Abstract
We isolated PTD, a member of the DEFICIENS (DEF) family of MADS box transcription factors, from the dioecious tree, black cottonwood (Populus trichocarpa). In females, in situ hybridization experiments showed that PTD mRNA was first detectable in cells on the flanks of the inflorescence meristem, before differentiation of individual flowers was visually detectable. In males, the onset of PTD expression was delayed until after individual flower differentiation had begun and floral meristems were developing. Although PTD was initially expressed throughout the inner whorl meristem in female and male flowers, its spatial expression pattern became sex-specific as reproductive primordia began to form. PTD expression was maintained in stamen primordia, but excluded from carpel primordia, as well as vegetative tissues. Although PTD is phylogenetically most closely related to the largely uncharacterized TM6 subfamily of the DEF/APETELA3(AP3)/TM6 group, its spatio-temporal expression patterns are more similar to that of DEF and AP3 than to other members of the TM6 subfamily.
One of the greatest challenges in plant developmental biology is ascertaining the genetic mechanisms responsible for the great variety of floral forms. We are studying floral developmental genetics in black cottonwood (Populus trichocarpa), a dioecious tree with unusual two-whorled flowers. Black cottonwood is a fast-growing tree native to the northwestern United States and Canada (Fig. 1A). It is the dominant species in many riparian ecosystems and is commonly used in breeding hybrids for use in fiber and energy plantations. Several factors render cottonwoods (black cottonwood and Populus deltoides) attractive as model tree species: their small genome size (approximately 1.1 pg per diploid cell and approximately 500 mbp per haploid genome [Bradshaw and Stettler, 1993]), the existence of an efficient transformation system (Han et al., 1996), ease of propagation, and a relatively short time from germination to flowering (≤5 years).
Figure 1.
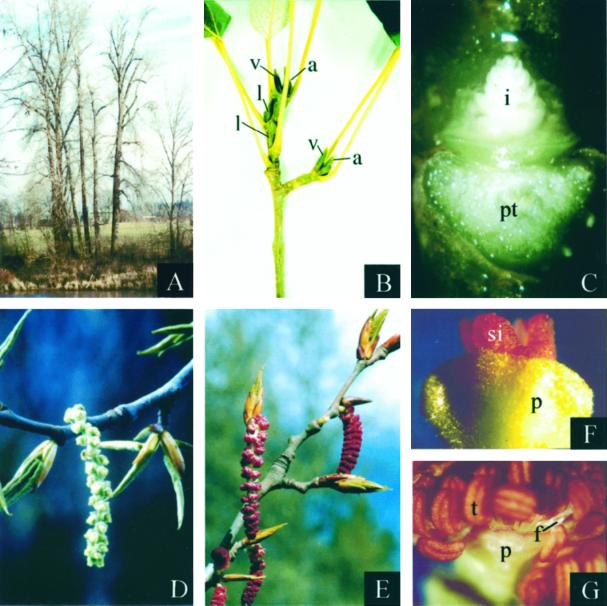
Black cottonwood floral morphology. Images are digitized. A, Mature black cottonwood trees on a riverbank during winter in Corvallis, Oregon. B, Developing inflorescence and vegetative buds in early summer. C, Developing inflorescence bud in early summer. Bud scales and the adjacent leaf petiole were removed. D, Mature female catkin in early spring. E, Mature male catkins in early spring. F, Mature female flower in early spring. G, Mature male flower in early spring. a, Axillary accessory inflorescence bud; f, filament of stamen; i, developing inflorescence; l, lateral axillary inflorescence bud; p, perianth cup; pt, petiole attachment (petiole removed); t, stamen; si, stigma; v, vegetative bud.
The flowers of black cottonwood develop on pendulous inflorescences (aments and catkins) that begin to develop in spring (late April to early May in Corvallis, OR) of the year before the spring in which they will open. Inflorescence buds develop as axillary accessory buds on short shoots and as axillary lateral buds near branch tips (Fig. 1, B and C). After anthesis, the new inflorescences begin to elongate rapidly within the bud scales. Concurrently, bract primordia develop acropetally along the inflorescence flanks. As the bract primordia enlarge, cells in the axils of the bracts become organized into flattened floral discs (Boes and Strauss, 1994). Continued growth at the perimeter of each floral disc produces a raised ring of tissue that will develop into an unusual structure known as the perianth cup. Vascular traces and developmental morphology indicate that the perianth cup is derived from the fusion of perianth parts (Fisher, 1928; Stoehr et al., 1988; Kaul, 1995). However, it remains unclear if the perianth cup is derived from fused sepals, fused petals, or adnation of petals and sepals. In females, three carpel primordia arise from the floral meristem and later unite to form a unilocular ovary. The mature female inflorescence bears 20 to 40 flowers, with each flower consisting of a stigma surrounded by a perianth cup (Figs. 1, D and F, and 2A). In males, stamen primordia begin to arise in the center of the floral disc and organogenesis proceeds centrifugally. The mature male flower consists of 40 to 60 stamens surrounded by a perianth cup (Figs. 1, E and G, and 2B).
Figure 2.
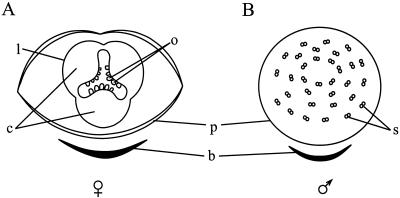
Diagrams of female and male black cottonwood flowers. Each flower consists of only two whorls: the perianth cup and either stamens or a pistil. A, Female flower with three fused carpels. The double line denoting the perianth cup indicates infolding that occurs at the rim. B, Male flower with 40 to 60 stamens. b, Bract; c, carpel; l, pistil; o, ovule; p, perianth cup; s, stamen.
Dioecy is estimated to occur in less than 4% of the angiosperms and is achieved by different means (for review, see Ainsworth et al., 1998). Most commonly, male and female organs are initiated, and then production of either ovules or pollen is blocked at a later stage. More rarely, only organs of a single sex are initiated and the floral meristems are thus unisexual. Detailed light and electron microscopy studies indicate that cottonwoods belong to the latter group in which initiation of stamen development does not occur in female flowers and initiation of carpel development does not occur in male flowers (Boes and Strauss, 1994; Kaul, 1995; Sheppard, 1997). The cottonwood flower thus consists of only two whorls of organs: the perianth cup and either stamens or a pistil.
Bulked segregant analyses of randomly amplified polymorphic DNA data indicate that sex determination in cottonwoods may be controlled by multiple loci acting in an additive or epistatic manner (McLetchie and Tuskan, 1994). However, perfect flowers and flowers of inappropriate sex have been noted on rare occasions (Larsen, 1970; Stettler, 1971) and environmental factors such as infestation by Eriophyid mites have been implicated in sex modification (Heslop-Harrison, 1924). The exact mode of sex determination in cottonwoods remains unclear.
