Abstract
Species and populations are disappearing at an alarming rate as a direct result of human activities. Loss of genetic diversity associated with population decline directly impacts species’ long-term survival. Therefore, preserving genetic diversity is of considerable conservation importance. However, to assist in conservation efforts, it is important to understand how genetic diversity is spatially distributed and how it changes due to anthropogenic pressures. In this study, we use historical museum and modern faecal samples of two critically endangered eastern gorilla taxa, Grauer’s (Gorilla beringei graueri) and mountain gorillas (Gorilla beringei beringei), to directly infer temporal changes in genetic diversity within the last century. Using over 100 complete mitochondrial genomes, we observe a significant decline in haplotype and nucleotide diversity in Grauer’s gorillas. By including historical samples from now extinct populations we show that this decline can be attributed to the loss of peripheral populations rather than a decrease in genetic diversity within the core range of the species. By directly quantifying genetic changes in the recent past, our study shows that human activities have severely impacted eastern gorilla genetic diversity within only four to five generations. This rapid loss calls for dedicated conservation actions, which should include preservation of the remaining peripheral populations.
Introduction
The rate at which species and populations disappear due to human activities has dramatically accelerated over the last centuries1–3, leading to what has been considered the onset of the sixth mass extinction4–6. The major contributors to species extinctions, changes in species assemblages and decrease in population size are hunting and habitat degradation due to climate change, agricultural and urban developments, and armed conflict3,7,8. However, direct quantitative assessments of the impact of human activities on natural populations remain limited, causing continued discussion of the magnitude of anthropogenic effects9.
Recently, attention has been drawn to the dramatic rate of population extinctions and population decline6. Reduction in population size frequently leads to a decrease in genetic diversity10. Populations with low genetic diversity have limited capacities to adapt to fast changing environments11, display lower fertility12, and are prone to infectious diseases13. Consequently, genetic diversity is considered one of the three forms of biodiversity that deserves global conservation attention14. However, species with long-term low genetic diversity may have developed adaptations to mitigate the negative effects of inbreeding or have experienced genetic purging, which removes strongly deleterious variants15,16. In addition, genetic diversity is strongly influenced by species’ life history and demography17,18. Hence, simply quantifying genetic diversity is insufficient to evaluate the conservation needs of a species. Therefore, it is imperative to distinguish long-term from short-term processes and to examine temporal changes in genetic diversity. Here, museum collections play an essential role, as they frequently span the critical period of the last few hundred years during which human influence on natural ecosystems has increased. By providing a window into the past, they thus help to disentangle long-term population processes from rapid, anthropogenically driven changes, offering a unique opportunity to directly quantify human impact19–21.
Here we focus on two critically endangered eastern gorilla taxa, the Grauer’s (Gorilla beringei graueri) and mountain gorillas (Gorilla beringei beringei). All four recognized gorilla taxa have experienced a continued long-term population decline over the last 100,000 years. However, this decline was particularly pronounced in the eastern species22, which is reflected in low estimates of genetic diversity in eastern compared to western gorillas23. Furthermore, the two eastern subspecies differ from each other in current population sizes24,25. Field-based studies have estimated up to 90% population decline in Grauer’s gorillas in the last two decades due to habitat loss and poaching, with reported current population size of less than 4,000 individuals26. The mountain gorilla population of the Virunga Volcanoes Massif counted only 250 individuals in the early 1980s27, but has recovered to around 480 individuals today24,28 thanks to major international conservation effort. Therefore, eastern gorillas represent an intriguing case of potentially rapid population decline over the last few generations that happened against the backdrop of slow long-term reduction in population size.
Demographic analyses of gorillas have so far only considered modern samples and therefore have limited power to resolve how genetic diversity has changed in recent times29. We aim to fill this gap by using historical museum and modern faecal samples that span the last 110 years, thus providing a direct past-to-present comparison. To quantify anthropogenic effects on genetic diversity (and hence evolutionary potential) of eastern gorillas, we focus on complete mitochondrial genomes, a powerful and widely used marker in conservation genetics that is well suited to reflect recent demographic changes30–32. Due to the low effective population size of mtDNA (1/4th of the nuclear genome), nucleotide variants rapidly become fixed through genetic drift, allowing accurate resolution of demographic processes, even when the sampling period encompasses only a few generations33–35. Changes in genetic diversity can rapidly be detected as emergence or loss of unique genetic variants for this maternally inherited, non-recombining locus, whereas longer time periods (more generations) are needed before these processes are reflected in the nuclear genome36–38.
The aims of our study are two-fold. First, we want to quantitatively assess how genetic diversity has changed over the course of only a few generations in two critically endangered gorilla subspecies that differ in their historical population sizes. Second, we aim to evaluate the relative contribution of decline in population size versus local population extinction to the genetic diversity of Grauer’s gorillas.
Methods
An extensive description of methods for sample collection, extraction, hybridisation capture of complete mitochondrial genomes and sequencing procedures is presented in39. Briefly, 69 historical Grauer’s gorilla samples (median collection year = 1950 [1910–1980]), 64 modern Grauer’s gorilla fecal samples (collected in 2014), and 22 historical mountain gorilla samples (median collection year = 1921 [1913–1956]) were included in this study (Table S1). Historical Grauer’s and mountain gorilla samples were obtained from mainly adult individuals (age class estimates were based on skull morphology, Table S1). Therefore their expected birth date predates the time when major anthropogenic factors started to affect these subspecies26,27. Fecal samples were collected in 2014 in two locations in the Democratic Republic of Congo (DRC) from two social groups: one social group from the Nkuba Research and Conservation Area (NK) in the Walikale territory, and one from the high altitude sector of the Kahuzi-Biega National Park (KBNP)40 (Table S1). Approximate individual age class was determined by dung bolus size41 and we excluded juvenile samples with dung diameter <5.5 cm from further analyses (N = 8) to reduce confounding effects of having pre-dispersed maternally related individuals that share the same mitochondrial haplotype.
