Abstract
Otitis media (OM) is one of the most common pediatric infections worldwide, but the complex microbiology associated with OM is poorly understood. Previous studies have shown an association between OM and gastroesophageal reflux (GER) in children. Therefore, in order to bridge the gap in our current understanding of the interaction between GER and OM, we investigated the nasopharyngeal and middle ear microbiota of children suffering from GER-associated OM and OM only, using culture-independent 16S rRNA gene sequencing. Middle ear fluid, nasopharyngeal swabs, and clinical data were collected as part of a prospective pilot study conducted at the Department of Otorhinolaryngology of the Erasmus MC-Sophia Children’s Hospital, Rotterdam, the Netherlands. A total of 30 children up to 12 years of age who suffered from recurrent acute otitis media (AOM) (5), chronic otitis media with effusion (OME) (23), or both (2), and who were listed for tympanostomy tube placement, were included in the study. Nine children were included in the GER-associated OM cohort and 21 in the OM-only cohort. We found no obvious effect of GER on the nasopharyngeal and middle ear microbiota between the two groups of children. However, our results highlight the need to assess the true role of Alloiococcus spp. and Turicella spp. in children presenting with a high prevalence of recurrent AOM and chronic OME.
Introduction
Respiratory tract infections are the leading causes of morbidity and death in children and adults worldwide, with otitis media (OM) being one of the most prevalent pediatric respiratory tract infections [1]. OM causes severe pain and can lead to serious complications, such as meningitis, mastoiditis, and hearing loss. As an example, OM-related hearing impairment has a prevalence of 30.82 per 10,000, and, each year, 21,000 people die due to complications of OM (estimated from 21 World Health Organization [WHO] regional areas) [2]. Importantly, the pathogenesis of OM is multifactorial, involving genetic, microbiological, and environmental factors [3].
A potentially important, though relatively unknown, factor to be associated with OM is gastroesophageal reflux (GER). GER, also known as acid reflux, is a clinical manifestation in which the stomach’s contents return back up into the esophagus and mouth. Reflux is a normal process that occurs in healthy infants, children, and adults. Most episodes are brief and do not cause bothersome symptoms or complications. However, earlier studies have shown a strong association between GER and OM [4–6]. The mean prevalence of GER in children with (chronic) OM with effusion (OME) is 48% and in children with (recurrent) acute OM (AOM) 63%, while the mean prevalence of GER in healthy children is less than 10% [7]. The data suggest a role for GER in the multifactorial etiology of OM in children, a hypothesis strengthened by the detection of pepsin and bile acids in the middle ears of children presenting with OM [4–6], as well as the possible role of Helicobacter pylori in the etiology of OM [8–11]. However, to date, no one has used culture-independent techniques to investigate the possible influence of GER on the nasopharyngeal and middle ear microbiota in GER-prone children. In this respect, differences in the presence or absence of (specific) microorganisms in the middle ear of GER-prone versus GER non-prone children could help further establish a role for GER in the pathogenesis of OM.
Therefore, in order to bridge the gap in our current understanding of the interaction between GER and OM, we investigated the nasopharyngeal and middle ear microbiota of children suffering from GER-associated OM and OM only, using culture-independent 16S rRNA gene sequencing.
Materials and methods
Middle ear fluid (MEF), nasopharyngeal swabs (NPS), and clinical data were collected according to Klokkenburg et al. [4] and Stol et al. [12], as part of a prospective pilot study conducted at the Department of Otorhinolaryngology of the Erasmus MC-Sophia Children’s Hospital, Rotterdam, the Netherlands. Children with a history of craniofacial malformations/syndromes, or primary or acquired immune deficiency were excluded from participation. Questionnaires were used to obtain relevant patient characteristics from the parents of the GER-associated OM cohort and the OM-only cohort. The GER-associated cohort was defined via questionnaires based on the questions “Has your child been diagnosed with reflux in the previous three months?” and “Has your child been diagnosed with reflux in the past?” The study was approved by the Erasmus MC Medical Ethical Committee (MEC-2012-487). Written informed consent was obtained from parents or caregivers.
