide synthesis. Nucleic Acids Res. 2017;45:8392–8402. doi: 10.1093/nar/gkx532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SternJohn J, Hati S, Siliciano PG, Musier-Forsyth K. Restoring species-specific posttransfer editing activity to a synthetase with a defunct editing domain. Proc Natl Acad Sci U S A. 2007;104:2127–2132. doi: 10.1073/pnas.0611110104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fang P, Sachs MS. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol Cell Biol. 1998;18:7528–7536. doi: 10.1128/mcb.18.12.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Sachs MS. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EC, Kang KR, Kim YS, Park MH. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids. 2007;33:341–350. doi: 10.1007/s00726-007-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Guydosh NR, Green R, Buskirk AR. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 2015;11:13–21. doi: 10.1016/j.celrep.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xaplanteri MA, Petropoulos AD, Dinos GP, Kalpaxis DL. Localization of spermine binding sites in 23S rRNA by photoaffinity labeling: parsing the spermine contribution to ribosomal 50S subunit functions. Nucleic Acids Res. 2005;33:2792–2805. doi: 10.1093/nar/gki557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Palam LR, Wu C, Sachs MS, Wek RC. Ribosome Elongation Stall Directs Gene-specific Translation in the Integrated Stress Response. J Biol Chem. 2016;291:6546–6558. doi: 10.1074/jbc.M115.705640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Willy JA, Wu C, Sachs MS, Wek RC. Ribosome Reinitiation Directs Gene-specific Translation and Regulates the Integrated Stress Response. J Biol Chem. 2015;290:28257–28271. doi: 10.1074/jbc.M115.693184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


