Abstract
Replication of positive-strand RNA viruses [(+)RNA viruses] takes place in membrane-bound viral replication complexes (VRCs). Formation of VRCs requires virus-mediated manipulation of cellular lipid synthesis. Here, we report significantly enhanced brome mosaic virus (BMV) replication and much improved cell growth in yeast cells lacking PAH1 (pah1Δ), the sole yeast ortholog of human LIPIN genes. PAH1 encodes Pah1p (phosphatidic acid phosphohydrolase), which converts phosphatidate (PA) to diacylglycerol that is subsequently used for the synthesis of the storage lipid triacylglycerol. Inactivation of Pah1p leads to altered lipid composition, including high levels of PA, total phospholipids, ergosterol ester, and free fatty acids, as well as expansion of the nuclear membrane. In pah1Δ cells, BMV replication protein 1a and double-stranded RNA localized to the extended nuclear membrane, there was a significant increase in the number of VRCs formed, and BMV genomic replication increased by 2-fold compared to wild-type cells. In another yeast mutant that lacks both PAH1 and DGK1 (encodes diacylglycerol kinase converting diacylglycerol to PA), which has a normal nuclear membrane but maintains similar lipid compositional changes as in pah1Δ cells, BMV replicated as efficiently as in pah1Δ cells, suggesting that the altered lipid composition was responsible for the enhanced BMV replication. We further showed that increased levels of total phospholipids play an important role because the enhanced BMV replication required active synthesis of phosphatidylcholine, the major membrane phospholipid. Moreover, overexpression of a phosphatidylcholine synthesis gene (CHO2) promoted BMV replication. Conversely, overexpression of PAH1 or plant PAH1 orthologs inhibited BMV replication in yeast or Nicotiana benthamiana plants. Competing with its host for limited resources, BMV inhibited host growth, which was markedly alleviated in pah1Δ cells. Our work suggests that Pah1p promotes storage lipid synthesis and thus represses phospholipid synthesis, which in turn restricts both viral replication and cell growth during viral infection.
Author summary
Phosphatidate (PA) plays crucial roles in lipid metabolism because it is the shared precursor for major membrane component phospholipids and for storage lipid triacylglycerols (TAGs). Phosphatidate phosphatase (PAP) is involved in converting PA to TAG via diacylglycerol and directs the lipid flux from membrane synthesis for cell growth to lipid storage during inactive growth stages. PAP is structurally and functionally conserved among eukaryotes, from Pah1p in yeast to lipins in mammals. Inactivation of PAP substantially affects lipid composition and causes human diseases, such as lipodystrophy and insulin resistance, whereas overexpression of PAP leads to obesity. We show that viruses compete with host cells for limited lipids, inhibiting host growth as a consequence. In yeast cells with inactive PAP, total phospholipids, sterol esters, and free fatty acids accumulated to high levels at the expense of TAG. Altered lipid composition substantially promotes cell growth during viral replication. However, this simultaneously enhances genomic replication of brome mosaic virus (BMV), a well-studied model for examining positive-strand RNA viruses. These results suggest that PAP restricts virus replication by limiting phospholipid synthesis, which is required for viral replication. Our data strengthen the notion that viruses may take advantage of disrupted PAP or other nutrient homeostasis for infection.
Introduction
Positive-strand RNA viruses [(+)RNA viruses] are the largest of all virus classes and cause numerous important diseases in humans, animals, and plants. All of the well-studied (+)RNA viruses have been shown to remodel host intracellular membranes to build viral replication complexes (VRCs) for genomic replication [1–4]. Because cellular lipids are the major building blocks of membranes, their metabolism and/or composition are crucial for virus-induced membrane rearrangements [4–6].
Brome mosaic virus (BMV) is the type member of the family Bromoviridae and a representative member of the alphavirus-like superfamily [7]. BMV induces spherular VRCs at the perinuclear endoplasmic reticulum (nER) membrane in the yeast Saccharomyces cerevisiae and in barley cells [8–11]. BMV has three capped genomic RNAs and a subgenomic mRNA, RNA4. For viral replication, RNA1- and RNA2-encoded replication proteins 1a and 2a polymerase (2apol) are necessary and sufficient for BMV replication in barley and Nicotiana benthamiana [7,12,13] as well as in yeast [11]. With a central RNA-dependent RNA polymerase (RdRp) domain, 2apol serves as the replicase. In addition, the N-terminus of 2apol interacts with the C-terminal domain of 1a [14–16]. 1a has an N-terminal RNA capping domain that adds a cap to the 5’ end of viral RNAs [17–19] and a C-terminal ATPase/helicase-like domain that is required for translocating viral genomic RNAs into VRCs [20]. 1a localizes to the nER membrane, which is the nuclear membrane or nuclear envelop, where it invaginates the outer nER membrane into the ER lumen to form spherules that have an overall negative membrane curvature [11,21]. Spherules become VRCs when 2apol and viral genomic RNAs are recruited by 1a during viral replication [11]. Several properties of 1a are required for this process, including its membrane association domain, an amphipathic α-helix (1a amino acids 392–407) [22], and its ability to self-interact [23].
Lipids play crucial roles in BMV replication, similar to other (+)RNA viruses [5,6]. In yeast, an ~30% increase of accumulated total fatty acids (FAs) per cell was induced by the expression of 1a along with the formation of spherules [24]. A mild decrease in unsaturated FAs (UFAs) inhibited BMV RNA replication more than 20-fold [24,25]. It was further shown that the decreased UFAs particularly affected the membranes surrounding VRCs, indicating that the lipid environment of VRC membranes is different from the rest of the nER membrane [24,25]. BMV replication also requires host ACB1-encoded acyl-Coenzyme A (acyl-CoA) binding protein, which binds to long-chain fatty acyl-CoAs and is important in maintaining lipid homeostasis. In the ACB1 deletion mutant, BMV RNA replication is inhibited by more than 10-fold and spherules are smaller in size but greater in number than those in wild-type (wt) cells [26]. Enhanced accumulation of phosphatidylcholine (PC) is also associated with BMV replication sites [27]. In addition, cellular PC synthesis enzyme Cho2p (phosphatidylethanolamine (PE) methyltransferase) (Fig 1A) is recruited to BMV replication sites by 1a via a specific 1a-Cho2p interaction, suggesting an enhanced PC synthesis at the viral replication sites. As expected, deletion of CHO2 significantly inhibits BMV replication, raising the possibility of controlling the viral replication by blocking the 1a-mediated Cho2p recruitment [27].
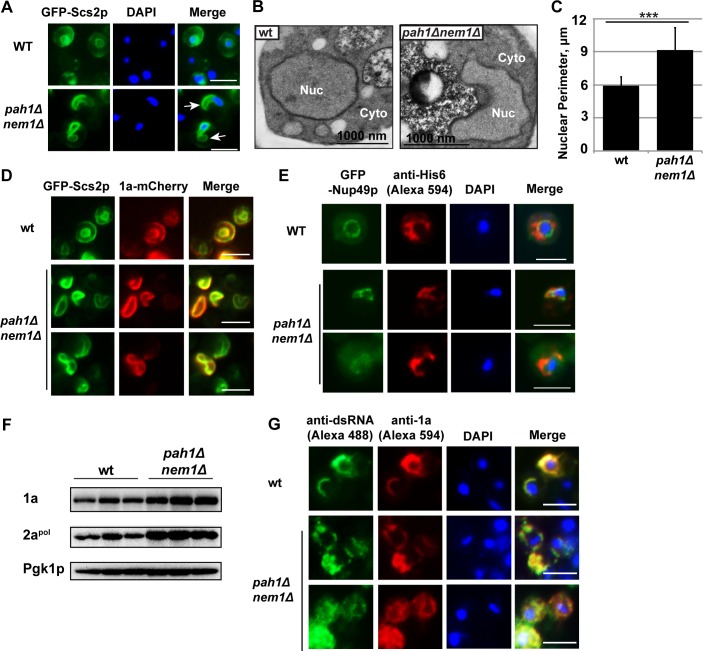
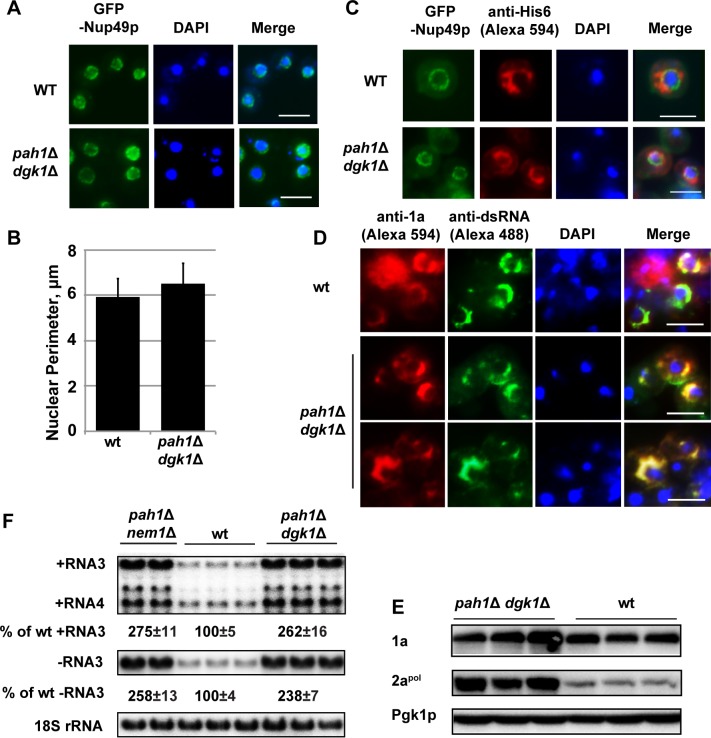
Fig 1. The inactivation or disruption of Pah1p promotes BMV genomic replication.
(A) Diagram of lipid metabolism in yeast. Key enzymes are shown. PA serves as a substrate for phospholipids and TAG. PA and Scs2p bind to and sequester Opi1p, keeping it from reaching to the nucleus, where Opi1p interacts with Ino2p and represses transcription of CHO2, OPI3 and other genes involved in phospholipid synthesis. Pah1p*** represents the hyperphosphorylated inactive Pah1p. (B) Accumulated BMV RNAs in wt and mutant cells with PAH1 deleted or Pah1p inactivated. Positive- and negative-strand viral RNAs were detected by using BMV RNA strand-specific probes. 18S rRNA was included as a control to eliminate loading variations. All experiments shown in the figure and in subsequent figures have been repeated multiple times and a representative figure is shown. (C) BMV replication in wt cells overexpressing wt or a defective mutant D177A of Dgk1p. (D) BMV replication in wt cells overexpressing PAH1, NEM1, SPO7, or SKI8. SKI8, a well-known antiviral gene, serves as a positive control.
Phosphatidate (PA) is a common precursor for both phospholipids and storage lipids. PA is produced de novo from glycerol-3-phosphate [28–30] and can be converted to CDP-diacylglycerol (CDP-DAG) [31–33], which is subsequently used to produce phospholipids, including PC, PE, phosphatidylinositol (PI), and phosphatidylserine (PS) (Fig 1A). PA can also be converted to diacylglycerol (DAG) by PAH1-encoded Pah1p, which is an Mg2+-dependent phosphatidate phosphatase, and further to TAG, the major storage lipid [34]. In yeast, PA also regulates the expression of lipid synthesis genes by sequestering a transcription repressor, Opi1p (overproduction of inositol1), at the nER membrane [35]. When PA levels are low, Opi1p is released from the nuclear membranes and translocated to the nucleus to repress transcriptions of many genes involved in phospholipid synthesis, including CHO2 and OPI3 (Fig 1A) [35,36].