Microbiota profiles were analyzed using next-generation sequencing of hypervariable V5-V6 regions of the 16S rRNA gene, as previously described [13]. Prior to 16S rRNA gene sequencing, the total number of 16S rRNA gene copies within each DNA extraction was measured using a 16S quantitative polymerase chain reaction (PCR) according to Yang et al. [14]. This was performed in order to remove any specimens from the analysis where the contaminating bacterial DNA that is already present within chemicals and consumables used in the experimental procedure could be responsible for the results obtained. In this respect, all specimens that contained < 1000 16S rRNA gene copies/μL were not sequenced [15]. All sequence data obtained from specimens containing > 1000 16S rRNA gene copies/μL was processed using bioinformatics modules present in the mothur v.1.33.0 software package [16].
Statistical analyses were performed using IBM SPSS Statistics 21. Fisher’s exact test was used to calculate the statistical differences between patient baseline characteristics, differences between antibiotic use and specimen exclusion, antibiotic use, and pathogen detection, and to calculate the statistical differences of prominent middle ear microbiota between GER-prone and GER non-prone children. A p-value of ≤ 0.05 was considered to be statistically significant.
Data availability
The datasets generated and analyzed during the current study are available in the NCBI Sequence Read Archive (SRA) repository with the accession number SRP099862, https://www.ncbi.nlm.nih.gov/sra/?term=SRP099862.
Results and discussion
16S rRNA gene sequencing
Thirty children up to 12 years of age who suffered from recurrent AOM (5), chronic OME (23), or both (2), and who were listed for tympanostomy tube placement, were included in the study (Table 1). Children were diagnosed based on the surgeon’s report.
Table 1.
Demographic and clinical parameters stratified by the gastroesophageal reflux (GER)-associated otitis media (OM) cohort (n = 21) and the OM-only cohort (n = 9)
| No GER, n = 21 (%) | GER, n = 9 (%) | p-Value | |
|---|---|---|---|
| Demographic parameters | |||
| Male sex | 13 (61.9) | 5 (55.6) | 1 |
| Age, years | 5.3 [1.3–6.0] | 3.7 [0.8–12.8] | |
| Antibiotics ≤ 6 months preceding inclusion | 12 (57.1) | 5 (55.6) | 1 |
| Siblings | 16 (76.2) | 5 (55.6) | 0.39 |
| Day-care | 7 (33.3) | 7 (77.8) | 0.05 |
| Smoking | 14 (66.7) | 5 (55.6) | 0.69 |
| Breastfeeding | 11 (52.4) | 8 (88.9) | 0.10 |
| Diagnosis | |||
| OME | 16 (76.2) | 7 (77.8) | 1 |
| rAOM + (OME) | 5 (23.8) | 2 (22.2) | 1 |
| Ventilation tube placement in past | 11 (52.4) | 1 (11.1) | 0.05 |
| Adenotonsillectomy in past | 7 (33.3) | 0 (0) | 0.07 |
In total, 11/30 MEF specimens and 11/30 NPS specimens (not, by definition, paired specimens) were excluded from 16S rRNA gene sequencing due to low DNA content (< 1000 16S rRNA gene copies/μL). Antibiotic use ≤ 6 months prior to inclusion in the study did not affect the exclusion of samples due to low DNA content (Table 2; MEF p = 0.13, NPS p = 0.45). The microbial composition of the remaining 38 specimens were sequenced and the data were processed using mothur. To reduce the effects of uneven sequencing depths, sequences were rarefied to 1000 sequences per specimen, which was sufficient to obtain a high degree of sequence coverage with an average Good’s coverage value of 0.98 (± 0.01).
Table 2.
Inclusion and exclusion of samples related to antibiotic use ≤ 6 months preceding inclusion
| MEF | p-Value | NPS | p-Value | |||
|---|---|---|---|---|---|---|
| Inclusion | Exclusion | Inclusion | Exclusion | |||
| Antibiotics | 13 | 4 | 0.13 | 12 | 5 | 0.45 |
| No antibiotics | 6 | 7 | 7 | 6 | ||
Sequencing of the MEF microbiota resulted in the identification of 29 bacterial operational taxonomic units (OTUs) in 19 MEF specimens with an abundance > 1%, with an average of 4 OTUs (± 3) per specimen. Sequencing the nasopharyngeal microbiota identified 39 bacterial OTUs in 19 NPS specimens with > 1% abundance, with an average of 9 OTUs (± 4) per specimen.
