Visual Abstract
Key Words: cardiac precursor cells, differentiation, heart disease, lncRNAs, miRNAs, NOTCH
Abbreviation and Acronyms: CARMEN, (CAR)diac (M)esoderm (E)nhancer-associated (N)oncoding RNA; CPC, cardiac precursor cell; DLL1, Delta-like1; GO, gene ontology; J1, Jagged1; lncRNA, long noncoding RNA; miRNA, microRNA; NICD, NOTCH intracellular domain; RT-PCR, reverse transcription polymerase chain reaction
Highlights
-
•
Human CPCs produce predominantly smooth muscle cells.
-
•
CPCs can be redirected to the cardiomyocyte fate by transient activation followed by inhibition of NOTCH signaling.
-
•
Inhibition of NOTCH signaling during differentiation represses MIR-143/145 expression and blocks smooth muscle differentiation.
-
•
Expression of the microRNAs is under control of CARMEN, a long noncoding RNA associated with an enhancer located in the MIR-143/145 locus and target of NOTCH signaling.
-
•
The CARMEN/MIR-145/143 locus represents a promising therapeutic target to favor production of cardiomyocytes in cell replacement therapies.
Summary
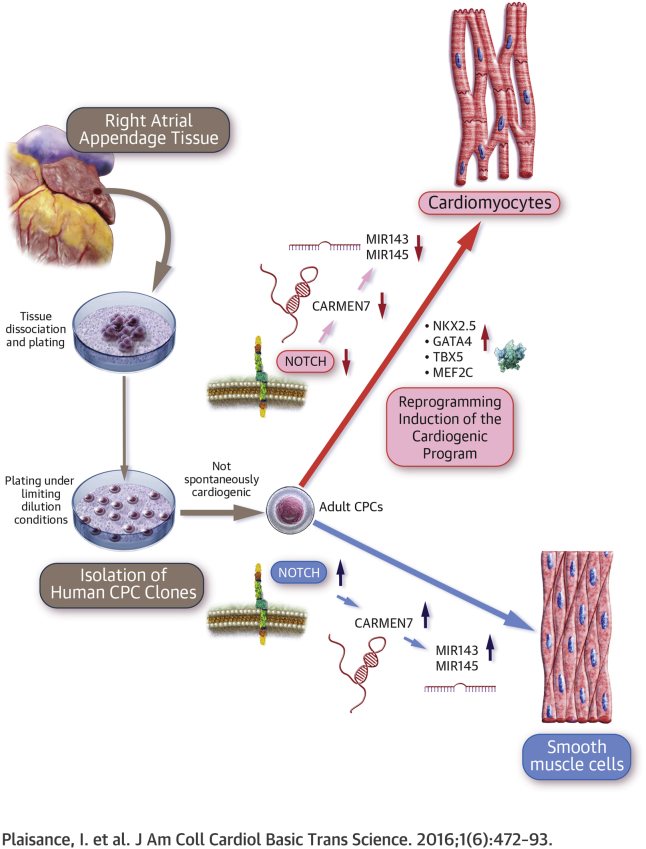
The mechanisms controlling differentiation in adult cardiac precursor cells (CPCs) are still largely unknown. In this study, CPCs isolated from the human heart were found to produce predominantly smooth muscle cells but could be redirected to the cardiomyocyte fate by transient activation followed by inhibition of NOTCH signaling. NOTCH inhibition repressed MIR-143/145 expression, and blocked smooth muscle differentiation. Expression of the microRNAs is under control of CARMEN, a long noncoding RNA associated with an enhancer located in the MIR-143/145 locus and target of NOTCH signaling. The CARMEN/MIR-145/143 axis represents, therefore, a promising target to favor production of cardiomyocytes in cell replacement therapies.
The adult human heart has poor regenerative potential, and heart failure gradually develops following injury 1, 2. Ultimately, heart transplantation represents the only therapeutic option for end-stage heart failure. Within this context, stimulation of cardiomyocyte production in the damaged heart to promote regeneration represents an attractive therapeutic approach 3, 4. In particular, cell replacement therapy via injection of precursor cells into the damaged heart represents an interesting therapeutic avenue. The main challenge for transferring cell therapies for heart disease into a clinical setting is to identify a suitable source of human cardiac precursor cells (CPCs). Direct isolation of CPCs from the heart of cardiac patients would represent a great advantage by the autologous nature of the isolated cells. This would indeed reduce the problems associated with immune rejection. The existence of resident CPCs in the adult mammalian heart, including the human heart, capable of differentiating into functional cardiomyocytes, has been demonstrated 3, 4. However, the number of CPCs in the adult myocardium is quite low, and isolation of these cells is a challenging procedure. Indeed, no truly specific markers are currently available to distinguish CPCs from other cell types 3, 4. Multipotent mesenchymal stromal cells expressing cardiac transcription factors such as GATA4, NKX2.5, and MEF2C, but no proteins expressed by fully differentiated cardiomyocytes such as proteins of the sarcomere, could therefore be operationally defined as CPCs. Nevertheless, the effective generation of new cardiomyocytes from transferred CPCs is still a matter of intense debate, and restoration of function has been attributed to paracrine mechanisms mediated by factors secreted from the transferred cells 2, 3, 4. Therefore, a clear understanding of the regulatory networks controlling mobilization and differentiation of endogenous CPCs toward the cardiac lineage is required in order to facilitate the ultimate goal of cardiac regeneration.
Several pathways that are important during cardiac morphogenesis are reactivated in the damaged myocardium. Among these, the NOTCH pathway plays crucial roles in the developing and adult heart 5, 6. NOTCH is an evolutionarily conserved cell-to-cell communication system that takes place between 2 adjacent cells (7). The signal-sending cell expresses a membrane-bound ligand such as Jagged (J)1, J2, Delta-like1 (DLL1), DLL3, and DLL4, and the signal-receiving cell expresses a NOTCH receptor such as NOTCH (N)1, N2, N3, and N4. Receptor engagement results in its cleavage and liberation of the NOTCH intracellular domain (NICD). NICD translocates into the nucleus, where it interacts with co-activators, in particular a transcription factor known as RBPJ, to activate target gene expression. NOTCH target genes include repressors of the Hairy enhancer of split (HES) and the related HEY families (8). During development, NOTCH regulates trabeculation, myocyte proliferation, and valve formation. In the neonatal heart, NOTCH controls cardiac precursor expansion and differentiation (9). In the adult heart, NOTCH signaling is activated in cardiomyocytes, CPCs, and fibroblasts 10, 11, 12, 13, 14. Interestingly, NOTCH appears to prevent premature cardiogenic differentiation in precursor cells, and to favor proliferation in this transient amplifying cell compartment (14). Consistent with this observation, blockade of the NOTCH pathway in embryonic stem cells favors commitment into the cardiac mesoderm, and subsequently, into cardiomyocytes, at the expense of the neuroectodermal lineage (15). NOTCH signaling has also been reported to induce early cardiac commitment in embryonic and induced pluripotent stem cells, supporting a biphasic role of NOTCH in cardiogenesis (16). Accordingly, NOTCH signaling was suggested to promote cardiogenesis in the post-natal heart 17, 18. Furthermore, NOTCH has been implicated in the differentiation of cardiosphere-derived cells into smooth muscle cells (19). This finding is reminiscent of the role of NOTCH in vascular smooth muscle cells, in which Jagged1-activated NOTCH signaling promotes a differentiated phenotype (20). Interestingly, NICD, associated with RBPJ, binds to an enhancer within the locus encoding the small regulatory noncoding RNAs MIR-143/145, 2 microRNAs (MIRs) that were previously shown to regulate smooth muscle cell phenotype and behavior 20, 21.
In recent years, it has become apparent that the noncoding genome is pervasively transcribed, generating thousands of long noncoding RNAs (lncRNAs). These transcripts, which are defined as being >200 nucleotides in length with no apparent protein coding potential, are typically expressed in a highly cell- and context-specific manner. LncRNAs are emerging, therefore, as master regulators of gene expression and important mediators of lineage-specific commitment during development 22, 23. In the developing and adult hearts, lncRNAs are dynamically expressed and exert control of the cardiac gene regulatory network 24, 25, 26. A growing body of evidence suggests that they could be pivotal during the pathophysiological response in the damaged heart 27, 28. LncRNAs efficiently operate in cis, at the site of transcription, and in trans, at remote locations in the genome. In particular, lncRNAs, which are transcribed from active enhancers, contribute to neighboring gene activation via cis mechanisms 29, 30. In this context, we recently identified CARMEN [(CAR)diac (M)esoderm (E)nhancer-associated (N)oncoding RNA], a lncRNA that is a crucial regulator of cardiac specification in human CPCs isolated from the fetal heart 31, 32. Interestingly, CARMEN is templated from the NOTCH-responsive enhancer element within the MIR-143/145 locus. In the present study, we aimed at evaluating the cardiogenic potential of CPCs isolated from adult human hearts. We show that human clonogenic CPCs can be readily obtained from atrial appendages, expanded in vitro, and induced to differentiate into either smooth muscle cells or cardiomyocytes. This binary cell fate decision depends on the state of activation or inhibition of the NOTCH pathway. Activation of NOTCH signaling promotes adoption of a smooth muscle lineage, whereas sequential activation and inhibition favor cardiomyocyte specification. NOTCH signaling appears to target CARMEN in differentiating CPCs. More precisely, we demonstrated that a particular CARMEN isoform regulates commitment into the smooth muscle fate. Therefore, NOTCH inhibition, via down-regulation of the smooth muscle cell–specific isoform of CARMEN, represses MIR-143/145 expression, and forces CPCs to adopt a cardiomyocyte fate.
Methods
Culture of human adult CPCs
Human atrial appendages were obtained from male patients (35 to 90 years old) undergoing cardiac surgery through donation. The protocol received authorization from the University Hospital Ethics Committee and the Cantonal Ethics Committee on research involving humans. Tissues were minced and enzymatically digested in a buffer containing 0.45 mg/ml collagenase (Worthington Biochemical Corporation, Lakewood, New Jersey) and 1 mg/ml pancreatin (Invitrogen, Carlsbad, California) (Supplemental Figure 1A). Following 24 hours in culture, cells that remained in suspension were discarded, and adherent cells were expanded in expansion medium (3:1 DMEM 1g/l glucose/Medium 199 [Invitrogen] supplemented with 10% horse serum [Serotec, Kidlington, United Kingdom], 5% fetal bovine serum [Serotec], 100 U/ml penicillin [Invitrogen], and 100 μg/ml streptomycin [Invitrogen]). For inducing differentiation, cells were switched to MEM alpha (Invitrogen) containing 2% horse serum, 1 μmol/l dexamethasone (Sigma-Aldrich, St. Louis, Missouri), 50 μg/ml ascorbic acid (Sigma-Aldrich), 10 mmol/l β-glycerophosphate (Sigma-Aldrich), 100 U/ml penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen) (differentiation medium [33]), and cultured for 2 to 3 weeks before analysis.
Experiments in SCID neonates
Immunodeficient SCID mice (S/B6.CB17-PrKdc scid Ma SN1913, The Jackson Laboratory, Bar Harbor, Maine) were used in cell transfer experiments. Animal experiments were approved by the Government Veterinary Office (Lausanne, Switzerland) and performed according to the guidelines from Directive 2010/63/EU of the European Parliament. Mice were maintained under specific pathogen-free conditions. CPCs were either not stimulated (None) or exposed to DLL1 for 24h. Cells (105 in 50 μl NaCl 0.09%) were injected into neonatal mice through the superficial temporal vein. Twenty-four hours post cell-injection, the test group was injected with DAPT [N-(N-(3,5-difluorophenacetyll)-L-alanyl)S-phenylglycine t-butyl ester] diluted in dimethyl sulfoxide (DMSO) (4 μl/g of body weight/day), whereas the control group received DMSO only (No DAPT group) for 3 consecutive days. Twelve days post-injection, hearts were collected and embedded in optimal cutting temperature compound. To identify heart sections containing human cells, every second cryosection was collected for reverse transcription polymerase chain reaction (RT-PCR) detection of the human α-satellite chromosome (8 sections per PCR tube). Sections were digested with Proteinase K (Sigma-Aldrich) in Direct PCR Tail Buffer (Viagen Biotech, Los Angeles, California). DNA was amplified using Taq NEB (New England Biolabs, Ipswich, Massachusetts). The primers are described in the Supplemental Methods. Immunostaining directed against human LAMIN and α-ACTININ was performed. The surface of the LAMIN area was measured using confocal images and the ImageJ application (Version 1.50B, National Institutes of Health, Bethesda, Maryland). Tridimensional volumes were reconstructed using IMARIS software (Version 7.7.1, Bitplane, Belfast, United Kingdom).