Pah1p is highly regulated given its important roles in directing PA for the synthesis of storage lipids and thus, away from phospholipid synthesis [37–39]. Primarily localized in the cytosol as a hyperphosphorylated inactive form, Pah1p is dephosphorylated by a phosphatase complex that is composed of the catalytic subunit Nem1p (nuclear envelop morphology1) and the regulatory partner Spo7p (sporulation7) [40–42]. The Nem1p-Spo7p complex also recruits Pah1p to ER membranes where the active Pah1p is associated with membranes via an insertion of an amphipathic α-helix [43,44]. Both Nem1p and Spo7p are required for protein phosphatase activity and absence of either subunit inactivates the protein phosphatase activity of the complex, and thus, Pah1p PA phosphatase activity [42]. In PAH1 deletion mutant (pah1Δ) cells, total phospholipid levels increase by ~2-fold while TAG levels decrease significantly and in addition, the nER membrane expanded compared to that of wt cells [40,45]. It has been shown that in pah1Δ cells, the nER membrane always expands at the site close to nucleolus, the site of ribosome biogenesis, and the chromosome DNA-occupied area remain the same as that in wt cells [46]. Pah1p shares structural and functional similarities to human lipins (lipin1, 2, and 3) as well as to AtPah1 and AtPah2 of Arabidopsis thaliana in that LIPINs or AtPAHs can complement phenotypical defects in yeast pah1Δ cells, including the decreased TAG and expanded nER membrane [47–49].
In a previous large-scale screening of a yeast deletion array, it was found that deleting NEM1 or SPO7 significantly enhanced BMV replication [50]. In addition, deletion of PAH1 facilitates robust RNA replication of tomato bushy stunt virus (TBSV). TBSV normally replicates in peroxisomes but assemble their VRCs at expanded ER membranes in pah1Δ cells. In addition, TBSV VRCs in pah1Δ cells are more active than those in wt cells [51,52].
Here, we report that disruption of PAH1 promotes BMV replication and results in the formation of VRCs that are 2-fold more abundant in number compared to those in wt cells, suggesting that a group of (+)RNA viruses could take advantage of the inactivation of Pah1p to promote their replication. We further demonstrate that the enhanced BMV replication phenotype is not due to the extended nER membrane but due to the increase in total phospholipid levels. In addition, we show that deleting PAH1 also alleviates BMV-inhibited yeast cell growth. We conclude that Pah1p, by targeting lipid flux away from phospholipid synthesis, constrains both viral replication and cell growth during BMV replication.
Results
There is a direct correlation between the inactivation of Pah1p and enhanced BMV RNA replication in yeast
In a previous genome-wide screen of yeast deletion mutants in which BMV RNA replication was measured by the expression of a Renilla luciferase reporter, there was a dramatic increase in BMV replication in yeast strains that had either NEM1 or SPO7 deleted [50]. The pah1Δ mutant, however, was missing from the library when the screen was performed. Since both Nem1p and Spo7p are required for Pah1p activation, deleting NEM1, SPO7 or PAH1 causes similar phenotypes in yeast [40,53]. To validate results of the screen and to determine the possible role of Pah1p in BMV replication, we tested BMV replication in nem1Δ, spo7Δ, and pah1Δ single mutants, as well as the double mutants pah1Δ nem1Δ and nem1Δ spo7Δ by performing Northern hybridization with viral RNA strand-specific probes. As shown in Fig 1B, in the nem1Δ or spo7Δ mutants, both negative- and positive-strand RNA accumulation increased by approximately 2-fold compared to those in wt cells. However, no further increase of BMV RNA replication was observed when both NEM1 and SPO7 were deleted, agreeing well with the notion that each is necessary to activate Pah1p. Providing further support that Pah1p restricts BMV replication, deleting PAH1 enhanced BMV positive- and negative-strand RNA3 accumulation by about 3-fold compared to that in wt cells (Fig 1B). It should be noted, however, that cells in which both PAH1 and NEM1 were deleted consistently supported the highest levels of BMV genomic replication; thus, we used this double mutant in the majority of experiments described below (Fig 1B). These data indicated that a lack of, or inactivation of, Pah1p promoted BMV replication in yeast, most likely through the increased production of PA and thus, increased total phospholipids and the expanded nER membrane.
To strengthen the notion that increased PA levels in pah1Δ cells is a major contributor to the enhanced BMV replication, we tested whether BMV replication was affected by overexpressing DGK1. DGK1 encodes DAG kinase, which converts DAG to PA in yeast (Fig 1A). Similar to deleting PAH1, overexpressing DGK1 leads to a decrease in TAG accumulation and an increase in PA levels, resulting in an expanded nER membrane in yeast cells [54]. As expected, overexpression of DGK1 also enhanced BMV RNA replication to levels comparable to that in pah1Δ cells (Fig 1C). To confirm that Dgk1p enzymatic activity was required for the effect, we used a Dgk1p mutant, D177A, which lacks DAG kinase activity and whose overexpression does not extend the nER membrane [54]. Indeed, overexpression of D177A did not promote BMV RNA replication (Fig 1C), consistent with the notion that redirecting lipid synthesis from TAG to phospholipids could enhance BMV replication.
In contrast to the above deletion mutants, overexpression of PAH1 inhibited BMV replication ~2-fold (Fig 1D). Similar inhibition in BMV replication was also observed in yeast cells overexpressing NEM1 or SPO7. These effects were comparable to that of SKI8 (superkiller8), a well-known antiviral gene [50,55] (Fig 1D). Taken together, our results indicate that there is a positive correlation between the inactivation or disruption of Pah1p function and enhanced BMV replication levels in yeast, indicating that Pah1p is a limiting factor for BMV replication.
BMV 1a localizes to the extended nuclear membrane in cells lacking PAH1
A dramatically extended nER membrane is present in cells lacking NEM1, SPO7, and/or PAH1 [40,53] or when DGK1 is overexpressed [54]. Since BMV 1a invaginates the outer nER membrane into the lumen to form spherules, the extended nER membrane in these mutant cells may provide an expanded surface area for VRC formation and thus, promote BMV replication [11].
We first examined whether the extended nER membrane was present in pah1Δ nem1Δ cells in the absence of BMV components using epifluorescence microscopy and transmission electron microscopy (TEM). ER membranes, which were identified using a GFP-tagged ER resident protein Scs2p (suppressor of choline sensitivity2, GFP-Scs2p), were observed as two-ring structures in wt cells (Fig 2A). The larger outer ring is the peripheral ER membrane, which is underneath the plasma membrane in yeast. The smaller inner ring indicates the nER membrane, which surrounds the DAPI-stained, round-shaped nucleus. Like the misshapen nER membrane in nem1Δ, spo7Δ, and pah1Δ mutants, the nER membrane was extended in the pah1Δ nem1Δ mutant (Fig 2A). Agreeing well with previous report, the extended nuclear membrane was away from the DAPI-stained chromosome DNA area (Fig 2A) and has been shown to be close to the nucleolus [46]. Consistent with what was observed by epifluorescence microscopy, the strikingly proliferated nER membrane was also confirmed in pah1Δ nem1Δ cells using TEM (Fig 2B). To further characterize the extended nuclear membrane in pah1Δ nem1Δ cells, we measured the perimeter of nER membranes in wt and pah1Δ nem1Δ cells. While the nuclear membrane perimeter in wt cells was ~5.9 μm, it increased to approximately 9.2 μm in the mutant, a 55% increase that was statistically significant (Fig 2C).
Fig 2. BMV 1a and double-stranded RNA localize to the extended nuclear membrane in pah1Δ nem1Δ cells.
(A) Epifluorescence microscopic images of the extended nuclear membrane observed in pah1Δ nem1Δ cells. GFP tagged Scs2p, an ER membrane protein, represents ER membranes. Nuclei were stained with DAPI. White arrows indicate the extended nER, which is away from DAPI-stained chromosome DNA (blue). (Scale bar, 5μm) (B) Morphology of the nuclear membrane in wt and pah1Δ nem1Δ cells under transmission electron microscope. (C) The perimeters of nuclei in wt and pah1Δ nem1Δ cells. ***, p < 0.001 (ANOVA single factor test). (D) Epifluorescence microscopic images showing the localization of 1a in wt and pah1Δ nem1Δ cells. The localization of 1a is indicated by mCherry, which is fused to the C-terminus of 1a. The yellow color in merged images represents the co-localization of 1a and Scs2p signals. (Scale bar, 5μm) (E) Immunofluorescence microscopic images showing the localization of 1a-His6 in wt and pah1Δ nem1Δ cells. Localization of 1a-His6 was detected using a polyclonal anti-His6 antibody and followed by a secondary anti-rabbit antibody conjugated to Alexa Fluor 594. GFP tagged Nup49p, a component of nuclear pore complexes, indicates the nuclear membrane. Nuclei were stained with DAPI. (Scale bar, 5μm) (F) Accumulated BMV 1a and 2apol in wt and pah1Δ nem1Δ cells. Total proteins were extracted from the same numbers of BMV replicating-yeast cells and analyzed by Western blotting using antibodies specific to 1a and 2apol. Pgk1p serves as a loading control. (G) Immunofluorescence microscopic images of the co-localization of 1a and dsRNA signals in wt and pah1Δ nem1Δ cells. BMV 1a was detected with anti-1a antiserum followed by a secondary anti-rabbit antibody conjugated to Alexa Fluor 594. dsRNA was detected by a dsRNA-specific monoclonal antibody (J2) and a secondary anti-mouse antibody conjugated to Alexa Fluor 488. The yellow color in merged images represents the co-localization of 1a and dsRNA signals. Note 1a and dsRNA appear as a half ring in wt but not in mutant cells. Nuclei were stained with DAPI. (Scale bar, 5μm).
The expression of 1a, without other BMV components, induces spherule formation in the nER membrane of yeast [11]. We tested whether the localization of 1a could be affected in pah1Δ nem1Δ cells. To visualize 1a, we first used an mCherry-tagged 1a, which primarily localized to the nER and partially localized to the peripheral ER in wt cells (Fig 2D, upper panels) [56]. In mutant cells, 1a-mCherry dominantly co-localized with GFP-Scs2p at the extended nER membrane (Fig 2D). To further confirm that 1a was associated with the nER membrane, we used a GFP tagged nuclear pore complex component, Nup49p (Nuclear Pore 49, GFP-Nup49p) [57]. His6-tagged 1a, when expressed alone, co-localized with the GFP-Nup49p-labeled nER membrane in both wt and pah1Δ nem1Δ cells as determined by immunofluorescence microscopy (Fig 2E).
We next checked the accumulation and localization of BMV replication proteins during BMV replication. Both 1a and 2apol accumulated at higher levels in pah1Δ nem1Δ cells compared to those in wt cells based on Western blotting using anti-1a or 2apol antibodies (Fig 2F). The increased levels of 1a was consistent with the localization of 1a-mCherry and 1a-His6 along the expanded nER membrane (Fig 2D and 2E). To determine the site of BMV replication in pah1Δ nem1Δ cells, we tested the distribution of double-stranded RNA (dsRNA) using a dsRNA-specific monoclonal antibody J2. As a replication intermediate, dsRNA is considered a hallmark of viral VRCs [58] and the J2 antibody has been commonly used to confirm localization of viral replication sites [58,59]. In wt cells, dsRNA signal co-localized nicely with that of 1a, as determined by immunofluorescence microscopy (Fig 2G). Moreover, both signals showed a half-ring structure surrounding the nucleus. However, at least two alterations were noticed in the majority of pah1Δ nem1Δ cells (Fig 2G): 1) Both dsRNA and 1a signals were not detected as a half-ring but localized at the extended nER membrane, and 2) Both signals extended away from the nucleus in many cells.
