Abstract
The human microbiome is important for health and plays a role in essential metabolic functions and protection from certain pathogens. Conversely, dysbiosis of the microbiome is seen in the context of various diseases. Recent studies have highlighted that a complex microbial community containing hundreds of bacteria colonizes the healthy urinary tract, but little is known about the human urinary viruses in health and disease. To evaluate the human urinary virome in the context of kidney transplantation (tx), variations in the composition of the urinary virome were evaluated in urine samples from normal healthy volunteers as well as patients with kidney disease after they had undergone kidney tx. Liquid chromatography-mass spectrometry/mass spectrometry analysis was undertaken on a selected cohort of 142 kidney tx patients and normal healthy controls, from a larger biobank of 770 kidney biopsy matched urine samples. In addition to analysis of normal healthy control urine, the cohort of kidney tx patients had biopsy confirmed phenotype classification, coincident with the urine sample analyzed, of stable grafts (STA), acute rejection, BK virus nephritis, and chronic allograft nephropathy. We identified 37 unique viruses, 29 of which are being identified for the first time in human urine samples. The composition of the human urinary virome differs in health and kidney injury, and the distribution of viral proteins in the urinary tract may be further impacted by IS exposure, diet and environmental, dietary, or cutaneous exposure to various insecticides and pesticides.
Keywords: kidney transplantation, virome, urine, proteomics, biomarkers
Introduction
The human microbiome has been studied extensively in various biofluids, such as blood (1), urine (2–4), saliva (5, 6), cerebrospinal fluid (7), and bronchoalveolar lavage (8, 9), for its influence on human health and disease (10). Several reports on the assessment of microbiomes in different regions of human body have been reported including lung and gut (11–15). However, only little is known about the urinary microbiome and its changes in the context of kidney injury after kidney transplantation (tx) (16, 17). Most human microbiome studies have mapped 16S rRNA for bacterial profiling (18), and there is only a small body of data examining the urine virome (19, 20), and a single published study has used next-generation sequencing to map viral genomic components in the urine of kidney transplant patients (19).
Dysbiosis of the microbiome is associated with multiple diseases such as inflammatory bowel disease, colon cancer, obesity and pulmonary disease (21–25), but little is known about alterations of the virome in human body fluids, such as saliva, bronchoalverolar lavage and urine. It is recognized that various components of the microbiome perform essential functions including biosynthesis of cofactors and vitamins, metabolism of essential compounds, and barrier protection from pathogens (26–28). Similar dysbiotic and protective functions may also be ascribed to the urine virome; hence, it is important to ascertain the composition of the human virome in health and disease. For purposes of this study, we chose to conduct these studies in urine in health and in the context of IS exposure after kidney tx, during stable kidney function and in the context of kidney tx injury. The focus of the study was to evaluate the repertoire of viruses as determined by the peptides present in human urine and their perturbations after IS exposure and various causes of acute, chronic, and infectious kidney injury.
The microbiome changes over time and correlates with organism diversity. Microbiome analysis can be segregated into both structural analysis based on operational taxonomic units based on sequence phylogeny, and functional analysis based on metagenomics sequencing and proteomics (29, 30). Identified microbiota in different diseases can be further studied by cultivation, functional metagenomics, and multiplexed immunofluorescence and in situ hybridization. The NIH human microbiome project has published the human microbiome in 15 body sites from 300 individuals (31).
Materials and Methods
A total of 142 unique samples were evaluated from a biorepository containing 2016 collected by IRB approved informed consent from adult and pediatric samples from the kidney tx programs at Stanford University and University of California San Francisco, between urine samples of which 770 were accompanied with matched kidney tx bx with centralized pathology histology reads and compartment scores using the standardized Banff schema (32) for scoring kidney tx bx injury (Figure 1). The study was approved by The Human Research Protection Program of the University of California, San Francisco. The urine samples were phenotyped based on the matched kidney bx pathology into five groups: healthy control (HC; n = 9), Stable graft (STA; n = 40), acute rejection (AR; n = 37), chronic allograft nephropathy (CAN; n = 39), and BK virus nephritis (BKVN; n = 17). Urine was centrifuged 2,000 × g at 4°C for 20 min to get rid of urine sediments. The supernatant was passed through a filer membrane of 10 kDa to remove native peptides from intact proteins larger than 10 kDa in size. The total protein was then trypsin digested and the resulting tryptic peptides were analyzed by LC-MS platform (Orbitrap Velos MS). The detail methods of protein preparation and analyses are reported elsewhere (33).
Figure 1.