We are studying floral homeotic genes in black cottonwood to better understand the genetic factors involved in floral development and to facilitate genetic engineering of reproductively sterile trees for the purpose of transgene containment. Most floral homeotic genes isolated to date belong to the MADS box family of transcription factors (for review, see Riechmann and Meyerowitz, 1998). Because MADS box genes are known to play fundamental roles in floral development, variations in floral MADS box gene sequences are likely to play a significant role in specifying the great diversity of floral forms. Recent phylogenetic analyses based on DNA sequences (Soltis et al., 1999), fossil evidence, and traditional classifications based on morphology (for review, see Eckenwalder, 1996) suggest that the two-whorled black cottonwood flowers evolved from a four-whorled flower typically found among the higher eudicots. The establishment of the four organ types of the typical eudicot flower are largely explained by the ABC model of flower development, which describes the combinatorial action of three classes of homeotic genes (for review, see Riechmann and Meyerowitz, 1998). From the standpoint of understanding how regulation of cottonwood floral development deviates from this model, the B-class organ identity genes are of particular interest. The combined action of A and B class genes specify petals, whereas B and C class genes specify stamens. The study of B-class genes in cottonwood may therefore provide insight into the derivation of the perianth cup and unisexuality. In Arabidopsis and snapdragon, two MADS box genes belonging to sister clades are necessary to specify B-function. The Arabidopsis genes are AP3 and PISTILLATA (PI), whereas the corresponding snapdragon orthologs are DEF and GLOBOSA (GLO).
We describe the isolation of PTD, a MADS box gene from black cottonwood that has homology with the floral homeotic transcription factors DEF (Sommer et al., 1990) and AP3 (Jack et al., 1992). Although phylogenetic analyses indicate that PTD is a member of the largely uncharacterized TM6 clade of the DEF/AP3/TM6 family, we report that the expression patterns of PTD are more similar to those of DEF and AP3 than to other members of the TM6 group. PTD is not expressed in vegetative tissues and its spatial and temporal expression patterns are sex-specific.
RESULTS
cDNA and Genomic Clone Isolation and Structure
To identify black cottonwood MADS box genes potentially involved in floral development, an early female inflorescence cDNA library was probed at low stringency with the MADS box region of the Arabidopsis AGL1 gene. One of the cDNA sequences subsequently isolated had high homology with the DEF gene from snapdragon and is referred to here as PTD (black cottonwood DEF-like). The PTD cDNA was 917 bp in length with an open reading frame corresponding to 224 amino acid residues. An initiating Met was not present in the cDNA clone.
The PTD genomic clone was isolated from a black cottonwood genomic library. The genomic sequence (GenBank accession no. AF057708) matched the cDNA sequence exactly. An ATG was present 10 bases upstream from the truncated 5′ end of the cDNA thus the deduced PTD protein sequence consists of 228 amino acid residues.
Gel blots of male and female genomic DNA probed with the 3′ region of PTD revealed one major band in lanes containing DNA digested with DraI or EcoRI, and two bands in lanes containing DNA digested with HindIII or XbaI, indicating that PTD is a single copy gene (data not shown). No differences were observed between the hybridization patterns of DNA from male and female trees.
The PTD gene consists of seven exons, and the number and positions of introns are nearly identical with those of the DEF gene. PTD introns have a slightly larger average size than those of AP3 (Schwarz-Sommer et al., 1992), but are smaller than those of DEF (Irish and Yamamoto, 1995) and ST-DEF (Garcia-Maroto et al., 1993).
A TATA-like sequence [TATTTA] was present 30 bases upstream from an Inr motif [TTCACCCTT], and CCAAT motifs were present at −215 and −245 relative to the putative translational start site. A homeobox protein binding consensus site [ATTTAATTGA] is present 878 bases upstream from the putative translational start site. Sequences matching the CArG box consensus sequence [CC(A/T)6A/GG] are present at −1,031 to −1,021 [CCTATAATAG], −920 to −910 [CCTTTTAAAG], and −159 to −149 [CCTTATTTAG]. The latter site encompasses the TATA motif. Statistical inference (see “Materials and Methods”) indicated that the −807 to −407 region has high similarity to known matrix attachment regions.
Phylogenetic Analyses
Forty-four deduced amino acid sequences belonging to the DEF/AP3/TM6 and GLO/PI groups (Table I) were analyzed by three methods: neighbor-joining (NJ) with Poisson correction, NJ using a Dayhoff-weighted genetic distance matrix, and maximum parsimony (MP; see “Materials and Methods”). All three methods were employed in the analysis of two different data sets: the complete deduced amino acid sequences and a slightly edited data set, for a total of six analyses. We applied a successive approximations approach to character weighting in the MP analysis.
Table I.
Genes included in the phylogenetic analyses
| Gene | Species | Accession No. or Reference |
|---|---|---|
| AP3 | Arabidopsis thaliana | A42095 |
| BobAP3 | Brassica oleraceae var. botrytis | BOU67456 |
| Boi1AP3 | B. oleraceae var. italica | BOU67453 |
| Boi2AP3 | B. oleraceae var. italica | BOU67455 |
| CMB2 | Dianthis caryophyllus | L40405 |
| CUM26 | Cucumis sativus | AAD02250 |
| DeAP3 | Dicentra exima | AF052875 |
| DEF | Antirrhinum majus | S12378 |
| GDEF1 | Gerbera hybrida | CAA08802 |
| GDEF2 | G. hybrida | CAA08803 |
| GGM13 | Gnetum gnemon | CAB44459 |
| GLO | A. majus | S28062 |
| HPI2 | Hyacinthus orientalis | AF134115 |
| LeAP3 | Lycopersicon esculentum cv Celebrity | AF052868 |
| LtAP3 | Liriodendrum tulipifera | AF052878 |
| MADS 16 | Oryza sativa | AAD19872 |
| MfAP3 | Michelia figo | AF052877 |
| NMH7 | Medicago sativa | L41727 |
| NTDEF | Nicotiana tabacum | X96428 |
| NTGLO | N. tabacum | X67959 |
| OsMADS2 | O. sativa | AAB52709 |
| PcAP3 | Papover californicum | AF052872 |
| PD2 | Solanum tuberosum | Garcia-Maroto et al., 1993 |
| PhAP3 | Peperomia hirta | AF052879 |
| PI | A. thaliana | D30807 |
| PMADS1 | Petunia hybrida | X69946 |
| PnAP3-1 | Papover nudicaule | AF052873 |
| PnAP3-2 | P. nudicaule | AF052874 |
| PtAP3-1 | Pachysandra terminalis | AF052870 |
| PtAP3-2 | P. terminalis | AF052871 |
| PTD | Populus trichocarpa | AAC13695 |
| RAD1 | Rumex acetosa | X8913 |
| RAD2 | R. acetosa | X89108 |
| RbAP3 | Ranunculus bulbosus | AF052876 |
| RbAP3-2 | R. bulbosus | AAD31697 |
| RfAP3-1 | R. ficaria | AF052854 |
| RfAP3-2 | R. ficaria | AF130870 |
| RfPI2 | R. ficaria | AAD31700 |
| ScPI | Sanguinaria canadensis | AAD31699 |
| SLM3 | Silene latifolia | X80490 |
| STDEF | S. tuberosum | X67511 |
| SvAP3 | Syringa vulgaris | AF052869 |
| TAMADS51 | Triticum aestivum | AB007506 |
| TM6 | L. esculentum cv Tiny Tim | X60759 |
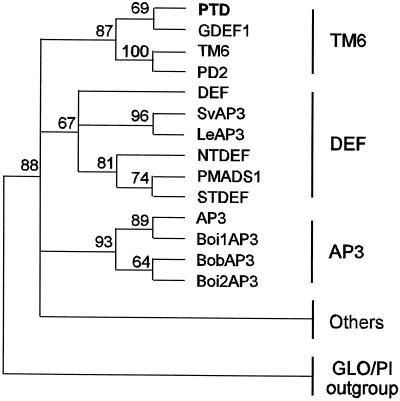
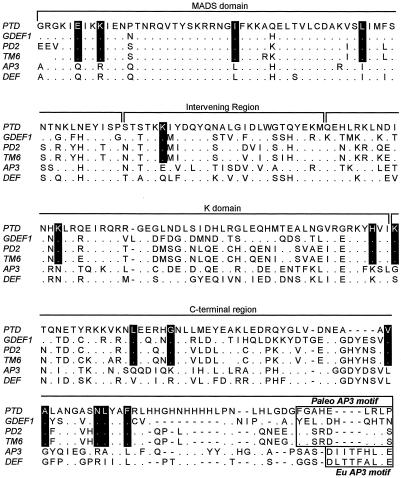
Three major, moderately well-supported groups within the DEF/AP3/TM6 family were revealed by our phylogenetic analyses: the TM6 group, the DEF group, and the AP3 group (Fig. 3). In all tests, PTD resolved as a member of the TM6 group containing GDEF1 from Gerbera hybrida, TM6 from tomato, and PD2 from potato. The TM6 group had an 87% bootstrap value in all MP trees. We used PAUP (Phylogenetic Analysis Using Parsimony, version 4.0b, Sinaur Associates, Sunderland, MA) to further test support of the PTD/TM6 group by imposing a PTD/TM6-constrained tree, then calculating the MP trees and keeping only trees that were not compatible with the constrained tree. Then these trees were compared with the strict consensus unconstrained tree. The results of this test confirmed that the PTD/TM6 group is well supported: all of the best trees that were not compatible with the PTD/TM6-constrained tree were always longer by at least two steps. The amino acid residues present at 15 positions are diagnostic of the TM6 clade, and five amino acids of the PaleoAP3 motif described by Kramer et al. (1998) are common among PTD, PD2, and TM6, but not GDEF1 (Fig. 4).