Double-barcoded, double-indexed sequencing libraries were generated from 40 historical Grauer’s gorilla samples, all 22 mountain gorilla samples, and three extraction blanks following42. The libraries were subjected to 125 bp paired-end shotgun sequencing on the Illumina Hiseq2500 platform (High Output Mode). In addition, we captured complete mitochondrial genomes from all adult fecal samples (N = 56) and historical Grauer’s gorilla samples that were not shotgun-sequenced or for which we obtained insufficient mitochondrial genome coverage after shotgun sequencing (N = 59)39.
Sequence data processing
Sequences were demultiplexed, merged, mapped to the gorilla reference genome (GorGor3.1), and quality filtered as described in39. Complete mitochondrial sequences were obtained requiring at least 3x coverage for each nucleotide position39. We removed the mitochondrial D-loop before further analysis, as this hypervariable region is problematic to assemble in great apes, especially from degraded samples43,44, and frequently represents a major source of assembly error from shotgun data45. After combining data from identical individuals represented by multiple fecal samples (as determined by their multilocus microsatellite genotypes, see40), and removing genomes with more than 25% of missing sites, the newly generated dataset contained a total of 115 mitochondrial genomes: 68 historical Grauer’s individuals, 29 unique modern Grauer’s individuals, and 18 historical mountain gorilla individuals sequenced at an average depth of 120x (Table S1, Fig. S1).
Sequence reliability
Sequence data from the historical samples showed low rates of typical ancient DNA damage patterns (see39), as is expected for archived museum samples46,47. As fecal samples were sonicated prior to library preparation and sequencing, no DNA damage analysis was performed for them39. Human contamination in all samples was low (0.94% ± 0.04, 0–2.4%) and mtDNA sequences of all replicate samples (N = 48) were identical, demonstrating the reliability of the used method (see39). For all polymorphic sites in Grauer’s (N = 37) and mountain gorillas (N = 4) we manually confirmed that no stop-codons were introduced by these variants. In Grauer’s gorillas we identified one polymorphic site within a non-coding region, five polymorphic sites within rRNA and tRNA genes, with the remaining 31 variants residing within coding regions. Most of the identified polymorphisms in the coding regions were either synonymous (13 out of 31) or coded for an amino acid change commonly observed in nature (15 out of 31, based on the PAM250 matrix, score ≥0). Transition accounted for the majority of substitutions (33 out of 37). In mountain gorillas, all polymorphic sites were transition within coding regions encoding for an amino acid change commonly observed in nature (PAM250 matrix ≥0).
Mitochondrial genome analyses
We combined our data with published Grauer’s (N = 7) and mountain (N = 8) gorilla mitochondrial genomes22,44 (Table S2). Additionally, we included published mitochondrial genomes from western lowland gorillas (Gorilla gorilla gorilla, N = 43), a subspecies of the western gorillas with a large population size of ~140.000 individuals48 (Table S2). This was done to allow cross-species comparison of genetic diversity. Birth year of individuals from published samples ranged from after 1990 to 2006 (Table S2). Geographic coordinates of the sampling locations were obtained from the museum labels (historical samples), field records (fecal samples), or derived from publications (published sequences). All mitochondrial genomes were aligned with Clustal omega 1.2.449. Minimum spanning haplotype networks were constructed in PopART V1.7 (http://popart.otago.ac.nz). We grouped all mitochondrial genomes by sub-species and sample type (historical or modern sample) and calculated haplotype statistics, molecular diversity indices, and neutrality tests in Arlequin3.550. We assessed the effect of unequal sample size by using permutation tests. To this end, we randomly subsampled mitochondrial genomes from each population to match the population with the smallest sample size (modern mountain gorillas, n = 8) and repeatedly (n = 1000) recalculated haplotype and nucleotide diversity.
To accurately model eastern gorilla demography, we estimated the gorilla specific mitochondrial mutation rate using BEAST 2.4.651. We used the estimated split date of western and eastern gorillas of ~1 million years ago obtained from nuclear data as calibration point52,53. To exclude confounding effects of different sample ages, only modern mitochondrial genomes were used. We first estimated genome wide mutation rate using the HKY-site model and enforcing a strict molecular clock. Birth-rate and clock-rate priors were set as gamma distribution with α = 0.001 and β = 1000. The prior for the split time between western and eastern gorillas was set as a log-normal distribution with M = 0.5 and S = 0.1, which corresponds to a divergence time of 0.848–1.18 Mya (95% CI). The Bayesian model was run for an MCMC length of 500 million and we used Tracer 1.654 to confirm run convergence and obtain probability distributions. We then partitioned the aligned mitochondrial genomes into coding genes (splitting triplets into 1st + 2nd and 3rd position), rRNAs, tRNAs, and non-coding regions in Geneious 10.1.2 using the annotated gorilla reference. The same BEAST model was then used to estimate partition-specific mutation rates. The obtained mutation rates were highly similar to those published for the human mitochondrial genome55,56 (Fig. S2, Table S3).
We used the Bayesian skyline model in BEAST 2.4.651 to infer demographic changes within the last centuries in Grauer’s and mountain gorillas. The best partition scheme and the best fitting model for each partition were identified with PartitionFinder 257, only considering models available in the BEAST 2 software package. The skyline analysis was then run for both subspecies separately under a strict molecular clock model using the inferred gorilla mutation rates for the different partitions and MCMC length of 500 million.
To account for the possibility of more recent demographic changes that could be bypassed by the skyline model, we also employed an approximate Bayesian computation approach58. Since initial exploratory analyses failed to detect the onset of demographic change within the last 2000 years due to the lack of power over this large time period, we narrowed down the time period to the last 110 years. This time frame reflects the temporal distribution of our historical samples and encompasses the documented population size decline within the last four decades in Grauer’s and mountain gorillas26,27. We divided our dataset into two temporal groups: present (modern samples) and historical (museum specimens).
We performed two inferences: (1) a model choice analysis with three competing models in which effective population size was constant, decreased or increased in a single recent event (30–110 years ago); and (2) a parameter estimation of two effective population sizes—present and historical. We ran 30 million simulations (10 million per model) for the model choice analysis and employed 10 acceptance proportions (0.005–0.05%) for assessing consistency of the estimated model likelihoods. For the parameter estimation analysis, we ran 1 billion simulations and applied rejection with a linear regression adjustment over the 0.001% (=10,000) accepted simulations. In a dedicated analysis, we selected the following summary statistics: number of haplotypes, number of private haplotypes and genetic diversity for each of the two temporal groups as well as pairwise differences and FST between the two temporal groups.