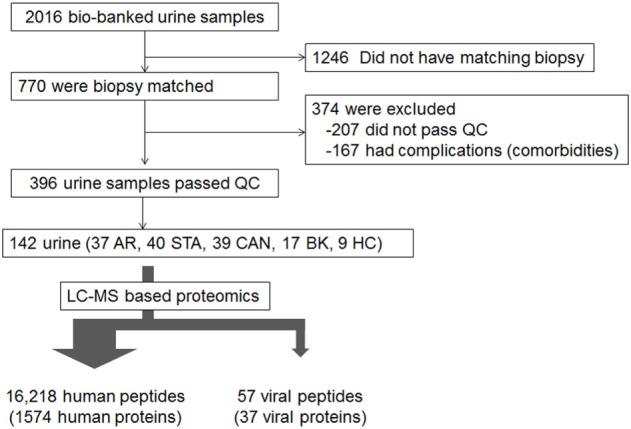
Source of samples. LC-MS based proteomics was performed on the 142 samples chosen: 37 with acute rejection (AR), 40 stable (STA), 39 with chronic allograft nephropathy (CAN), 17 with BK virus nephritis, and 9 healthy controls.
The MSGF plus customized algorithm generated by our group (https://omics.pnl.gov/software/ms-gf), was used to search MS/MS spectra against the combined human protein sequence database and the NCBI viral database. Peptides were initially identified from database searching applying the following criteria: MSGF spectrum E-value (a probability value of the peptide to MS/MS spectrum match with the lower value the higher probability to be correct match) to be <10-10, Peptide level Q-value (false discovery rate estimated by targeted-decoy database search) to be <0.01, and mass measurement error <10 ppm (±5 ppm). The decoy database searching methodology was used to confirm the final false discovery rate at the unique peptide level to be <1%. Due to the anticipated higher false discovery rate for peptides from viral proteins, a more stringent filtering criteria with MSGF spectrum E value to be <1E-13 was applied. The false discovery rate was estimated to nearly 0% based on the well-accepted target-decoy searching strategy because no decoy hits were observed following this stringent cutoff. Data are shown as percentages and mean ± SD. Comparisons of different categories are done using ANOVA and p values of <0.05 are considered significant.
Results
Our group has previously published a detailed analysis of biologically relevant human proteins in these urine samples collected from kidney transplant recipients with different graft injury phenotypes, as confirmed by matched kidney transplant histopathology on the biopsy, collected at the same time as the urine sample; this data has been deposited in the proteomic MassIVE repository (accession MSV000079262) and in the ProteomeXchange repository (accession PXD002761) (33). In this study, we only focused on the identification and analysis of viral proteins in the same cohort of kidney transplant patients, with the inclusion also of age- and gender-matched healthy control human urine samples to evaluate viral proteins in both health and kidney injury. It is important to note that of the total analyzed human and viral proteins in urine, viral proteins alone constitute <0.2% of the total identified proteins, highlighting the very low abundance of rather rare viral proteins in human urine, irrespective of the type of kidney injury, and irrespective of baseline immunosuppression usage.
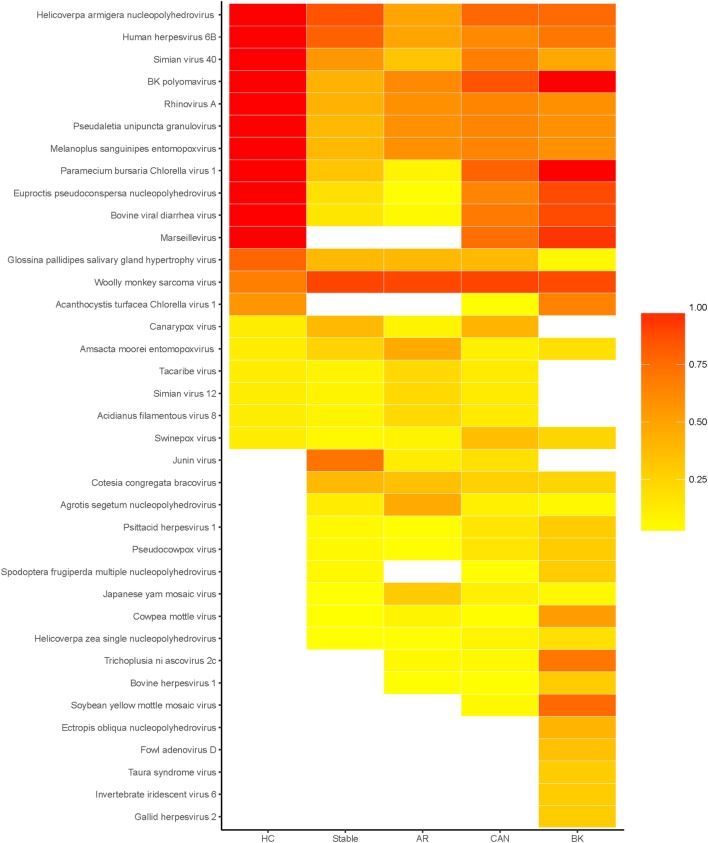
The results presented in this paper come from viral peptide mapping, unlike genomic sequencing of viral components as reported by previous publications (19). As an initial analysis, we focus on the prevalence of urinary viral proteins specific to each kidney transplant phenotype of STA, AR, CAN, and BKVN, and the variations noted over the healthy control urine virome. Urinary viral protein data for each sample was evaluated for sample abundance relative to the mean level of that virus in the entire sampled population. We found that on an average, 22% (range 4–67%) of kidney transplant patients had different viral proteins detected in their urine, excluding patients with BKVN, where detection of the BK virus in urine is to be expected, as these patients have infection with BK virus in their urinary tract and in the kidney transplant. The distribution of different viral proteins in different tx injury states and normal health are shown in the prevalence heat map (Figure 2). Overall, a total of 57 viral proteins from 37 unique viruses were identified, many of these being unexpected and previously not described as “commensals” in human urine. Of these unique viruses, 8 viruses were single-stranded RNA viruses, 1 virus was a single-stranded RNA-RT virus, and 28 viruses were double-stranded DNA viruses (Table 1). Out of the 142 patients, all patients had at least one viral protein in their urine, with an average of 10.23 ± 4.57 viral proteins/sample.