Substantially increased numbers of viral replication complexes are formed in pah1Δ nem1Δ cells compared to wild-type cells
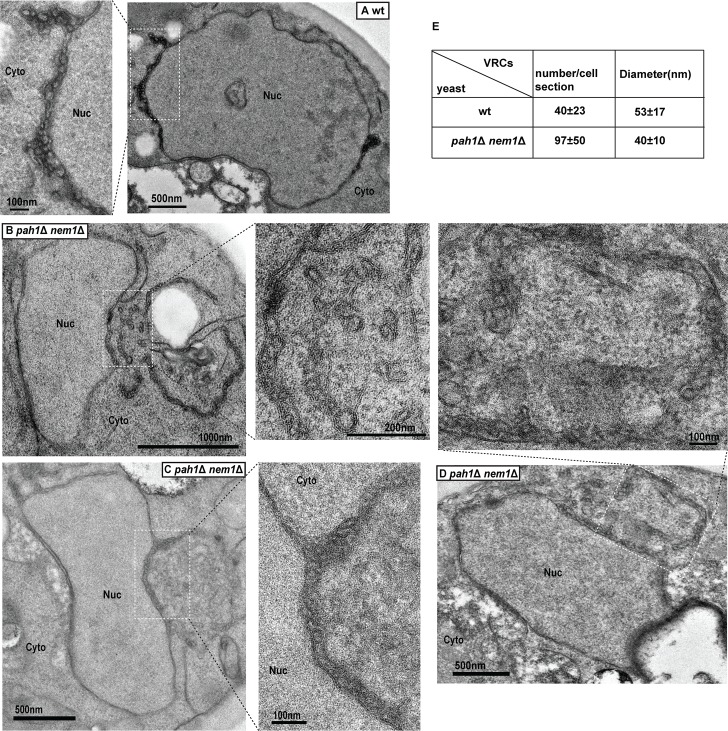
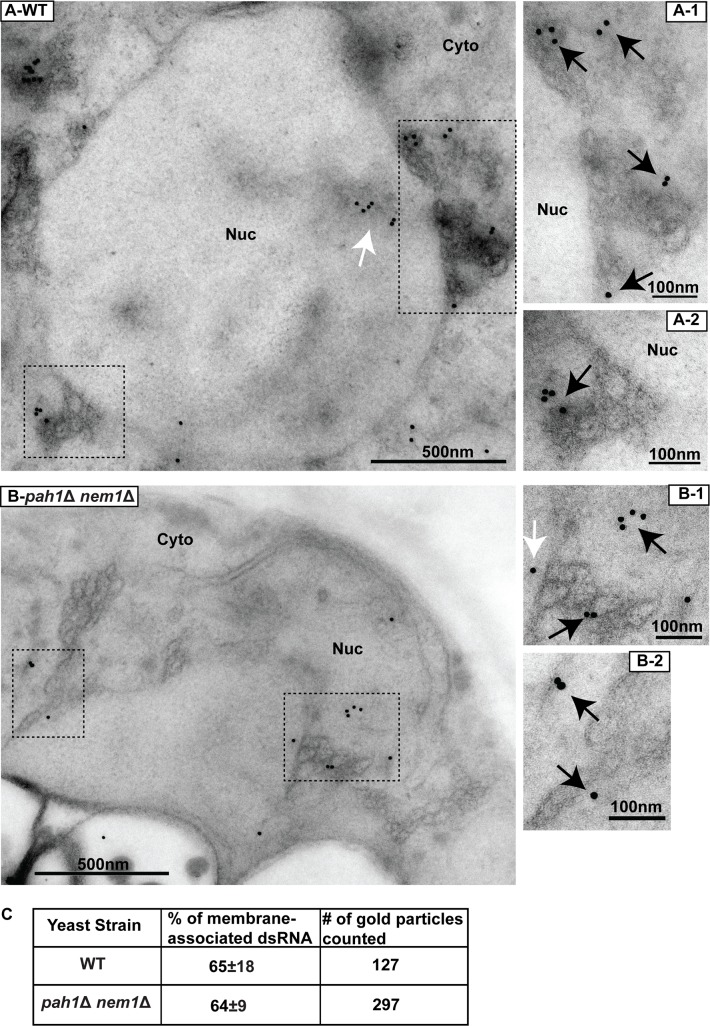
To determine whether VRC assembly was affected in the pah1Δ nem1Δ mutant, we checked the morphology of spherular VRCs using TEM in both wt and mutant cells during BMV replication. In wt cells, viral spherular VRCs were found in the lumen of the nER membrane. In wt cells of the RS453 background, the average number of spherular VRCs per cell section was approximately 40 (40 ± 3, [mean±SD]) with an average diameter of ~53 nm (53 ± 17 nm, Fig 3A). In BMV-replicating pah1Δ nem1Δ cells, an extended nER membrane was clearly observed (Fig 3B and 3C), similar to what was seen in mutant cells without BMV components (Fig 2C). We found VRCs that were 24% smaller in diameter (40 ± 10 nm, Fig 3B–3E) but about 2.4-fold more abundant in number (97 ± 50, Fig 3B–3E) compared to those in wt cells. These spherular VRCs were generated from membranes connected to the nER membrane. The increased numbers of VRCs is consistent with higher accumulation of both BMV 1a and 2apol (Fig 2F). To confirm that these smaller VRCs were active in viral replication, we performed immunogold electron microscopy analysis (IEM) using the J2 antibody [59]. About 65% of the gold particles were associated with viral VRCs in BMV-replicating wt cells (65%±18, n = 127) (Fig 4A). A similar ratio was observed (64%±9, n = 297) in pah1Δ nem1Δ cells (Fig 4B). We have similarly detected BMV 1a in VRCs in wt and pah1Δ nem1Δ cells with similar ratios, 71% (n = 110) and 75% (n = 206), respectively (S1 Fig). Given the fact that spherular VRCs are the site of RNA synthesis and that there was an increase in the accumulation of both positive- and negative-strand RNA in the mutant cells (Fig 1B), these results suggest that the smaller spherular VRCs in mutant cells support efficient viral RNA synthesis.
Fig 3. Number of spherular VRCs is substantially increased in pah1Δ nem1Δ cells.
Electron micrographs of spherular VRCs formed in wt (A) and pah1Δ nem1Δ cells (B-D) are shown. Micrographs at a higher magnification of boxed areas are also shown. Note spherular VRCs are in membranes extended from the nER membrane. (E) Average number of VRCs per cell section and diameter of VRCs in wt and pah1Δ nem1Δ cells. Nuc, nucleus; Cyto, cytoplasm.
Fig 4. BMV replication sites are localized at the expanded nuclear ER membranes in pah1Δ nem1Δ cells.
Immunogold labeling of dsRNA in wt (A) and pah1Δ nem1Δ (B) cells during BMV replication. Images at a higher magnification of the boxed areas (A-1, A-2, B-1, and B-2) are also shown. Black arrows indicate the gold particles that were associated with membranes or spherular VRCs. White arrows indicate the gold particles that were not associated with membranes. (C) Number of total particles counted and the percentage of particles that were localized to the nER membrane and spherular structures among total counted particles in wt and pah1Δ nem1Δ cells. Particles within 20 nm of the nER membrane or spherular VRCs, the distance spanned by primary and secondary antibodies, were counted as positive [98]. Nuc, nucleus; Cyto, cytoplasm.
In addition to smaller spherular VRCs, we also observed more dramatic membrane rearrangements in pah1Δ nem1Δ cells replicating BMV, usually multiple layers of bilayer membrane surrounding the nucleus (S2 Fig). These layers of membrane are likely generated during BMV replication because such structures have not been previously reported and were not observed in the absence of BMV replication (Fig 2B). However, the nature of and the relationship of the layers to viral replication is currently unclear and is under further investigation.
Total phospholipid levels increase in cells lacking PAH1 in the presence of BMV replication
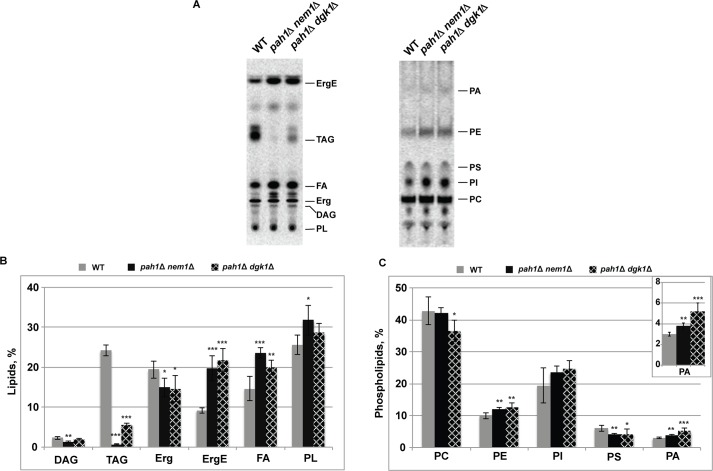
As reported previously, levels of total phospholipids, ergosterol esters (ErgE) and free FAs increased at the expense of TAG in pah1Δ mutant cells [54]. To confirm that similar altered lipid composition was still present in pah1Δ nem1Δ cells in the presence of BMV replication, we measured lipids of wt and mutant cells grown in the presence of [2-14C] acetate to radiolabel neutral lipids and phospholipids (Fig 5A). The mol percentages of each measured lipid was reported in Fig 5B and 5C. The mol percentage of both DAG (p<0.01) and TAG (p<0.001) decreased significantly while total phospholipid levels increased (p<0.05) in pah1Δ nem1Δ mutant cells compared to those in wt cells (Fig 5B). Moreover, there was a significant decrease in ergosterol levels but a substantial increase in ErgE and free FAs levels in the presence of BMV (Fig 5B). The similar compositional changes of all aforementioned lipids, in the absence of BMV, have been previously reported [54], indicating that BMV did not alter the trend of lipid compositional changes in mutant cells.
Fig 5. Increased total phospholipid levels in yeast cells lacking PAH1 in the presence of BMV replication.
WT, pah1Δ nem1Δ and pah1Δ dgk1Δ cells with BMV components were grown at 30°C in SC-Ura-Leu medium in the presence of galactose as the carbon source and [2-14C] acetate (1μCi/ml). Lipids were extracted, separated by the one-dimensional thin-layer chromatography system for phospholipids or neutral lipids, visualized by phosphoimaging and analyzed by ImageQuant software. (A) Chromatograms of neutral lipid composition and total phospholipids (left), and phospholipid composition (right). The chromatograms shown in the panel are representative of three independent experiments. (B) and (C) The mol percentages shown for the individual neutral lipids and phospholipids were normalized to the total 14C-labeled chloroform fraction, which also contained the unidentified neutral lipids and phospholipids shown in (A). Each data point represents the average of three experiments ± S.D.. *, P<0.05; **, P<0.01; ***, p < 0.001 (based on single factor ANOVA test).
The phospholipid composition was also altered in pah1Δ nem1Δ cells during BMV replication. Levels of PA (p<0.01) and PE (p<0.01) increased while there was a decrease in PS levels (p<0.01) in the mutant compared to wt (Fig 5C). However, there were no statistically significant changes in PC or PI levels (Fig 5C). Thus, our data agrees with a previous report [54], which showed that total phospholipid levels, PA in particular, increase upon deletion of PAH1, even in the presence of BMV replication.
