Our understanding of new genes is often built upon the knowledge of well-characterized genes. One avenue toward revealing such connections involves creation of strains with mutations in two or more defined genes to permit genetic interaction analysis. Strain manipulations can yield unexpected mutations at loci outside the defined targeted genes. In this report, we describe a method for rapid validation of multigene mutants, thus allowing an appraisal of the contribution of the defined targeted genes to the strain’s phenotype.
KEYWORDS: CRISPR, Candida albicans, genetics
ABSTRACT
The biological function of a gene is often probed through its interactions with other genes. This general approach has been especially useful to build knowledge about poorly understood genes upon the bedrock of well-characterized genes. Genetic interaction analysis requires the construction of strains with mutations in two or more genes. Single-gene mutants of microbial pathogens are generally validated through introduction of a wild-type copy of the affected gene to create a complemented or reconstituted strain, followed by testing for restoration of a wild-type phenotype. This practice, formalized as one of Falkow’s “molecular Koch’s postulates” ensures that the phenotype of the mutant depends upon the known mutation. However, multigene mutants are seldom validated because of the labor required to assemble multiple genomic segments into a vector that can be introduced into the mutant strain. We present here an approach, concatemer assembly for rescue of mutant abilities (CARMA), that circumvents this impediment through an in vivo recombinational assembly strategy that does not require cloning at all. Our results show that CARMA allows genetic rescue of two double-gene mutant strains of the fungal pathogen Candida albicans.
IMPORTANCE Our understanding of new genes is often built upon the knowledge of well-characterized genes. One avenue toward revealing such connections involves creation of strains with mutations in two or more defined genes to permit genetic interaction analysis. Strain manipulations can yield unexpected mutations at loci outside the defined targeted genes. In this report, we describe a method for rapid validation of multigene mutants, thus allowing an appraisal of the contribution of the defined targeted genes to the strain’s phenotype.
INTRODUCTION
For many organisms, our understanding of gene function is based upon analysis of engineered mutations at predetermined genomic sites. The manipulations that yield engineered mutations can also yield adventitious mutations elsewhere in the genome. For this reason, when characterizing a defined mutant strain, there is a concern that its phenotype may be due to adventitious mutations, rather than the engineered mutation. For microbial pathogens, mutant strain characterization can be extensive, involving, for example, animal infection models. Moreover, strain phenotypes may appear to be complex because many mechanistic underpinnings of virulence have yet to be discovered. For these organisms, the possible contributions of adventitious mutations to the phenotype are often assessed through analysis of a complemented or reconstituted strain, in which a wild-type copy of the gene that had been altered by engineering is introduced at a distant genomic site (complementation) or at the native locus (reconstitution). The importance of including complemented or reconstituted strains in pathogen phenotypic characterization was emphasized as the third postulate among the cornerstone “molecular Koch’s postulates” coined by Falkow (1).
Once there is a panel of well-characterized genes in an organism, it is useful to build upon that knowledge in the discovery or characterization of new genes. The approach of genetic interaction analysis, also called epistasis analysis, is a widely used strategy that can connect the functions of two genes. In premier genetic organisms like Saccharomyces cerevisiae, systematic gene interaction analysis has been applied on a large scale to reveal new gene functions and novel connections among seemingly distinct biological processes (2). Our organism of interest, Candida albicans, is not as amenable to genetic manipulation as S. cerevisiae, but nonetheless, it has a growing panel of characterized genes that provide the foundation for genetic interaction analysis. In fact, the use of double and multigene mutants in C. albicans to draw functional inferences has a long history (3–11). Studies of C. albicans double and multigene mutants have provided key insights into functional redundancy, pathway relationships, and major effectors that mediate pathway outputs. The analysis of double and multigene mutants has been of exceptional value for the understanding of C. albicans, as it has for diverse other organisms.
It is likely that analysis of C. albicans multigene mutants will have an increasingly prominent role in elucidation of the basis for key traits such as pathogenicity and drug resistance. This expectation is based upon the value of the information from these strains as well as the increasing ease with which multigene mutants can be created. Strain construction in C. albicans has been accelerated through implementation of clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9-based methods (12), and recently, a gene drive system designed specifically for creation of double mutant strains (10). Unfortunately, the development of multigene complementation or reconstitution strategies, which we refer to together as genetic rescue strategies, has not kept pace with these developments. Strain validation through genetic rescue generally requires vector-based cloning, an often tedious procedure when multiple genes must be introduced together.