Figure 3.
Phylogenetic tree of the DEF/AP3/TM6 gene family derived from MP analysis. Major groups are indicated at the right. The numbers above the nodes are percentages of bootstrap confidence levels from 1,000 replicates. Nodes with values less than 60% are collapsed. “Others” consists of sequences associated with the DEF/AP3/TM6 family (bootstrap value 88%) that are not members of any well-defined group within the family. Sequences included in “others” are CMB2, DEAP3, GDEF2, GGM13, LtAP3, MADS16, MfAP3, NMH7, PcAP3, PhAP3, PnAP3–1, PnAP3–2, PtAP3–1, PtAP3–2, RAD1, RAD2, RbAP3, RbAP3–2, RfAP3–1, RfAP3–2, SLM3, and TaMADS51. Sequences included in the GLO/PI outgroup are CUM26, GLO, HPI2, NTGLO, OsMADS2, PI, RfPI-2, and ScPI.
Figure 4.
Alignment of the deduced amino acid sequence of PTD with GDEF1, PD2, TM6, AP3, and DEF. Fifteen residues diagnostic of the TM6 group are shown with a black background. The Paleo AP3 and Eu AP3 motifs (Kramer et al., 1998) are boxed. Dots indicate identical residues. Dashes represent gaps introduced to maximize the alignment.
A second group (DEF) with weaker bootstrap support (67%) consisted of DEF, SvAP3, LeAP3, NTDEF, PMADS1, and STDEF. A third, well-supported group (AP3; 93%) contained the brassicaceous genes AP3, BoiAP3, BobAP3, and Boi2AP3. Twenty-two additional genes resolved within the DEF/AP3/TM6 family, but did not associate strongly with any of the three groups just described nor did they form significant relationships with each other except for several strongly associated pairs from species of close taxonomic affinity.
RNA Gel Blot
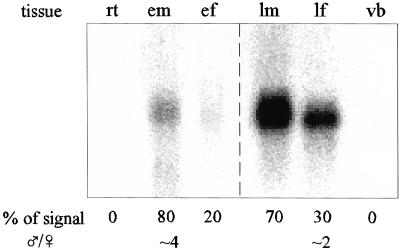
Inflorescences were collected from wild black cottonwood trees at two stages of maturity: in late spring when the inflorescences that will bear mature flowers the following year are developing, and in early spring when the flowers are nearly mature just prior to anthesis. Black cottonwood flowers mature acropetally on inflorescences, thus the early inflorescences bore flowers in various stages of development: the least mature “flowers” were cells on the flanks of the inflorescence meristem that would develop into floral meristems; the most mature flowers (at the proximal end of the inflorescence) had carpel or stamen primordia emerging from the floral meristem. A 1.1-kb PTD transcript was present in developing and mature inflorescence buds of both sexes, but not in vegetative buds or roots (Fig. 5). Quantitation of PhosphorImager scans (see “Materials and Methods”) showed that the signal intensity in early male inflorescences is approximately four times greater than that of early female inflorescences. In mature inflorescences, the signal from male inflorescences was approximately twice that of female inflorescences.
Figure 5.
PTD mRNA expression in various tissues. The RNA gel blot was probed with the NdeI/XhoI fragment of PTD and visualized using a phosphorimager as described in “Materials and Methods.” The first three lanes each contained 30 μg of total RNA. The last three lanes each contained 2 μg of poly(A)+ RNA. Values in the first row are the percentage of total signal contributed by each tissue after correction following ribosomal RNA hybridization (see “Materials and Methods”). Values in the final row are the proportion of signal from male inflorescences as compared with female inflorescences at the same stage of maturity. rt, Root; em, early male inflorescence; ef, early female inflorescence; lm, late male inflorescence; lf, late female inflorescence; vb, vegetative bud.
In Situ Hybridization Analyses
We used in situ hybridization analysis to determine the cell-specific expression patterns of PTD in developing and mature inflorescences of both sexes. Black cottonwood tissue sections were probed with a fragment from the non-conserved 3′ region of the PTD cDNA. In developing male inflorescences, PTD expression was strong at its onset, which occurred well after the floral meristem had differentiated from the inflorescence meristem, as the perianth cup differentiated, but prior to any indications of stamen organogenesis (Fig. 6, A–C). At this stage, PTD signal was present across the entire male floral meristem excluding the perianth cup (Fig. 6, D and E). In slightly more mature male flowers with developing stamens, hybridization signal was visible in the stamens, but appeared more diffuse (Fig. 6F). In mature male flowers a strong hybridization signal was present in the sporophytic tissues of the stamen, particularly the tapetum and filament (Fig. 6, G and H). No signal was detected in the perianth cup, pedicel, bracts, or peduncle of either sex at any stage of development.
Figure 6.