Characterization of middle ear bacterial communities
The most commonly detected OTUs were Alloiococcus spp. and Turicella spp., present in 12 and 11 MEF specimens, respectively. There has been some debate as to whether members of the bacterial genera Alloiococcus and Turicella actually play a role in the pathogenesis of OM in children or if they are part of the commensal microbiota (and invading the middle ear following perforation) [17–23]. Though A. otitidis and T. otitidis are frequently detected in OM children, it is questionable whether these organisms have enough pathogenic potential to induce OM, although studies have shown that A. otitidis may have enough immunogenic potential to modulate a host immune response [24–26]. In addition, A. otitidis is able to form both single- and multispecies biofilms with Haemophilus influenzae. When present in polymicrobial biofilms, A. otitidis can promote H. influenzae growth and survival by increasing biofilm production in adverse growth conditions and by altering antimicrobial resistance [27]. In this study, the use of antibiotics ≤ 6 months prior to inclusion in the study did not have an effect on the detection of Alloiococcus spp., Haemophilus spp., Streptococcus spp., or Moraxella spp. (Table 3). These findings are clinically important, as antibiotics overuse or misuse can promote the development of bacterial resistance to antibiotics. Alloiococcus spp. and Turicella spp. were not detected by 16S rRNA gene sequencing in any nasopharyngeal swab. Previous PCR- and culture-based studies from other populations have reported A. otitidis in 7–12% of nasopharyngeal swabs, in contrast to the study of Marsh et al., who found no A. otitidis in paired nasopharyngeal swabs [17, 18, 28, 29]. Turicella otitidis is isolated almost exclusively from middle ear exudates [20, 23]. However, our study was limited to 30 children and it is possible that Alloiococcus spp. and Turicella spp. would have been detected in a larger cohort of children. Failure to detect Alloiococcus spp. and Turicella spp. in nasopharyngeal swabs from any of the children with positive ear MEF samples suggests that they are unlikely to be primary otopathogens in this population, since nasopharyngeal colonization is considered to be the antecedent of OM [30]. To better understand the potential role of Alloiococcus spp. and Turicella spp. in OM, further studies into host interactions and colonization of adjacent sites in healthy and diseased children will be necessary. Ultimately, the findings of this study will need to be further validated in the future with larger cohort sizes.
Table 3.
Antibiotic use and pathogen detection in middle ear fluid (MEF)
| Pathogen | No. of specimens | |||||
|---|---|---|---|---|---|---|
| GER (n = 6) | p-Value | GER (n = 13) | p-Value | |||
| Without antibiotics (n = 2) | With antibiotics (n = 4) | Without antibiotics (n = 4) | With antibiotics (n = 9) | |||
| Alloiococcus spp. | 2 | 0 | 0.07 | 3 | 7 | 1 |
| Streptococcus spp. | 0 | 1 | 1 | 1 | 4 | 1 |
| Haemophilus spp. | 1 | 1 | 1 | 1 | 6 | 0.27 |
| Moraxella spp. | 0 | 0 | 1 | 0 | 1 | 1 |
OTUs consistent with classical otopathogens such as Haemophilus spp., Streptococcus spp., and Moraxella spp. were detected in 9, 6, and 1 MEF specimens, respectively (Fig. 1). In addition, staphylococci were also found in the MEF of seven children, with an abundance up to 100%. Our findings are consistent with several other studies; for example, Haemophilus (influenzae) has been found to be the most common pathogen isolated in the context of widespread conjugate pneumococcal vaccination [31, 32]. Further, the presence of Haemophilus (influenzae) has been associated with an increased risk of OM, whereas the presence of Corynebacterium spp. and Dolosigranulum spp. have been associated with a decreased risk of OM [33, 34].
Fig. 1.