Figure 2.
Heatmap of prevalence by disease status. Heatmap of the percentage of patients, within each disease status, in which each virus was detected. Total number of unique viruses in each disease status: healthy control (HC) 20, stable 27, acute rejection (AR) 28, chronic allograft nephropathy (CAN) 32, and BK 32.
Table 1.
Breakdown of unique viruses.
| Virus | Virus family | Virus type | Host | Described in humans | Used as pesticide/insecticide |
|---|---|---|---|---|---|
| Acanthocystis turfacea Chlorella virus 1 | Phycodnaviridae | dsDNA | Algae | Yes | No |
| Acidianus filamentous virus 8 | Lipothrixviridae | dsDNA | Sulfo bacteria | No | No |
| Agrotis segetum nucleopolyhedrovirus | Baculoviridae | dsDNA | Lepidoptera | No | No |
| Amsacta moorei entomopoxvirus | Poxviridae | dsDNA | Butterfly | No | No |
| BK polyomavirus | Polyomaviridae | dsDNA | Humans | Yes | No |
| Bovine herpesvirus 1 | Herpesviridae | dsDNA | Cow | No | No |
| Bovine viral diarrhea virus | Flaviviridae | (+)ssRNA | Cow | No | No |
| Canarypox virus | Poxviridae | dsDNA | Canary | No | No |
| Cotesia congregata bracovirus | Polydnaviridae | dsDNA | Wasp | No | No |
| Cowpea mottle virus | Tombusviridae | (+)ssRNA | Cowpeas | No | No |
| Ectropis obliqua nucleopolyhedrovirus | Baculoviridae | dsDNA | Butterfly | No | Yes |
| Euproctis pseudoconspersa nucleopolyhedrovirus | Baculoviridae | dsDNA | Butterfly | No | Yes |
| Fowl adenovirus D | Adenoviridae | dsDNA | Birds | No | No |
| Gallid herpesvirus 2 | Herpesviridae | dsDNA | Chicken | No | No |
| Glossina pallidipes salivary gland hypertrophy virus | Hytrosaviridae | dsDNA | Tsetse fly | No | No |
| Helicoverpa armigera nucleopolyhedrovirus | Baculoviridae | dsDNA | Butterfly | No | Yes |
| Helicoverpa zea single nucleopolyhedrovirus | Baculoviridae | dsDNA | Butterfly | No | Yes |
| Human herpesvirus 6B | Herpesviridae | dsDNA | Humans | Yes | No |
| Invertebrate iridescent virus 6 | Iridoviridae | dsDNA | Insects | No | No |
| Japanese yam mosaic virus | Potyviridae | (+)ssRNA | Yam | No | No |
| Junin virus | Arenaviridae | (−)ssRNA | Mice | Yes | No |
| Marseillevirus | Marseilleviridae | dsDNA | Amoeba | Yes | No |
| Melanoplus sanguinipes entomopoxvirus | Poxviridae | dsDNA | Grasshoppers | No | No |
| Paramecium bursaria Chlorella virus 1 | Phycodnaviridae | dsDNA | Algae | No | No |
| Pseudaletia unipuncta granulovirus | Baculoviridae | dsDNA | Moth | No | No |
| Pseudocowpox virus | Poxviridae | dsDNA | Cow | No | No |
| Psittacid herpesvirus 1 | Herpesviridae | dsDNA | Parrots | No | No |
| Rhinovirus A | Picornaviridae | (+)ssRNA | Humans | Yes | No |
| Simian virus 12 | Polyomaviridae | dsDNA | Baboons | No | No |
| Simian virus 40 | Polyomaviridae | dsDNA | Humans | Yes | No |
| Soybean yellow mottle mosaic virus | Tombusviridae | (+)ssRNA | Soybean | No | No |
| Spodoptera frugiperda multiple nucleopolyhedrovirus | Baculoviridae | dsDNA | Lepidoptera | No | No |
| Swinepox virus | Poxviridae | dsDNA | Boars | No | No |
| Tacaribe virus | Arenaviridae | (−)ssRNA | Rodents | Yes | No |
| Taura syndrome virus | Dicistroviridae | (+)ssRNA | Shrimp | No | No |