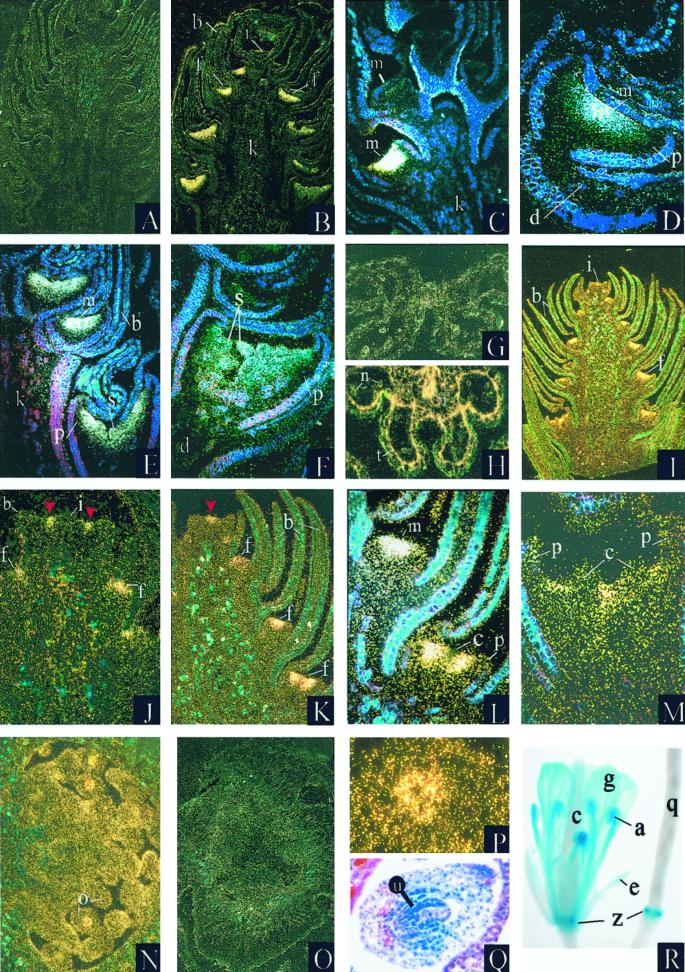
Analysis of PTD expression. A through Q, In situ hybridization analysis of PTD expression in black cottonwood. R, Expression pattern of pPTD:GUS1 in transgenic Arabidopsis. A, G, and O were hybridized with sense control probe; B through F, H, I through N, and P were hybridized with antisense PTD probe. G and H depict cross sections. All others are longitudinal sections. White and gold dots indicate areas of PTD hybridization. Images are digitized. A and B, Developing male inflorescences. Developing flowers along the inflorescence represent a gradient of maturational stages with the most (Legend continues on facing page.)mature flowers at the proximal end (lower portion of photograph); C, distal (i.e. upper) portion of male inflorescence. Four floral meristems are visible; D, developing male flower; E, medial portion of male inflorescence showing three developing flowers; F, developing male flower with stamens beginning to differentiate. Expression is evident in the stamen primordia; G and H, mature anther; I, developing female inflorescence; J, female inflorescence meristem and the two distal-most developing flowers. The area of expression in the cells at the flanks of the inflorescence meristem is perpendicular to the plane of the section and is indicated by arrows; K, distal portion of female inflorescence. The area of hybridization in the inflorescence meristem is parallel to the plane of the section and is indicated by an arrow; L, medial portion of developing female inflorescence; M, female flower with developing carpels; N and O, ovary of mature female flower; P, mature ovule. Hybridization signal is concentrated in the nucellus; Q, mature ovule, brightfield micrograph. R, Arabidopsis mature flower (left) and developing silique (right). a, Anther; b, bract; c, carpel; d, pedicel; e, sepal; f, flower; g, petal; i, inflorescence meristem; k, peduncle; l, filament; m, floral meristem; n, pollen; o, ovule; p, perianth cup; q, developing silique; s, developing stamens; t, tapetum; u, nucellus; z, floral abscission zone.
In contrast to male inflorescences, female inflorescences first showed a weak hybridization signal in the cells on the flanks of the inflorescence meristem that would develop into floral meristems (Fig. 6I). A weak signal was present in the earliest floral meristems and became stronger as the perianth cup differentiated (Fig. 6, J and K). As carpel organogenesis began, PTD mRNA became excluded from the cells differentiating into carpel primordia (Fig. 6L). Later, when carpel primordia emerged, hybridization occurred in the areas of the floral meristem between and around the carpel primordia, but was excluded from the carpel primordia (Fig. 6M). In mature female flowers, hybridization signal was present in the nucellus of the ovules (Fig. 6, N–Q). No expression was detected in vegetative buds and seedlings of either sex (not shown).
Histochemical Analysis of pPTD:GUS1 Expression in Transgenic Arabidopsis
pPTD:GUS1 was generated by fusing approximately 1.9 kb of sequence upstream of the PTD coding region, including 30 bp of 5′-untranslated sequence, with the uidA reporter gene. T1 seeds of transformed Arabidopsis were selected on kanamycin-containing medium and grown to maturity under 16-h days. Leaf, root, pedicel, and floral tissues of 10 independent lines were evaluated for β-glucuronidase (GUS) activity by histochemical analysis. Visible GUS activity was detected only in floral tissues, even when entire plants were cleared and stained (Fig. 6R). In flowers, strong expression was observed in the petals and stamens. Two lines displayed expression in sepals; however, it was weak. Staining was typically stronger in stamens than in petals, and strongest in the anthers. In older flowers, expression was maintained in a discrete band at the base of the developing silique, probably corresponding to the floral abscission zone.
DISCUSSION
PTD Is a Member of the TM6 Subgroup
We performed phylogenetic analysis of PTD along with 31 other members of the DEF/AP3/TM6 family and eight members of the GLO/PI family. Our analyses indicate that PTD belongs to a subgroup of the DEF/AP3/TM6 family that includes the largely uncharacterized genes TM6, PD2, and GDEF1 (Fig. 3). The four genes formed a well-supported clade (87%) in all of our MP trees. Placement of PTD within the TM6 group was further supported by PAUP analysis. Black cottonwood is placed in the rosid clade (Soltis et al., 1999) thus PTD is the first nonasterid gene in the TM6 subgroup to be described.
Two other groups were resolved: one (DEF) comprised largely of genes from the Solanaceae; the other (AP3) comprised of four Brassicaceae genes. Twenty-two other genes were clearly members of the DEF/AP3/TM6 family, but did not form strong relationships with the TM6, AP3, or DEF subgroups nor with each other. The existence of two paralogous lineages within the DEF/AP3/TM6 family, one containing DEF and AP3 and their orthologs, the other containing TM6 and its orthologs, has been suggested (Doyle, 1994; Kramer et al., 1998). It is possible that two DEF-like genes are present in black cottonwood and that PTD is orthologous to PD2, TM6, and GDEF1, whereas an as yet unidentified gene is orthologous to DEF and AP3. However, the results of our RNA gel-blot and in situ hybridization studies indicate that although PTD is a member of the TM6 group, its expression pattern is unlike that of GDEF1 and TM6, and is more similar to that of DEF and AP3. Whereas GDEF1 is expressed primarily in leaves, bracts, and scapes (Yu et al., 1999), PTD is not expressed in vegetative tissues. In addition, although PTD is expressed early in floral development (discussed above), expression of GDEF1 is not detectable at this early stage. Although GDEF1 is expressed only weakly in the corolla and stamens of mature flowers, PTD, like DEF (Schwarz-Sommer et al., 1992) and AP3 (Jack et al., 1992), is strongly expressed in the portion of the floral meristem that gives rise to stamen primordia.