Finally, to discriminate between negative results that were due to lack of demographic change versus those due to limited information content of the data, we employed pseudo-observed datasets (PODs) to assess the statistical power of our samples and methodology. All analyses were performed separately for Grauer’s and Mountain gorilla mitochondrial genomes in the software BaySICS v1.959 using the estimated gorilla mutation rate (1.28 × 10−8), no gamma parameter, and a Ts/Tv bias of 0.75 and 0.933 in mountain and Grauer’s gorillas, respectively (as identified by the selection model of molecular evolution carried out in MEGA 760).
Data accessibility
Sequence reads are available for download at the European Nucleotide Archive (ENA) under accession number PRJEB21370.
Mitochondrial genome sequences are available at Genbank under accession numbers MH177628 - MH177754.
Used python scripts are available upon request.
Results
Haplotype network
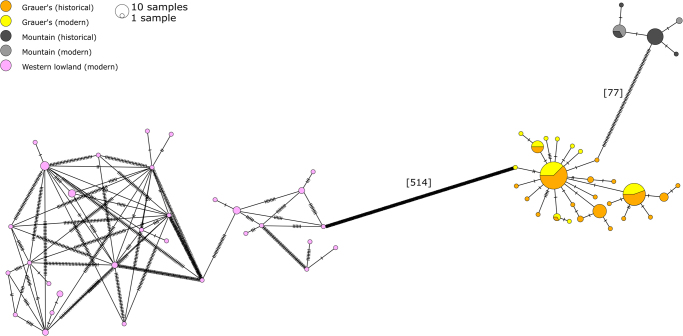
The minimum spanning haplotype network clearly differentiated the three studied gorilla subspecies into separate clusters (Fig. 1). Based on the complete mitochondrial genomes (without the D-loop), we detected 32 haplotypes in western lowland gorillas, with on average 66.4 nucleotides differences between them (Fig. 1). Combining historical and modern eastern gorilla samples, we found 27 Grauer’s and five mountain gorilla haplotypes and significantly lower average number of nucleotide difference between haplotypes than in western lowland gorillas (1.6 for Grauer’s and 0.775 for mountain gorillas, Pwestern-Grauer’s < 0.001, Pwestern-mountain < 0.001). Consequently, haplotype diversity and nucleotide diversity for both eastern gorilla species was significantly lower than in western lowland gorillas (Table S4). To evaluate the effect of using only parts of the complete mitochondrial genome, we sub-sampled the genomes to cytochrome c oxidase (subunit 1–3), cytochrome b, and 16 S rRNA, regions commonly used as markers in populations genetics and phylogenetics. To this end, the target genes were extracted from the alignment based on the annotated reference (gorGor3.1) using Geneious 10.1.261. After subsampling, most of the unique haplotypes could not be identified (Fig. S3), showing the importance of obtaining complete mitochondrial genomes when studying species with extremely low diversity.
Figure 1.
Minimum spanning haplotype network based on complete mitochondrial genomes. Haplotypes are coloured by (sub)species and sample age (historical or modern). Ticks on the haplotype edges correspond to the number of substitution. The total number of substitutions between subspecies is shown in square brackets.
Temporal changes in genetic diversity
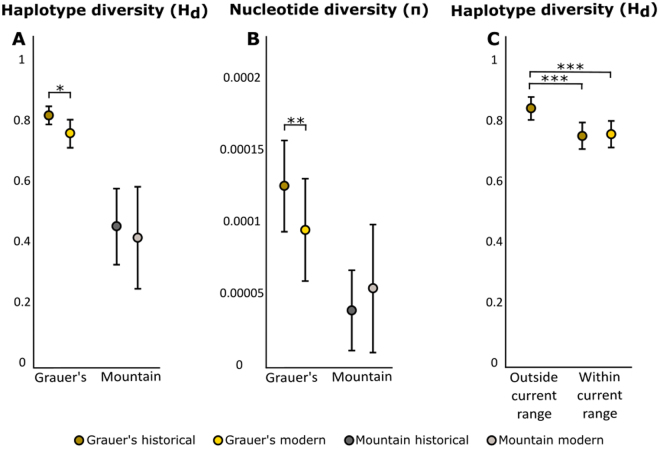
In Grauer’s gorillas, we identified 20 and 11 haplotypes among historical and modern samples, respectively, and a significant decline in haplotype and nucleotide diversity between the two age categories (PHd = 0.0169, Pπ = 0.0062, 2-sample t-test, Fig. 2A,B, Table S4), which could not be explained by differences in sample size (Fig. S4, Table S5).
Figure 2.
(A) Haplotype and (B) nucleotide diversity in both eastern gorilla subspecies. (C) Haplotype diversity in Grauer’s gorilla, grouped by geographic region and sampling period. Error bars depict 95% CI.
In mountain gorillas, we found that genetic diversity was already extremely low prior to the 1980s, with only four unique haplotypes found in historical samples that predate the reported population low. However, we found only two haplotypes in the modern samples (median birth year 1997 [1982–2012]), one that was shared with the historical samples and one unique to the modern samples. We detected no significant difference in nucleotide and haplotype diversity between modern and historical samples (Pπ = 0.60, PHd = 0.47, 2-sample T-test), a result that is independent of sample size (Fig. S4), although the haplotype diversity mean was lower in the modern data compared to the historical data (Fig. 2A, Table S4). Historical and modern mountain gorillas were genetically less diverse than historical and modern Grauer’s gorillas, respectively (Phistorial-Hd < 0.001, Pmodern-Hd < 0.001, Phistorial-π < 0.001, Pmodern-π = 0.0046, 2-sample-T-test, Fig. 2A,B, Table S4).