RNA sequencing and analysis
Total RNA was isolated from proliferating and differentiated CPCs using the RNeasy isolation kit (Qiagen, Valencia, California). Sequencing libraries were prepared according to Illumina RNA Seq library kit instructions with Poly(A) selection (Illumina, San Diego, California). Libraries were sequenced with the Illumina HiSeq2000 (2 × 100 bp).
Statistical analysis
All data were collected from at least 3 independent experiments, performed in triplicate. Data throughout the paper are expressed as mean ± SEM. Data were processed using GraphPad Prism (version 7.00, GraphPad Software, La Jolla, California), and analyzed by the Kolmogorov-Smirnov test to check for normal distribution. Analysis was performed using analysis of variance with post hoc Tukey test. For non-normally distributed data, Kruskal-Wallis analysis with the Dunn multiple comparison test were used, and median values were calculated. Values of p < 0.05 were considered significant.
For transcriptomic data, statistical analysis was performed in R version 3.0.2 (R Foundation for Statistical Computing, Vienna, Austria). Raw counts were normalized using TMM (EdgeR, R version 3.4.0 [34]) for genes with 1 count per million (cpm) in at least 1 sample. Log transformation was applied on the normalized counts using Voom function (limma package, R version 3.18.2 [35]). Differential expression was computed with limma (36), and a moderated t test was used for each comparison. Adjusted p values were computed by the Benjamini-Hochberg method, controlling for a false discovery rate.
An expanded methods section is also available in the Supplemental Appendix.
Results
Characterization of cardiac precursor cells isolated from adult human atrial appendages
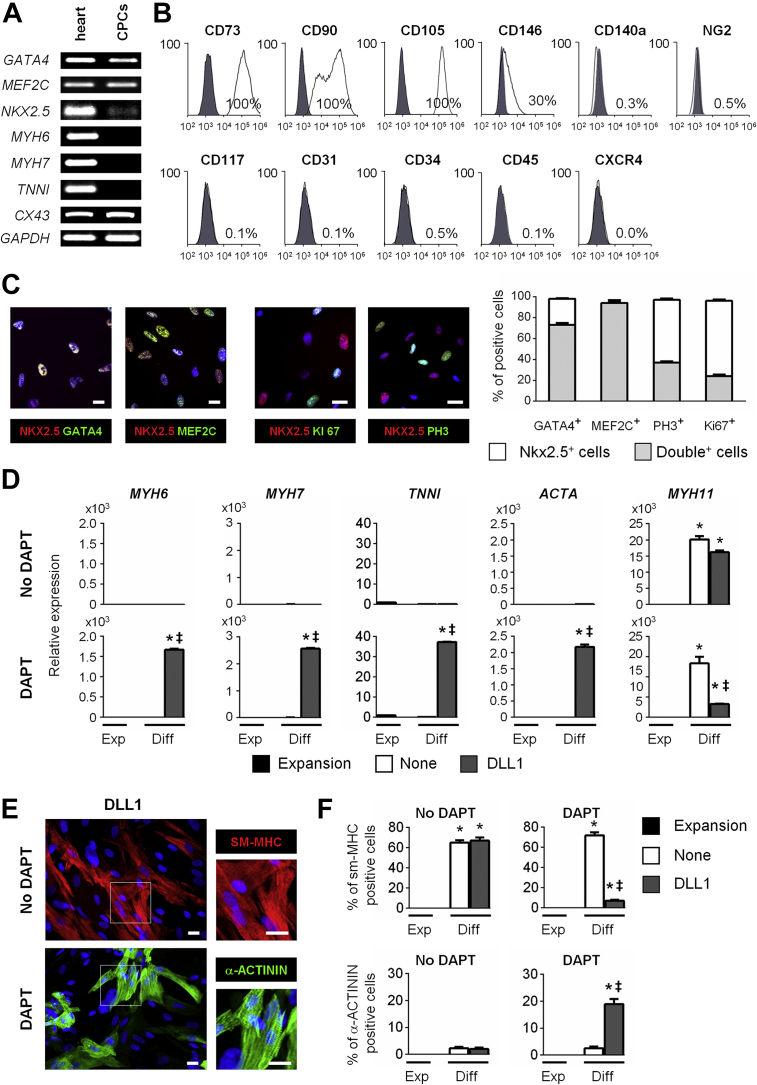
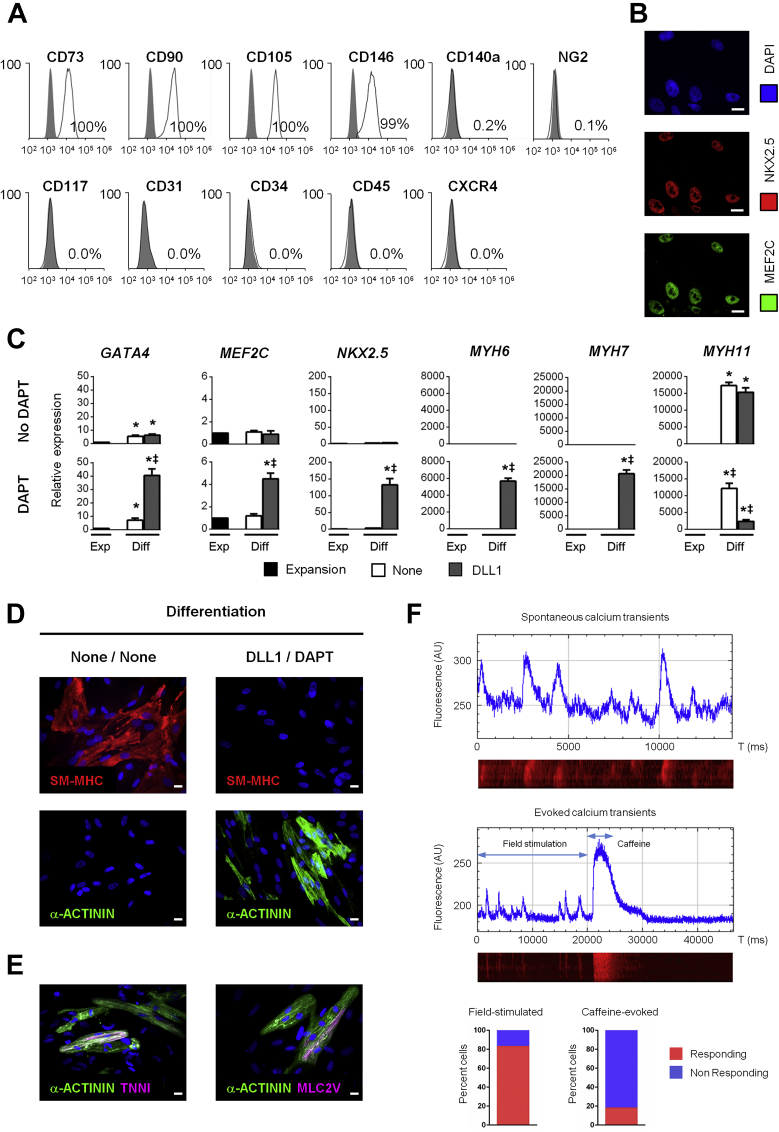
Human CPCs were isolated from right atrial appendages of male patients (35 to 86 years old) undergoing cardiac surgery (Supplemental Figure 1A). Following expansion in vitro, we routinely obtained several 100 thousand cells from each biopsy (Supplemental Table 1). Cells could be stored frozen, thawed, and retained a proliferative capacity for more than 15 passages. We first measured the expression of markers of cardiac specification and differentiation by RT-PCR (Figure 1A). Under expansion conditions, these cells expressed early cardiac markers such as GATA4, NKX2.5, and MEF2C, but no markers of differentiated cardiomyocytes such as α-myosin heavy chain (MYH6), β-myosin heavy chain (MYH7), cardiac actin (ACTA), or cardiac troponin I (TNNI). We examined cell surface marker expression by flow cytometry (Figure 1B). The cells uniformly expressed mesenchymal stem cell markers such as CD73, CD90, and CD105, whereas endothelial and hematopoietic markers (CD31, CD34, CD45), CD117 (KIT), CXCR4, and CD140 (PDGFRα) were not expressed. A significant proportion expressed CD146, suggesting that they could derive from pericytes (37). However, these cells did not express NG2, another pericyte marker. The characteristics of the isolated cells were further investigated by immunostaining. All cells were found positive for NKX2.5 (Figure 1C). In addition, NKX2.5-positive (NKX2.5POS) cells were MEF2CPOS. The vast majority of NKX2.5POS cells (73%) also expressed GATA4. Proliferation was examined using immunostaining directed against the proliferation markers Ki67 and phospho-Histone 3 (PH3). Respectively, 37% and 24% of NKX2.5POS were found Ki67POS and pH3POS. Altogether, these data indicated that the isolated population was composed of proliferative CPCs from mesenchymal origin, which are referred throughout this paper as adult CPCs. Interestingly, adult CPCs exhibited similar characteristics as CPCs previously isolated from the human fetal heart, which were shown to produce fully differentiated cardiomyocytes (31).
Figure 1.
Characterization and Differentiation of Adult CPCs
(A) Expression of early (GATA4, MEF2C, NKX2.5) and late cardiac markers (MYH6, MYH7, TNNI, CX43) in the human adult heart and in isolated cardiac precursor cells (CPCs). (B) Flow cytometry analysis of CD31, CD34, CD45, CD73, CD90, CD105, CD117, CXCR4, CD140a, CD146, and NG2 expression in adult CPCs. Histograms show control (gray) and specific fluorescent intensity signal (white). (C) Analysis of cardiac transcription factor and proliferation marker expression by immunostaining. Nuclei were stained with DAPI (blue). Scale bar: 20 μm. Quantification of NKX2.5POS/GATA4POS, NKX2.5POS/MEF2CPOS, NKX2.5POS/PH3POS, and NKX2.5POS/KI67POS CPCs. Data represent mean ± SEM (minimum 4,000 cells in 50 different fields at a magnification of 40× per quantification). (D) Gene expression of MYH6, MYH7, TNNI, ACTA, and MYH11 in proliferating CPCs (Expansion; black) and in adult CPCs exposed to DLL1 (gray), and differentiated in the absence (upper panels) or presence of DAPT (lower panels). Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating unstimulated (None; white) CPCs in absence of DAPT (n = 5). (E) Analysis of SM-MHC and α-ACTININ expression by immunostaining in CPCs exposed or not to DLL1 and differentiated in the absence or presence of DAPT. Nuclei were stained with DAPI (blue). Scale bars: 20 μm. (F) Quantification of SM-MHCPOS and α-ACTININPOS in adult CPCs exposed to DLL1 (gray), and differentiated in the absence or presence of DAPT. Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating unstimulated (None) (white) CPCs in absence of DAPT (minimum 2,000 cells in 30 different fields at a magnification of 40× per quantification). Diff = differentiation medium; Exp = expansion medium.