In the hermaphroditic floral meristems of snapdragon and Arabidopsis, this area excludes the central fourth whorl where carpel primordia will develop, whereas in male black cottonwood it encompasses the entire floral meristem excluding the perianth cup. In mature stamens, PTD expression, like DEF, is localized to the tapetum and filament. In developing carpels, PTD expression, like AP3 and DEF, is absent or low, whereas TM6 expression is high (Pneuli et al., 1991). PTD and AP3 are expressed in ovules (Jack et al., 1992). Indeed, the expression patterns of all three members of the TM6 phylogenetic group that have been studied differ from each other significantly. Thus although they belong to the same phylogenetic group, it is doubtful that members of the TM6 subfamily are orthologs with similar functions. Given the similar expression patterns of PTD, DEF, and AP3, it seems more likely that PTD performs a function similar to that of DEF and AP3.
The Temporal and Spatial Expression Patterns of PTD Are Sex Specific
Black cottonwood is well suited to the study of sex-specific developmental programs. In black cottonwood, no morphological indications of carpel initiation occur in male flowers; stamen primordia first appear in the center of the floral meristem and their emergence proceeds centrifugally (Boes and Strauss, 1994; Kaul, 1995). Likewise, no morphological indications of stamen initiation are present in developing female flowers. The unisexual nature of black cottonwood floral meristems allows sex-specific differences in gene expression patterns to be discerned early in floral development.
The temporal expression pattern of PTD in male black cottowood is similar to that observed in DEF and AP3: expression is not detected until after the floral meristem has formed and the initiation of sepal (or perianth cup, in black cottonwood) primordia have occurred. However in black cottonwood females, PTD expression is first detectable earlier in cells on the flanks of the inflorescence meristem. The hybridization signal in female black cottonwood became stronger as the floral meristem expanded, then more diffuse as carpel primordia developed. The area of expression initially encompassed the entire floral meristem exclusive of the perianth cup, but later became excluded from those cells that were differentiating into carpel primordia. When carpel primordia developed, PTD was excluded from them, yet remained present in the cells of the meristem that surrounded them. The presence of PTD mRNA in stamen primordia and the lack of PTD expression in carpel primordia are consistent with the expression pattern predicted for B-function genes by the ABC model. In several hermaphroditic species, one B-class gene is initially expressed in the whorl that will form carpels. In Arabidopsis, PI is initially expressed in the fourth whorl, whereas in snapdragon and the Solanaceae, DEF or DEF orthologs are initially expressed (for review, see Irish, 1999). Studies indicate that PI or DEF expression is not maintained in the fourth whorl because the second B-function gene (AP3 or GLO) is not expressed in this whorl, and both gene activities are needed for maintenance of expression. The similarity of PTDs expression pattern suggests the possibility that an analogous B-class gene autoregulatory mechanism may prevent the maintenance of PTD expression in developing carpels. Isolation and analysis of additional poplar B-class gene homologs would indicate if this is the case.
The early onset of PTD expression in female versus male inflorescence meristems and the initially high intensity of expression in developing female floral meristems suggest that PTD may play a role in female floral organogenesis, perhaps in cell proliferation. In snapdragon, the initiation of fourth whorl organogenesis in L1 chimeras indicated that DEF activity in epidermal cells promotes cell proliferation in the center of the meristem (Perbal et al., 1996). These studies also suggested that a high level of DEF expression in the L1 cell layer allows the growth and expansion of petal lobes by stimulating L1 cell division and/or cell shape and elongation. In a similar manner in the Arabidopsis floral meristem, AP3 and PI appear to have a role in cellular proliferation. Ectopic expression of AP3 and PI results in the production of extra whorls of stamens, and ectopic expression of AP3/PI rescues missing second whorl organs in class A mutants (Krizek and Meyerowitz, 1996). In petunia, ectopic expression of the DEF homolog PMADS1 causes delayed petal fusion and additional lateral growth of petal tissues, suggesting that PMADS1 might be involved in stimulation of directed cell proliferation (Halfter et al., 1994). Alternatively, PTD may have no function in the black cottonwood female inflorescence; PMADS1 is present in the third whorl of petunia flowers, yet plays no discernable role there (van der Krol et al., 1993; Angenent et al., 1995).
We observed no consistent differences between the banding patterns of DNA isolated from male and female trees on our genomic blots, thus it seems unlikely that PTD is directly involved in sex determination. However it is possible that sex-specific regulation of PTD expression is involved in sex determination as a consequence of the action of upstream sex determination genes.
Evidence indicates that the perianth cup of black cottonwood is derived from a fusion of perianth parts (Fisher, 1928; Stoehr et al., 1988; Kaul, 1995), however, it remains unclear which particular parts were incorporated into the structure. If we assume that the perianth cup is derived from fused sepals, then the lack of PTD expression in the cup is consistent with the predicted expression patterns of B-function genes such as DEF and AP3. On the other hand, it is possible that the perianth cup is derived from fused petals, or from adnation of petals and sepals, and that the sepal-like nature of the black cottonwood perianth results, at least in part, from the evolutionary loss of PTD expression in that organ. Experiments to determine the exact function of PTD by suppression of PTD expression in transgenic cottonwood trees are under way.
The PTD Promoter Contains Consensus Sites for Binding Regulatory Proteins
MADS box gene products are believed to regulate gene expression by binding to sites present in the promoters of target genes (Riechmann et al., 1996). These binding sites, known as CArG boxes, are found in the promoter regions of plant MADS box genes, including DEF (Schwarz-Sommer et al., 1990; Tröbner et al., 1992) and AP3 (Riechmann et al., 1996), and are thought to be targets for autoregulation. The AP3 promoter contains three CArG boxes that regulate different aspects of AP3 expression (Hill et al., 1998; Tilly et al., 1998). We found three sites in the PTD promoter region that match the CArG box consensus motif. One site encompasses the TATA box; the other two are approximately 1 kb upstream from the initial ATG. When a construct consisting of the PTD 5′-flanking region fused with the uidA coding region was introduced into Arabidopsis, we observed GUS expression in the organs where AP3 is normally expressed, namely petals and stamens. This conserved B-function expression pattern suggests that the transcription factors responsible for regulation of AP3 expression in Arabidopsis are able to bind to sites present in the PTD promoter region and supports the hypothesis that PTD performs a function similar to that of AP3.
The PTD gene promoter also contains a motif that exactly matches the consensus site for binding homeodomain proteins. Homeodomain proteins are known to be expressed in inflorescence and floral meristems (Jackson et al., 1994) and in ovules (Reiser et al., 1995), and are involved in the regulation of floral homeotic MADS box gene expression (Ray et al., 1994). The interaction of homeodomain and MADS domain transcription factors has been reported in animals (for review, see Duprey and Lesens, 1994). It is possible that a homeodomain protein binds to the consensus site present in the PTD promoter and is involved in regulating PTD gene expression. Consensus sites for binding homeodomain proteins are not present in the promoters of DEF, AP3, or the three ST-DEF promoters.