Geographic distribution of genetic diversity within Grauer’s gorilla
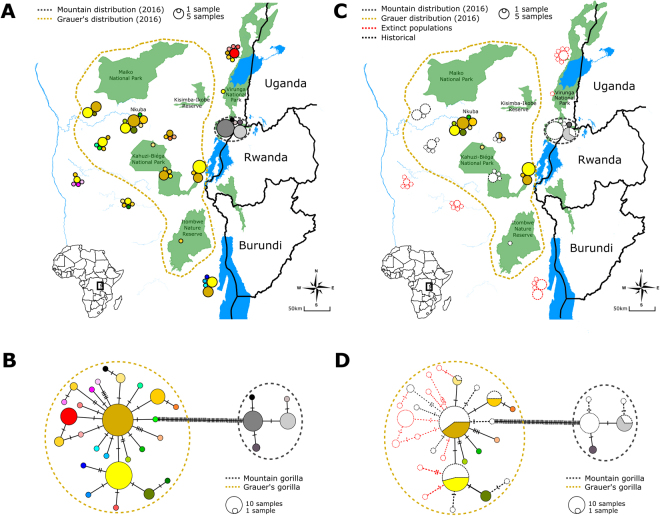
Historical Grauer’s gorilla samples were obtained throughout the current species distribution range and also included areas where this species is extinct today26,62 (Fig. 3). Modern fecal sample collection was limited to two regions: the high-altitude sector of the KBNP on the eastern border of the species range and NK, a population from the centre of the subspecies’ range (Fig. 3). The provenance of most published Grauer’s gorilla sequences is recorded as the confiscation location, frequently a trading center22,44 (Table S2). Hence, in most cases, the exact geographic origin of published sequences is uncertain.
Figure 3.
Haplotype map and haplotype network showing the geographic and genetic placement of haplotypes for both historical and modern samples. Each haplotype is marked by a unique colour. Dashed line in panels A and C designates the currently estimated distribution range of Grauer’s gorillas based on expert knowledge26. Distribution of studied mountain gorilla samples is limited to the Virunga Massif, but is presented larger on the map for clarity. (A) Geographic location of all samples and (B) the corresponding mtDNA haplotype network. (C) Geographic location and (D) the corresponding mtDNA haplotype network of modern samples (colored) and historical samples (shown as outlines). Red outlines designate historical samples from locations outside the current distribution range, where Grauer’s gorillas are extinct today. Maps were obtained from ©OpenStreetMap contributors83 and modified in Inkscape 0.92 (https://inkscape.org/)
Unique haplotypes among the historical Grauer’s gorilla samples were predominantly found in the peripheral populations (Fig. 3). As result, haplotype diversity for historical samples outside of the current distribution was significantly higher than for historical and modern samples within the range (Phist. outside vs. hist. inside < 0.001, Phist. outside vs. modern inside < 0.001, Table S6, Fig. 2C). For historical samples, this result is unlikely the consequence of differences in sample size, as these were highly similar (n = 31 outside the current range and n = 37 within the current range, Table S6). Especially the southernmost (Baraka Sibatwa forest) and the northernmost (around Lubero and Butembo) populations, both extinct today, contained many unique haplotypes (n = 8). We note that six Grauer’s gorillas, of which only one is a female that is already beyond reproductive age, still remain in a highly isolated forest fragment on Mt. Tshiaberimu in the north-eastern part of the historical distribution range63. However, since this population has no means of long-term survival, we excluded the site from the current Grauer’s distribution range. Within the current distribution range, we did not observe a significant difference in nucleotide and haplotype diversity between modern and historical samples (PHd = 0.72, Pπ = 0.76, 2-sample T-test, Fig. 2C, Table S6), despite more limited modern sampling. This pattern persisted after geographically limiting the distribution of historical samples to close proximity of modern sampling locations (Fig. S5). However, when including the nearby extinct historical populations, we again observed higher genetic diversity in historical samples (Fig. S5).
Eastern gorilla population demography
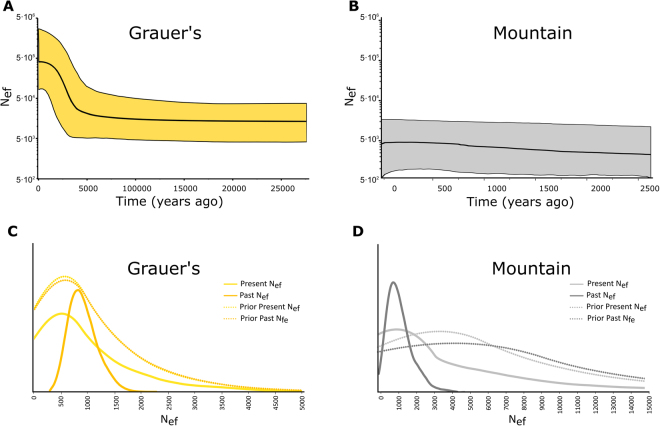
The significantly negative Tajima’s D (Table S4) and the star-like haplotype topology observed in Grauer’s gorillas (Fig. 1) indicate a relatively recent population expansion. This scenario is also supported by the Bayesian skyline plot analysis, which suggests a population expansion 5000 ± 2500 years ago (Fig. 4A). However, since the assumption of a single panmictic population that underlies this approach is likely violated, this result has to be interpreted with caution. In mountain gorillas, Bayesian skyline plot analysis suggested a relatively constant population size within the last 2500 years (Fig. 4B). However, this analysis is based on only four variable sites, the confidence intervals are wide and the observed pattern is therefore little informative.
Figure 4.
(A,B) Extended Bayesian skyline plots for both eastern gorilla subspecies. Time is presented on the x-axis (years ago, note different time scales in Grauer’s and mountain gorillas), the effective female population size, assuming a generation time of 20 years84, is shown on the y-axis (log-transformed). Black line shows the estimate of the mean and the colored areas corresponds to the 95% highest probability density interval. (C,D) Estimates of female effective population size in historical and modern Grauer’s and mountain gorillas, as inferred by approximate Bayesian computation. Probability is shown on the y-axis (unit-less). Solid lines show the inferred posterior probability distribution of present and past female effective population sizes, dotted lines depict the priors.