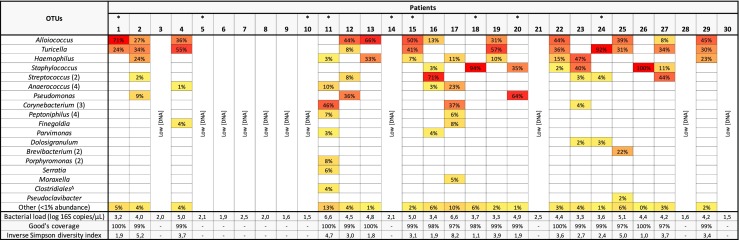
Heatmap demonstrating the relative abundance of bacterial taxa in individual middle ear fluid (MEF) specimens. Bacterial operational taxonomic units (OTUs) are grouped and shown at the genus level, unless the OTU was not identified below the taxonomic order level (Δ). A cut-off of 1% abundance was used for visual differentiation between specimens. The number of merged OTUs is shown in parentheses. Children that suffered from gastroesophageal reflux (GER) are indicated with an asterisk
No upper gastrointestinal tract microbiota previously associated with OM (e.g., H. pylori) was detected in the middle ear. Further, we did not find any microbial difference in the MEF of children with or without GER (Table 4), suggesting that GER was not associated with the translocation of stomach microbiota to the middle ear in our cohort.
Table 4.
Effect of GER on the presence of prominent middle ear microbiota
| Pathogen | No. of specimens | ||
|---|---|---|---|
| GER (n = 6) | No GER (n = 13) | p-Value | |
| Alloiococcus spp. | 2 | 10 | 0.13 |
| Haemophilus spp. | 2 | 7 | 0.66 |
| Streptococcus spp. | 1 | 5 | 0.60 |
| Moraxella spp. | 0 | 1 | 1 |
Co-occurrence of OM-related pathogens
To study the co-occurrence of OM-related pathogens in the nasopharynx and middle ear, we compared the microbiota obtained from 12 paired NPS and MEF specimens (Fig. 2). Nine OTUs were present in both specimen types, of which four OTUs, including Haemophilus spp., Streptococcus spp., Parvimonas spp., and Staphylococcus spp. were present in both paired specimen types from 10/12 children. Importantly, Haemophilus spp. and Streptococcus spp. were only detected in MEF when the nasopharynx of the child was concurrently colonized with these bacterial genera. This finding is in agreement with previous studies, and suggests that the more easily accessible nasopharyngeal microbiota could be used to predict the presence of pediatric otopathogens in the middle ears of children suffering from OM. However, it should be noted that the majority of OTUs, including the dominant genera Alloiococcus spp. and Turicella spp., were unique to the sampling site and, therefore, caution is necessary when using the nasopharyngeal microbiota as a proxy for the MEF microbiota, as discussed previously by van Dongen et al. [35].
Fig. 2.

Co-occurrence model showing bacterial taxa present within 12 paired nasopharyngeal (NPS) and middle ear fluid (MEF) specimens. Data (Cytoscape v3.2.1) [37] are presented for 12 children, and include a single NPS and MEF specimen per patient. Taxonomic assignment for each OTU is at the genus level, unless the OTU was not identified below the taxonomic order level (*). A cut-off of 1% abundance was used for visual differentiation between both specimen types. Bracketed numbers [NPS/MEF] and node sizes indicate the number of specimens that contain a specified OTU. Shared OTUs are visualized in the middle and connected by a line if the presence of an OTU in both specimen types was derived from the same patient (solid) or from different children (dashed). Specimen-specific OTUs are grouped at the genus or order levels to ease visualization
Strengths and limitations of the study
The strength of this publication is that it is the first study to investigate the possible association between the microbial composition of the nasopharynx and middle ear in GER-prone and GER non-prone children using culture-free 16S rRNA gene sequencing. However, several limitations in the study protocol should be recognized.
Firstly, an actual diagnosis of GER is difficult to prove, as most cases of pediatric GER are diagnosed based solely on the clinical presentation and parental observation (and not, for example, on the presence of pepsin and bile acids in the middle ear). There are no recognized classic physical signs of GER in the pediatric population, and, in general, a doctor diagnoses GER by reviewing an infant’s symptoms and medical history. In this study, only parental-reported clinical information was collected. Essentially, we asked parents retrospectively if their child was diagnosed with reflux in the previous three months and if their child had been diagnosed with reflux at any time in the past. To date, the most thoroughly evaluated questionnaire for infant symptoms is the Infant Gastroesophageal Reflux Questionnaire Revised (I-GERQ-R) [36]. We designed our own questionnaire, as even an established questionnaire such as the I-GERQ-R was not validated to be adequately specific to differentiate GER infants from other symptomatic, but non-GER, infants, particularly if the treatments being tested are directed at acid reflux. For that purpose, additional inclusion criteria, such as esophageal pH monitoring or histology, are currently necessary to ensure adequate diagnostic specificity [36]. In the current study, we did not confirm the parental-reported diagnosis of GER by the identification of bile acids/pepsin in MEF or measurement of the MEF pH levels. These tools have proven to be useful for determining the presence of pathological GER [4–6]. The absence of these tools in our study may have influenced the fact that we found no statistically significant association between the microbiota profiles of GER-prone and GER non-prone children.