| Trichoplusia ni ascovirus 2c | Ascoviridae | dsDNA | Moths | No | No |
| Wooly monkey sarcoma virus | Retroviridae | ssRNA-RT | Wooly Monkeys | No | No |
Prevalence of Human Urine Viral Proteins
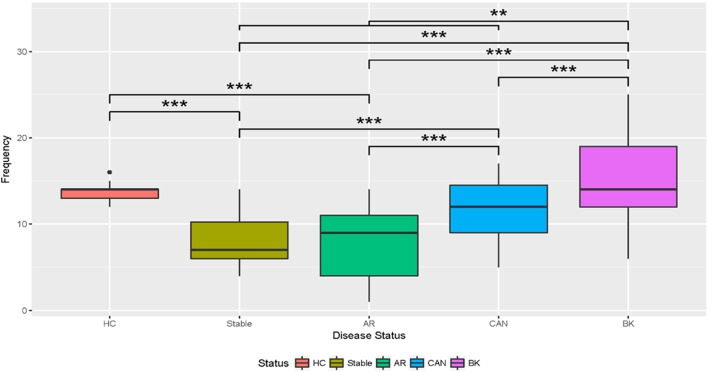
The healthy control group examined had 20 unique viral proteins with a group average of 13.67 ± 1.32 viral proteins. Kidney tx patients with maintenance IS and stable kidney tx function had a statistically significant lower number of unique viruses in the group (8.08 ± 2.78; p = 7e-10) when compared to the healthy cohort (normal kidney function and a normal immune system), without any IS exposure. Nine new viral proteins are instead detected in the STA cohort compared to healthy controls, belonging to the following viruses: Junin virus, Cotesia congregata bracovirus, Agrotis segetum nucleopolyhedrovirus, Psittacid herpesvirus 1, Pseudocowpow virus, Spodoptera frugiperda multiple nucleopolyhedrovirus, Japanese yam mosaic virus, Cowpea mottle virus, and Helicoverpa zea single nucleopolyhedrovirus. Kidney injury after tx (despite continued IS exposure), results in an overall increase in the number of unique viral proteins over the STA tx cohort, with a significant increase in urine viral proteins in the CAN patient group (11.46 ± 3.52; p = 8e-10), and the BKVN patient group (15.76 ± 5.65; p = 13.7e-10). The repertoire of urine viral proteins appears to be quite distinct in different tx injury categories (Figure 3). The prevalence of BKV viral proteins in urine increases to 60–70% in AR, 70–80% in patients in CAN, and 100% in patients with BKVN (Figure 2), highlighting that increasing abundance of the BK virus in urine may result from augmentation of IS, as seen in the AR category, and with greater time post-tx, as seen in the CAN category. BKVN urine samples show the maximal divergence of urinary viral proteins, as expected. Four viral proteins that are consistently present in all other samples, inclusive of normal healthy controls (Canarypox virus, Tacaribe virus, Simian virus 12, Acidianus filamentous virus 8), are no longer observed in the BKVN cohort. As BKV is a DNA virus, we examined if the changes in the urinary virome are largely related to an emergence of new DNA viruses in BKVN. Five new viral proteins are noted only in the BKVN cohort (Ectropis obliqua nucleopolyhedrovirus, Fowl adenovirus D, Taura syndrome virus, Invertebrate iridescent virus 6, Gallid herpesvirus 2). 4/5 of BKVN cohort viruses were double-stranded DNA viruses (Table 1).
Figure 3.
Boxplots of the number of unique viruses discovered by disease status. The boxplots show that the relative abundance of unique viruses discovered in stable and acute rejection (AR) patients is lower than the number discovered in healthy control (HC). The number of viruses discovered increased in chronic allograft nephropathy (CAN) above stable and AR. The number of viruses increased in BK over all immunosuppressed categories. T-Tests with Bonferroni corrections were used for multiple testing. *Significant at p < 0.05; **significant at p < 0.005; ***significant at p < 0.001.