Skyline plots are well known for their limitation in detecting recent or short-lasting and sudden demographic changes64. Therefore, to explore changes in gorilla demography within the last several decades, we employed an approximate Bayesian computation approach. We detected population size reduction in both eastern gorilla species, but the support for this model was weak, with 62% and 69% of simulations displaying a recent population decline in Grauer’s and mountain gorillas, respectively. Best-supported simulations showed on average a 4.5x (95% CI 0.3x to 16.0x) and 2.5x (95% CI 0.2x–68x) population size reduction in Grauer’s and mountain gorillas, respectively (Fig. 4C,D). However, the magnitude of reduction has to be interpreted with caution due to the wide confidence intervals (Table S7). These results are consistent with the PODs inferences, which suggested low power for inferring the direction and magnitude of population size change with our data (Fig. S6).
Discussion
As ecosystems worldwide are facing rapid species and population loss, the importance of understanding how human activities impact natural populations has increased. However, distinguishing between recent anthropogenic and long-term demographic factors is not always trivial. Here we used a temporal approach to study recent changes in mitochondrial genetic diversity in the critically endangered eastern gorillas. By comparing temporally spaced samples, we can clearly differentiate between long-term processes that took place during the last few thousand years and short-term changes that happened in the last few decades, corresponding to only four-five gorilla generations. By including historical samples from outside of the current distribution range of Grauer’s gorillas, we were able to evaluate the importance of peripheral populations for maintenance of species-wide genetic diversity and to distinguish between signatures of population loss and general decline in species abundance.
Historical sampling uncovers changes in genetic diversity in eastern gorillas
Our demographic modelling of the eastern gorilla populations suggested that Grauer’s gorillas experienced a period of population growth 7500 to 2500 years ago (Fig. 4A). This result is concordant with previous inferences about Grauer’s gorilla evolutionary history based on morphological analyses, and coincides with the expansion of forest habitat in central Africa65,66. In mountain gorillas, the extremely low levels of genetic diversity, even after including historical samples, precluded meaningful demographic inferences over the time scale of the last several thousand years (Fig. 4B).
By juxtaposing over 100 complete eastern gorilla mitochondrial genomes from modern and historical samples, we could demonstrate pronounced loss of genetic diversity in Grauer’s gorillas within the last century (Fig. 2). The use of mitochondrial markers to describe population-level processes is justified in gorillas, a species that is characterized by female dispersal67,68. Combined with our relatively large sample size, we therefore likely captured a reasonable representation of the Grauer’s gorilla population as a whole, even with relatively limited modern sampling.
We detected only four different haplotypes in the historical mountain gorilla population, suggesting that mitochondrial diversity was already extremely low historically. The number of unique haplotypes was limited to only two in the modern samples. Such low levels of diversity in the datasets make it impossible to derive any statistically meaningful conclusions about temporal changes in haplotype and nucleotide diversity. However, ABC modelling supports the scenario of a recent population size decrease in both eastern gorilla subspecies (Fig. 4C,D). The best supported models suggested a 25–75% reduction in effective female population size in both eastern gorilla subspecies, albeit with wide confidence intervals and low statistical power (Fig. 4, Fig. S6). This observation is also concordant with signs of very recent inbreeding in both eastern gorillas22. It is furthermore corroborated by known historical events that impacted Grauer’s and mountain gorillas during the last few decades, including human encroachment, civil war, and on-going political unrest in the DRC, which are associated with high rates of bushmeat hunting and deforestation8,27,69,70. For instance, the only closely monitored Grauer’s gorilla population of KBNP suffered an estimated 50% loss in 199971. Bushmeat hunting linked to artisanal mining and the activity of numerous armed militia remains a major threat to wildlife throughout Grauer’s gorilla range26. As a result, Grauer’s gorilla populations continue to decline dramatically and total population loss has been estimated to be up to 90% in the last two decades26. In contrast, intense conservation efforts have allowed the Virunga Massif mountain gorilla population to almost double in size since the early 1980s24.
Present-day mitochondrial diversity in the eastern gorillas is currently the lowest of all great apes: It is over 20 times lower than in Homo sapiens72, almost 50 times lower than in eastern chimpanzee (Pan troglodytes schweinfurthii) and the bonobo (Pan paniscus)45, and around 100 times lower than in Sumatran orangutan (Pongo abelli)73,74. We also detected pronounced differences in mitochondrial diversity among gorilla species: western lowland gorillas harbor many highly diverse haplotypes, whereas only a few, closely related, haplotypes are found in the eastern species, particularly in mountain gorillas. These estimates are consistent with previous studies based on the mitochondrial hypervariable region75–77, nuclear data22,25,44, and match both current and historical census population size estimates22,48.
Our temporal approach and the use of complete mitochondrial genomes constitute a powerful strategy for detecting recent population size changes in threatened and endangered species. The extremely low levels of genetic diversity found in eastern gorillas would make sequencing of only a small part of the mitochondrial genome prohibitively uninformative (Fig. S3).
Extirpation of peripheral populations is the main cause for the loss of genetic diversity
Grauer’s gorillas have not only experienced reduction in population size and increased fragmentation throughout their range26,70, but also a general range contraction, accompanied by local population extirpation62. Because our historical sampling contains populations from both within and outside of the current species’ distribution range, it allowed us to effectively evaluate the effect of population size reduction compared to population loss. We find that genetic diversity within the Grauer’s gorilla core distribution range remained relatively stable over the study period (Fig. 2C), despite reported decline in encounter rates26. However, genetic diversity in the species overall declined significantly as the result of extirpation of populations that are outside the current species range. This result holds also after accounting for differences in geographic distribution between historical and modern samples (Fig. S5). We found a particularly large number of unique mitochondrial haplotypes in these peripheral historical populations and detect multiple unique haplotypes even within relatively small geographic regions (Fig. 3). The observed pattern is possibly a result of long-term historical isolation of peripheral populations, which, by limiting (female) dispersal, may have allowed multiple unique variants to drift to high frequency. This interpretation is in line with early surveys that reported pronounced habitat fragmentation within the Grauer’s gorilla range already in the 1950s78.
Conclusions
The significant loss of mitochondrial diversity in eastern gorillas over a short time period is alarming, as it likely also holds for autosomal variation (albeit to a lesser extent due to differences in effective population size), which is directly linked to the species’ evolutionary potential and hence long-term survival. Populations with low levels of standing genetic variation struggle to adapt in fast changing environments79,80, and it is this rapid, human-induced change that many species, including gorillas, are facing today. The recent declines in effective population size suggested by the ABC analyses may also have consequences for genetic load in eastern gorillas. Whereas genetic purging under slow inbreeding may remove deleterious variants, rapid loss of genetic diversity has been shown to have detrimental effects15.