Human adult CPCs differentiate primarily into smooth muscle cells
To examine the potential of adult CPCs for producing a differentiated progeny, cells were exposed to a differentiation medium shown to induce robust cardiomyocyte differentiation in human fetal CPCs 31, 33. Under these conditions, however, adult CPCs produce exclusively smooth muscle cells (Supplemental Figures 1B and 1C). Three weeks after inducing differentiation, expression of smooth muscle myosin heavy chain (MYH11) was significantly increased. In addition, 80% of the cells were smooth muscle-myosin heavy chain (SM-MHC)POS by immunostaining, whereas <1% stained positive for the cardiomyocyte-specific marker α-ACTININ. Cardiac transcription factors (GATA4, MEF2C, NKX2.5) and differentiation markers (MYH6 and MYH7) were minimally induced. No endothelial cells were detected in differentiating cell cultures (data not shown). Therefore, adult CPCs displayed limited cardiogenic potential under these conditions, and differed markedly from fetal CPCs in their capacity to produce cardiomyocytes (31). In order to investigate the underlying mechanisms controlling adult CPC differentiation, we first compared expression of various cardiac markers in adult vs. fetal CPCs under basal and differentiation conditions. Cardiac transcription factors (GATA4, MEF2C, NKX2.5, and TBX5) were significantly more expressed in fetal than in adult CPCs, suggesting that fetal CPCs are more committed to the cardiac lineage than adult CPCs (Supplemental Figure 1D). Furthermore, MYH6 and MYH7 were significantly induced in fetal CPCs upon differentiation, whereas these genes were not activated in adult CPCs (Supplemental Figure 1E). On the contrary, MYH11 expression was induced in adult CPCs during the differentiation process.
NOTCH activation stimulates cardiac transcription factor expression and proliferation in adult CPCs
Because of the role of the NOTCH pathway in cardiac precursor renewal and differentiation during development and in the adult heart, we measured expression of NOTCH receptors and ligands in fetal and adult CPCs (Supplemental Figure 2A). Although expression of all NOTCH receptors was comparable between fetal and adult CPCs, NOTCH ligands, i.e., J1, J2, DLL1, DLL3, and DLL4 were significantly down-regulated in adult CPCs. The levels of HES1 and HEY1 were also significantly lower in adult CPCs, whereas HEY2 was more expressed. These results led us to postulate that activation of NOTCH signaling could contribute to sustaining cardiogenesis in fetal CPCs. Therefore, we investigated the possibility of restoring a cardiogenic potential in adult CPCs via manipulation of the NOTCH pathway. Adult CPCs were cultured on immobilized DLL1 ligand to activate NOTCH signaling (Supplemental Figure 2B). The efficiency of this procedure was demonstrated using the hN1 reporter cell line, in which YFP expression is driven by N1ICD/RBPJ (Supplemental Figure 2C) (38). Activation of NOTCH signaling in adult CPCs exposed to immobilized DLL1 was verified by the induction of HES1, HEY1, and HEY2 transcription (Supplemental Figure 2D). This manipulation had no significant effect on expression of NOTCH receptors and ligands. Exposure to DLL1 markedly stimulated proliferation in adult CPCs as judged by expression of several genes encoding positive (CCND1, C-MYC, and PCNA) and negative (CDKI1A) regulators of the cell cycle (Supplemental Figure 2E). Consistently, EdU (5-ethynyl-2′-deoxyuridine) incorporation was also higher in NOTCH-induced CPCs (Supplemental Figure 2F). We next evaluated the impact of NOTCH stimulation in adult CPCs on the expression of cardiac transcription factors. DLL1-mediated NOTCH activation induced a marked expression of GATA4, NKX2.5, MEF2C, and TBX5 (Supplemental Figures 2G and 2H). Importantly, exposure to DLL1 increased the number of NKX2.5POS, GATA4POS, MEF2CPOS CPCs (Supplemental Figure 2I).
Sequential NOTCH activation and inhibition in adult CPCs favors differentiation into cardiomyocytes
To investigate whether the increase in cardiac transcription factor expression following NOTCH activation restored a cardiogenic potential in adult CPCs, DLL1-stimulated CPCs were cultured under differentiation conditions. However, NOTCH-stimulated CPCs produced again exclusively smooth muscle cells (Figures 1D [upper panel] to 1F). Indeed, these cells demonstrated high MYH11 expression and no expression of cardiomyocyte markers such as MYH6 and MYH7. As a consequence, the vast majority of differentiated cells were SM-MHCPOS, whereas none expressed α-ACTININ. The action of NOTCH as an inducer of cardiac commitment and proliferation is in accordance with previous observations in CPCs and immature cardiomyocytes 10, 14, 39, 40. However, NOTCH also exerts inhibitory actions on terminal cardiac differentiation 10, 15, 40. Therefore, while promoting CPC specification, NOTCH could at the same time block cardiomyocyte production from committed CPCs. To examine this possibility, adult CPCs were exposed to DLL1 and induced to differentiate in the presence of DAPT, an inhibitor of gamma secretase and thereby of the NOTCH pathway (Supplemental Figure 3A). DAPT efficiently blocked NOTCH signaling during differentiation as judged by the blunted expression of NOTCH target genes in treated CPCs (Supplemental Figure 3B). In sharp contrast to what was observed in the absence of DAPT, a marked expression of cardiac transcription factors and genes expressed in terminally differentiated cardiomyocytes were readily observed in NOTCH-inhibited CPCs, which therefore produced α-ACTININPOS cardiomyocytes and very few smooth muscle cells (Supplemental Figure 3C, Figures 1D to 1F). Importantly, exposure to DLL1 was a prerequisite for restoring the cardiogenic potential in differentiating CPCs. Indeed, in the absence of this sensitization step, adult CPCs gave rise to smooth muscle cells even if treated with DAPT. These results underlined the importance of a transient NOTCH activation to promote cardiomyocyte specification. Moreover, we formally demonstrated the dependence on the NOTCH1 signaling pathway for specifying CPCs in the cardiomyocyte lineage by taking advantage of selective inhibitory antibodies directed against N1 (namely NRR1 [41]). Specific blockade of NOTCH1-mediated pathways in differentiating DLL1-sensitized CPCs was sufficient for producing cardiomyocytes (Supplemental Figure 3D). These experiments rules out nonspecific effects of DAPT in differentiating CPCs. Finally, it is important to note that CPCs exposed to a different NOTCH ligand, i.e., J1, were equally able to produce cardiomyocytes following NOTCH inhibition (Supplemental Figure 4). However, J1-activated CPCs did not demonstrate increased proliferation. Therefore, DLL1 was chosen for further experiments.
Adult CPC–derived cardiomyocytes exhibit functional electrical properties
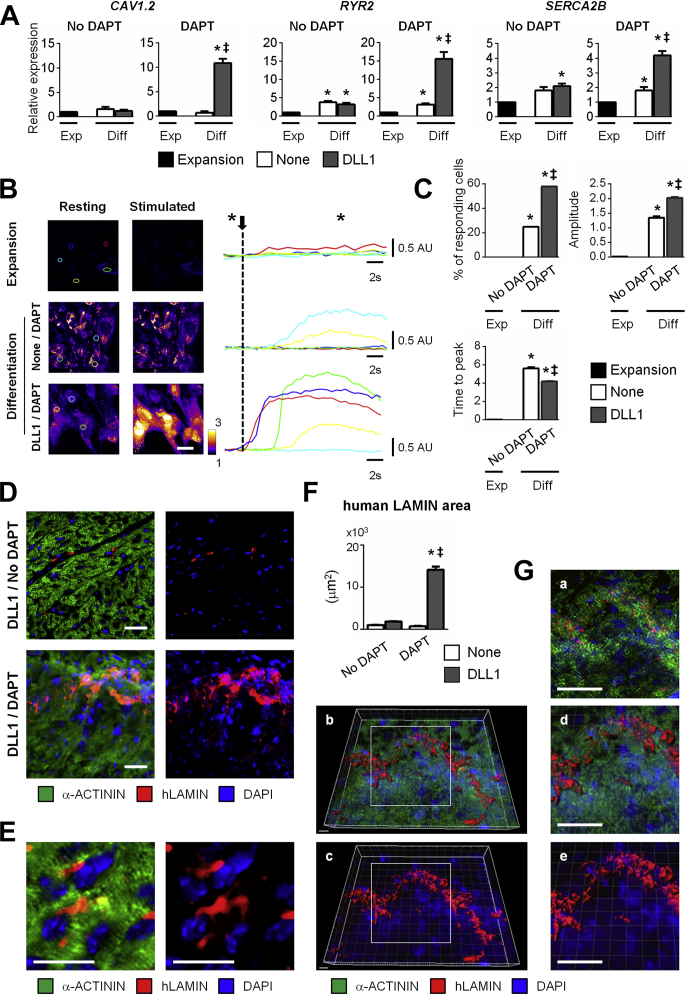
To functionally characterize CPC-derived cardiomyocytes, we first measured expression of several calcium handling proteins (Figure 2A). Expression of the L-type calcium channel (CAV1.2), ryanodine receptor (RYR2), and sarcoplasmic reticulum calcium ATPase (SERCA2B) was induced in adult CPCs differentiating into cardiomyocytes. Marginal expression was observed in situations giving rise to smooth muscle cells. To evaluate whether differentiating CPCs exhibited electrophysiological properties, we assessed Ca2+ signaling in resting and field-stimulated cells (Figure 2B). Under resting conditions or pacing (0.5 Hz), undifferentiated proliferating CPCs did not exhibit any calcium transients. Similarly, the 2 groups of differentiated cells did not show significant spontaneous calcium response. By contrast, electrical pacing triggered a marked and synchronized increase in cytosolic Ca2+ in CPC-derived cardiomyocytes produced by exposure to DLL1 and the presence of DAPT during differentiation. These findings were consistent with cardiomyocytes exhibiting functional calcium-induced calcium release when electrically stimulated. These calcium signals appear to last longer than those present in fully differentiated cardiomyocytes, similar to observations made in differentiating induced pluripotent stem cells (42). Although some calcium transients were detected in CPC-derived smooth muscle cells, the 2 differentiating populations differed markedly in their capacity to respond to electrical stimulation (Videos 1 and 2). Quantification of the response, as measured by determining the number of responding cells, amplitude of the calcium transients, and time to reach maximal Ca2+ release, supported electrical competence in CPC-derived cardiomyocytes as compared with smooth muscle cells (Figure 2C). These data demonstrated that the CPC-derived cardiomyocytes developed electrical properties similar to those of mature cardiomyocytes.
Figure 2.
Adult CPCs Produce Functional Cardiomyocytes In Vitro and In Vivo
(A) Expression of calcium handling proteins (CAV1.2, RYR2, SERCA2B) in proliferating CPCs (Expansion; black) and in adult CPCs exposed to DLL1 (gray), and differentiated in the absence or presence of DAPT. Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating unstimulated (None; white) CPCs in absence of DAPT (n = 4). (B) Cytosolic Ca2+ signals were recorded in adult CPCs in expansion (upper panels), and in differentiating CPCs following exposure or not to DLL1 in the presence of DAPT. Cells were either evaluated under resting conditions or electrical stimulated (10 s at 0.5 Hz). Scale bar: 50 μm. Representative tracing of normalized Ca2+ changes obtained following induction of pacing (arrow). Colored lines correspond to regions of recording as indicated in B. Images depicted in B were taken at the indicated time (*). See Videos 1 and 2. (C) Quantification of calcium transient parameters. Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating unstimulated (None) (white) CPCs in absence of DAPT (n = 55 cells per quantification). (D) Analysis of h-LAMIN (red) and α-ACTININ expression (green) in human CPC-derived cardiomyocytes in SCID mouse heart sections by immunostaining. DLL1-stimulated CPCs were injected in neonatal SCID mice. Mice were injected with DAPT or vehicle alone. Nuclei were stained with DAPI (blue). Scale bars: 20 μm. (E) High magnification of human hLAMINPOS α-ACTININPOS cardiomyocytes in the mouse myocardium. Scale bars: 20 μm. (F) Quantification of human hLAMINPOS cardiomyocyte patches in SCID mouse hearts. Data represent mean ± SEM; *p < 0.05 as compared with control group (unstimulated CPCs [None]; No DAPT administration); ‡p < 0.05 as compared with the group of animals injected with CPCs exposed to DLL1 (gray), but not injected with DAPT (minimum 3,000 cells in 40 different fields at a magnification of 40× per quantification, 3 animals per condition). (G) Confocal analysis and tridimensional reconstruction of a representative patch of human cardiomyocytes in the mouse myocardium. (a) Confocal image of a representative patch. Scale bars: 20μm. (b and c) Tridimensional reconstruction of the representative human cardiomyocyte depicted in a. hLAMIN (red), α-ACTININ expression (green), DAPI (blue). Scale bar: 5 μm. (d and e) High magnifications of areas indicated in b and c. Scale bar: 20 μm. Abbreviations as in Figure 1.