Extension of the nuclear membrane is not the major contributor to the increase in BMV genomic replication in cells lacking PAH1
In pah1Δ cells, several alterations may account for the enhanced BMV replication: 1) Since BMV assembles its VRCs at the nER membrane, the extended nER membrane will provide a larger surface area for the formation of BMV VRCs; 2) Since phospholipids are major components of membranes, the increased total phospholipid levels may provide building materials to form more VRCs. To determine which or both of these are the major contributor(s), we took advantage of the pah1Δ dgk1Δ mutant, in which both PAH1 and DGK1 are deleted. It was reported that the mutant has similar lipid compositional changes as those in the pah1Δ mutant but the nER membrane is normal [54]. We first checked the morphology of GFP-Nup49p-tracked nER membrane and confirmed that the nER membrane was indeed round shaped (Fig 6A) and that the size of nuclei in pah1Δ dgk1Δ cells was similar to that of wt cells (Fig 6B). The average perimeter of the nER membrane in pah1Δ dgk1Δ cells were 6.5 μm (n = 136), a 10% increase over that of wt cells at 5.9 μm (Fig 6B). However, this increase is not statistically significant. In addition, we confirmed that 1a-His6 co-localized with GFP-Nup49p in the nER membrane (Fig 6C). Consistent with the localization of 1a-His6, 1a and dsRNA were all localized at the round-shaped nER membrane during BMV replication in pah1Δ dgk1Δ cells (Fig 6D). In addition, as seen in wt cells, both 1a and dsRNA localized as a half-ring in pah1Δ dgk1Δ cells. Surprisingly, 1a and 2apol still accumulated at much higher levels compared to wt cells, even the nER membrane was not extended (Fig 6E). In addition, BMV replication increased up to ~2.5-fold, similar to that in the pah1Δ nem1Δ mutant (Fig 6F). We also observed smaller but many more abundant spherular VRCs in pah1Δ dgk1Δ cells during BMV replication compared to those in wt cells (Fig 7A and 7B). The average size of spherular VRCs was 42 ± 9 nm and the number of VRCs was about 79 ± 44 per cell section (Fig 7C). Lipid analysis indicated that the pah1Δ dgk1Δ and pah1Δ nem1Δ mutants shared similar trends in lipid compositional changes, including decreased DAG, TAG, and Erg but increased ErgE, free FAs, and total phospholipids (Fig 5B). These data indicate that the altered lipid composition, but not the extended nuclear membrane, is responsible for the enhanced BMV genomic replication in cells with disrupted Pah1p activity.
Fig 6. The extended nuclear membrane is not responsible for the enhanced BMV replication in cells that lack PAH1.
(A) Epifluorescence microscopic images of the round-shaped nuclear membrane observed in wt and pah1Δ dgk1Δ cells. GFP-Nup49p was used as a nuclear membrane marker. Nuclei were stained with DAPI. (Scale bar, 5μm) (B) Nuclei perimeter measurements in wt and pah1Δ dgk1Δ cells. (C) BMV 1a localization in wt and pah1Δ dgk1Δ cells. GFP-Nup49p was used as a nuclear membrane marker. (Scale bar, 5μm) Note the wt cell (upper panel) is the same one in Fig 2E (upper panel). (D) Immunofluorescence microscopic images showing localization of dsRNA and 1a in wt and pah1Δ dgk1Δ cells. Note 1a and dsRNA appear as a half-ring structure in both wt and mutant cells. (Scale bar, 5μm) (E) Accumulated BMV 1a and 2apol in wt and pah1Δ dgk1Δ cells. Protein extraction and Western blotting were done as in Fig 2. (F) BMV replication in wt, pah1Δ nem1Δ, and pah1Δ dgk1Δ cells. Viral RNAs and 18S rRNA were detected as in Fig 1.
Fig 7. BMV replication complexes are associated with the perinuclear ER membrane in pah1Δ dgk1Δ cells.
(A) and (B) Representative TEM images of spherular VRCs formed in pah1Δ dgk1Δ cells. Micrographs at a higher magnification of boxed areas (A-1, A-2, B-1, and B-2) are also shown. (C) Number and diameter of VRCs in pah1Δ dgk1Δ cells are shown. Nuc, nucleus; Cyto, cytoplasm.
The contribution of enhanced phospholipids in the promoted BMV replication in cells lacking PAH1
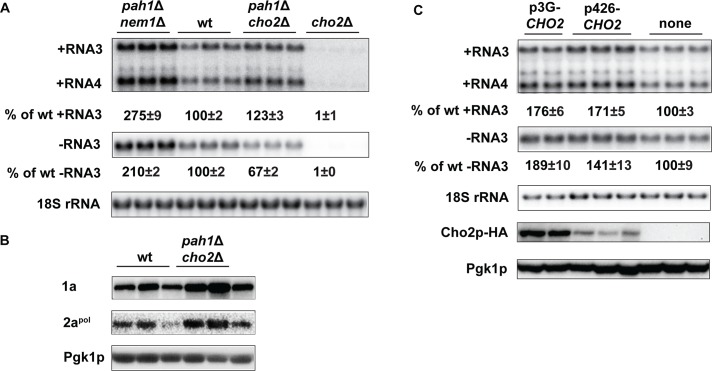
We further tested whether increased total phospholipid levels, among lipid compositional changes, could play an important role in promoting VRC formation and BMV replication because phospholipids are major membrane components and both ErgE and free FAs are not present in membranes. We have previously shown that a pool of PC is synthesized in the site of viral replication by recruiting host Cho2p [27], which is involved in converting PE to PC (Fig 1A) [60]. We first deleted CHO2 and found that BMV replication was hardly detectable in the cho2Δ mutant in the RS453 background (Fig 8A). When CHO2 and PAH1 were simultaneously deleted, positive-strand RNA3 accumulation increased by 23% but negative-strand RNA3 levels decreased by 33% compared to those in wt cells. However, comparing to that in the pah1Δ nem1Δ mutant, BMV replication significantly reduced (Fig 8A). Of note, 1a and 2apol proteins increased in the pah1Δ cho2Δ mutant background compared to those in wt cells (Fig 8B).
Fig 8. The active synthesis of phosphotidylcholine is required for the enhanced BMV replication in pah1Δ cells.
(A) BMV replication in wt, cho2Δ, pah1Δ cho2Δ and pah1Δ nem1Δ cells. (B) BMV 1a and 2apol accumulation in wt and pah1Δ cho2Δ cells. Protein extraction and Western blotting were done as in Fig 2F. (C) Over-expression of CHO2 promotes BMV replication in wt cells. Low or high levels of Cho2p-HA was expressed from p426-CHO2 (a high-copy-number plasmid) or p3G-CHO2 (a low-copy-number plasmid) under the control of the CHO2 or GAL1 promoter, respectively. The bottom panel shows accumulated Cho2p that was expressed from different vectors. Pgk1p serves as a loading control.
As a result of the increase in PA levels in the pah1Δ mutant, transcription of phospholipid synthesis genes increases due to the sequestration of the transcription repressor Opi1p (Fig 1A) [36]. To simulate those conditions, we tested whether enhanced CHO2 expression would promote BMV replication. To achieve different levels of overexpression in wt cells, CHO2 was expressed from a high-copy-number plasmid under its endogenous promoter (p426-CHO2) or from the strong GAL1 promoter (p3G-CHO2), respectively (Fig 8C). An increase of 40% or 90% of negative-strand RNA3 over that in wt cells was associated with different levels of overexpressed Cho2p (Fig 8C). An approximate 70% increase in positive-strand RNA was also noticed when CHO2 was overexpressed (Fig 8C). However, these increases in positive- and negative-strand RNA synthesis was not as significant as that in pah1Δ cells, suggesting other phospholipids besides PC contribute to the enhanced BMV replication phenotype in pah1Δ cells (Fig 1B).
Deleting PAH1 improves yeast cell growth during BMV replication
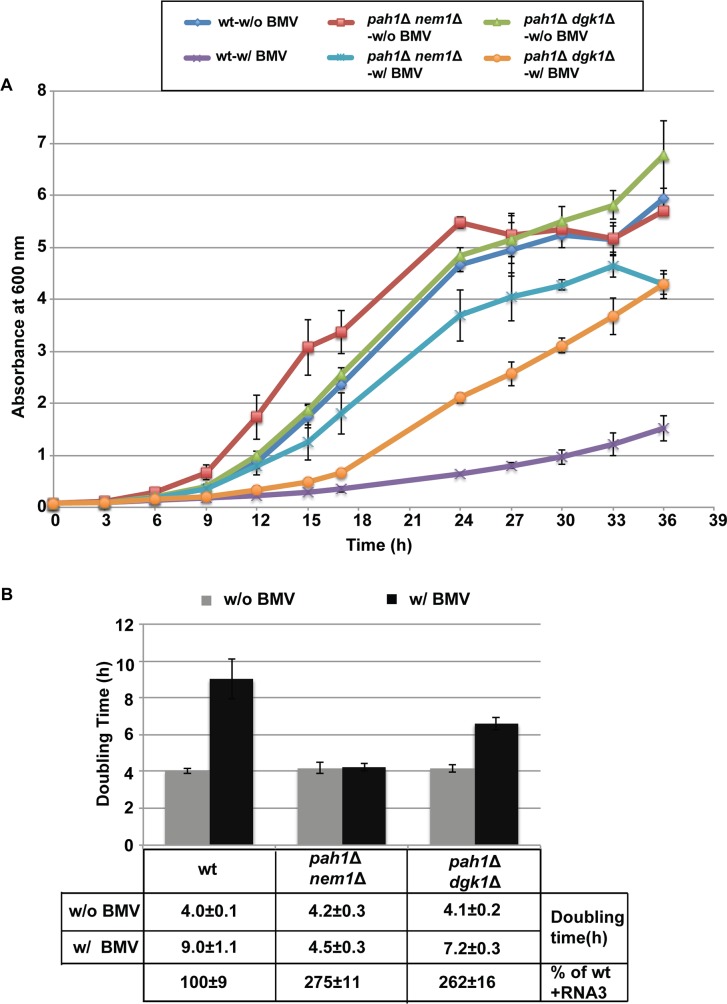
Phospholipids are the major components of cellular membranes [60] and are utilized by various viruses for infection [5,27,61–63]. Viruses compete with their hosts for limited resources and, as a direct result, viral infections usually affect cell growth. We measured cell growth and calculated doubling times (based on growth during the exponential stage) of wt, pah1Δ nem1Δ, and pah1Δ dgk1Δ cells in the absence or presence of BMV in the galactose medium, which is to induce BMV replication (Fig 9A). BMV replication substantially slowed down the growth of wt cells. In wt cells, the doubling time increased from ~4 hours/generation in the absence of BMV to ~9 hours/generation in the presence of BMV, an approximately 2-fold increase (Fig 9B). In addition, the cell density of the culture expressing BMV components never reached to that of cells without BMV. Deleting PAH1 profoundly improved the growth of cells with BMV replication (Fig 9). The doubling times of pah1Δ nem1Δ and pah1Δ dgk1Δ mutants in the presence of BMV replication were approximately 4.5 and 7.2 hours/generation, respectively. It should be noted that these mutant cells grew at the same rate as wt cells in the absence of BMV components (Fig 9), indicating that the growth differences between wt cells and the above mutants are directly related to BMV replication.