Secondly, our study included only children primarily diagnosed with OME, as these children are mainly listed for tympanostomy tube placement. Most children with AOM experience a self-limiting illness and lack the presence of fluid in the middle ear. Many will not present to a doctor. This means that an association between GER and AOM disease is much more difficult to prove than between GER and OME disease, but does not affect the hypothesis that GER may be associated with increased OM disease in GER-prone children.
Thirdly, as previously discussed, using the more easily accessible nasopharyngeal microbiota to predict the presence of pediatric otopathogens in the middle ears of children suffering from OME and AOM is questionable. Because of the size of the population studied and lack of data from other studies, data interpretation should be made with caution and regarded as preliminary. Further, we were unable to analyze the microbiota to the species level (a common problem with microbiota analysis [13]) and were, therefore, limited to bacterial taxonomic identification at the genus level.
Acknowledgements
The authors would like to thank the staff of the operating rooms of the Department of Otorhinolaryngology of the Erasmus MC-Sophia Children’s Hospital, the parents who consented, and the children who participated in this study.
Funding
None to declare.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the Erasmus MC Medical Ethical Committee (MEC-2012-487).
Informed consent
Written informed consent was obtained from parents or caregivers.
References
- 1.Marchisio P, Bianchini S, Capaccio P, Esposito S, Fusi M, Nazzari E, Torretta S, Principi N. Insights into infectious otitis media. Int J Immunopathol Pharmacol. 2010;23(1 Suppl):20–23. [PubMed] [Google Scholar]
- 2.Monasta L, Ronfani L, Marchetti F, Montico M, Vecchi Brumatti L, Bavcar A, Grasso D, Barbiero C, Tamburlini G. Burden of disease caused by otitis media: systematic review and global estimates. PLoS One. 2012;7(4):e36226. doi: 10.1371/journal.pone.0036226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rovers MM, Schilder AG, Zielhuis GA, Rosenfeld RM. Otitis media. Lancet. 2004;363(9407):465–473. doi: 10.1016/S0140-6736(04)15495-0. [DOI] [PubMed] [Google Scholar]
- 4.Klokkenburg JJ, Hoeve HL, Francke J, Wieringa MH, Borgstein J, Feenstra L. Bile acids identified in middle ear effusions of children with otitis media with effusion. Laryngoscope. 2009;119(2):396–400. doi: 10.1002/lary.20115. [DOI] [PubMed] [Google Scholar]
- 5.He Z, O’Reilly RC, Bolling L, Soundar S, Shah M, Cook S, Schmidt RJ, Bloedon E, Mehta DI. Detection of gastric pepsin in middle ear fluid of children with otitis media. Otolaryngol Head Neck Surg. 2007;137(1):59–64. doi: 10.1016/j.otohns.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 6.He Z, O’Reilly RC, Mehta D. Gastric pepsin in middle ear fluid of children with otitis media: clinical implications. Curr Allergy Asthma Rep. 2008;8(6):513–518. doi: 10.1007/s11882-008-0094-7. [DOI] [PubMed] [Google Scholar]
- 7.Miura MS, Mascaro M, Rosenfeld RM. Association between otitis media and gastroesophageal reflux: a systematic review. Otolaryngol Head Neck Surg. 2012;146(3):345–352. doi: 10.1177/0194599811430809. [DOI] [PubMed] [Google Scholar]
- 8.Sudhoff H, Rajagopal S, Baguley DM, Ebmeyer J, Schmelzer A, Schreiber S, Moffat DA. A critical evaluation of the evidence on a causal relationship between Helicobacter pylori and otitis media with effusion. J Laryngol Otol. 2008;122(9):905–911. doi: 10.1017/S0022215107000989. [DOI] [PubMed] [Google Scholar]