Electrical stimulation of CPC-derived smooth muscle cells. Related to Figure 2. Cytosolic Ca2+ signals were recorded in differentiated adult CPCs, not exposed to DLL1 in the absence of DAPT. Cells were evaluated following electrical stimulation (10 s at 0.5 Hz).
Electrical stimulation of CPC-derived cardiomyocytes. Related to Figure 2. Cytosolic Ca2+ signals were recorded in differentiated adult CPCs, following exposure to DLL1 in the presence of DAPT. Cells were evaluated following electrical stimulation (10 s at 0.5 Hz).
Adult human CPCs produce cardiomyocytes in vivo
To determine whether adult CPCs could produce cardiomyocytes in vivo, cells were injected into SCID mouse neonates intravenously. This protocol was previously demonstrated to result in significant engraftment and differentiation of CPCs in the post-natal heart (33). Cells were, therefore, either exposed or not to DLL1, and injected into neonatal mice, which then were administered with DAPT to favor cardiac differentiation (Supplemental Figure 5A). Twelve days thereafter, hearts were sectioned entirely and analyzed to determine the degree of CPC engraftment and differentiation. We first used PCR to detect human satellite DNA in cryosections. Provided that CPCs were exposed to DLL1, DAPT treatment promoted engraftment as shown by the increased number of PCR-positive sections in the relevant group (Supplemental Figure 5B). To further quantify engraftment and differentiation of human CPCs within the host myocardium, we performed immunostaining directed against human (h)LAMIN (human-specific antigen) and α-ACTININ (cardiac marker). Occasional undifferentiated human CPCs were detected in the hearts of mice receiving no DAPT (Figure 2D). Similarly, in mice treated with DAPT but injected with CPCs that were not exposed to DLL1, hLAMINPOS cells did not differentiated into cardiomyocytes (Supplemental Figure 5C). Strikingly, in the group receiving DLL1-sensitized CPCs and treated with DAPT, large clusters of hLAMINPOS human cells expressing α-ACTININ were readily detected (Figures 2D and 2E). Quantification of hLAMIN area confirmed that numerous human cells engrafted in the mouse myocardium under these conditions (Figure 2F). To ascertain the degree of differentiation of CPC-derived cardiomyocytes, we performed confocal microcopy. Z-stack analysis across hLAMINPOS human cardiomyocytes confirmed colocalization of hLAMIN and α-ACTININ (Supplemental Figure 5D). Sarcomeric organization confirmed that transferred CPCs gave rise to mature cardiomyocytes. Finally, we generated a tridimensional reconstruction of areas containing human cardiomyocytes, which supported functional integration of human cardiomyocytes in the mouse myocardium (Figure 2G). Altogether, these data indicated that NOTCH signaling could be modulated in vivo to enhance the specification of adult CPCs towards the cardiomyocyte fate.
NOTCH-dependent MIR-143/145 expression controls differentiation of adult CPCs into smooth muscle cells
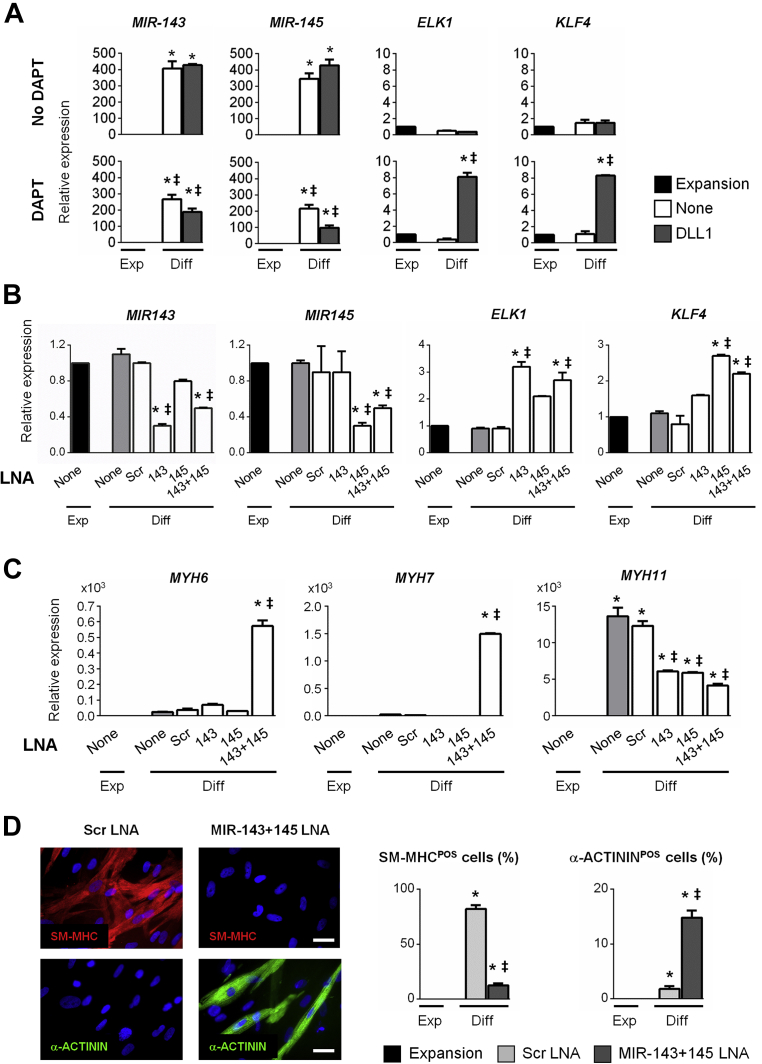
Proliferation and differentiation of smooth muscle cells has been shown to be regulated by MIR-143/145 20, 21. We therefore measured MIR-143/145 expression in fetal and adult CPCs (Supplemental Figure 6A). The expression levels of both microRNAs (miRNAs) were strongly down-regulated in fetal CPCs during spontaneous differentiation into cardiomyocytes. In sharp contrast, expression was markedly induced during differentiation of adult CPCs, consistent with their preferential commitment toward the smooth muscle cell lineage. We next evaluated whether MIR-143/145 expression was modulated in adult CPCs according to their differentiation into either cardiomyocytes or smooth muscle cells. Adult CPCs were, therefore, induced to differentiate following exposure to DLL1 in the presence or absence of DAPT. MIR-143/145 expression was measured under these different conditions (Figure 3A). Expression of MIR-143/145 was robustly increased in all situations giving rise to smooth muscle cells. On the contrary, expression of the two miRNAs was significantly blunted in CPCs giving rise to cardiomyocytes. Target genes of MIR-143/145, negatively controlling smooth muscle cells differentiation, have been previously identified (21). Expression of ETS-domain protein 1 (ELK1) and Krueppel-like factor 4 (KLF4) was, therefore, measured. ELK1 and KLF4 were found to be up-regulated in adult CPCs differentiating into cardiomyocytes, concomitantly with MIR-143/145 down-regulation (Figure 3A). These results demonstrated that MIR-143/145 were under control by the NOTCH pathway in adult CPCs.
Figure 3.
NOTCH-Mediated Regulation of Cell Fate Decision Between the Smooth Muscle Cell and Cardiomyocyte Lineages Depends on MIR-143/145 Expression
(A) Expression of MIR-143 and MIR-145 in proliferating CPCs (Expansion; black) and in adult CPCs exposed to DLL1 (gray), and differentiated in the absence (upper panels) or presence of DAPT (lower panels). Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating unstimulated (None; white) CPCs in absence of DAPT (n = 5). (B) Expression of MIR-143, -145, and miRNA targets (ELK1, KLF4) in proliferating CPCs (Expansion; black) and in differentiated adult CPCs (without prior DLL1 exposure, in the absence of DAPT). Cells were either transfected (white) or not (None; gray) with the indicated miRNA LNA inhibitor. Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating CPCs transfected with scrambled miRNA LNA inhibitor (n = 3). (C) Expression of sarcomeric proteins (MYH6, MYH7, MYH11) in proliferating CPCs (Expansion; black) and in differentiated adult CPCs (without prior DLL1 exposure, in the absence of DAPT). Cells were transfected (white) or not (None; gray) with the indicated miRNA LNA inhibitor. Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating CPCs transfected with scrambled miRNA LNA inhibitor (Scr) (n = 3). (D) Analysis of SM-MHC and α-ACTININ expression by immunostaining in differentiated CPCs (without prior DLL1 exposure, in the absence of DAPT), transfected or not with the indicated miRNA LNA inhibitor. Nuclei were stained with DAPI (blue). Scale bars: 50 μm. Quantification of the numbers of α-ACTININPOS cardiomyocytes or sm-MHCPOS smooth muscle cells. Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating CPCs transfected with scrambled miRNA LNA inhibitor (Scr) (minimum 2,000 cells in 30 different fields at a magnification of 40× per quantification). Abbreviations as in Figure 1.
To further investigate the importance of MIR-143/145 in lineage determination in adult CPCs, we used anti-MIR LNA inhibitors and miRNA mimics to modulate MIR-143/145 activity during adult CPC differentiation. We first cultured CPCs under conditions that favor smooth muscle cell differentiation (i.e., no exposure to DLL1 and no DAPT) (Supplemental Figure 6B), and transfected these cells with anti–MIR-143 and anti–MIR-145 LNA inhibitors, alone or in combination (Figures 3B and 3C). Efficient MIR blockade was demonstrated by the reduced mature MIR levels observed in transfected adult CPCs and the up-regulation of the corresponding target genes in these cells. CPCs receiving no inhibitors or transfected with a scrambled LNA inhibitor produced smooth muscle cells, as judged by high MYH11 expression and low expression of MYH6 and MYH7 in differentiating cells. Furthermore, CPCs transfected with either the anti–MIR-143 or the anti–MIR-145 LNA demonstrated a reduced capacity to produce smooth muscle cells. These cells, however, did not give rise to cardiomyocytes. In sharp contrast, CPCs transfected with a combination of anti–MIR-143 and anti–MIR-145 were characterized by up-regulation of MYH6 and MYH7, down-regulation of MYH11, and therefore, massively differentiate into cardiomyocytes (Figure 3D). We subsequently evaluated whether MIR-143/145 expression were sufficient to force CPCs to differentiate into smooth muscle cells. For this, CPCs were exposed to DLL1 and cultured in the presence of DAPT, a protocol inducing cardiomyocyte production (Supplemental Figure 6C). Untransfected CPCs or CPCs transfected with a scrambled miRNA mimic demonstrated high MYH6 and MYH7, and low MYH11 expression, indicating that these cells differentiated into cardiomyocytes (Supplemental Figures 6D and 6E). On the contrary, when CPCs were transfected with either a MIR-143 or a MIR-145 mimic, MIR-143/145 target genes were down-regulated, MYH6 and MYH7 expression was significantly blunted, and MYH11 expression was markedly induced, showing that CPCs were redirected into the smooth muscle cell lineage. Altogether, these data indicated that MIR-143 and MIR-145 were indispensable for specifying adult CPCs into the smooth muscle fate. It further demonstrated that concomitant inhibition of the 2 miRNAs was sufficient to promote cardiogenesis in adult CPCs.
Binary cell fate decision between the cardiomyocyte and smooth muscle cell fates in adult CPC clones
In order to demonstrate the potential of individual CPCs to adopt a cardiomyocyte and a smooth muscle cell fate, we derived a series of clones from proliferating adult CPCs. Figure 4 depicts results obtained with 1 clone, representative of 15 sharing a similar phenotype in 3 independent experiments. This clone expressed cardiac transcription factors and demonstrated a surface phenotype identical to that observed for the bulk population (Figures 4A and 4B). Although CD140a (PDGFRα) and NG2 were not detected on the surface of the different clones, these populations expressed homogenously CD146, confirming their probable pericyte origin. Furthermore, all clones were found positive for NKX2.5, GATA4, and MEF2C, and maintain a high proliferative capacity. Importantly, provided that they were subjected to DLL1 and DAPT during differentiation, the clones produced large amounts of cardiomyocytes (Figures 4D and 4E). Cardiomyocytes stained positive for α-ACTININ, troponin I (TNNI), and myosin light chain 2v (MLC2V), showing their relative mature phenotype. Furthermore, spontaneous and evoked calcium transients demonstrated electrical competence (Figure 4F). The differentiated cells also responded to caffeine, suggesting that calcium-induced calcium release operated normally. In the absence of this sequential NOTCH activation and inhibition, these clones produced exclusively smooth muscle cells. These results demonstrated the bipotential of single adult CPCs to differentiate into either smooth muscle cells or cardiomyocytes.