Abundance of Human Urine Viral Proteins
We next evaluated the abundance of different viral proteins across all tx patients and HC urine samples. Among the 37 unique viruses examined, the following viruses were the most abundant in STA IS patients (Acidianus filamentous virus 8, Agrotis segetum nucleopolyhedrovirus, BK polyomavirus, Canarypox virus, Cotesia congregata bracovirus, Cowpea mottle virus, Glossina pallidipes salivary gland, hypertrophy virus, Simian virus 12, Tacaribe virus), and increased abundance of the Marsellavirus is seen in both CAN and BKVN. Despite intrarenal infection with the BKV virus in BKVN patients, we observe that low levels of the BK virus are detected also in all examined tx categories and healthy controls, likely as the BK virus is a normal human urine commensal, with its persistence in the human uroepithelium at low levels in almost all examined HC and kidney tx patients is not unexpected.
Discussion
A study the role of the microbiome in organ transplantation (13, 17, 34) takes in to account of the impact of food intake, digestion, metabolism, and modulation; however, dysbiosis of microbiota due to the tx and IS medicines is a contributing factor that decreases in the baseline predominant microbes and also results in a loss of overall diversity, and the emergence of few new dominant microbial populations (35). A very interesting finding from this study is that only 8 out of the 37 viruses identified in this dataset have been previously described in humans, and many of these have also been described to be pathogenic. Human infections have been described with some of the identified viruses, suggesting that their presence in urine may be relate to the pathogenesis of general systemic or the underlying renal injury. Junin virus (an arenavirus) infections can result in clinical human disease inclusive of fever, as well as an entity known as Argentine hemorrhagic fever (36). Tacaribe virus is an arenavirus that can also cause human fevers and hemorrhagic disease (36). The HHV-6 virus is a highly prevalent virus in children and causes fever, diarrhea, and rashes (37). Rhinoviruses are single-stranded RNA viruses that are the most infectious agent in humans and the most likely culprit for the common cold (38). Acanthocystis turfacea chlorella virus 1 has been in 44% of healthy humans (39) and has only been described in the oropharynx and is not known to be pathogenic to humans. The Marseille virus is a nucleocytoplasmic large DNA virus that has been described in blood and stool of patient with nonfebrile lymphadenopathy (40).
The SV40 virus is a DNA polyoma virus that was likely introduced into the human population through contaminated vaccines (41) and infection with this virus is frequently co-infected with BK virus infection, this being an almost invariant finding in the tx population. Recent studies have also highlighted the role of chronic SV40 infection with human cancers (42). Antibodies against the T antigen of SV40 cross reacts with the T antigens of BK and JC viruses, which are all in the polyoma virus family, and the SV40 stain is used for diagnosis of BK virus infection in BKVN. BK virus exposure, in the kidney, is seen in 90% of the normal population, and our data suggests that the urinary virome has traces of BK viral proteins in all the healthy controls sampled. BK virus replication is seen in 10–60% of kidney tx patients have been described to shed BK virus in their urine, which is confirmed by our data (Figure 2). BK virus is latent in renal tubular epithelial cells and its prevalence is known to increase in immune dysfunction and immunosuppression exposure (43). In this study, we could not identify any urinary virome specific differences that could distinguish the AR and BKVN groups, despite these conditions being immunologically distinct and requiring very different treatment approaches, i.e., minimization of immunosuppression in BKVN and increased immunosuppression in AR. The AR category was bx confirmed and there was negative SV40 stain in the bx tissue examined. Nevertheless, this data suggests that the detection of BKVN may be under-reported in kidney tx patients, and histological changes may be patchy resulting in BKVN disease underdiagnosis.
Of the remaining 19 viruses that have not been previously described in humans, the following have been noted to be used as pesticides/insecticides (E. obliqua nucleopolyhedrovirus, Euproctis pseudoconspersa nucleopolyhedrovirus, Helicoverpa armigera nucleopolyhedrovirus, H. zea single nucleopolyhedrovirus). Pesticides in public health use are intended to limit the potential for disease but they have been known to make their way into human tissues/bodily fluids if not properly handled and that organic diets limit the amount of pesticides in children urine (44, 45). The finding of these viral proteins in human urine samples raises a question of environmental exposure to these previously undiscovered, possible contaminants.