Fig 9. Deleting PAH1 alleviates BMV-repressed host cell growth.
Cells of wt or mutants were grown in media using galactose as the carbon source in the absence or presence of BMV replication. (A) Growth curves of wt, pah1Δ nem1Δ, and pah1Δ dgk1Δ cells in the absence or presence of BMV replication in 36 hours. (B) Doubling time of yeast strains in the absence or presence of BMV replication during exponential phase. Doubling time was calculated using the following equation: Doubling time = [hours cells grown * Ln(2)]/[Ln(A600 nm at the end / A600 nm at the start)].
Expression of plant PAH1 orthologs inhibits BMV genomic replication in yeast and Nicotiana benthamiana plants
The PAP enzyme is present in yeast, plants and humans [64]. The two PAH1 orthologs in Arabidopsis thaliana (AtPAH1 and AtPAH2) could complement the phenotypical defects in pah1Δ cells [47–49] even though Arabidopsis and yeast Pah proteins share only ~14% identity at the protein level (Fig 10A). We have additionally identified five putative PAH genes in the genome of Nicotiana benthamiana based on the sequence similarity to Arabidopsis AtPAH1 and AtPAH2: NbPAH1A (Niben101Scf01009g01015.1), NbPAH1B (Niben101Scf05306g01007.1), NbPAH1C (Niben101Scf07223g03002.1), NbPAH2A (Niben101Scf05628g01019.1), and NbPAH 2B (Niben101Scf08200g05005.1). They can be classified into two clades, NbPAH1A, 1B, and 1C as one clade and NbPAH2A and 2B as the other one, based on their sequence similarity to AtPAH1 and AtPAH2 and among themselves (Fig 10A). To test the role of plant PAHs in BMV genomic replication, we expressed NbPAH1A, NbPAH2A, AtPAH1 or AtPAH2 in yeast cells to test whether their expression could inhibit BMV replication in a similar manner to that of yeast PAH1 (Fig 1D). All genes were expressed from a high-copy-number plasmid under the control of the GAL1 promoter [49]. Like yeast PAH1, the expression of plant orthologs inhibited BMV replication by ~40–50% (Fig 10B).
Fig 10. Expression of plant PAH1 homologs inhibits BMV replication in yeast and Nicotiana benthamiana.
(A) Homology analysis and phylogenetic tree of Pah1 proteins from yeast (ScPah1p), Arabidopsis (AtPah1 and AtPah2), and N. benthamiana (NbPah1A, 1B, 1C, 2A, and 2B). (B) BMV replication in wt yeast cells expressing ScPAH1, AtPAH1, AtPAH2, NbPAH1A or NbPAH2A. Positive-strand viral RNAs as well as 18S rRNA were detected as in Fig 1. Genome replications of BMV (C), TMV (D), or TRV (E) in N. benthamiana leaves expressing AtPAH2, NbPAH1A or NbPAH2A. BMV, TMV, and TRV were launched by agroinfiltration in N. benthamiana leaves 2 days after agroinfiltration to express the plant PAH1 orthologs. Virus-infected leaves were harvested 3 days after agroinfiltration. RNA was extracted and viral positive-strand RNA was detected by using virus strand-specific probes as in Fig 1.
We next tested how BMV genome replication was affected when plant PAH1 orthologs were highly expressed in N. benthamiana, which is a systemic host for BMV [13,65,66] and serves as a universal host for plant viruses [67]. AtPAH2, NbPAH1A and NbPAH2A were expressed from an enhanced cauliflower mosaic virus (CaMV) 35S promoter by agroinfiltration. BMV genome replication was inhibited by 40–50% based on the accumulation of positive-strand RNA3 when AtPAH2 or NbPAH2A was expressed. However, the expression of NbPAH1A only inhibited BMV replication by ~25%, suggesting that different plant PAH1 orthologs may play different roles in plants (Fig 10C).
To determine whether the inhibition of viral replication by plant PAH1 orthologs was specific to replication of BMV or if it was a general effect on other plant (+)RNA viruses, we also tested tobacco mosaic virus (TMV) and tobacco rattle virus (TRV). TMV replicates in association with ER membranes while TRV replicates on mitochondrial membranes [68,69]. We previously included both viruses and showed that a dominant negative mutant of AtSNF7-2 (sucrose nonfermenting7) did not affect the replication of TMV and TRV but specifically affected BMV [70]. Here we found that TMV genome replication was inhibited by ~50% when AtPAH2 or NbPAH2A was expressed in N. benthamiana. However, TMV replication was only slightly inhibited by the NbPAH1A expression (Fig 10D), which similarly inhibited BMV replication at a lesser degree compared to AtPAH2 and NbPAH2A (Fig 10C). On the contrary, based on two genomic RNAs, RNA1 and 2, TRV genome replication was not significantly affected by any of plant orthologs in N. benthamiana (Fig 10E). However, the accumulation of its subgenomic RNA1a levels were lower than that in untreated plants (Fig 10E). It is unclear why the accumulation of different TRV RNAs was differently affected by the overexpression of NbPAHs.
Discussion
We report here that the host enzyme phosphatidic acid phosphohydrolase restricts BMV RNA replication by limiting the phospholipid synthesis and that BMV takes advantage of altered lipid composition, including the increased total phospholipids, in yeast cells lacking PAH1 to assemble many more VRCs and substantially promote its genomic replication. Although deleting PAH1 leads to several phenotypes that could facilitate BMV replication, our data suggest that the increased levels of total phospholipids but not the proliferated nER membrane is the primary contributor (Figs 5 and 6). The rise in levels of total phospholipids in cells lacking PAH1, possibly with other altered lipids, also significantly improved cell growth during viral replication. It has been reported that in pah1Δ cells TBSV assembles its VRCs and replicates robustly in the extended ER membranes [52], but the improved replicase activity is primarily responsible for the enhanced TBSV replication [51]. Thus, our work complements and expands the current understanding of Pah1p’s role in balancing cellular phospholipid and storage lipid synthesis as well as in replication of various viruses.
Enhanced levels of total phospholipids, not the extended nuclear membrane, are primarily responsible for the increased BMV replication in cells that lack PAH1
Although both BMV and TBSV replicate at much higher levels in pah1Δ cells compared to wt cells, the mechanisms by which each virus takes advantage of PAH1 deletion to improve its replication are different. Under normal conditions TBSV forms spherular VRCs in peroxisomes, however, it preferentially assembles VRCs in association with extended ER membranes in pah1Δ nem1Δ cells [51,52]. It is not specified whether more VRCs are formed in pah1Δ nem1Δ cells [52], but it is clear that TBSV VRCs isolated from the mutant cells are more efficient in supporting viral RNA synthesis in vitro than those from wt cells [51]. Our TEM data showed that in pah1Δ nem1Δ cells, viral spherules were present in the extended nER membrane and were about 2.4-fold more abundant in number than those in wt cells (Fig 3B–3D). Despite the fact that the nuclear membrane was normal in pah1Δ dgk1Δ cells (Figs 6 and 7) [54], BMV replication levels and the number of VRCs were similar in the pah1Δ dgk1Δ and pah1Δ nem1Δ mutants (Figs 6F and 7), indicating that the expanded nER membrane is not the major factor in promoting the VRC formation and viral genomic replication.
Two lines of evidence support that increased total phospholipid levels played an important role in the enhanced BMV replication in pah1Δ cells: (1) Reducing PC synthesis by deleting CHO2 in the pah1Δ mutant diminished the substantially enhanced BMV replication (Fig 8A); (2) Overexpressing CHO2 in wt cells enhanced BMV replication (Fig 8C). However, the enhancement in BMV genomic replication was not as significant as that in pah1Δ cells because total phospholipids, not just PC, increased in pah1Δ cells.
There are increases in three major lipids in cells lacking PAH1, either in the absence [54] or presence of BMV (Fig 5B), total phospholipids, ErgE, and free FAs. We focused on the roles of increased total phospholipids in this work because ErgE is not present in membranes and its involvement in genomic replication of any viruses has not been reported. Although it was reported previously that the expression of BMV 1a enhanced the accumulation of total FAs by 33% per yeast cell [24], it should point out that what we measured here was free FAs, which were not incorporated into phospholipids and not present in membranes. How are free FAs involved in BMV replication is not clear. Nevertheless, it merits further investigation whether ErgE or free FAs is involved in BMV replication. Although phospholipids are major building blocks of membranes and our work suggested that increased total phospholipids play an important role in the enhanced BMV replication (Fig 8), the possible contribution of other significantly altered lipids, such as ErgE and free FAs, cannot be totally ruled out.
Deleting PAH1 facilitates both BMV replication and host cell growth
Lipid-containing cellular membranes are the sites where (+)RNA virus replication invariably takes place, although different viruses exploit specific organelle membranes and require different lipid microenvironment for their replication [1,4–6]. For example, TBSV requires a PE-enriched microenvironment [61,71]. Phosphatidylinostol-4-phosphate (PI4P) is produced in VRCs by hepatitis C virus (HCV)-recruited phosphatidylinositol-4-kinase III α (PI4KIIIα) [72,73] or Coxsackievirus B3-engaged PI4KIIIβ [74] either for the assembly or proper function of the VRCs, respectively. Our prior work showed that PC content is enriched at the viral replication sites of a number of (+)RNA viruses, including BMV, HCV, and poliovirus [27].
It has recently been reported that pah1Δ cells are susceptible to abiotic stresses and have a short chronological life span [75]. Our data showed that deleting PAH1 substantially improved host cell growth during viral replication. Yeast mutants that lack PAH1, either by itself or in combination with a second mutation, divided at a faster rate than wt cells during BMV replication (Fig 9). It should be noted that the mutant cells divided at a similar rate to wt cells in the absence of BMV replication (Fig 9). Both pah1Δ nem1Δ and pah1Δ dgk1Δ mutants grew faster than wt, suggesting that both of the increased viral replication and host cell growth were likely due to changes in lipid composition, possibly to the increase in total phospholipid levels. These results are consistent with the notion that BMV competes for the limited intracellular phospholipid resources with host cells, and that increasing total phospholipid levels could satisfy the requirements for both viral replication and host cell growth. As such, Pah1p serves as a limiting factor for BMV and possibly other (+)RNA viruses by directing lipid synthesis away from phospholipid synthesis, via converting PA to storage lipids. It is also possible that Pah1p promotes storage lipid synthesis at the onset of viral replication as a host reaction to stresses imposed by viral infection and in turn, limits cell growth.
Possible roles of PA in regulating nuclear membrane morphology and viral replication
In eukaryotic cells, besides serving as a key intermediate in lipid synthesis, PA is involved in multiple biological processes as a signaling molecule, such as cell growth and proliferation, secretion, endocytosis, and vesicular trafficking in mammalian cells [76–79] as well as responses to biotic and abiotic stress and seed germination in plants [78–83].