Figure 4.
Analysis of Representative Human CPC Clones
(A) Flow cytometric analysis of CD31, CD34, CD45, CD73, CD90, CD105, CD117, CXCR4, CD140a, CD146, and NG2 expression in 1 CPC clone. Histograms show control (gray) and specific fluorescent intensity signal (white). (B) Analysis of cardiac transcription factor expression by immunostaining. NKX2.5 (red); MEF2C (green). Nuclei were stained with DAPI (blue). (C) Expression of cardiac transcription factors (GATA4, MEF2C, NKX2.5), and sarcomeric proteins (MYH6, MYH7, MYH11) in proliferating adult CPCs (Expansion; black) and in adult CPCs exposed to DLL1 (gray), and differentiated in the absence (upper panels) or presence of DAPT (lower panels). Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating unstimulated (None; white) CPCs in absence of DAPT (n = 3). (D) Analysis of SM-MHC and α-ACTININ expression by immunostaining in CPCs exposed or not to DLL1 and differentiated in the absence or presence of DAPT. Nuclei were stained with DAPI (blue). Scale bars: 20 μm. (E) Analysis of α-ACTININ (green) and TNNI (magenta), or MLC2V (magenta) expression by immunostaining in CPCs-derived cardiomyocytes. Nuclei were stained with DAPI (blue). Scale bars: 20 μm. (F) Cytosolic Ca2+ were recorded in CPCs-derived cardiomyocytes. Cells were either evaluated under resting conditions, electrical stimulation (30 V, 2 ms at 5 Hz), or perfusion with caffeine. Spontaneous (top panel) and evoked (middle panel) Ca2+ oscillations were detected. Quantification shows the percentage of cells with spontaneous Ca2+ activity or responding to caffeine. Abbreviations as in Figure 1.
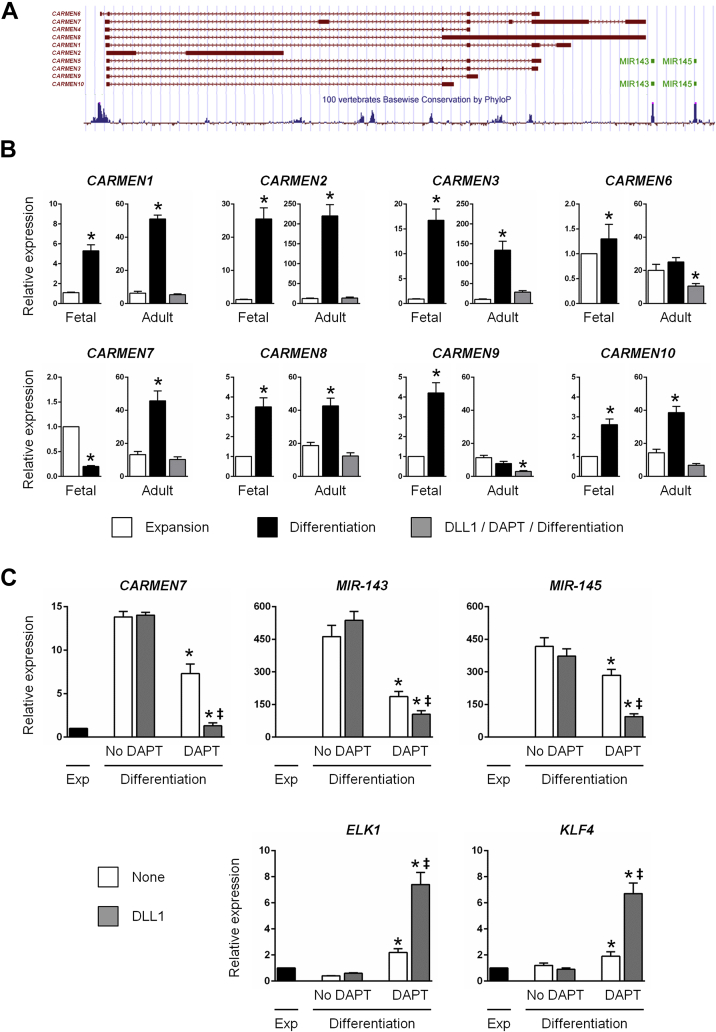
CARMEN, an enhancer-associated lncRNA, controls MIR-143/145 expression and the smooth muscle cell fate in adult CPCs
Using transcriptome profiling to identify lncRNAs that were differentially expressed in human fetal CPCs during differentiation into cardiomyocytes, we recently discovered CARMEN, an enhancer-associated lncRNA (32). Importantly, CARMEN is located in the genome immediately upstream of MIR-143/145, and corresponds to the enhancer that has been shown to be the target of NOTCH signaling in smooth muscle cells 20, 32. We therefore investigated whether CARMEN was implicated in lineage determination in adult CPCs. Ten different CARMEN isoforms have been identified. We thus named these isoforms CARMEN1 to 10 (Figure 5A). Primers were designed to measure individual expression of CARMEN1, 2, 3, 6, 7, 8, 9, and 10. Specifically, we measured the expression of each isoform in differentiating fetal CPCs that gave rise to cardiomyocytes, and in adult clonal CPCs differentiating into either smooth muscle cells or cardiomyocytes (Figure 5B). In adult CPCs differentiating into smooth muscle cells, all CARMEN isoforms were either up-regulated or not modulated. By contrast, in fetal CPCs, a single isoform, CARMEN7, was significantly down-regulated during differentiation into cardiomyocytes. In addition, expression of all isoforms including CARMEN7 was abolished in adult CPCs subjected to sequential NOTCH activation and inhibition to promote a cardiomyocyte fate. In order to determine whether CARMEN7 and MIR-143/145 were indeed coregulated in adult CPCs, we measured their expression in different situations favoring the emergence of a smooth muscle cell or a cardiomyocyte lineage (Figure 5C). In adult CPCs producing smooth muscle cells, CARMEN7 and MIR-143/145 were highly expressed. As a consequence, KLF4 and ELK1 expression was minimal. By contrast, CARMEN7 and MIR-143/145 were down-regulated in adult CPCs specified into the cardiomyocyte fate. In this case, the levels of KLF4 and ELK1 were elevated.
Figure 5.
CARMEN Transcripts Are Differentially Regulated During Fetal and Adult Differentiation Into Smooth-Muscle Cells Or Cardiomyocytes
(A) UCSC screenshot of the human CARMEN locus. The 10 CARMEN isoforms, that is, CARMEN1, 2, and 3, 4, 5, 6, 7, 8, 9, and 10 are indicated in red. The 2 miRNAs, MIR-143/145, are indicated in green. (B) Expression of CARMEN transcripts (CARMEN1, 2, 3, 6, 7, 8, 9, 10) in proliferating (Expansion; white), and in differentiated fetal and adult CPCs without prior DLL1 stimulation and cultured in the absence of DAPT (Differentiation; black) or after stimulation with DLL1 and cultured in presence of DAPT (DLL1/DAPT/Differentiation; light gray). Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs (n = 4). (C) Expression of CARMEN7, MIR-143, MIR-145, ELK1, and KLF4 transcripts in proliferating CPCs (Expansion [Exp]; black) and in differentiated adult CPCs (white and light gray), exposed or not to DLL1 in the absence or presence of DAPT. Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating unstimulated (None; white) CPCs in absence of DAPT (n = 3).
Targeting CARMEN7 inhibits smooth muscle commitment and promotes a cardiomyocyte fate in adult CPCs
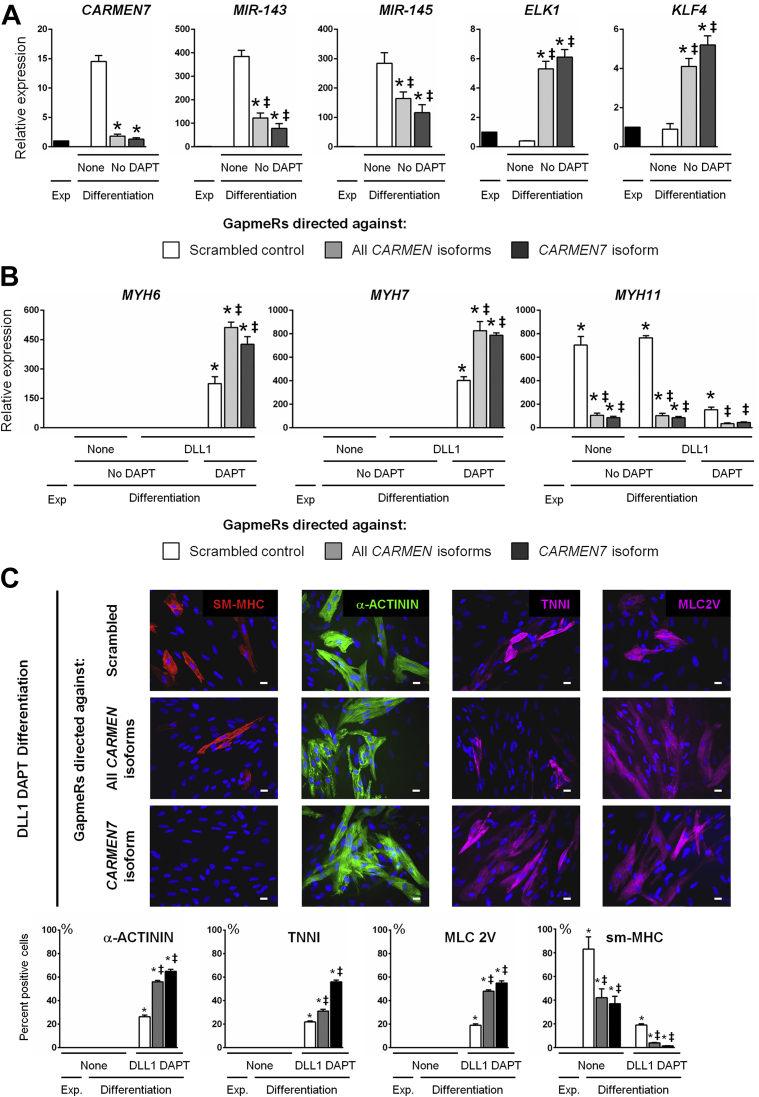
Since a positive correlation existed between CARMEN7 and MIR-143/145 expression, we postulated that CARMEN7 could directly control MIR-143/145 expression via cis-acting regulation, a mechanism that is common for enhancer-derived lncRNAs 30, 43. To test this hypothesis, we designed modified antisense oligonucleotides (GapmeRs) targeting either all CARMEN isoforms or specifically CARMEN7, that is, in the latter case, the unique exon that is specific of this particular isoform. Adult CPCs were induced to differentiate into smooth muscle cells (no DLL1 exposure, no DAPT), a situation in which all CARMEN isoforms are significantly expressed. Cells were in addition transfected with GapmeRs directed against either all CARMEN isoforms or uniquely CARMEN7. The experiment demonstrated that CARMEN7 was the only isoform specifically down-regulated by CARMEN7 GapmeRs, whereas the pan-CARMEN GapmeRs abolished expression of all isoforms (Supplemental Figure 7A). We next used these 2 different GapmeRs to evaluate the significance of CARMEN7 in the regulation of the smooth muscle cell versus cardiomyocyte fate in differentiating adult CPCs. We first evaluated the effects of CARMEN7 blockade on expression of MIR-143/145 and their respective target genes. Specific CARMEN7 knockdown resulted in a marked down-regulation of MIR-143/145 expression, and a simultaneous up-regulation of ELK1 and KLF4 expression (Figure 6A).
Figure 6.