The PTD Promoter Should Be Useful for Engineering Reproductive Sterility in Trees
Genetic engineering shows potential for the improvement of qualitative and quantitative traits in trees (Tzfira et al., 1998). Transgenic cottonwoods with engineered insect and herbicide resistance have been produced (Meilan et al., 2000). However, there is a high potential for escape of transgenes in trees into wild populations because of their long distance movement of seeds and pollen and their ubiquitous wild relatives. Genetically engineered sterility has been proposed as the best method for transgene containment in trees (Strauss et al., 1995). Fusion of reproductive tissue-specific promoters with coding regions from genes that encode ablative proteins has been used to engineer male and female sterility in a variety of agronomic and model plants (e.g. Mariani et al., 1990; Goldman et al., 1994). If deleterious effects on vegetative growth are to be avoided, this approach requires a promoter that limits expression specifically to reproductive tissues. Because PTD is expressed early in the floral meristems of male and female trees and does not appear to be expressed in vegetative buds, seedlings, or roots, its promoter should be useful for engineering of the complete male and female sterility desirable for gene containment. Studies to test the effectiveness of this approach in transgenic trees are under way.
MATERIALS AND METHODS
Plant Materials
Plant materials were collected from wild black cottonwood (Populus trichocarpa) trees near Corvallis, Oregon. Cut branches were partially submersed in aerated water to produce the roots used in the RNA gel blot. Seedlings used for in situ hybridization experiments were sprouted in potting soil in the greenhouse.
Nucleic Acid Isolation
RNA was isolated using the method of Hughes and Galau (1988) with the following modifications: Tissues were ground in liquid nitrogen in a blender then homogenized in homogenization buffer using a polytron (Brinkman Instruments, Westbury, NY); after thawing the homogenates, 0.5 volume of 5 m potassium acetate, pH 6.5, was added; the phenol/chloroform extraction was performed prior to the LiCl precipitation.
Poly(A)+ RNA was isolated using oligo(dT) spin columns (CLONTECH, Palo Alto, CA). Genomic DNA was isolated from vegetative buds using the CTAB method of Wagner et al. (1987).
cDNA and Genomic Clone Isolation
Floral buds were collected from a native female black cottonwood tree in late May. The bud scales were removed and poly(A)+ RNA isolated from the developing inflorescences was used to construct a cDNA library in λ Zap (Stratagene, La Jolla, CA). The PTD cDNA was obtained by probing 1 × 106 clones of the amplified library at low stringency with an EcoRI fragment of pCIT2241 that contained the highly conserved MADS box region of AGL1 (Ma et al., 1991). Plasmids containing putative MADS box cDNA clones were excised from λ ZAP as suggested by the supplier.
Construction of the black cottonwood genomic library has been described (Rottmann et al., 2000). Isolation of the PTD genomic clone was accomplished by screening 6 × 105 genomic clones with a BSU36I/XhoI restriction fragment that contained the 3′ portion of the PTD cDNA, but not the highly conserved MADS box region.
DNA Sequencing and Analysis
Sequencing was performed by the Oregon State University Central Services Laboratory using fluorescent primer dye/dideoxy chain termination and an automated DNA sequencer (ABI, Sunnyvale, CA). Both strands of the templates were sequenced. Sequence analysis was conducted using the database of motifs in the GeneRunner version 3.0 program (Hastings Software, Hastings, NY) supplemented with the CArG box consensus sequence [CC(A/T)6A/GG]. The location of a putative matrix attachment region was statistically inferred using the computer program MAR-Finder (Kramer et al., 1997).
Sequence Alignment and Phylogenetic Analyses
The sequence of PD2 was obtained from Garcia-Maroto et al. (1993). All other sequences were obtained from GenBank (Table I). Alignment of protein sequences was accomplished using the GeneDoc software program (Nicholas and Nicholas, 1997). GLO and its homologs were chosen as outgroup (least related) sequences because as members of the DEF sister-clade (Doyle, 1994) their sequences were similar enough to allow alignment within the minimally conserved C-terminal region.
A total of six phylogenetic trees were derived from two different data sets using three different methods. One data set employed the complete deduced amino acid sequences of all genes. For the other, sequences were edited to exclude those residues where corresponding positions in all other sequences were gaps. The NJ method (Saitou and Nei, 1987) of the MEGA computer program (Molecular Evolutionary Genetic Analysis, version 1.0, Pennsylvania State University, University Park) was used for heuristic distance-based phylogenetic analyses of both data sets. Poisson distribution-correction distance was employed to estimate the number of amino acid substitutions per site. Two additional distance-based trees were constructed, based on the weighted genetic distance matrix computed from deduced protein sequences using the PROTDIST program in the PHYLIP software package (Phylogeny Inference Package, version 3.57, Department of Genetics, University of Washington, Seattle), where amino acid substitutions were scaled using Dayhoff's PAM 001 empirical matrix of mutation rates (Dayhoff et al., 1978). Consensus trees and estimates of statistical confidence were inferred from 1,000 bootstrapped (randomly sampled with replacement) data sets.
Parsimony analysis (derivation of a phylogenetic tree requiring the smallest number of evolutionary changes) was performed using the PROTPARS program in the PHYLIP package (Felsenstein, 1995). The SEQBOOT program was used to generate 1,000 data sets. Majority-rule and strict consensus trees were generated using CONSENSE. The computer program PAUP was used to apply a successive approximations approach to character weighting in the MP analysis (Farris, 1969). To begin, a heuristic search with 10 randomizations of sequence input order was used to find the most parsimonious trees. A strict consensus tree was calculated from the equally most parsimonious trees obtained in the search. This tree was used in successive approximations weighting with the characters reweighted by maximum value of rescaled consistency indices in consecutive successive approximations weighting runs until identical trees were found in two consecutive searches. We used PAUP to test support of the PTD/TM6 group by imposing the PTD/TM6-constrained tree, then calculating the maximum parsimony trees and keeping only those trees that were not compatible with the constrained tree. These trees were then compared with the strict consensus unconstrained tree.
DNA and RNA Gel Blots
For the RNA gel blot, 2 μg of poly(A)+ RNA from vegetative buds and mature male and female catkins of black cottonwood collected in February, or 30 μg of total RNA from roots and from developing male and female floral buds collected in May, were applied to a formaldehyde agarose gel, subjected to electrophoresis, blotted onto a nitrocellulose membrane, hybridized, and washed at high stringency (0.1× SSC; 1% [w/v] SDS, 69°C). For the DNA gel blot, 10 μg of black cottonwood genomic DNA were digested with restriction enzymes, blotted onto a nylon membrane, hybridized, and washed according to established procedures (Sambrook et al., 1989). The blots were probed with a 595-bp NdeI/XhoI fragment that contained only the 3′ portion of the PTD-coding region to avoid cross-hybridization with other MADS box sequences present in the black cottonwood genome. The RNA gel blot was stripped and reprobed with 18S rDNA from P. deltoides (D'Ovidio, 1992) as control. Both RNA and DNA gel blots were exposed to a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) plate and digitized using a SI PhosphorImager (Molecular Dynamics). The values in Figure 5 were obtained by quantifying signals on digitized images of an RNA gel blot probed sequentially with PTD and an18s rDNA from P. deltoides (D'Ovidio, 1992) as a control using ImageQuant software (version 4.2, Molecular Dynamics), correcting for background, and adjusting for variation on the assumptions that rRNA was present at equal amounts in all tissues and that the oligo(dT) columns removed equal proportions of rRNA from the three poly(A)+ samples.