Increased PA may be involved in viral replication through several nonexclusive mechanisms. One is through the extension of the nER membrane, which could provide more room for VRC assembly, such as the substantially enhanced replication of TBSV and related viruses [51]. Another possible mechanism is that higher PA levels may recruit PA-dependent effectors. As a signaling lipid, PA executes its function by binding to effector proteins and recruiting them to a specific membrane [79]. Because such binding is dependent on the concentration of PA in the bilayer, higher levels of PA in the nER membrane may recruit its effectors more efficiently [79]. Some of these effectors may play positive, yet unclear, roles in (+)RNA virus replication. This is supported by the enhanced BMV replication in the pah1Δ dgk1Δ mutant (Fig 6), which has wt nER membrane but enhanced levels of total phospholipids including a high level of PA [54]. A third possible option is that the enhanced accumulation of PA and other phospholipids may affect protein conformation and stability. This is supported by increased accumulations of 1a and 2apol in both pah1Δ nem1Δ and pah1Δ dgk1Δ mutants (Figs 2F and 6E). It is also possible that PA may play a direct role in the formation of VRCs because the incorporation of PA, a cone-shaped lipid, promotes the formation of negative curvature [84–86]. BMV spherules are formed by invaginating the outer nER membrane away from the cytoplasm, thus inducing a negative curvature. Higher PA levels may facilitate the formation of viral spherules, which may explain significantly increased numbers of viral VRCs formed in pah1Δ nem1Δ (Fig 3) and pah1Δ dgk1Δ cells (Fig 7).
Besides de novo synthesis, PA can be produced by phospholipase D (PLD)-catalyzed removal of the choline head group from PC. PLD-generated PA plays an important role in supporting replication of plant (+)RNA viruses [87]. The replication protein p27 of red clover necrotic mosaic virus (RCNMV) in the Tombusviridae family binds to PA directly. Knocking down the expression or inactivation of PLD severely inhibited RCNMV replication [87]. It should be noted that inhibition of PLD activity by addition of n-butanol in tobacco protoplasts also inhibited BMV replication, indicating an important role of PA in the replication of a group of plant (+)RNA viruses [87]. Our data in yeast agree with the important role of PA in BMV replication in plants, although the sources of the increased PA are different.
In summary, our work suggests that altered lipid composition, likely through the enhanced total phospholipids, is the major factor not only for promoting BMV genomic replication but also for alleviating the virus-repressed cell growth in cells lacking Pah1p. Our data complement and extend prior findings on the role of PA in lipid metabolism and virus infections.
Materials and methods
Yeast strains and growth condition
All yeast strains used in this study are listed in Table 1 and were derived from the strain RS453 (MATa ade2-1, his3-11, 15 leu2-3, ura3-52, 112 trp1-1). The spo7Δ mutant was generated by replacing SPO7 with a HIS3MAX6 cassette. The pah1Δ nem1Δ mutant was made by replacing NEM1 with a KanMAX4 cassette in the pah1::TRP1 background. In the majority of experiments presented, the pah1Δ nem1Δ mutant was used.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Ref./source |
|---|---|---|
| RS453 | MATa ade2-1, his3-11, ura3-52, 15 leu2-3, 112 trp1-1 | [54] |
| nem1Δ | RS453 nem1::HIS3 | [53] |
| pah1Δ | RS453 pah1::TRP1 | [54] |
| spo7Δ | RS453 spo7::HIS3 | This study |
| cho2Δ | RS453 pah1::TRP1 cho2::KanMX4+YCplac33-URA3-PAH1 | [54] |
| nem1Δ spo7Δ | RS453 nem1::HIS3 spo7::HIS3 | [53] |
| pah1Δ cho2Δ | RS453 pah1::TRP1 cho2::KanMX4 | [54] |
| pah1Δ nem1Δ | RS453 pah1::TRP1 nem1::KanMX4 | This study |
| pah1Δ dgk1Δ | RS453 pah1::TRP1 dgk1::HIS3 | [54] |
Yeast cells were grown at 30°C in synthetic complete (SC) medium containing 2% galactose as the carbon source. Histidine, leucine, uracil, or combinations of them were omitted from the medium depending on the selection markers of plasmids [26]. After two passages (24~48 hours) in SC medium, cells were harvested when the absorbance at 600 nm (A600 nm) reached between 0.4–1.0 [26].
Plasmids and antibodies
The plasmids used in this study are shown in Table 2. To launch BMV replication in yeast, plasmids pB12VG1 and pB3VG128 were used in the experiments as described before [26]. In the pB12VG1 plasmid, 1a is controlled by the GAL1 promoter while 2apol is under the control of the GAL10 promoter. RNA3 is under the control of the copper-inducible CUP1 promoter but no copper was purposely included in the medium. DGK1, DGK1-D177A are overexpressed from a low-copy number plasmid YCplac33 under the control of the GAL1 promoter and tagged with HA. The plasmid pB1YT3-mCherry was used to express mCherry-tagged BMV 1a. Rabbit anti-1a antiserum (a gift from Dr. Paul Ahlquist at University of Wisconsin-Madison), mouse anti-His6 (Genescript, 6G2A9), mouse anti-dsRNA J2 antibody (English and Scientific Consulting, Hungary), and rabbit anti-HA (Thermo Fisher Scientific, 71–5500) were used at 1:100 dilution for Immunofluorescence microscopy and 1:10,000 or 1:3,000 for Western blotting. For Western blotting, we also used mouse anti-BMV 2apol at 1:3,000 dilution, and mouse anti-Pgk1p (Invitrogen, 459250) at 1:10,000 dilution.
Table 2. Plasmids used in this study.
| Plasmid | Description | Ref./source |
|---|---|---|
| pB12VG1 | BMV 1a and 2apol are driven by GAL1 or GAL10 promoter respectively in a CEN/LEU vector | [27] |
| pB3VG128-U | BMV RNA3 is under control of CUP1 promoter in a CEN/URA vector | [27] |
| pB3VG128-H | BMV RNA3 is under control of CUP1 promoter in a CEN/HIS vector | [27] |
| pB1YT3 | BMV 1a is under control of GAL1 promoter in a CEN/URA vector | [27] |
| pB1YT3-mCherry-L | 1a-mCherry is under control of GAL1 promoter in a CEN/LEU vector | [56] |
| pUN100-GFP-NUP49 | GFP-Nup49p is constructed in pUN100, a CEN/LEU vector | [88] |
| p3G-DGK1-HA | DGK1 is under control of GAL1 promoter in a CEN/URA vector | This study |
| p3G-DGK1-D177A-HA | dgk1-D177A is under control of GAL1 promoter in a CEN/URA vector | This study |
| pBG1805-SPO7 | SPO7 is under control of GAL1 promoter in the pBG1805, 2μ/URA vector | [89] |
| pBG1805-NEM1 | NEM1 is under control of GAL1 promoter in the pBG1805, 2μ/URA vector | [89] |
| pBG1805-PAH1 | PAH1 is under the control of GAL1 promoter in the pBG1805, 2μ/URA vector | [89] |
| pBG1805-SKI8 | SKI8 is under control of GAL1 promoter in the pBG1805, 2μ/URA vector | [89] |
| p3G-CHO2-HA | CHO2 is under control of GAL1 promoter in a CEN/URA vector | [27] |
| p426-CHO2-HA | CHO2 is under control of CHO2 endogenous promoter in a 2μ/URA vector | This study |
| YCplac33-PAH1 | PAH1 is under control of its endogenous promoter in a CEN/URA vector | [54] |
| p3G-PAH1-HA | PAH1 is under control of GAL1 promoter in a CEN/URA vector | This study |
| pYes2-AtPAH1 | AtPAH1 is under control of GAL1 promoter in the pYES2.1/NT 2μ/URA vector | [49] |
| pYes2-AtPAH2 | AtPAH2 is under control of GAL1 promoter in the pYES2.1/NT 2μ/URA vector | [49] |
| pYes2-NbPAH1A | NbPAH1A is under control of GAL1 promoter in the pYES2.1/NT 2μ/URA vector | This study |
| pYes2-NbPAH2A | NbPAH2A is under control of GAL1 promoter in the pYES2.1/NT 2μ/URA vector | This study |
| pPWHT- NbPAH1A | NbPAH1A is under control of an enhanced CaMV 35S promoter | This study |
| pPWHT- NbPAH2A | NbPAH2A is under control of an enhanced CaMV 35S promoter | This study |
| pAG2p-AtPAH2 | AtPAH2 is under control of an enhanced CaMV 35S promoter | This study |
RNA extraction and Northern blotting
Total RNA was extracted using a hot phenol method [90]. Equal amounts of total RNA were used for Northern blotting analysis. P32-labled probes specific to BMV positive- or negative-strand RNA or 18S rRNA were used in the hybridization. Radioactive signals were scanned using a Typhoon FLA 7000 phosphoimager and the intensity of radioactive signals were quantified by using ImageQuant TL (GE healthcare). The 18S rRNA signal was used to normalize BMV RNA signals to eliminate loading variations [26].
Western blotting
Two A600 nm units of yeast cells were harvested and total proteins were extracted as described previously [25]. Equal volumes of total proteins were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membrane. Rabbit anti-BMV 1a (1:10,000 dilution), mouse anti-BMV 2apol (1:3,000 dilution), rabbit anti-HA (1:5,000 dilution), and mouse anti-Pgk1p (1:10,000 dilution) were used to detect 1a, 2apol, HA, and Pgk1p [26]. Pgk1p was used as a loading control. Horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse antibodies (Thermo Fisher Scientific 32460 or 32430, 1:5,000 dilution) together with Supersignal West Femto substrate (Thermo Fisher Scientific, 34096) were used for signal detection.
Electron microscopy
Samples were prepared as described previously [26]. Briefly, 10 A600 nm units of cells were fixed with 4% paraformaldehyde and 2% glutaraldehyde for 1 hour followed by secondary fixation in 1% osmium tetroxide for another 1 hour. After dehydration through an ethanol gradient, yeast cells were embedded in Spurr’s resin (Electron Microscopy Sciences) for overnight. The sample sections were stained with uranyl acetate and lead citrate and observed under a JEOL JEM 1400 TEM at the Virginia-Maryland College of Veterinary Medicine.
For immunogold labeling, 4% paraformaldehyde and 0.5% glutaraldehyde were used to fix 10 A600 nm units of cells for 1 hour and followed by 0.1% osmium tetroxide secondary fixation for another 15 minutes. After dehydration through an ethanol gradient, yeast cells were embedded in LR White resin (Electron Microscopy Sciences) for overnight. Embedded samples were sectioned and nickel grids were used to hold the samples. After treated with blocking solution (AURION) for 30 minutes, grids were incubated with primary antibody diluted in incubation buffer (PBS, pH7.4, 0.15% AURION BSA-c and 15mM NaN3) and secondary antibody conjugated with colloidal gold particles (10nm or 15nm particles were conjugated to anti-mouse or anti-rabbit secondary antibody, AURION) diluted in incubation buffer. The primary antibodies were rabbit anti-1a antiserum (1:50), mouse-anti dsRNA monoclonal antibody J2 (1:50). Secondary antibodies were diluted at 1:20. Sections were counterstained with uranyl acetate (10 minutes) and lead citrate (3 minutes) and observed under a JEOL JEM 1400 TEM at 80KV at the Virginia-Maryland College of Veterinary Medicine.
Immunofluorescence microscopy
Yeast cells were harvested and fixed with 4% formaldehyde for 30 minutes. To prepare spheroplasts, the cell wall was removed by lyticase. After permeabilization with 0.1% Triton X-100 for 15 minutes, the spheroplasts were incubated with primary antibodies (1:100 dilution) overnight at 4°C followed by incubation with secondary antibodies (1:100 dilution) for 1 hour at 37°C. Finally, the nucleus was stained with DAPI (Vector laboratories) for 10 minutes. Samples were observed using a Zeiss epifluorescence microscope (Observer.Z1) at the Fralin microscopy facility, VT.
Measurement of yeast nuclear membrane perimeters
Measurements were performed with ImageJ software. Briefly, the scale was set based on the scale bar in images. The color threshold was adjusted to allow the spot to fit the nucleus perfectly and adding the target spot to the ROI (Region of Interest) manager by using the wand (tracing) tool. The perimeter was measured by performing the “measure” in the ROI manager tool.