CARMEN Down-Regulation Promotes CPC Differentiation Into Cardiomyocytes
(A) Expression of CARMEN7, MIR-143 MIR-145, ELK1, and KLF4 transcripts in proliferating CPCs (Expansion [Expansion]; black) and in differentiated (Differentiation) adult CPCs without prior DLL1 stimulation and cultured in the absence of DAPT (None No DAPT). Cells were transfected with either scrambled GapmeRs (white), GapmeRs directed against all CARMEN isoforms (light gray), or GapmeRs directed specifically against CARMEN7(dark gray). Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating CPCs (white) transfected with scrambled GapmeRs (n = 3). (B) Expression of sarcomeric proteins (MYH6, MYH7, MYH11) in proliferating CPCs (Expansion; black) and in differentiated adult CPCs exposed or not to DLL1 in the absence or presence of DAPT. Cells were transfected with either scrambled GapmeRs (white), GapmeRs directed against all CARMEN isoforms (light gray), or GapmeRs directed specifically against CARMEN7(dark gray). Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating CPCs (white) transfected with scrambled GapmeRs (n = 3). (C) Analysis of SM-MHC, α-ACTININ, TNNI, and MLC2V expression by immunostaining in differentiated CPCs exposed to DLL1 in the presence of DAPT, transfected with either scrambled GapmeRs (white), GapmeRs directed against all CARMEN isoforms (light gray), or GapmeRs directed specifically against CARMEN7(dark gray). Scale bar: 25 μm. Quantification of the numbers of α-ACTININPOS, TNNIPOS, or MLC2VPOS cardiomyocytes and sm-MHCPOS smooth muscle cells. Data represent mean ± SEM; *p < 0.05 as compared with proliferating CPCs; ‡p < 0.05 as compared with differentiating CPCs (white) transfected with scrambled GapmeRs (minimum 1,500 cells in 20 different fields at a magnification of 40× per quantification).
The role of CARMEN7 in the adoption of a particular fate was then investigated in adult CPCs. CPC clones were therefore cultured under differentiating conditions after no or previous exposure to DLL1, and in the absence or presence of DAPT (Supplemental Figure 7B). Cells were transfected with either control scrambled GapmeRs, GapmeRs targeting all CARMEN isoforms or CARMEN7-specific GapmeRs. Specification into the smooth muscle fate was completely blocked by GapmeRs directed against either all CARMEN isoforms or CARMEN7, as judged in particular by the down-regulation of MYH11 expression and the marked decrease in the number of SM-MHCPOS cells in all experimental conditions (Figures 6B and 6C). These findings confirmed the dependence of the smooth muscle lineage on CARMEN7 expression. The 2 GapmeRs alone did not, however, impact significantly cardiogenesis. Indeed, in transfected CPCs that were not exposed to DLL1 and not cultured in DAPT-containing medium, MYH6 and MYH7 expression was not different than under controlled conditions. Nevertheless, CARMEN knockdown strikingly reinforce the cardiogenic program in committed cells. Large amounts of cardiomyocytes were detected in this case. Specifically, up to 60% of CPC clones adopted a cardiomyocyte fate. In particular, these cells demonstrated expression of the cardiac proteins α-ACTININ, TNNI, and myosin light chain 2 (MLC2V) (Figure 6C). Moreover, most of the cells are characterized by the presence of organized sarcomeres.
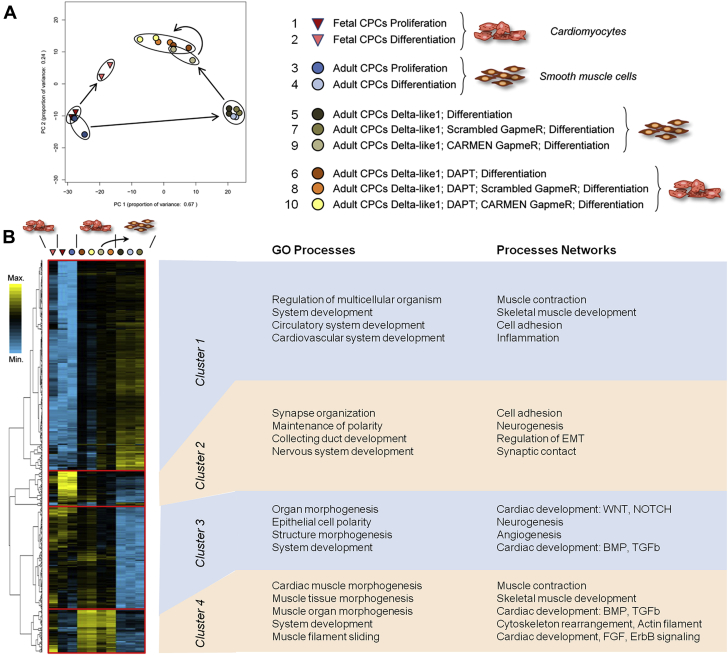
Gene regulatory network controlling CPC differentiation
In order to elucidate the transcriptional events associated with CPC differentiation, we profiled the transcriptome using RNA sequencing in differentiating fetal and adult CPCs under various conditions (Supplemental Table 2). We first performed a principal component analysis of all genes significantly modulated in either CPCs differentiating into cardiomyocytes or CPCs differentiating into smooth muscle cells (adjusted p value <0.05) (Figure 7A). Ninety-one percent of total variation can be attributed to the first 2 principal components. Strikingly, proliferating undifferentiated fetal and adult CPCs clustered together, consistent with the notion that these cells demonstrated a similar phenotype, and giving further support to previous analysis of surface marker expression (Figure 1) (31). In sharp contrast, differentiated fetal and adult CPCs clearly differed in this analysis, in accordance with their respective capacity to produce either cardiomyocytes or smooth muscle cells. However, adult CPCs exposed to DLL1 and induced to differentiate in the presence of DAPT, that is, producing cardiomyocytes, showed a shift along the PC1 and PC2 axes, and were similar to fetal CPC-derived cardiomyocytes. Hierarchical clustering confirmed that proliferating CPCs from fetal and adult hearts share an identical transcriptional signature (Figure 7B, 2nd and 3rd columns). Clustering analysis then demonstrated that genes activated in each situations giving rise to cardiomyocytes were down-regulated during differentiation into smooth muscle cells. Differentiation into cardiomyocytes was associated with relevant gene ontology (GO) terms and networks, such as cardiac development and morphogenesis, coupled to activation of specific cardiogenic pathways implicating important modulators of cardiogenesis, in particular NOTCH, WNT, BMP, FGF, and TGFβ (Clusters 3 and 4). Conversely, adult CPCs differentiating into smooth muscle cells expressed a transcriptional program associated with blood vessel development (Cluster 1).
Figure 7.
Transcriptional Signature Activated During CPC Differentiation Into Cardiomyocytes
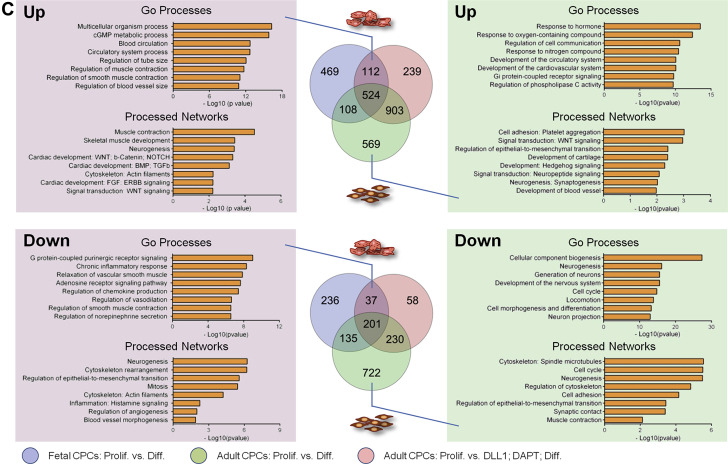
(A) Principal component analysis of genes differentially expressed during differentiation of CPCs into either cardiomyocytes or smooth muscle cells. Groups are indicated in the figure. Numbers refer to Supplemental Table 2. (B) Hierarchical clustering of genes differentially expressed during differentiation of CPCs into either cardiomyocytes or smooth muscle cells. Groups are indicated in the Figure. Gene ontology (Go) terms and biological processes annotated by Gene Go are indicated. (C) Venn diagram of up- and down-regulated genes common or unique to fetal CPCs differentiating in cardiomyocytes (blue), adult CPCs differentiating into smooth muscle cells (green), and adult CPCs differentiating into cardiomyocytes following exposure to DLL1 and DAPT treatment (red). Gene ontology terms and biological processes annotated by Gene Go are indicated for sets of genes common to cells differentiating into cardiomyocytes, and for set of genes unique to cells differentiating into smooth muscle cells. Abbreviations as in Figure 1.
In order to define a core transcriptional program controlling differentiation into cardiomyocytes, we first identified genes commonly modulated during fetal CPC differentiation and during differentiation of adult CPCs exposed to DLL1 and DAPT. Genes that were differentially expressed in adult CPCs differentiating into smooth muscle cells were subtracted from those that were common to both datasets (Figure 7C). We next determined GO category enrichment for up- and down-regulated genes. GO terms and networks associated with up-regulated genes (112 genes) were representative of cardiac differentiation and morphogenesis, and muscle contraction. On the contrary, negatively modulated genes (37 genes) were associated with smooth muscle cell differentiation, blood vessel morphogenesis, and angiogenesis. We also analyzed GO categories in up- and down-regulated genes, which were found uniquely modulated in adult CPCs differentiating into smooth muscle cells. Consistently, we found that up-regulated genes (569 genes) were associated with GO terms defining development of blood vessels. Interestingly, down-regulated genes (722 genes) were implicated primarily in neurogenesis.
To identify processes under control of NOTCH signaling, we next compared adult CPCs induced to differentiate following DLL1 exposure in the absence or presence of DAPT (Supplemental Figure 8A). GO term enrichment in genes that were up- and down-regulated exclusively in DLL1-sensitized CPCs cultured in DAPT-containing medium were determined. As expected, up-regulated genes (359 genes) were found to be implicated in cardiac development. Consistent with a control of smooth muscle cell differentiation by NOTCH-mediated expression of MIR-143/145, down-regulated genes (98 genes) were associated with blood vessel morphogenesis. Importantly, neurogenesis was also a prime target of NOTCH inhibition. A similar approach was used to determine processes under the control by the CARMEN. We compared differentiating adult CPCs following exposure to DLL1, transfected with either scrambled control GapmeRs or GapmeRs directed against all CARMEN isoforms in the absence of DAPT (Supplemental Figure 8B). Up-regulated genes (258 genes) were involved in cardiac muscle differentiation. This is in accordance with the transcriptional signature observed in the hierarchical clustering analysis (Figure 7B). Indeed, DLL1-activated adult CPCs treated with GapmeRs targeting CARMEN produced a signature similar to that of cardiogenic cells despite the fact that CARMEN-depleted adult CPCs produced smooth muscle cells. CARMEN down-regulation appeared to favor, therefore, the emergence of a cardiogenic program, even if not sufficient to redirect CPCs into a cardiomyocyte fate.
Several important factors and transcriptional regulators were identified among the top modulated genes in cardiogenic CPCs. We confirmed their differential expression in various situations by quantitative RT-PCR (Supplemental Figure 8C). In particular, HOPX, a transcription factor implicated in cardiomyocyte proliferation and differentiation, was markedly up-regulated (44). Hopx was recently shown to control cardiogenic commitment of cardiovascular progenitors in the developing mouse heart (45). Specifically, Hopx acts as a co-repressor in the BMP4 signaling pathway to modulate WNT and promote cardiogenesis. BMP2 and BMP4 expression were indeed induced in CPCs differentiating into cardiomyocytes. In addition, several WNT ligands were modulated, in particular WNT5A and WNT11, demonstrating a pattern of expression known to favor cardiogenic differentiation in cardiac progenitors (46). Key transcription factors playing crucial roles in cardiogenesis were found also up-regulated in adult CPCs giving rise to cardiomyocytes, such as ISL1, IRX1, IRX2, and IRX4 4, 47. Finally, to identify potential factors regulating specification into cardiomyocytes, we performed a predictive analysis of upstream regulators using the gene set common to fetal and adult CPCs producing cardiomyocytes (Figure 7C, Supplemental Table 3). Among cytokines and growth factors that were predicted to regulate cardiogenic commitment, several factors were involved in cardiomyocyte differentiation and maintenance of cell identity, such as BMP4, FGF2, and CXCL12 33, 48, 49, and as inducers of cell division in cardiac myocytes such as neuregulin (NRG1), oncostatin M (OSM), and interleukin (IL)13 50, 51.