- 9.Yılmaz MD, Aktepe O, Çetinkol Y, Altuntaş A. Does Helicobacter pylori have role in development of otitis media with effusion? Int J Pediatr Otorhinolaryngol. 2005;69(6):745–749. doi: 10.1016/j.ijporl.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Boronat-Echeverría N, Aguirre-Mariscal H, Carmolinga-Ponce M, Sevilla-Delgado Y, Miceli-Flores R, Kennedy-Padilla A, Mejía-Aranguré JM. Helicobacter pylori detection and clinical symptomatology of gastroesophageal reflux disease in pediatric patients with otitis media with effusion. Int J Pediatr Otorhinolaryngol. 2016;87:126–129. doi: 10.1016/j.ijporl.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 11.Yılmaz T, Ceylan M, Akyön Y, Özçakýr O, Gürsel B. Helicobacter pylori: a possible association with otitis media with effusion. Otolaryngol Head Neck Surg. 2006;134(5):772–777. doi: 10.1016/j.otohns.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Stol K, Verhaegh SJ, Graamans K, Engel JA, Sturm PD, Melchers WJ, Meis JF, Warris A, Hays JP, Hermans PW. Microbial profiling does not differentiate between childhood recurrent acute otitis media and chronic otitis media with effusion. Int J Pediatr Otorhinolaryngol. 2013;77(4):488–493. doi: 10.1016/j.ijporl.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, Bruin J, Montijn R, Bonten M, Sanders E. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One. 2011;6(2):e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S, Lin S, Kelen GD, Quinn TC, Dick JD, Gaydos CA, Rothman RE. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J Clin Microbiol. 2002;40(9):3449–3454. doi: 10.1128/JCM.40.9.3449-3454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biesbroek G, Sanders EA, Roeselers G, Wang X, Caspers MP, Trzciński K, Bogaert D, Keijser BJ. Deep sequencing analyses of low density microbial communities: working at the boundary of accurate microbiota detection. PLoS One. 2012;7(3):e32942. doi: 10.1371/journal.pone.0032942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tano K, von Essen R, Eriksson PO, Sjöstedt A. Alloiococcus otitidis—otitis media pathogen or normal bacterial flora? APMIS. 2008;116(9):785–790. doi: 10.1111/j.1600-0463.2008.01003.x. [DOI] [PubMed] [Google Scholar]
- 18.Harimaya A, Takada R, Somekawa Y, Fujii N, Himi T. High frequency of Alloiococcus otitidis in the nasopharynx and in the middle ear cavity of otitis-prone children. Int J Pediatr Otorhinolaryngol. 2006;70(6):1009–1014. doi: 10.1016/j.ijporl.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 19.Chan CL, Wabnitz D, Bardy JJ, Bassiouni A, Wormald PJ, Vreugde S, Psaltis AJ. The microbiome of otitis media with effusion. Laryngoscope. 2016;126(12):2844–2851. doi: 10.1002/lary.26128. [DOI] [PubMed] [Google Scholar]
- 20.Jervis-Bardy J, Rogers GB, Morris PS, Smith-Vaughan HC, Nosworthy E, Leong LE, Smith RJ, Weyrich LS, De Haan J, Carney AS, Leach AJ, O’Leary S, Marsh RL. The microbiome of otitis media with effusion in Indigenous Australian children. Int J Pediatr Otorhinolaryngol. 2015;79(9):1548–1555. doi: 10.1016/j.ijporl.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Holzmann D, Funke G, Linder T, Nadal D. Turicella otitidis and Corynebacterium auris do not cause otitis media with effusion in children. Pediatr Infect Dis J. 2002;21(12):1124–1126. doi: 10.1097/00006454-200212000-00007. [DOI] [PubMed] [Google Scholar]
- 22.von Graevenitz A, Funke G. Turicella otitidis and Corynebacterium auris: 20 years on. Infection. 2014;42(1):1–4. doi: 10.1007/s15010-013-0488-x. [DOI] [PubMed] [Google Scholar]