Radiolabeling and analysis of lipids
The steady-state labeling of lipids with [2-14C] acetate was performed as described previously [91]. Briefly, equal number of cells (2.5 x 105 cells/ml) were inoculated into SC-Ura-Leu with galactose as carbon source along with the [2-14C] acetate. The cells were grown to exponential phase (A600nm = ~0.5) and harvested. Lipids were extracted [92] from the radiolabeled cells, and then separated by one-dimensional TLC for neutral lipids [93] or phospholipids [94]. The resolved lipids were visualized by phosphorimaging and quantified by ImageQuant software using a standard curve of [2-14C] acetate. The identity of radiolabeled lipids was confirmed by comparison with the migration of authentic standards visualized by staining with iodine vapor. The mol percentage of each neutral lipid or phospholipid was normalized to the total 14C-labeled chloroform fraction. Single factor ANOVA was used for statistical analysis of lipid differences between wt and mutants.
BMV replication assay in Nicotiana benthamiana
Replication analysis of BMV, TRV, and TMV [95] in N. benthamiana was performed as previously reported [96]. Arabidopsis thaliana AtPAH2, N. benthamiana NbPAH1A and NbPAH2A were expressed in N. benthamiana leaves by agroinfiltration following a protocol described before [97]. The AtPAH2 was cloned into pAG2p vector between an enhanced CaMV 35S promoter and a terminator [70]. The NbPAH1A and NbPAH2A were cloned into pPWHT vector between an enhanced CaMV 35S promoter and a terminator through gateway cloning. N. benthamiana leaves were first infiltrated with Agrobacteria (GV3101) harboring pAG2p-AtPAH2, pPWHT-NbPAH1A or pPWHT-NbPAH2A plasmid. Two days later, the same leaves were infiltrated with the mixed Agrobacteria cultures harboring plasmids that launch BMV RNA 1, 2, and 3, or TRV1 and 2, or TMV. The infiltrated leaves were harvested 3 days post viral inoculation. Total RNA was extracted following the hot phenol method. Viral RNA accumulation was analyzed by Northern blotting with BMV-, TRV-, or TMV-specific probes as described before [70].
Supporting information
Immunogold labeling of BMV 1a in wt (A), pah1Δ nem1Δ (B) or pah1Δ dgk1Δ (C) cells in the presence of BMV replication. Anti-1a antiserum was used as a primary antibody and a 15-nm gold particle-conjugated anti-rabbit antibody was used as a secondary antibody. Micrographs at a higher magnification (A-1, B-1, B-2, C-1 and C-2) are also shown. (D) Percentage of gold particles localized in or near spherular structures in wt, pah1Δ nem1Δ or pah1Δ dgk1Δ cells. The total number of gold particles counted in each strain is also included.
(TIF)
(A) and (B) Micrographs showing BMV-replicating pah1Δ nem1Δ cells with proliferated membranes. The micrographs of boxed areas at a higher magnification are shown in A-1, B-1, and B-2. Arrows indicate the dramatically proliferated membranes.
(TIF)
Acknowledgments
We thank Haijie Liu, Jianhui Li, Elizabeth Barton, Nicholas Todd, and Nancy Kalaj for general assistance, Dr. Symeon Siniossoglou at University of Cambridge (UK) for providing yeast strains, Drs. Elzbieta Mietkiewska and Randall J. Weselake at University of Alberta (Canada), and Dr. Valérie Doye from Institute Jacques Monod (France) for providing plasmids, Drs. Li-Ka Liu and William Prinz from National Institute of Diabetes and Digestive and Kidney Diseases for help in lipid work, Drs. Tero Ahola and Janet Webster for critical reading of the manuscript. We appreciate the help from Ms. Kathy Lowe at Virginia-Maryland College of Veterinary Medicine (Virginia Tech) and Ms. Shannon Modla at the Delaware Biotechnology Institute for electron microscopy work, and Dr. Kristi DeCourcy at the Fralin Life Science Institute (Virginia Tech) for fluorescence microscopy work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
ZZ. and GH were supported by the China Scholarship Council. Work at the GMC laboratory is supported by National Institute of Health grant GM028140. This work is partially supported by National Science Foundation grant IOS-1645740 to AD and XW. Work in the XW laboratory is also supported by the Virginia Agricultural Experiment Station and Hatch Program of National Institute of Food and Agriculture, United States Department of Agriculture. Microscopy access in Delaware Biotechnology Institute was supported by grants from the NIH-NIGMS (P20 GM103446), the NSF (IIA-1301765) and the State of Delaware. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang A. Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu Rev Phytopathol. 2015;53:45–66. doi: 10.1146/annurev-phyto-080614-120001 [DOI] [PubMed] [Google Scholar]
- 2.Paul D, Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol. 2013;2(2):32–48. doi: 10.5501/wjv.v2.i2.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laliberte JF, Zheng H. Viral Manipulation of Plant Host Membranes. Annu Rev Virol. 2014;1(1):237–59. doi: 10.1146/annurev-virology-031413-085532 [DOI] [PubMed] [Google Scholar]
- 4.Hyodo K, Okuno T. Pathogenesis mediated by proviral host factors involved in translation and replication of plant positive-strand RNA viruses. Curr Opin Virol. 2016;17:11–8. doi: 10.1016/j.coviro.2015.11.004 [DOI] [PubMed] [Google Scholar]
- 5.Chukkapalli V, Heaton NS, Randall G. Lipids at the interface of virus-host interactions. Curr Opin Microbiol. 2012;15(4):512–8. doi: 10.1016/j.mib.2012.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belov GA, van Kuppeveld FJ. (+)RNA viruses rewire cellular pathways to build replication organelles. Curr Opin Virol. 2012;2(6):740–7. doi: 10.1016/j.coviro.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Ahlquist P. Brome mosaic virus. Encyclopedia of virology; doi: 10.1016/B978-012374410-4.00560–42008 p. 381–6. [Google Scholar]
- 8.Restrepo-Hartwig M, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol. 1999;73(12):10303–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovanni P, Martelli, Marcrello R. Virus-host relationships: symptomatological and ultrastructural aspects. In: RIB F, editor. The plant viruses; doi: 10.1007/978-1-4684-4937-2_61985 p. 163–205. [Google Scholar]
- 10.Diaz A, Wang X. Bromovirus-induced remodeling of host membranes during viral RNA replication. Curr Opin Virol. 2014;9:104–10. doi: 10.1016/j.coviro.2014.09.018 [DOI] [PubMed] [Google Scholar]
- 11.Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell. 2002;9(3):505–14. doi: 10.1016/S1097-2765(02)00474-4 [DOI] [PubMed] [Google Scholar]
- 12.Annamalai P, Rao AL. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology. 2005;338(1):96–111. doi: 10.1016/j.virol.2005.05.013 [DOI] [PubMed] [Google Scholar]
- 13.Gopinath K, Dragnea B, Kao C. Interaction between Brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J Virol. 2005;79(22):14222–34. doi: 10.1128/JVI.79.22.14222-14234.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Ahlquist P. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J Virol. 2000;74(9):4310–8. doi: 10.1128/JVI.74.9.4310–4318.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao CC, Ahlquist P. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol. 1992;66(12):7293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Reilly EK, Paul JD, Kao CC. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J Virol. 1997;71(10):7526–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahola T, Ahlquist P. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J Virol. 1999;73(12):10061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahola T, den Boon JA, Ahlquist P. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J Virol. 2000;74(19):8803–11. doi: 10.1128/JVI.74.19.8803–8811.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong F, Sivakumaran K, Kao C. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology. 1999;259(1):200–10. doi: 10.1006/viro.1999.9763 [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Lee WM, Watanabe T, Schwartz M, Janda M, Ahlquist P. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J Virol. 2005;79(21):13747–58. doi: 10.1128/JVI.79.21.13747-13758.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Restrepo-Hartwig MA, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70(12):8908–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L, Westler WM, den Boon JA, Wang X, Diaz A, Steinberg HA, et al. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 2009;5(3):e1000351 doi: 10.1371/journal.ppat.1000351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz A, Gallei A, Ahlquist P. Bromovirus RNA replication compartment formation requires concerted action of 1a's self-interacting RNA capping and helicase domains. J Virol. 2012;86(2):821–34. doi: 10.1128/JVI.05684-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee WM, Ahlquist P. Membrane Synthesis, Specific Lipid Requirements, and Localized Lipid Composition Changes Associated with a Positive-Strand RNA Virus RNA Replication Protein. Journal of Virology. 2003;77(23):12819–28. doi: 10.1128/JVI.77.23.12819-12828.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee WM, Ishikawa M, Ahlquist P. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J Virol. 2001;75(5):2097–106. doi: 10.1128/JVI.75.5.2097-2106.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Diaz A, Mao L, Ahlquist P, Wang X. Host acyl coenzyme A binding protein regulates replication complex assembly and activity of a positive-strand RNA virus. J Virol. 2012;86(9):5110–21. doi: 10.1128/JVI.06701-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Zhang Z, Chukkapalli V, Nchoutmboube JA, Li J, Randall G, et al. Positive-strand RNA viruses stimulate host phosphatidylcholine synthesis at viral replication sites. Proc Natl Acad Sci U S A. 2016;113(8):E1064–73. doi: 10.1073/pnas.1519730113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Athenstaedt K, Daum G. Biosynthesis of phosphatidic acid in lipid particles and endoplasmic reticulum of Saccharomyces cerevisiae. J Bacteriol. 1997;179(24):7611–6. doi: 10.1128/jb.179.24.7611–7616.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Athenstaedt K, Weys S, Paltauf F, Daum G. Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181(5):1458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Z, Zou J. The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J Biol Chem. 2001;276(45):41710–6. doi: 10.1074/jbc.M104749200 [DOI] [PubMed] [Google Scholar]
- 31.Rattray JB, Schibeci A, Kidby DK. Lipids of yeasts. Bacteriol Rev. 1975;39(3):197–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carman GM, Henry SA. Phospholipid biosynthesis in yeast. Annu Rev Biochem. 1989;58:635–69. doi: 10.1146/annurev.bi.58.070189.003223 [DOI] [PubMed] [Google Scholar]
- 33.Paltauf F, Kohlwein S, Henry S. Regulation and compartmentalization of lipid synthesis in yeast. Cold Spring Harbor Monograph Archive. 1992;415(21B):415–500. doi: 10.1101/087969365.21B.415 [Google Scholar]
- 34.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem. 2002;277(46):44507–12. doi: 10.1074/jbc.M207712200 [DOI] [PubMed] [Google Scholar]
- 35.Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, et al. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304(5677):1644–7. doi: 10.1126/science.1096083 [DOI] [PubMed] [Google Scholar]
- 36.Carman GM, Han GS. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu Rev Biochem. 2011;80:859–83. doi: 10.1146/annurev-biochem-060409-092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci. 2006;31(12):694–9. doi: 10.1016/j.tibs.2006.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carman GM, Han GS. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J Biol Chem. 2009;284(5):2593–7. doi: 10.1074/jbc.R800059200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez-Murray JP, McMaster CR. Lipid synthesis and membrane contact sites: a crossroads for cellular physiology. J Lipid Res. 2016;57(10):1789–805. doi: 10.1194/jlr.R070920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24(11):1931–41. doi: 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi HS, Su WM, Han GS, Plote D, Xu Z, Carman GM. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J Biol Chem. 2012;287(14):11290–301. doi: 10.1074/jbc.M112.346023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han GS, Siniossoglou S, Carman GM. The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J Biol Chem. 2007;282(51):37026–35. doi: 10.1074/jbc.M705777200 [DOI] [PubMed] [Google Scholar]