Discussion
The rate of cardiomyocyte renewal in the adult mammalian heart is low, and not sufficient to replenish the heart with newly formed myocytes following injury (2). The reasons for this relative incapacity of the heart to induce effective regeneration are still obscure. Many laboratories have identified various kinds of CPCs, which demonstrate ability to generate cardiomyocytes in vitro 3, 4. However, these cells usually have a poor capacity to produce a functional myocardium in vivo. In the present study, we have isolated a subpopulation of multipotent clonogenic CPCs from human atrial appendages. These cells demonstrate a unique pattern of surface marker expression. Indeed, CPC clones express uniformly CD73, CD90, and CD105, which characterize these cells as multipotent mesenchymal stromal cells. In addition, the expression of CD146 suggest a possible pericyte origin (37). By contrast, clones express neither NG2 nor PDGFRα (CD140a). Cardiac-resident mesenchymal stromal cells that occupy a perivascular adventitial niche have been described (52). However, these cells express PDFGRα, contrary to the adult CPCs described in our study. Moreover, this also distinguishes them from pericytes recently isolated from human ventricles (53). Finally, these cells do not express CD117, CD31, CD34, CD45, and CXCR4. More importantly, human adult CPCs coexpressed NKX2.5, GATA4, and MEF2C. Furthermore, cardiac transcription factors such as TBX5 and HAND2 are up-regulated in CPCs differentiating into cardiomyocytes. In the normal heart, 25% of resident cardiac non-myocyte cells were found to be positive for GATA4, MEF2C, and TBX5 (54). Interestingly, forced expression of cardiac transcription factors, in combination with activators of Wnt and JAK/STAT signaling, was able to reprogram cardiac non-myocyte cells into cardiac progenitors, and finally, relatively mature cardiomyocytes 55, 56, 57. Nevertheless, intrinsic pre-specification in vivo is not sufficient, and cardiogenic non-myocyte cells do not contribute new cardiomyocytes post-myocardial infarction (54). The reason why pre-specified cells do not spontaneously complete cardiogenic differentiation is currently unknown. However, our results provide a possible scenario. It appears that adult CPCs preferentially produce smooth muscle cells over cardiomyocytes. This is in line with observations suggesting that increased perfusion more than de novo cardiogenesis produces beneficial effects in cell therapy for heart disease. Moreover, paracrine factors released from the transferred cells are thought to stimulate angiogenesis and promote repair in the injured heart (58). Therefore, reprogramming toward the cardiomyocyte fate might be needed in order to reveal the full potential of adult CPCs in vivo. Sequential activation and inhibition of NOTCH signaling is sufficient to redirect CPCs from a default smooth muscle commitment to a differentiation into cardiomyocytes. Importantly, results using clonal CPC populations formally demonstrate their capacity for development in either of 2, apparently mutually exclusive, lineages. Prior activation of NOTCH signaling via exposure to NOTCH ligand is a prerequisite for revealing the dormant capacity of CPCs to produce cardiomyocytes. The NOTCH pathway has been shown to promote CPC commitment and expansion in the developing and postnatal hearts 9, 17, 18. Accordingly, we show that key cardiac transcription factors such as NKX2.5, GATA4, MEF2C, and TBX5 are up-regulated in human adult CPCs exposed to DLL1, and proliferation is stimulated. This is also reminiscent of a situation observed in the heart of transgenic mice overexpressing Jagged1, in which the number of NKX2.5-positive CPCs is significantly increased (14). Of note, NOTCH also stimulates proliferation in immature cardiomyocytes 39, 40. Moreover, in embryonic stem cells and in undifferentiated precursors, NOTCH is known to inhibit cardiogenesis and maturation 10, 15. Our observation supports, therefore, the notion that NOTCH activates a cardiogenic program in adult CPCs while preventing terminal differentiation to promote expansion. In addition, one target of NOTCH signaling in adult CPCs is the locus encoding MIR-143/145, 2 miRNAs that are known to promote smooth muscle differentiation 20, 21. Therefore, NOTCH activates simultaneously a smooth muscle cell program. As a consequence, NOTCH inhibition is, on one hand, mandatory for alleviating the inhibitory action of NOTCH on cardiogenesis and, on the other hand, for blocking the activation of the smooth muscle program.
In the MIR-143/145 locus, NOTCH activates a proximal enhancer element that is associated with different isoforms of an enhancer-associated lncRNA. We recently named this enhancer-associated lncRNA CARMEN, and demonstrated its importance as a regulator of cardiac differentiation 20, 32. Enhancers are cis-regulatory genomic elements that integrate chromatin state transitions to elicit appropriate transcriptional and cellular responses. In recent years, it has become evident that pervasive transcription occurs at active enhancer sequences during different cellular contexts, in particular during specification and differentiation 29, 30. Interestingly, the vast majority of differentially modulated heart–enriched lncRNAs following myocardial infarction were derived from active enhancers 24, 27. Enhancer-associated lncRNAs activate target gene expression primarily via cis mechanisms involving chromatin looping between the enhancer sequences and their target genes within neighboring or distal domains 30, 43. Several studies, including ours, demonstrated that degradation of particular enhancer-associated lncRNAs is sufficient to reduce expression of their target coding genes 24, 27. In the present study, we show that specification toward the smooth muscle cell fate in adult CPCs depends on CARMEN expression. More specifically, among the different CARMEN isoforms that are produced upon enhancer activation, CARMEN7 is of crucial importance. A positive correlation exists in adult CPCs between CARMEN7 and MIR-143/145 expression. Moreover, CARMEN7 is down-regulated following sequential NOTCH activation and inhibition resulting in the reactivation of the cardiogenic program. Accordingly, CARMEN7 knockdown results in MIR-143/145 down-regulation and blocks differentiation of adult CPCs into smooth muscle cells. Altogether, these data are consistent with a cis regulation of MIR-143/145 expression by CARMEN via CARMEN7 production in adult CPCs. Targets of MIR-143/145 that regulate smooth muscle cell proliferation and differentiation have been identified (21). In particular, the pluripotency factor KLF4, a MIR-145 target, inhibits terminal differentiation and promotes expansion of smooth muscle cells following injury. Furthermore, the MIR-143 target ELK-1 is known as a competitive inhibitor of MYOCARDIN. Finally, other MIR-143/145 targets can be implicated in maintaining smooth muscle identity. For instance, MIR-145 has been recently shown to regulate TGFβ receptor II expression in smooth muscle cells (59). TGFβ is known to drive smooth muscle differentiation (60).
The situation observed in adult CPCs is in contrast to what is seen in fetal CPCs, in which CARMEN7 is not significantly expressed. In fetal CPCs spontaneously differentiating into cardiomyocytes, MIR-143/145 expression is inversely correlated with expression of other CARMEN isoforms, which are indispensable for the induction of cardiogenesis (32). It is also important to note that none of these CARMEN isoforms represent the precursor of the 2 miRNAs. Indeed, because a negative correlation exists in differentiating fetal CPCs between CARMEN and MIR-143/145 expression, CARMEN induction is associated to MIR-143/145 down-regulation, and CARMEN depletion induces MIR-143/145 expression. Moreover, CARMEN boundaries have been well defined by analysis of promoter-specific histone modification and polyA signal frequency 61, 62. Isoforms of CARMEN, and in particular CARMEN7, terminate at a position situated upstream of the 2 miRNAs. Finally, CARMEN regulates cardiogenesis independently of MIR-143/145 expression in fetal CPCs (32). Altogether, this suggests also that CARMEN transcripts other than CARMEN7 exert trans regulatory function to initiate the cardiogenic program. Indeed, many characterized lncRNAs operate in trans as decoys or recruiters for transcription factors and chromatin-remodeling complexes. LncRNAs act efficiently at remote locations in the genome via their particular affinity with RNA-binding proteins such as components of the Trithorax and Polycomb complexes to activate or silence specific expression programs (63).
Gene clustering analysis supports the view that smooth muscle cell specification in adult CPCs prevents expression of the cardiogenic program. GO analysis demonstrates that cardiac differentiation is not induced in adult CPCs not exposed to DLL1 or in NOTCH-activated cells without following inhibition. In these instances, MIR-143/145 expression is activated and smooth muscle cells are produced. Interestingly, CARMEN down-regulation alone is not sufficient to implement cardiomyocyte production. Strikingly, however, adult CPCs exposed to DLL1 and transfected with anti-CARMEN GapmeRs are characterized by a gene expression pattern similar to that observed in adult CPCs differentiating into cardiomyocytes. Therefore, cardiogenesis is initiated by concomitant activation of the NOTCH pathway and CARMEN down-regulation but is not productively efficient. This observation emphasizes the strength of NOTCH-mediated inhibition on cardiac differentiation and maturation. This also suggests that NOTCH blockade exerts pro-cardiogenic effects beyond CARMEN down-regulation and subsequent modulation of MIR-143/145 expression. Our transcriptional data indicate that neurogenesis might need to be inhibited before the cardiogenic program can be initiated. Along this line, a binary cell fate decision between the cardiac mesoderm and the neuroectoderm has been shown to be controlled by NOTCH in embryonic stem cells (15). Remarkably, under conditions of NOTCH and CARMEN inhibition, up to 60% of differentiated cells are terminally differentiated cardiomyocytes, and practically none are smooth muscle cells. Altogether, these data suggest that NOTCH and CARMEN cooperate to control specification into the cardiomyocyte and smooth muscle lineages. Within this gene regulatory network controlling CPC differentiation, MIR-143/145 down-regulation is a mandatory step that permits cardiomyocyte specification. The visual abstract proposes a schematic of the molecular pathways implicated in the control of fate in human adult CPCs. Interestingly, HOPX is highly induced in CPCs specified to the cardiac lineage. This transcriptional repressor has been recently shown to be expressed in cardiogenic cell intermediates in the developing mouse heart (45). Hopx is necessary for BMP4-mediated repression of Wnt signaling. Importantly, Hopx-positive cells give rise exclusively to cardiomyocytes. It is therefore possible that reactivation of this developmental pathway in adult CPCs forces the cells to adopt a cardiomyocyte fate.
Appropriately specified CPCs produce functional cardiomyocytes in vivo and in vitro. Cardiomyocytes demonstrate spontaneous and triggered calcium signals. In particular, rapid synchronized calcium transients are readily observable in CPC-derived cardiomyocytes following electrical stimulation, indicating that cardiomyocytes develop electrical competence similar to that of mature cardiomyocytes. Furthermore, CPCs differentiate into cardiomyocytes in vivo. These cells engraft in the mouse post-natal heart and form structures that appear functionally integrated in the host myocardium. These findings indicate that the atrial appendage–derived CPCs described in the present study could represent a valuable source of precursors in cell replacement therapy for heart disease. Provided that NOTCH is inhibited following CPC injection into the damaged myocardium, cardiomyocytes could be produced at the site of injury. In this regard, although unspecific γ-secretase inhibitors can produce toxicity in vivo, in particular in the gut, this difficulty can be overcome using selective blockers of NOTCH receptor paralogs (41). We have indeed tested this possibility, and demonstrated that NRR1 antibodies can substitute for DAPT during production of CPC-derived cardiomyocytes. Finally, we recently demonstrated that CARMEN is induced in human cardiac pathologies and in mouse models of cardiac disease (32). In this context, it would be interesting to evaluate whether manipulation of endogenous cardiogenic mesenchymal precursors in vivo via modulation of the NOTCH-CARMEN-MIR-143/145 axis is able to favor cardiomyocyte renewal in the damaged heart, and promotes true heart regeneration.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Cellular replacement therapies for heart disease represent promising therapeutic approaches. However, even if improvement in cardiac function is sometime observed, the generation of a new myocardium in the human heart has never been demonstrated. This has been attributed to a relative incapacity of cardiac precursor cells to produce cardiomyocytes. Long noncoding (lnc)RNAs are emerging as master regulator of cell specification and differentiation. Within this context, these molecules play crucial roles in generating cell-specific programs, which regulate cell identity. These properties can be utilized to program precursor cells to adopt a cardiomyocyte fate.