In Situ Hybridizations
In situ hybridizations were performed as described by Kelly et al. (1995) with the following modifications. Eight- to 10-μm sections were probed with either antisense PTD cRNA transcribed using the T7 promoter in the pBlueScript vector or with sense (control) PTD cRNA transcribed from the T3 promoter. The antisense cDNA template was digested with NdeI at a site 380 bases from the 5′ end to terminate transcription within the K box, yielding a probe that lacked the highly conserved MADS box region. Control sections were probed with a sense transcript generated from a T3 promoter flanking the 5′ end of the cDNA. The sense template was digested with XhoI directly at the 5′ end of the cDNA (thus the control probe included the entire PTD cDNA sequence). Probe was applied at a concentration of 10 to 30 ng/mL of hybridization solution; higher probe concentrations resulted in unacceptable levels of non-specific hybridization. Silver grains were visualized with a Axioskop (Zeiss, Jena, Germany) microscope equipped with a darklight illuminator and/or a darkfield stop in the substage condenser. Tissue sections were photographed with a camera (Contax, UK) mounted on the microscope using slide film (Sensia ISO 100, Fuji Phot Film, Tokyo). The ovule depicted in the brightfield micrograph was stained with hematoxylin and eosin. The images in Figures 1 and 6 were digitized using a Microtek Scanmaker III. Images were adjusted to match the original slides and prints using Adobe Photoshop 4.0.1 (Adobe Systems Inc., Mountain View, CA, http://www.adobe.com) and composites were assembled using Presentations 3 (Corel, Ottawa, ON, Canada).
pPTD:GUS1 Construction and Analysis of Expression in Arabidopsis
pPTD:GUS1 was constructed by subcloning a approximately 1.9-kb HindIII/AvrII fragment of the PTD gene upstream region into HindIII/XbaI-digested pBI101 (CLONTECH) that had been modified by replacing the existing uidA gene with an intron-containing version of uidA (Vancanneyt et al., 1990). The AvrIII site, which constituted the 3′-terminus of the subcloned PTD fragment, occurred approximately 90 bp upstream of the presumed translational start site. Transformation of Arabidopsis ecotype Columbia with the pPTD:GUS1 fusion construct as well as selection and growth of transgenic plants were performed as previously described (Rottmann et al., 2000). Histochemical GUS staining was accomplished as described by Jefferson et al. (1987). Digital images of stained tissues were produced using a digital documentation system (UVP Inc., Upland, CA).
ACKNOWLEDGMENTS
We thank Alan Kelly and Ry Meeks-Wagner (University of Oregon, Eugene) for their advice on in situ hybridizations, Elliot Meyerowitz (California Institute of Technology, Pasadena) for providing pCIT2241, Renato D'Ovidio (Universita' della Tuscia, Viterbo, Italy) for providing the P. deltoides 18S rDNA, and Don Copes (U.S. Department of Agriculture Forest Service, Corvallis, OR) for allowing us to use his microscope.
Footnotes
This research was supported in part by the National Science Foundation, by the U.S. Department of Agriculture National Research Initiative (grant no. 93–37301–9425), by members of the Tree Genetic Engineering Research Cooperative based at Oregon State University (Alberta Pacific, Boise Cascade, Department of Energy Biofeedstocks Program, Electric Power Research Institute, Fort James, Georgia Pacific, International Paper, MacMillan Bloedel, Monsanto, Potlatch, Shell, Union Camp, Westvaco, and Weyerhaeuser), and by an endowment from the late Conrad Wessela.
LITERATURE CITED
- Ainsworth C, Parker J, Buchanan-Wollaston V. Sex determination in plants. In: Moscano AA, Monroy A, editors. Current Topics in Developmental Biology. Vol. 38. San Diego: Academic Press; 1998. pp. 167–223. [DOI] [PubMed] [Google Scholar]
- Angenent GC, Busscher M, Franken J, Dons HJM, van Tunen AJ. Functional interaction between the homeotic genes fbp1 and pMADS1during petunia floral organogenesis. Plant Cell. 1995;7:507–516. doi: 10.1105/tpc.7.5.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boes TK, Strauss SH. Floral phenology and morphology of black cottonwood, Populus trichocarpa(Salicaceae) Am J Bot. 1994;81:562–567. [Google Scholar]
- Bradshaw HD, Jr, Stettler RF. Molecular genetics of growth and development in Populus: I. Triploidy in hybrid poplars. Theor Appl Genet. 1993;86:301–307. doi: 10.1007/BF00222092. [DOI] [PubMed] [Google Scholar]
- Dayhoff M, Schwartz RM, Orcutt BC. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of Protein Sequence and Structure. Vol. 5 1978. , Supplement 3. National Biomedical Research Foundation, Silver Spring, MD, pp 345–352. [Google Scholar]
- D'Ovidio R. Nucleotide sequence of a 5.8S rDNA gene and of the internal transcribed spacers from Populus deltoides. Plant Mol Biol. 1992;19:1069–1072. doi: 10.1007/BF00040539. [DOI] [PubMed] [Google Scholar]
- Doyle JJ. Evolution of a plant homeotic multigene family: toward connecting molecular systematics and molecular developmental genetics. Syst Biol. 1994;43:307–328. [Google Scholar]
- Duprey P, Lesens C. Control of skeletal muscle-specific transcription: involvement of paired homeodomain and MADS domain transcription factors. Intl J Dev Biol. 1994;38:591–604. [PubMed] [Google Scholar]
- Eckenwalder JE. Systematics and evolution of Populus. In: Stettler RF, Bradshaw HD, Heilman PE, Hinckley TM, editors. Biology of Populus and Its Implications for Management and Conservation. Ottawa, Canada: NRC Research Press, National Research Council of Canada; 1996. pp. 7–32. [Google Scholar]
- Felsenstein J. PHYLIP: A Phylogeny Interface Package Version 3.57C. Seattle: Department of Genetics, University of Washington; 1995. [Google Scholar]
- Farris J. A successive approximations approach to character weighting. Syst Zool. 1969;18:374–385. [Google Scholar]
- Fisher MJ. The morphology and anatomy of the flowers of the SalicaceaeII. J Bot. 1928;15:372–394. [Google Scholar]
- Garcia-Maroto F, Salamini F, Rohde W. Molecular cloning and expression patterns of three alleles of the Deficiens-homologous gene St-Deficiens from Solanum tuberosum. Plant J. 1993;4:771–780. doi: 10.1046/j.1365-313x.1993.04050771.x. [DOI] [PubMed] [Google Scholar]
- Goldman MHS, Goldberg RB, Mariani C. Female sterile tobacco plants are produced by stigma-specific cell ablation. EMBO J. 1994;13:2976–2984. doi: 10.1002/j.1460-2075.1994.tb06596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ali N, Stockhaus J, Ren L, Chua N-H. Ectopic expression of a single homeotic gene, the Petunia gene green petal, is sufficient to convert sepals to petaloid organs. EMBO J. 1994;13:1443–1449. doi: 10.1002/j.1460-2075.1994.tb06398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K-H, Gordon MP, Strauss SH. Cellular and molecular biology of Agrobacterium-mediated transformation of plants and its application to genetic transformation of Populus. In: Stettler R, Bradshaw Jr H, Heilman P, Hinckley TM, editors. Biology of Populus and Its Implication for Management and Conservation. Ottawa, Canada: National Research Council; 1996. pp. 201–222. [Google Scholar]
- Heslop-Harrison JW. Sex in the Salicaceae and its modification by eriophyid mites and other influences. Br J Exp Biol. 1924;1(4):445–472. [Google Scholar]