- 43.Choi HS, Su WM, Morgan JM, Han GS, Xu Z, Karanasios E, et al. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), AND SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J Biol Chem. 2011;286(2):1486–98. doi: 10.1074/jbc.M110.155598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karanasios E, Han GS, Xu Z, Carman GM, Siniossoglou S. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci U S A. 2010;107(41):17539–44. doi: 10.1073/pnas.1007974107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fakas S, Qiu Y, Dixon JL, Han GS, Ruggles KV, Garbarino J, et al. Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J Biol Chem. 2011;286(33):29074–85. doi: 10.1074/jbc.M111.258798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell JL, Lorenz A, Witkin KL, Hays T, Loidl L, Cohen-Fix O. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol Biol Cell. 2006;17(4):1768–78. doi: 10.1091/mbc.E05-09-0839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, et al. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci U S A. 2009;106(49):20978–83. doi: 10.1073/pnas.0907173106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimsey N, Han GS, O'Hara L, Rochford JJ, Carman GM, Siniossoglou S. Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J Biol Chem. 2008;283(43):29166–74. doi: 10.1074/jbc.M804278200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mietkiewska E, Siloto RM, Dewald J, Shah S, Brindley DN, Weselake RJ. Lipins from plants are phosphatidate phosphatases that restore lipid synthesis in a pah1Delta mutant strain of Saccharomyces cerevisiae. FEBS J. 2011;278(5):764–75. doi: 10.1111/j.1742-4658.2010.07995.x [DOI] [PubMed] [Google Scholar]
- 50.Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A. 2003;100(26):15764–9. doi: 10.1073/pnas.2536857100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chuang C, Barajas D, Qin J, Nagy PD. Inactivation of the host lipin gene accelerates RNA virus replication through viral exploitation of the expanded endoplasmic reticulum membrane. PLoS Pathog. 2014;10(2):e1003944 doi: 10.1371/journal.ppat.1003944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez de Castro I, Fernandez JJ, Barajas D, Nagy PD, Risco C. Three-dimensional imaging of the intracellular assembly of a functional viral RNA replicase complex. J Cell Sci. 2017;130(1):260–8. doi: 10.1242/jcs.181586 [DOI] [PubMed] [Google Scholar]
- 53.Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17(22):6449–64. doi: 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han GS, O'Hara L, Carman GM, Siniossoglou S. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J Biol Chem. 2008;283(29):20433–42. doi: 10.1074/jbc.M802903200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickner RB. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60(1):250–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Fuchs S, Zhang J, Wellford S, Schuldiner M, Wang X. An unrecognized function for COPII components in recruiting the viral replication protein BMV 1a to the perinuclear ER. J Cell Sci. 2016;129(19):3597–608. doi: 10.1242/jcs.190082 [DOI] [PubMed] [Google Scholar]
- 57.Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119(4):705–23. doi: 10.1083/jcb.119.4.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng X, Deng P, Cui H, Wang A. Visualizing double-stranded RNA distribution and dynamics in living cells by dsRNA binding-dependent fluorescence complementation. Virology. 2015;485:439–51. doi: 10.1016/j.virol.2015.08.023 [DOI] [PubMed] [Google Scholar]
- 59.Cao X, Jin X, Zhang X, Li Y, Wang C, Wang X, et al. Morphogenesis of Endoplasmic Reticulum Membrane-Invaginated Vesicles during Beet Black Scorch Virus Infection: Role of Auxiliary Replication Protein and New Implications of Three-Dimensional Architecture. J Virol. 2015;89(12):6184–95. doi: 10.1128/JVI.00401-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012:190(2):317–349. doi: 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu K, Nagy PD. RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes. Proc Natl Acad Sci U S A. 2015;112(14):E1782–91. doi: 10.1073/pnas.1418971112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belov GA. Dynamic lipid landscape of picornavirus replication organelles. Curr Opin Virol. 2016;19:1–6. doi: 10.1016/j.coviro.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altan-Bonnet N. Lipid Tales of Viral Replication and Transmission. Trends Cell Biol. 2017;27(3):201–13. doi: 10.1016/j.tcb.2016.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Csaki LS, Dwyer JR, Fong LG, Tontonoz P, Young SG, Reue K. Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog Lipid Res. 2013;52(3):305–16. doi: 10.1016/j.plipres.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mise K, Allison RF, Janda M, Ahlquist P. Bromovirus movement protein genes play a crucial role in host specificity. J Virol. 1993;67(5):2815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Annamalai P, Rao ALN. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: A functional role for viral replicase in RNA packaging. Virology. 2005;338(1):96–111. doi: 10.1016/j.virol.2005.05.013 [DOI] [PubMed] [Google Scholar]
- 67.Goodin MM, Zaitlin D, Naidu RA, Lommel SA. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact. 2008;21(8):1015–26. doi: 10.1094/MPMI-21-8-1015 [DOI] [PubMed] [Google Scholar]
- 68.Mas P, Beachy RN. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol. 1999;147(5):945–58. doi: 10.1083/jcb.147.5.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harrison BD, Kubo S, Robinson DJ, Hutcheson AM. Multiplication Cycle of Tobacco Rattle Virus in Tobacco Mesophyll Protoplasts. J Gen Virol. 1976;33(Nov):237–48. doi: 10.1099/0022-1317-33-2-237 [Google Scholar]
- 70.Diaz A, Zhang J, Ollwerther A, Wang X, Ahlquist P. Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog. 2015;11(3):e1004742 doi: 10.1371/journal.ppat.1004742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu K, Nagy PD. Enrichment of Phosphatidylethanolamine in Viral Replication Compartments via Co-opting the Endosomal Rab5 Small GTPase by a Positive-Strand RNA Virus. PLoS Biol. 2016;14(10):e2000128 doi: 10.1371/journal.pbio.2000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106(18):7577–82. doi: 10.1073/pnas.0902693106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9(1):32–45. doi: 10.1016/j.chom.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141(5):799–811. doi: 10.1016/j.cell.2010.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park Y, Han GS, Mileykovskaya E, Garrett TA, Carman GM. Altered Lipid Synthesis by Lack of Yeast Pah1 Phosphatidate Phosphatase Reduces Chronological Life Span. J Biol Chem. 2015;290(42):25382–94. doi: 10.1074/jbc.M115.680314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waggoner DW, Xu J, Singh I, Jasinska R, Zhang QX, Brindley DN. Structural organization of mammalian lipid phosphate phosphatases: implications for signal transduction. Biochim Biophys Acta. 1999;1439(2):299–316. doi: 10.1016/S1388-1981(99)00102-X [DOI] [PubMed] [Google Scholar]
- 77.Sciorra VA, Morris AJ. Roles for lipid phosphate phosphatases in regulation of cellular signaling. Biochim Biophys Acta. 2002;1582(1–3):45–51. doi: 10.1016/S1388-1981(02)00136-1 [DOI] [PubMed] [Google Scholar]
- 78.Testerink C, Munnik T. Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005;10(8):368–75. doi: 10.1016/j.tplants.2005.06.002 [DOI] [PubMed] [Google Scholar]
- 79.Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45(3):250–78. doi: 10.1016/j.plipres.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 80.Bargmann BO, Laxalt AM, Riet BT, Schouten E, van Leeuwen W, Dekker HL, et al. LePLDbeta1 activation and relocalization in suspension-cultured tomato cells treated with xylanase. Plant J. 2006;45(3):358–68. doi: 10.1111/j.1365-313X.2005.02631.x [DOI] [PubMed] [Google Scholar]
- 81.Kirik A, Mudgett MB. SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc Natl Acad Sci U S A. 2009;106(48):20532–7. doi: 10.1073/pnas.0903859106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Luit AH, Piatti T, van Doorn A, Musgrave A, Felix G, Boller T, et al. Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol. 2000;123(4):1507–16. doi: 10.1104/pp.123.4.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamaguchi T, Minami E, Ueki J, Shibuya N. Elicitor-induced activation of phospholipases plays an important role for the induction of defense responses in suspension-cultured rice cells. Plant Cell Physiol. 2005;46(4):579–87. doi: 10.1093/pcp/pci065 [DOI] [PubMed] [Google Scholar]
- 84.Kooijman EE, Chupin V, Fuller NL, Kozlov MM, de Kruijff B, Burger KN, et al. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 2005;44(6):2097–102. doi: 10.1021/bi0478502 [DOI] [PubMed] [Google Scholar]
- 85.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–6. doi: 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- 86.Burger KN. Greasing membrane fusion and fission machineries. Traffic. 2000;1(8):605–13. doi: 10.1034/j.1600-0854.2000.010804.x [DOI] [PubMed] [Google Scholar]
- 87.Hyodo K, Taniguchi T, Manabe Y, Kaido M, Mise K, Sugawara T, et al. Phosphatidic acid produced by phospholipase D promotes RNA replication of a plant RNA virus. PLoS Pathog. 2015;11(5):e1004909 doi: 10.1371/journal.ppat.1004909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belgareh N, Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. Journal of Cell Biology. 1997;136(4):747–59. doi: 10.1083/jcb.136.4.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Gene Dev. 2005;19(23):2816–26. doi: 10.1101/gad.1362105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-G [DOI] [PubMed] [Google Scholar]
- 91.Morlock KR, Lin YP, Carman GM. Regulation of phosphatidate phosphatase activity by inositol in Saccharomyces cerevisiae. J Bacteriol. 1988;170(8):3561–6. doi: 10.1128/jb.170.8.3561–3566.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–7. doi: 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 93.Henderson R, Tocher D. Thin-layer chromatography Hamilton R, Hamilton S, editors. New York: IRL Press; 1992. [Google Scholar]
- 94.Vaden DL, Gohil VM, Gu Z, Greenberg ML. Separation of yeast phospholipids using one-dimensional thin-layer chromatography. Anal Biochem. 2005;338(1):162–4. doi: 10.1016/j.ab.2004.11.020 [DOI] [PubMed] [Google Scholar]
- 95.Lindbo JA. TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 2007;145(4):1232–40. doi: 10.1104/pp.107.106377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31(6):777–86. doi: 10.1046/j.1365-313X.2002.01394.x [DOI] [PubMed] [Google Scholar]
- 97.Bendahmane A, Querci M, Kanyuka K, Baulcombe DC. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 2000;21(1):73–81. doi: 10.1046/j.1365-313x.2000.00654.x [DOI] [PubMed] [Google Scholar]
- 98.Hayat M. Colloidal gold: principles, methods, and applications Hayat M, editor. San Diego: Academic Press; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunogold labeling of BMV 1a in wt (A), pah1Δ nem1Δ (B) or pah1Δ dgk1Δ (C) cells in the presence of BMV replication. Anti-1a antiserum was used as a primary antibody and a 15-nm gold particle-conjugated anti-rabbit antibody was used as a secondary antibody. Micrographs at a higher magnification (A-1, B-1, B-2, C-1 and C-2) are also shown. (D) Percentage of gold particles localized in or near spherular structures in wt, pah1Δ nem1Δ or pah1Δ dgk1Δ cells. The total number of gold particles counted in each strain is also included.
(TIF)
(A) and (B) Micrographs showing BMV-replicating pah1Δ nem1Δ cells with proliferated membranes. The micrographs of boxed areas at a higher magnification are shown in A-1, B-1, and B-2. Arrows indicate the dramatically proliferated membranes.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.