TRANSLATIONAL OUTLOOK: Future research should take advantage of the high potential of lncRNAs to regulate cell-specific patterns of gene expression to control the fate of precursor cells used in cell therapies and improved cardiomyocyte production in the damaged heart.
Conclusions and Limitations
With the advent of reprogramming technologies, a novel approach to regeneration has emerged. In the context of our study, reprogramming particularly pertains to the controlled differentiation of CPCs into specific cardiac cells. Our study demonstrates that the subclass of lncRNAs that are associated to enhancer sequences can be manipulated to specify human CPCs into particular fates. Moreover, our findings suggest that the different lncRNA isoforms, produced by a single locus, regulate distinct gene programs, producing therefore diverse cellular responses. LncRNA isoform-specific effects certainly warrant further investigation. Altogether, these findings open new perspectives for the treatment of heart disease, and more specifically in cell replacement therapies. Several points should be however considered. MIR-143/145 are most likely not the sole targets of CARMEN transcripts. Within the frame of our study, we have not investigated trans mechanisms of actions. LncRNAs are known to regulate gene transcription at remote locations in the genome, different from their own site of transcription. In addition, LncRNAs can translocated into the cytoplasm whether they can associate with specific modulators of cell identity and behavior. Furthermore, NOTCH signaling exerts also a variety of actions that should be investigated. This integrated network implicating a signaling pathway, lncRNAs and miRNAs provide nevertheless a fascinating example of the complexity of the molecular mechanisms that regulate cell plasticity in organs such as the heart.
Acknowledgments
The authors are grateful to Andrée Porret for her technical assistance. The authors thank Sandra Calderon and Sylvain Pradervand, Genomic Technologies Facility, University of Lausanne, Switzerland, for helping with transcriptomic data analysis, and Yannick Kremp, Cellular Imaging Facility, University of Lausanne, Switzerland, for providing expertise in confocal microscopy. The authors also thank Dr. Michael B. Elowitz, California Institute of Technology, Pasadena, California, for making the hN1 reporter cell line available to them. The NRR1 antibodies were a kind gift from Christian W. Siebel, Genentech, South San Francisco, California.
Footnotes
This project was supported in part by grants to Dr. Pedrazzini from the Swiss National Science Foundation, Bern, Switzerland (grants no 33CM30-124090 and no 406340-128129) and by the Novartis Foundation for Medical-Biological Research, Basel, Switzerland (grant no 15A048). The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Appendix
References
- 1.Towbin J.A., Bowles N.E. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- 2.Garbern J.C., Lee R.T. Cardiac stem cell therapy and the promise of heart regeneration. Cell Stem Cell. 2013;12:689–698. doi: 10.1016/j.stem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Berlo J.H., Molkentin J.D. An emerging consensus on cardiac regeneration. Nat Med. 2014;20:1386–1393. doi: 10.1038/nm.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahara M., Santoro F., Chien K.R. Programming and reprogramming a human heart cell. EMBO J. 2015;34:710–738. doi: 10.15252/embj.201490563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nemir M., Pedrazzini T. Functional role of Notch signaling in the developing and postnatal heart. J Mol Cell Cardiol. 2008;45:495–504. doi: 10.1016/j.yjmcc.2008.02.273. [DOI] [PubMed] [Google Scholar]
- 6.de la Pompa J.L., Epstein J.A. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopan R. Notch signaling. Cold Spring Harb Perspect Biol. 2012;4:a011213. doi: 10.1101/cshperspect.a011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iso T., Kedes L., Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 9.Urbanek K., Cabral-da-Silva M.C., Ide-Iwata N. Inhibition of notch1-dependent cardiomyogenesis leads to a dilated myopathy in the neonatal heart. Circ Res. 2010;107:429–441. doi: 10.1161/CIRCRESAHA.110.218487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croquelois A., Domenighetti A.A., Nemir M. Control of the adaptive response of the heart to stress via the Notch1 receptor pathway. J Exp Med. 2008;205:3173–3185. doi: 10.1084/jem.20081427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gude N.A., Emmanuel G., Wu W. Activation of Notch-mediated protective signaling in the myocardium. Circ Res. 2008;102:1025–1035. doi: 10.1161/CIRCRESAHA.107.164749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kratsios P., Catela C., Salimova E. Distinct roles for cell-autonomous Notch signaling in cardiomyocytes of the embryonic and adult heart. Circ Res. 2010;106:559–572. doi: 10.1161/CIRCRESAHA.109.203034. [DOI] [PubMed] [Google Scholar]
- 13.Russell J.L., Goetsch S.C., Gaiano N.R., Hill J.A., Olson E.N., Schneider J.W. A dynamic notch injury response activates epicardium and contributes to fibrosis repair. Circ Res. 2011;108:51–59. doi: 10.1161/CIRCRESAHA.110.233262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemir M., Metrich M., Plaisance I. The Notch pathway controls fibrotic and regenerative repair in the adult heart. Eur Heart J. 2014;35:2174–2185. doi: 10.1093/eurheartj/ehs269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemir M., Croquelois A., Pedrazzini T., Radtke F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res. 2006;98:1471–1478. doi: 10.1161/01.RES.0000226497.52052.2a. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y., Li P., Liu K. Timely inhibition of Notch signaling by DAPT promotes cardiac differentiation of murine pluripotent stem cells. PLoS One. 2014;9:e109588. doi: 10.1371/journal.pone.0109588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boni A., Urbanek K., Nascimbene A. Notch1 regulates the fate of cardiac progenitor cells. Proc Natl Acad Sci U S A. 2008;105:15529–15534. doi: 10.1073/pnas.0808357105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Gude N., Joyo E., Toko H. Notch activation enhances lineage commitment and protective signaling in cardiac progenitor cells. Basic Res Cardiol. 2015;110:488. doi: 10.1007/s00395-015-0488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., Ashraf M., Wang Y. The role of notch 1 activation in cardiosphere derived cell differentiation. Stem Cells Dev. 2012;21:2122–2129. doi: 10.1089/scd.2011.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boucher J.M., Peterson S.M., Urs S., Zhang C., Liaw L. The miR-143/145 cluster is a novel transcriptional target of Jagged-1/Notch signaling in vascular smooth muscle cells. J Biol Chem. 2011;286:28312–28321. doi: 10.1074/jbc.M111.221945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordes K.R., Sheehy N.T., White M.P. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttman M., Donaghey J., Carey B.W. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ounzain S., Pezzuto I., Micheletti R. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J Mol Cell Cardiol. 2014;76C:55–70. doi: 10.1016/j.yjmcc.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klattenhoff C.A., Scheuermann J.C., Surface L.E. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grote P., Wittler L., Hendrix D. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ounzain S., Micheletti R., Beckmann T. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J. 2015;36:353–368. doi: 10.1093/eurheartj/ehu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boon R.A., Jae N., Holdt L., Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67:1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 29.Orom U.A., Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ounzain S., Pedrazzini T. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends Cardiovasc Med. 2015;25:592–602. doi: 10.1016/j.tcm.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Gonzales C., Ullrich N.D., Gerber S., Berthonneche C., Niggli E., Pedrazzini T. Isolation of cardiovascular precursor cells from the human fetal heart. Tissue Eng Part A. 2012;18:198–207. doi: 10.1089/ten.tea.2011.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ounzain S., Micheletti R., Arnan C. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol. 2015;89:98–112. doi: 10.1016/j.yjmcc.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Rosenblatt-Velin N., Lepore M.G., Cartoni C., Beermann F., Pedrazzini T. FGF-2 controls the differentiation of resident cardiac precursors into functional cardiomyocytes. J Clin Invest. 2005;115:1724–1733. doi: 10.1172/JCI23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie M.E., Phipson B., Wu D. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.da Silva Meirelles L., Caplan A.I., Nardi N.B. In search of the in vivo identity of mesenchymal stem cells. Stem Cells. 2008;26:2287–2299. doi: 10.1634/stemcells.2007-1122. [DOI] [PubMed] [Google Scholar]
- 38.Sprinzak D., Lakhanpal A., Lebon L. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campa V.M., Gutierrez-Lanza R., Cerignoli F. Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J Cell Biol. 2008;183:129–141. doi: 10.1083/jcb.200806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collesi C., Zentilin L., Sinagra G., Giacca M. Notch1 signaling stimulates proliferation of immature cardiomyocytes. J Cell Biol. 2008;183:117–128. doi: 10.1083/jcb.200806091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y., Cain-Hom C., Choy L. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 42.Lan F., Lee A.S., Liang P. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W., Notani D., Rosenfeld M.G. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 44.Kook H., Epstein J.A. Hopping to the beat. Hop regulation of cardiac gene expression. Trends Cardiovasc Med. 2003;13:261–264. doi: 10.1016/s1050-1738(03)00107-5. [DOI] [PubMed] [Google Scholar]
- 45.Jain R., Li D., Gupta M. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science. 2015;348:aaa6071. doi: 10.1126/science.aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gessert S., Kuhl M. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ Res. 2010;107:186–199. doi: 10.1161/CIRCRESAHA.110.221531. [DOI] [PubMed] [Google Scholar]
- 47.Kim K.H., Rosen A., Bruneau B.G., Hui C.C., Backx P.H. Iroquois homeodomain transcription factors in heart development and function. Circ Res. 2012;110:1513–1524. doi: 10.1161/CIRCRESAHA.112.265041. [DOI] [PubMed] [Google Scholar]
- 48.Cagavi E., Bartulos O., Suh C.Y. Functional cardiomyocytes derived from Isl1 cardiac progenitors via Bmp4 stimulation. PLoS One. 2014;9:e110752. doi: 10.1371/journal.pone.0110752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malliaras K., Ibrahim A., Tseliou E. Stimulation of endogenous cardioblasts by exogenous cell therapy after myocardial infarction. EMBO Mol Med. 2014;6:760–777. doi: 10.1002/emmm.201303626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bersell K., Arab S., Haring B., Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 51.O'Meara C.C., Wamstad J.A., Gladstone R.A. Transcriptional reversion of cardiac myocyte fate during Mammalian cardiac regeneration. Circ Res. 2015;116:804–815. doi: 10.1161/CIRCRESAHA.116.304269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chong J.J., Chandrakanthan V., Xaymardan M. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell. 2011;9:527–540. doi: 10.1016/j.stem.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen W.C., Baily J.E., Corselli M. Human myocardial pericytes: multipotent mesodermal precursors exhibiting cardiac specificity. Stem Cells. 2015;33:557–573. doi: 10.1002/stem.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furtado M.B., Costa M.W., Pranoto E.A. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res. 2014;114:1422–1434. doi: 10.1161/CIRCRESAHA.114.302530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qian L., Berry E.C., Fu J.D., Ieda M., Srivastava D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc. 2013;8:1204–1215. doi: 10.1038/nprot.2013.067. [DOI] [PubMed] [Google Scholar]
- 56.Song K., Nam Y.J., Luo X. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lalit P.A., Salick M.R., Nelson D.O. Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell. 2016;18:354–367. doi: 10.1016/j.stem.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hodgkinson C.P., Bareja A., Gomez J.A., Dzau V.J. Emerging concepts in paracrine mechanisms in regenerative cardiovascular medicine and biology. Circ Res. 2016;118:95–107. doi: 10.1161/CIRCRESAHA.115.305373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao N., Koenig S.N., Trask A.J. MicroRNA miR145 regulates TGFBR2 expression and matrix synthesis in vascular smooth muscle cells. Circ Res. 2015;116:23–34. doi: 10.1161/CIRCRESAHA.115.303970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Owens G.K., Kumar M.S., Wamhoff B.R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 61.Derrien T., Johnson R., Bussotti G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H., Hu J., Recce M., Tian B. PolyA_DB: a database for mammalian mRNA polyadenylation. Nucleic Acids Res. 2005;33:D116–D120. doi: 10.1093/nar/gki055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.