- Hill TA, Daly CD, Zondio S, Thackeray AG, Irish VF. Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development. 1998;125:1711–1721. doi: 10.1242/dev.125.9.1711. [DOI] [PubMed] [Google Scholar]
- Hughes DW, Galau G. Preparation of RNA from cotton leaves and pollen. Plant Mol Biol Rep. 1988;6:253–257. [Google Scholar]
- Irish VF. Petal and stamen development. In: Pederson RA, Schatten GP, editors. Current Topics in Developmental Biology. Vol. 41. San Diego: Academic Press; 1999. pp. 133–161. [DOI] [PubMed] [Google Scholar]
- Irish VF, Yamamoto YT. Conservation of floral homeotic gene function between Arabidopsis and Antirrhinum. Plant Cell. 1995;7:1635–1644. doi: 10.1105/tpc.7.10.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETELA3 of Arabidopsis thalianaencodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S. Expression of maize KNOTTED1related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–413. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul RB. Reproductive structure and organogenesis in a cottonwood, Populus deltoides(Salicaceae) Int J Plant Sci. 1995;156:172–180. [Google Scholar]
- Kelly AJ, Bonnlander MB, Meeks-Wagner DR. NFL, the tobacco homolog of FLORICAULA and LEAFY, is transcriptionally expressed in both vegetative and floral meristems. Plant Cell. 1995;7:225–234. doi: 10.1105/tpc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETELA3 and PISTILLATAMADS-box lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JA, Singh GB, Krawetz SA. Computer assisted search for sites of nuclear matrix attachment. Genomics. 1997;33:302–308. doi: 10.1006/geno.1996.0198. [DOI] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM. The Arabidopsis homeotic genes APETELA3 and PISTILLATAare sufficient to provide the B class organ identity function. Development. 1996;122:11–22. doi: 10.1242/dev.122.1.11. [DOI] [PubMed] [Google Scholar]
- Larsen CM. Recent advances in poplar breeding. Intl Rev Forestry Res. 1970;3:1–67. [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. AGL1-AGL6, an Arabidopsisgene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991;5:484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mariani C, De Beuckeleer M, Truettner J, Leemans J, Goldberg RB. Induction of male sterility in plants by a chimeric ribonuclease gene. Nature. 1990;347:737–741. [Google Scholar]
- McLetchie DM, Tuskan GA. Gender determination in Populus. Norw J Agric Sci Suppl. 1994;18:57–66. [Google Scholar]
- Meilan R, Ma C, Cheng S, Eaton JA, Miller LK, Crockett RP, DiFazio SP, Strauss SH. High levels of Roundup and leaf-beetle resistance in genetically engineered hybrid cottonwoods. In: Blatner KA, Johnson JJ, editors. Hybrid Poplars in the Pacific Northwest: Culture, Commerce and Capability. WA: Washington State University Cooperative Extension, Pullman; 2000. (in press) [Google Scholar]
- Nicholas K, Nicholas H (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. Distributed by the author. www.cris.com/∼ketchup/genedoc.shtml
- Perbal M-C, Haughn G, Saedler H, Schwarz-Sommer ZS. Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSAis exerted by their polar cell-to-cell trafficking. Development. 1996;122:3433–3441. doi: 10.1242/dev.122.11.3433. [DOI] [PubMed] [Google Scholar]
- Pneuli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer ZS, Lifschitz E. The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1991;1:255–266. [PubMed] [Google Scholar]
- Ray A, Robinson-Beers K, Ray S, Baker SC, Lang JD, Preuss D, Milligan SB, Gasser C. Arabidopsis floral homeotic gene BELL (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG) Proc Natl Acad Sci USA. 1994;91:5761–5765. doi: 10.1073/pnas.91.13.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haudhn GW, Fischer RL. The Bell1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsisovule primordium. Cell. 1995;83:735–742. doi: 10.1016/0092-8674(95)90186-8. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. MADS domain proteins in plant development. Biol Chem. 1998;378:1079–1101. [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM. DNA-binding properties of ArabidopsisMADS domain homeotic proteins APETELA1, APETELA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann WH, Meilan R, Sheppard LA, Brunner AM, Skinner JS, Ma C, Cheng S, Jouanin L, Pilate G, Strauss SH. Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J. 2000;22:235–245. doi: 10.1046/j.1365-313x.2000.00734.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig W-E, Saedler H, Sommer H. Characterization of the Antirrhinum floral homeotic MADS- box gene deficiens: evidence for DNA-binding and autoregulation of its persistent expression throughout floral development. EMBO J. 1992;11:251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science. 1990;250:931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- Sheppard L. PTD: a Populus trichocarpa gene with homology to floral homeotic transcription factors. PhD thesis. Corvallis, OR: Oregon State University; 1997. [Google Scholar]
- Soltis OS, Soltis DE, Chase MW. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature. 1999;402:402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- Sommer H, Beltran J-P, Huijser P, Pape H, Lönnig W-E, Saedler H, Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler RF. Variation in sex expression in black cottonwood and related hybrids. Silvae Genet. 1971;20:42–46. [Google Scholar]
- Stoehr MU, Zsuffa L, Eckenwalder JE. Anomalous solitary flowers on anther-derived plants of Populus maximowiczii. Am J Bot. 1988;745:594–597. [Google Scholar]
- Strauss SH, Rottmann WH, Brunner AM, Sheppard LA. Genetic engineering of reproductive sterility in forest trees. Mol Breed. 1995;1:5–26. [Google Scholar]
- Swofford DL, Begle DP. PAUP: Phylogenetic Analysis Using Parsimony, Version 3.1, User's Manual. IL: Illinois Natural History Survey, Champain; 1993. [Google Scholar]
- Tilly JJ, Allen DW, Jack T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3mediate diverse regulatory effects. Development. 1998;125:1647–1657. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig W-E, Saedler H, Sommer H, Schwarz-Sommer Z. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinumfloral organogenesis. EMBO J. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Zucker A, Altman A. Forest-tree biotechnology: genetic transformation and its application to future forests. Trends Biotechnol. 1998;16:439–445. [Google Scholar]
- Vancanneyt G, Schmidt R, O'Connor-Sanchez A, Willmitzer L, Rocha-Sosa M. Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet. 1990;220:245–250. doi: 10.1007/BF00260489. [DOI] [PubMed] [Google Scholar]
- van der Krol AR, Brunelle A, Tsuchimoto S, Chua N-H. Functional analysis of petunia floral homeotic MADS box gene pMADS1. Genes Dev. 1993;7:1214–1228. doi: 10.1101/gad.7.7a.1214. [DOI] [PubMed] [Google Scholar]
- Wagner DB, Furnier GR, Saghai-Maroof MA, Williams SM, Dancik BP, Allard RW. Chloroplast DNA polymorphisms in lodgepole pines and their hybrids. Proc Natl Acad Sci USA. 1987;84:2097–2100. doi: 10.1073/pnas.84.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Kotilainen M, Pöllänen E, Mehto M, Elomaa P, Helariutta Y, Albert VA, Teeri TH. Organ identity genes and modified patterns of flower development in Gerbera hybrida(Asteraceae) Plant J. 1999;17(1):51–62. doi: 10.1046/j.1365-313x.1999.00351.x. [DOI] [PubMed] [Google Scholar]