The finding of 100% prevalence of some viruses only in the healthy control group suggests that there are viral commensals that may exist either without harm or may even be hypothesized to assist in immune adaptation. Phylogentic tree analysis of these viruses may be interesting, as some viruses such as bacteriophages play an important role in health by removing pathogenic bacteria and helping boost innate immunity (16). A reduction in the protective viruses or phages may result in increased bacterial proliferation which is seen in the context of IS after kidney tx. Metagenomic sequencing of the new viral species found in tx patients under IS may identify if IS exposure has changed the resistance patterns or resistome of some of the viruses, especially those that are more prone to drive inflammation and immunity (16). Silver nanoparticle treatment has been shown to reduce gut viral and bacterial populations that are pro-inflammatory (46). Mucinophilic microbiomes may be more prone to provide non-host protective immunity, and the loss of any mucinophilic viruses/microbes with IS may also be a driver of the increased risk of cystitis and urinary tract infections seen in patients after tx.
The microbiome intersects with states of health and disease. The novel detection of a large number of viruses used as pesticides/insecticides is a surprising finding. Identification of viruses that are not expected to be present in the urine points to a possibility of their origin to enter bodily systems through ingestion (contamination of food, water, or other drinks such as milk) or cutaneous absorption (e.g., improper hand washing). In fact, according to federal law, a small residue of pesticides/insecticide contamination of human food is recognized and accepted (41, 47), even though no one to date has examined their presence in urine. It is also possible that these viruses colonize or invade the urinary tract by ascending infection, especially in the context of IS in a kidney tx patient, where there is lowered host immune defense.
Further validation of these results by evaluation of the human virome repertoire in different geographic and demographic cohorts of patients with different causes of kidney injury will better identify if the peculiar repertoire of viral proteins observed in this study is specific to a patient, their geography, their demography, or the kidney disease subtype. Additional validation studies by urine viral PCR assays can provide for rapid assessment of the clinical impact of these viruses in other kidney injury cohorts. Interestingly, in the repertoire of identified viral peptides, we did not observe any phages. This is attributed to the methodology used for peptide preparation from urine supernatant, which does not capture bacteria (which get pelleted out), the host of phage viruses (48). The virome report based on DNA sequence analysis also did not identify phages in the published report (19). The urine virome study that used purified bacteriophage isolation using cesium chloride density gradient has reported identification of phages (20). These articles all support that the methodology of sample prep influences the eventual findings.
Colonization, resistance, and microbial ecology has been described and well studied in the context of microbial infections (49), and it has been shown that antibiotic treatment (in the context of treating infections in the animals that form food products or as part of the antiviral prophylaxis for tx patients in the first 3–6 months post-tx) eliminates many commensal bacterial and viral species form the gut and other body fluids and reduces antimicrobial defense (50). Colonization by microbiota in early life shapes the immune system and create a window of opportunity for a homeostatic state, which if disrupted, in the context of tx and IS, can contribute to inflammation. Precision medicine approaches in the future may want to customize therapy from a “microbiota centered” and “host centered” approach to precision medicine. Until recently, we have not paid much attention to the food microbiome and the understanding that many emerging infections may be vectored by foods. Current efforts to develop a metagenome trackr tool that will evaluate the impact of different infections—bacterial, viral, and fungal and their fingerprints in the context of the food that we eat, that maybe influenced by ambient temperature, animal cohort poor survival causation, and agricultural practices inclusive of soil treatment (methyl bromide treatment of soil as a pesticide results in a huge increase in bacillus species), water sources [different microbial loads in pond vs. well water (51)], genetics of the food plant cultivar that may result in altered plant defense to microbes, plants, and pesticides (51). Viral and bacterial lipoproteins may be responsible for immune modulatory properties, and recent studies also suggest a role for the interaction of innate immunity for regulating microbiomal diversity and controlling infection, by specific cell lineages such as mucosal invariant T cells (MAIT cells). Hence, additional studies that focus on the causation of microbial diversity of human tissues and its impact on inflammation and immune responses will be paramount to conduct as part of future research.
Ethics Statement
All the study samples were collected from pediatric and young adult recipients transplanted between years 2000 and 2011 at Lucile Packard Children’s Hospital of Stanford University. The study was approved by the ethics committees of Stanford University Medical School and UCSF Medical Center. All adult patients and parents/guardians of non-adult patients provided written informed consent to participate in the research, in full adherence to the Declaration of Helsinki.
Author Contributions
MS and TS participated in the design of the study; SN, TS, NM, and MS participated in the writing of the article and the interpretation of the results; NM performed the statistical analyses and evaluation of results; CN, KB-J, and W-JQ generated and processed the raw data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer DP declared a shared affiliation, with no collaboration, with three of the authors NM, TS, and MS to the handling editor.
Footnotes
Funding. We acknowledge support from University of California San Francisco. The authors acknowledge the funding support from NIDDK R01DK083447 (to MS), DP3 DK110844 (W-JQ), and P41 GM103493 (W-JQ).