- 23.Gomez-Garces JL, Alhambra A, Alos JI, Barrera B, García G. Acute and chronic otitis media and Turicella otitidis: a controversial association. Clin Microbiol Infect. 2004;10(9):854–857. doi: 10.1111/j.1198-743X.2004.00965.x. [DOI] [PubMed] [Google Scholar]
- 24.Harimaya A, Takada R, Himi T, Yokota S, Fujii N. Evidence of local antibody response against Alloiococcus otitidis in the middle ear cavity of children with otitis media. FEMS Immunol Med Microbiol. 2007;49(1):41–45. doi: 10.1111/j.1574-695X.2006.00166.x. [DOI] [PubMed] [Google Scholar]
- 25.Tarkkanen J, Himi T, Harimaya A, Carlson P, Ylikoski J, Mattila PS. Stimulation of adenoidal lymphocytes by Alloiococcus otitidis. Ann Otol Rhinol Laryngol. 2000;109(10 Pt 1):958–964. doi: 10.1177/000348940010901010. [DOI] [PubMed] [Google Scholar]
- 26.Himi T, Kita H, Mitsuzawa H, Harimaya A, Tarkkanen J, Hendolin P, Ylikoski J, Fujii N. Effect of Alloiococcus otitidis and three pathogens of otitis media in production of interleukin-12 by human monocyte cell line. FEMS Immunol Med Microbiol. 2000;29(2):101–106. doi: 10.1111/j.1574-695X.2000.tb01511.x. [DOI] [PubMed] [Google Scholar]
- 27.Chan CL, Richter K, Wormald PJ, Psaltis AJ, Vreugde S. Alloiococcus otitidis forms multispecies biofilm with Haemophilus influenzae: effects on antibiotic susceptibility and growth in adverse conditions. Front Cell Infect Microbiol. 2017;7:344. doi: 10.3389/fcimb.2017.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur R, Adlowitz DG, Casey JR, Zeng M, Pichichero ME. Simultaneous assay for four bacterial species including Alloiococcus otitidis using multiplex-PCR in children with culture negative acute otitis media. Pediatr Infect Dis J. 2010;29(8):741–745. doi: 10.1097/INF.0b013e3181d9e639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsh RL, Binks MJ, Beissbarth J, Christensen P, Morris PS, Leach AJ, Smith-Vaughan HC. Quantitative PCR of ear discharge from indigenous Australian children with acute otitis media with perforation supports a role for Alloiococcus otitidis as a secondary pathogen. BMC Ear Nose Throat Disord. 2012;12:11. doi: 10.1186/1472-6815-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jervis-Bardy J, Carney AS, Duguid R, Leach AJ. Microbiology of otitis media in Indigenous Australian children: review. J Laryngol Otol. 2017;131(S2):S2–S11. doi: 10.1017/S0022215116009294. [DOI] [PubMed] [Google Scholar]
- 31.Casey JR, Adlowitz DG, Pichichero ME. New patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2010;29(4):304–309. doi: 10.1097/INF.0b013e3181c1bc48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unger SA, Bogaert D. The respiratory microbiome and respiratory infections. J Infect. 2017;74(Suppl 1):S84–S88. doi: 10.1016/S0163-4453(17)30196-2. [DOI] [PubMed] [Google Scholar]
- 33.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol. 2012;78(17):6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190(11):1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 35.van Dongen TM, van der Heijden GJ, van Zon A, Bogaert D, Sanders EA, Schilder AG. Evaluation of concordance between the microorganisms detected in the nasopharynx and middle ear of children with otitis media. Pediatr Infect Dis J. 2013;32(5):549–552. doi: 10.1097/INF.0b013e318280ab45. [DOI] [PubMed] [Google Scholar]
- 36.Orenstein SR. Symptoms and reflux in infants: Infant Gastroesophageal Reflux Questionnaire Revised (I-GERQ-R)—utility for symptom tracking and diagnosis. Curr Gastroenterol Rep. 2010;12(6):431–436. doi: 10.1007/s11894-010-0140-1. [DOI] [PubMed] [Google Scholar]
- 37.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the NCBI Sequence Read Archive (SRA) repository with the accession number SRP099862, https://www.ncbi.nlm.nih.gov/sra/?term=SRP099862.