Our findings stand in contrast to the common notion that small peripheral populations contain few unique genetic variants81,82 and instead identifies peripheral gorilla populations as important keepers of genetic diversity, highlighting their conservation importance. This observation is likely to apply to fragmented populations of many endangered species. Therefore, conservation actions aimed at preserving peripheral populations, improving habitat connectivity and hence facilitating gene flow, could have a disproportionally positive effect on maintaining overall species diversity.
Electronic supplementary material
Acknowledgements
We would like to thank the Dian Fossey Gorilla Fund International and Institut Congolais pour la Conservation de la Nature for permission and logistical support of research in the DRC. We are grateful to the Fossey Fund trackers and the ICCN park rangers who helped us collect samples in Nkuba and at Kahuzi-Biega NP. To Isabel Ordaz Németh and Zoltan T. Nagy for their support during the study of natural history collections; Nadin Rohland for help with laboratory protocols; Christian Roos from the Gene Bank of Primates for providing western lowland gorilla DNA; Pauline Baas for providing genotyping data for fecal samples. Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala and Stockholm. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The authors further acknowledge support from the Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. This research was supported by the following grants: Human Frontier Science Program Postdoctoral Fellowship to KG (LT000800/2011-L), the Jan Löfqvist and the Nilsson-Ehle Endowments of the Royal Physiographic Society of Lund to KG, FORMAS (project 2015–676) to LD, the Turner Foundation, the Daniel K. Thorne Foundation and the Margot Marsh Biodiversity Foundation to the Dian Fossey Gorilla Fund.
Author Contributions
T.v.d.V. and K.G. designed the study. T.v.d.V. and K.G. performed laboratory experiments. T.v.d.V. wrote scripts and performed data analyses. E.S. performed the ABC-modelling. L.D. and D.C. provided essential technical and logistical support and critical feedback throughout the study. D.C., U.N., E.B., R.N., E.G., G.S., P.S., D.C.K. provided samples and data. T.v.d.V. and K.G. wrote the article, with input from all co-authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24497-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tom van der Valk, Email: tom.vandervalk@ebc.uu.se.
Katerina Guschanski, Email: katerina.guschanski@ebc.uu.se.
References
- 1.Pimm SL, et al. The biodiversity of species and their rates of extinction, distribution, and protection. Science. 2014;344:1246752–1246752. doi: 10.1126/science.1246752. [DOI] [PubMed] [Google Scholar]
- 2.Lewis SL, Maslin MA. Defining the Anthropocene. Nature. 2015;519:171–180. doi: 10.1038/nature14258. [DOI] [PubMed] [Google Scholar]
- 3.Waters CN, et al. The Anthropocene is functionally and stratigraphically distinct from the Holocene. Science. 2016;351:aad2622. doi: 10.1126/science.aad2622. [DOI] [PubMed] [Google Scholar]
- 4.Barnosky AD, et al. Has the Earth’s sixth mass extinction already arrived? Nature. 2011;471:51–57. doi: 10.1038/nature09678. [DOI] [PubMed] [Google Scholar]
- 5.Ceballos G, et al. Accelerated modern human-induced species losses: Entering the sixth mass extinction. Sci. Adv. 2015;1:e1400253–e1400253. doi: 10.1126/sciadv.1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceballos G, Ehrlich PR, Dirzo R. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc. Natl. Acad. Sci. 2017;114:201704949. doi: 10.1073/pnas.1704949114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dirzo R, Raven PH. Global state of biodiversiy and loss. Annu. Rev. Environ. Resour. 2003;28:137–67. doi: 10.1146/annurev.energy.28.050302.105532. [DOI] [Google Scholar]
- 8.Daskin JH, Pringle RM. Warfare and wildlife declines in Africa’s protected areas. Nature. 2018;553:328–332. doi: 10.1038/nature25194. [DOI] [PubMed] [Google Scholar]
- 9.Dirzo R, et al. Defaunation in the Anthropocene. Science. 2014;345:401–406. doi: 10.1126/science.1251817. [DOI] [PubMed] [Google Scholar]
- 10.Frankham R. Relationship of genetic variation to population size in wildlife. Conserv Biol. 1996;10:1500–1508. doi: 10.1046/j.1523-1739.1996.10061500.x. [DOI] [Google Scholar]
- 11.Lande R, Shannon S. The Role of Genetic Variation in Adaptation and Population Persistence in a Changing Environment. Evolution (N. Y). 1996;50:434. doi: 10.1111/j.1558-5646.1996.tb04504.x. [DOI] [PubMed] [Google Scholar]
- 12.Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003;17:230–237. doi: 10.1046/j.1523-1739.2003.01236.x. [DOI] [Google Scholar]
- 13.Smith KF, Sax DF, Lafferty KD. Evidence for the role of infectious disease in species extinction and endangerment. Conservation Biology. 2006;20:1349–1357. doi: 10.1111/j.1523-1739.2006.00524.x. [DOI] [PubMed] [Google Scholar]
- 14.McNeely, A. J., Miller, K. R., Reid, A. R. & Werner, T. B. Conserving the world’s biological diversity. Conservation union (World Conservation Union, World Resources Institute, Conservation International, World Wildlife Fund–US, and the World Bank, Washington, D.C., 1990).