References
- 1.Potgieter M, Bester J, Kell DB, Pretorius E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol Rev (2015) 39(4):567–91. 10.1093/femsre/fuv013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Zhao F, Deng Y, Zhao Y, Ren H. Metagenomic and metabolomic analysis of the toxic effects of trichloroacetamide-induced gut microbiome and urine metabolome perturbations in mice. J Proteome Res (2015) 14(4):1752–61. 10.1021/pr5011263 [DOI] [PubMed] [Google Scholar]
- 3.Lewis DA, Brown R, Williams J, White P, Jacobson SK, Marchesi JR, et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol (2013) 3:41. 10.3389/fcimb.2013.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med (2012) 10:174. 10.1186/1479-5876-10-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim Y, Totsika M, Morrison M, Punyadeera C. The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Sci Rep (2017) 7(1):8523. 10.1038/s41598-017-07885-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nasidze I, Quinque D, Li J, Li M, Tang K, Stoneking M. Comparative analysis of human saliva microbiome diversity by barcoded pyrosequencing and cloning approaches. Anal Biochem (2009) 391(1):64–8. 10.1016/j.ab.2009.04.034 [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behav Immun (2014) 38:1–12. 10.1016/j.bbi.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer (2016) 102:89–95. 10.1016/j.lungcan.2016.10.016 [DOI] [PubMed] [Google Scholar]
- 9.Twigg HL, III, Morris A, Ghedin E, Curtis JL, Huffnagle GB, Crothers K, et al. Use of bronchoalveolar lavage to assess the respiratory microbiome: signal in the noise. Lancet Respir Med (2013) 1(5):354–6. 10.1016/S2213-2600(13)70117-6 [DOI] [PubMed] [Google Scholar]
- 10.Human Microbiome Project Consortium. A framework for human microbiome research. Nature (2012) 486(7402):215–21. 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroemer A, Elsabbagh AM, Matsumoto CS, Zasloff M, Fishbein TM. The microbiome and its implications in intestinal transplantation. Curr Opin Organ Transplant (2016) 21(2):135–9. 10.1097/MOT.0000000000000278 [DOI] [PubMed] [Google Scholar]
- 12.Weber D, Oefner PJ, Hiergeist A, Koestler J, Gessner A, Weber M, et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood (2015) 126(14):1723–8. 10.1182/blood-2015-04-638858 [DOI] [PubMed] [Google Scholar]
- 13.Vindigni SM, Surawicz CM. The gut microbiome: a clinically significant player in transplantation? Expert Rev Clin Immunol (2015) 11(7):781–3. 10.1586/1744666X.2015.1043894 [DOI] [PubMed] [Google Scholar]
- 14.Becker J, Poroyko V, Bhorade S. The lung microbiome after lung transplantation. Expert Rev Respir Med (2014) 8(2):221–31. 10.1586/17476348.2014.890518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, et al. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci U S A (2009) 106(40):17187–92. 10.1073/pnas.0904847106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rani A, Ranjan R, McGee HS, Andropolis KE, Panchal DV, Hajjiri Z, et al. Urinary microbiome of kidney transplant patients reveals dysbiosis with potential for antibiotic resistance. Transl Res (2017) 181:59–70. 10.1016/j.trsl.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fricke WF, Maddox C, Song Y, Bromberg JS. Human microbiota characterization in the course of renal transplantation. Am J Transplant (2014) 14(2):416–27. 10.1111/ajt.12588 [DOI] [PubMed] [Google Scholar]
- 18.Lee JR, Muthukumar T, Dadhania D, Toussaint NC, Ling L, Pamer E, et al. Gut microbial community structure and complications after kidney transplantation: a pilot study. Transplantation (2014) 98(7):697–705. 10.1097/TP.0000000000000370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rani A, Ranjan R, McGee HS, Metwally A, Hajjiri Z, Brennan DC, et al. A diverse virome in kidney transplant patients contains multiple viral subtypes with distinct polymorphisms. Sci Rep (2016) 6:33327. 10.1038/srep33327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santiago-Rodriguez TM, Ly M, Bonilla N, Pride DT. The human urine virome in association with urinary tract infections. Front Microbiol (2015) 6:14. 10.3389/fmicb.2015.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut (2004) 53(1):1–4. 10.1136/gut.53.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res (2012) 22(2):299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444(7122):1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 24.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature (2009) 457(7228):480–4. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla SD, Budden KF, Neal R, Hansbro PM. Microbiome effects on immunity, health and disease in the lung. Clin Transl Immunology (2017) 6(3):e133. 10.1038/cti.2017.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr (2011) 6(3):209–40. 10.1007/s12263-011-0229-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature (2011) 474(7351):327–36. 10.1038/nature10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med (2016) 375(24):2369–79. 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- 29.Morgan XC, Huttenhower C. Chapter 12: human microbiome analysis. PLoS Comput Biol (2012) 8(12):e1002808. 10.1371/journal.pcbi.1002808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ (2017) 356:j831. 10.1136/bmj.j831 [DOI] [PubMed] [Google Scholar]