- 15.Hedrick PW, Garcia-Dorado A. Understanding Inbreeding Depression, Purging, and Genetic Rescue. Trends in Ecology and Evolution. 2016;31:940–952. doi: 10.1016/j.tree.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Caballero A, Bravo I, Wang J. Inbreeding load and purging: implications for the short-term survival and the conservation management of small populations. Heredity (Edinb). 2017;118:177–185. doi: 10.1038/hdy.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lande R. Genetics and demography in biological conservation. Science. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- 18.Romiguier J, et al. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature. 2014;515:261–263. doi: 10.1038/nature13685. [DOI] [PubMed] [Google Scholar]
- 19.Wandeler P, Hoeck PEA, Keller LF. Back to the future: museum specimens in population genetics. Trends in Ecology and Evolution. 2007;22:634–642. doi: 10.1016/j.tree.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Holmes MW, et al. Natural history collections as windows on evolutionary processes. Molecular Ecology. 2016;25:864–881. doi: 10.1111/mec.13529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Díez-del-Molino, D., Sánchez-Barreiro, F., Barnes, I., Gilbert, M. T. P. & Dalén, L. Quantifying Temporal Genomic Erosion in Endangered Species. Trends in Ecology and Evolution (2017). [DOI] [PubMed]
- 22.Xue Y, et al. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science. 2015;348:242–5. doi: 10.1126/science.aaa3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhlwilm M, et al. Evolution and demography of the great apes. Curr. Opin. Genet. Dev. 2016;41:124–129. doi: 10.1016/j.gde.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Gray M, et al. Genetic census reveals increased but uneven growth of a critically endangered mountain gorilla population. Biol. Conserv. 2013;158:230–238. doi: 10.1016/j.biocon.2012.09.018. [DOI] [Google Scholar]
- 25.Roy J, et al. Recent divergences and size decreases of eastern gorilla populations. Biol. Lett. 2014;10:20140811–20140811. doi: 10.1098/rsbl.2014.0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plumptre AJ, et al. Catastrophic Decline of World’s Largest Primate: 80% Loss of Grauer’s Gorilla (Gorilla beringei graueri) Population Justifies Critically Endangered Status. PLoS One. 2016;11:e0162697. doi: 10.1371/journal.pone.0162697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalpers J, et al. Gorillas in the crossfire: population dynamics of the Virunga mountain gorillas over the past three decades. Oryx. 2003;37:326–337. doi: 10.1017/S0030605303000589. [DOI] [Google Scholar]
- 28.Gray M, et al. Censusing the mountain gorillas in the Virunga Volcanoes: Complete sweep method versus monitoring. Afr. J. Ecol. 2010;48:588–599. [Google Scholar]
- 29.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofman CA, et al. Mitochondrial Genomes Suggest Rapid Evolution of Dwarf California Channel Islands Foxes (Urocyon littoralis) PLoS ONE. 2015;10:e0118240. doi: 10.1371/journal.pone.0118240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gariboldi MC, et al. Patterns of population structure at microsatellite and mitochondrial DNA markers in the franciscana dolphin (Pontoporia blainvillei) Ecol. Evol. 2016;6:8764–8776. doi: 10.1002/ece3.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cassidy LM, et al. Capturing goats: documenting two hundred years of mitochondrial DNA diversity among goat populations from Britain and Ireland. Biol. Lett. 2017;13:20160876. doi: 10.1098/rsbl.2016.0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nyström V, Angerbjörn A, Dalén L. Genetic consequences of a demographic bottleneck in the Scandinavian arctic fox. Oikos. 2006;114:84–94. doi: 10.1111/j.2006.0030-1299.14701.x. [DOI] [Google Scholar]
- 34.Xenikoudakis G, et al. Consequences of a demographic bottleneck on genetic structure and variation in the Scandinavian brown bear. Mol. Ecol. 2015;24:3441–3454. doi: 10.1111/mec.13239. [DOI] [PubMed] [Google Scholar]
- 35.Tobler R, et al. Aboriginal mitogenomes reveal 50,000 years of regionalism in Australia. Nature. 2017;544:180–184. doi: 10.1038/nature21416. [DOI] [PubMed] [Google Scholar]
- 36.Moritz C, Dowling T. Evolution of animal mitochondrialDNA: relevance for population biology and systematics. Rev. Ecol. Syst. 1987;92:269–292. doi: 10.1146/annurev.es.18.110187.001413. [DOI] [Google Scholar]
- 37.Harrison RG. Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends in Ecology and Evolution. 1989;4:6–11. doi: 10.1016/0169-5347(89)90006-2. [DOI] [PubMed] [Google Scholar]
- 38.Desalle, R., Schierwater, B. & Hadrys, H. MtDNA: The small workhorse of evolutionary studies. Front. Biosci. (LandmarkEd22, 873–887 (2017). [DOI] [PubMed]
- 39.van der Valk T, Lona Durazo F, Dalén L, Guschanski K. Whole mitochondrial genome capture from faecal samples and museum-preserved specimens. Mol. Ecol. Resour. 2017;17:e111–e121. doi: 10.1111/1755-0998.12699. [DOI] [PubMed] [Google Scholar]
- 40.Baas, P. et al. Population-level assessment of genetic diversity and habitat fragmentation in Critically-Endangered Grauer’s gorillas. Am. J. Phys. Anthropol. 165, 565–575 (2018). [DOI] [PubMed]
- 41.McNeilage A, et al. Census of the mountain gorilla Gorilla beringei beringei population in Bwindi Impenetrable National Park, Uganda. Oryx. 2006;40:419–427. doi: 10.1017/S0030605306001311. [DOI] [Google Scholar]
- 42.Rohland N, Harney E, Mallick S, Nordenfelt S, Reich D. Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015;370:20130624. doi: 10.1098/rstb.2013.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guschanski K, et al. Next-generation museomics disentangles one of the largest primate radiations. Syst. Biol. 2013;62:539–554. doi: 10.1093/sysbio/syt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lobon I, et al. Demographic History of the Genus Pan Inferred from Whole Mitochondrial Genome Reconstructions. Genome Biol. Evol. 2016;8:2020–30. doi: 10.1093/gbe/evw124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dabney J, Meyer M, Pääbo S. Ancient DNA damage. Cold Spring Harb. Perspect. Biol. 2013;5:a012567. doi: 10.1101/cshperspect.a012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiß, C. L. et al. Temporal patterns of damage and decay kinetics of DNA retrieved from plant herbarium specimens. Open Sci. 3, (2016). [DOI] [PMC free article] [PubMed]
- 48.Williamson, E. A., Maisels, F. G. & Groves, C. P. In Handbook of the Mammals of the World Volume 3: Primates (eds Mittermeier, R. A., Rylands, A. B. & Wilson, D. E.) 792–854 (Lynx Edicions, 2013).
- 49.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 51.Bouckaert R, et al. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thalmann O, Fischer A, Lankester F, Paabo S, Vigilant L. The Complex Evolutionary History of Gorillas: Insights from Genomic Data. Mol. Biol. Evol. 2006;24:146–158. doi: 10.1093/molbev/msl160. [DOI] [PubMed] [Google Scholar]
- 53.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rambaut, A., Suchard, M., Xie, D. & Drummond, A. Tracer v1.6. (2014).
- 55.Soares P, et al. Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock. Am. J. Hum. Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rieux A, et al. Improved calibration of the human mitochondrial clock using ancient genomes. Mol. Biol. Evol. 2014;31:2780–2792. doi: 10.1093/molbev/msu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. Partitionfinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 58.Beaumont M. Approximate Bayesian computation in evolution and ecology. Annu. Rev. Ecol. Evol. Syst. 2010;41:379–406. doi: 10.1146/annurev-ecolsys-102209-144621. [DOI] [Google Scholar]
- 59.Sandoval-Castellanos E, Palkopoulou E, Dalén L. Back to BaySICS: A user-friendly program for Bayesian statistical inference from coalescent simulations. PLoS One. 2014;9:e98011. doi: 10.1371/journal.pone.0098011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maldonado, O. et al. Grauer’s Gorillas and Chimpanzees in Eastern Democratic Republic of Congo (Kahuzi-Biega, Maiko, Tayna and Itombwe Landscape): Conservation Action Plan 2012–2022. doi:ISBN: 978-2-8317-1535-3 (2012).
- 63.Sikubwabo CK. Can the gorillas of Mt Tshiaberimu survive? Gorilla J. 2015;3:3–5. [Google Scholar]
- 64.Shapiro B, et al. A bayesian phylogenetic method to estimate unknown sequence ages. Mol. Biol. Evol. 2011;28:879–887. doi: 10.1093/molbev/msq262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barks SK, et al. Variable temporoinsular cortex neuroanatomy in primates suggests a bottleneck effect in eastern gorillas. J. Comp. Neurol. 2014;522:844–860. doi: 10.1002/cne.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tocheri MW, et al. The evolutionary origin and population history of the grauer gorilla. Am. J. Phys. Anthropol. 2016;159:S4–S18. doi: 10.1002/ajpa.22900. [DOI] [PubMed] [Google Scholar]
- 67.Robbins AM, Stoinski T, Fawcett K, Robbins MM. Leave or conceive: natal dispersal and philopatry of female mountain gorillas in the Virunga volcano region. Anim. Behav. 2009;77:831–838. doi: 10.1016/j.anbehav.2008.12.005. [DOI] [Google Scholar]
- 68.Yamagiwa, J. & Kahekwa, J. In Mountain Gorillas: Three Decades of Research at Karisoke 89–122 (Cambridge University Press). 10.1017/CBO9780511661631.005 (2001).
- 69.Hart T, Hart JA. Conservation and Civil Strife: Two Perspectives from Central Africa Zaire. Conserv. Biol. 1997;11:308–314. doi: 10.1046/j.1523-1739.1997.011002308.x. [DOI] [Google Scholar]
- 70.Molinario G, et al. Quantification of land cover and land use within the rural complex of the Democratic Republic of Congo. Environ. Res. Lett. 2017;12:104001. doi: 10.1088/1748-9326/aa8680. [DOI] [Google Scholar]
- 71.Yamagiwa J. Slaughter of gorillas in the Kahuzi-Biega Park. Gorilla. Journal. 1999;19:4–6. [Google Scholar]
- 72.Carter RW. Mitochondrial diversity within modern human populations. Nucleic Acids Res. 2007;35:3039–3045. doi: 10.1093/nar/gkm207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Banes GL, Galdikas BMF. Effective characterisation of the complete orang-utan mitochondrial DNA control region, in the face of persistent focus in many taxa on shorter hypervariable regions. PLoS One. 2016;11:e0168715. doi: 10.1371/journal.pone.0168715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garner KJ, Ryder OA. Mitochondrial DNA Diversity in Gorillas. Mol. Phylogenet. Evol. 1996;6:39–48. doi: 10.1006/mpev.1996.0056. [DOI] [PubMed] [Google Scholar]
- 76.Jensen-Seaman MI, Kidd KK. Mitochondrial DNA variation and biogeography of eastern gorillas. Mol. Ecol. 2001;10:2241–2247. doi: 10.1046/j.0962-1083.2001.01365.x. [DOI] [PubMed] [Google Scholar]
- 77.Clifford SL, et al. Mitochondrial DNA phylogeography of western lowland gorillas (Gorilla gorilla gorilla) Mol. Ecol. 2004;13:1551–1565. doi: 10.1111/j.1365-294X.2004.02140.x. [DOI] [PubMed] [Google Scholar]
- 78.Emlen, J. & Schaller, G. B. Distribution and status of the mountain gorilla (Gorilla gorilla beringei). Zoologica 41–52 (1959).
- 79.O’Brien SJ. A role for molecular genetics in biological conservation. Proc. Natl. Acad. Sci. USA. 1994;91:5748–5755. doi: 10.1073/pnas.91.13.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Visser ME. Keeping up with a warming world; assessing the rate of adaptation to climate change Keeping up with a warming world; assessing the rate of adaptation to climate change. Proc. R. Soc. London B Biol. Sci. 2008;275:649–659. doi: 10.1098/rspb.2007.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lawton JH. Range, population abundance and conservation. Trends Ecol. Evol. 1993;8:409–413. doi: 10.1016/0169-5347(93)90043-O. [DOI] [PubMed] [Google Scholar]
- 82.Vucetich JA, Waite TA. Spatial patterns of demography and genetic processes across the species’ range: Null hypotheses for landscape conservation genetics. Conserv. Genet. 2003;4:639–645. doi: 10.1023/A:1025671831349. [DOI] [Google Scholar]
- 83.www.opendatacommons.org/licenses/odbl.
- 84.Langergraber KE, et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl. Acad. Sci. USA. 2012;109:15716–21. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence reads are available for download at the European Nucleotide Archive (ENA) under accession number PRJEB21370.
Mitochondrial genome sequences are available at Genbank under accession numbers MH177628 - MH177754.
Used python scripts are available upon request.