- 31.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486(7402):207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solez K, Colvin RB, Racusen LC, Haas M, Sis B, Mengel M, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant (2008) 8(4):753–60. 10.1111/j.1600-6143.2008.02159.x [DOI] [PubMed] [Google Scholar]
- 33.Sigdel TK, Gao Y, He J, Wang A, Nicora CD, Fillmore TL, et al. Mining the human urine proteome for monitoring renal transplant injury. Kidney Int (2016) 89(6):1244–52. 10.1016/j.kint.2015.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fricke WF, Bromberg JS. Checks and balances-microbiota shifts in immunosuppressed mice. Transplantation (2017) 101(1):26–7. 10.1097/TP.0000000000001493 [DOI] [PubMed] [Google Scholar]
- 35.Lu H, He J, Wu Z, Xu W, Zhang H, Ye P, et al. Assessment of microbiome variation during the perioperative period in liver transplant patients: a retrospective analysis. Microb Ecol (2013) 65(3):781–91. 10.1007/s00248-013-0211-6 [DOI] [PubMed] [Google Scholar]
- 36.Viral Hemorrhagic Fevers. Centers for Disease Control and Prevention; (2013). [Google Scholar]
- 37.Hall CB, Long CE, Schnabel KC, Caserta MT, McIntyre KM, Costanzo MA, et al. Human herpesvirus-6 infection in children. A prospective study of complications and reactivation. N Engl J Med (1994) 331(7):432–8. 10.1056/NEJM199408183310703 [DOI] [PubMed] [Google Scholar]
- 38.Love RA, Maegley KA, Yu X, Ferre RA, Lingardo LK, Diehl W, et al. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure (2004) 12(8):1533–44. 10.1016/j.str.2004.05.024 [DOI] [PubMed] [Google Scholar]
- 39.Petro C, et al. Herpes simplex type 2 virus deleted in glycoprotein D protects against vaginal, skin and neural disease. Elife (2015) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popgeorgiev N, Michel G, Lepidi H, Raoult D, Desnues C. Marseillevirus adenitis in an 11-month-old child. J Clin Microbiol (2013) 51(12):4102–5. 10.1128/JCM.01918-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martini F, Iaccheri L, Lazzarin L, Carinci P, Corallini A, Gerosa M, et al. SV40 early region and large T antigen in human brain tumors, peripheral blood cells, and sperm fluids from healthy individuals. Cancer Res (1996) 56(20):4820–5. [PubMed] [Google Scholar]
- 42.Vilchez RA, Butel JS. Emergent human pathogen simian virus 40 and its role in cancer. Clin Microbiol Rev (2004) 17(3):495–508. 10.1128/CMR.17.3.495-508.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mbianda C, El-Meanawy A, Sorokin A. Mechanisms of BK virus infection of renal cells and therapeutic implications. J Clin Virol (2015) 71:59–62. 10.1016/j.jcv.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fourth National Report on Human Exposure to Environmental Chemicals. Center for Disease Control; (2009). [Google Scholar]
- 45.Lu C, Toepel K, Irish R, Fenske RA, Barr DB, Bravo R. Organic diets significantly lower children’s dietary exposure to organophosphorus pesticides. Environ Health Perspect (2006) 114(2):260–3. 10.1289/ehp.8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams KM, Gokulan K, Cerniglia CE, Khare S. Size and dose dependent effects of silver nanoparticle exposure on intestinal permeability in an in vitro model of the human gut epithelium. J Nanobiotechnology (2016) 14(1):62. 10.1186/s12951-016-0214-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Summary of the Federal Food, Drug, and Cosmetic Act. 2002. United States Environmental Protection Agency; (2002). [Google Scholar]
- 48.Lavigne R, Ceyssens PJ, Robben J. Phage proteomics: applications of mass spectrometry. Methods Mol Biol (2009) 502:239–51. 10.1007/978-1-60327-565-1_14 [DOI] [PubMed] [Google Scholar]
- 49.Theriot CM, Young VB. Interactions between the gastrointestinal microbiome and clostridium difficile. Annu Rev Microbiol (2015) 69:445–61. 10.1146/annurev-micro-091014-104115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science (2016) 352(6285):544–5. 10.1126/science.aad9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ottesen AR, González Peña A, White JR, Pettengill JB, Li C, Allard S, et al. Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol (2013) 13:114. 10.1186/1471-2180-13-114 [DOI] [PMC free article] [PubMed] [Google Scholar]