Here, we present a genetic rescue strategy that relies on recombinational assembly in C. albicans of PCR products. Integration of the concatenated assembly into a mutant strain is augmented by CRISPR-Cas9 cleavage at the targeted locus. Then, the mutant strain that carries the concatenated assembly is tested to see whether a wild-type phenotype is restored. We report proof-of-principle studies for phenotypic rescue of auxotrophic markers and for mutants defective for biofilm formation and filamentation. This approach, which we call concatemer assembly for rescue of mutant abilities (CARMA), has the potential to accelerate multigene mutant validation in C. albicans and other organisms.
RESULTS
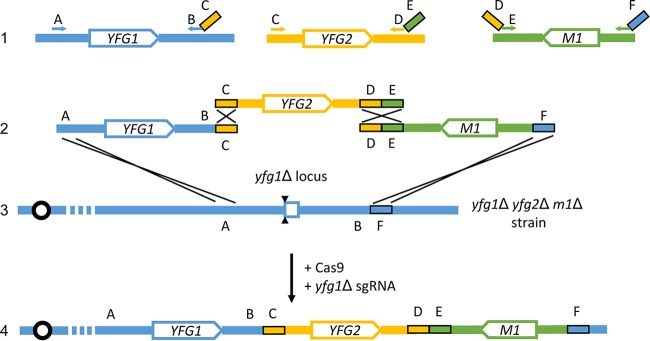
We developed a multigene rescue strategy that employs three DNA segments (Fig. 1). In this illustration, we seek to introduce functional YFG1 and YFG2 genes to assay phenotypic rescue of a yfg1Δ/Δ yfg2Δ/Δ double mutant. We include the marker M1 in order to select for transformation and integration of a concatemer of the three segments. Each segment is synthesized through a PCR templated by the genome or an existing plasmid, thus eliminating any need for custom cloning. The segments have homology at their ends to one another or to the genome (regions A to F in Fig. 1), so that cellular homologous recombination machinery can assemble the segments in a predictable order. We refer to an organization of homology that directs concatenation of the DNA segments as “concatenating homology.”
FIG 1 .
Strategy for concatemer assembly for rescue of mutant abilities (CARMA). Given a double mutant strain with the genotype yfg1Δ/Δ yfg2Δ/Δ with an available selectable marker M1, both YFG1 and YFG2 may conceptually be reintroduced at either the yfg1Δ (depicted in line 2 and line 3) or yfg2Δ locus. Cassettes containing YFG1 and YFG2 are generated by PCR from wild-type genomic DNA (line 1). The YFG1 cassette with sequences shown in blue is amplified with the primers indicated at the A and BC regions. It contains a segment of the YFG1 promoter, the YFG1 open reading frame (ORF), and terminator. Sequence C, containing homology to the YFG2 cassette, is introduced into the YFG1 cassette using the long primer indicated at the BC region. The YFG2 cassette with sequences shown in gold is amplified with the primers indicated at the C and DE regions. It contains a segment of the YFG2 promoter, the YFG2 ORF, and terminator. Sequence E, containing homology to the M1 marker cassette, is introduced into the YFG2 cassette using the long primer indicated at the DE region. The M1 marker cassette with sequences shown in green is amplified from a plasmid containing the M1 marker with the primers indicated at the DE and F regions. Sequence D, containing homology to the terminator region of YFG2, is introduced into the M1 cassette using the long primer indicated at the DE region. Sequence F, containing homology to a region downstream of YFG1 is introduced into the M1 cassette using the long primer indicated at the F region. All three cassettes are transformed into the yfg1Δ/Δ yfg2Δ/Δ m1Δ/Δ strain under M1 selection, along with DNA cassettes expressing CAS9 and a single guide RNA targeting the yfg1Δ locus. The double-strand break introduced by Cas9 complexed with the yfg1Δ targeting sgRNA is indicated by two black triangles. Expected homologous recombination events are depicted as single crosses, and together, they should yield a concatenated locus containing YFG1, YFG2, and M1 (line 4). Boxes C, D, E, and F each indicate segments of 80 bp. Where possible, homology between PCR cassettes is maximized. Sequences D and E together provide 160 bp of homology between the YFG2 and M1 cassettes. (In pilot experiments, we could not generate integrated concatemers with only 80 bp of homology between the YFG2 and M1 cassettes.) Sequences C and F provide 80 bp of homology to the YFG1 cassette and yfg1Δ locus, respectively. For construction of the ARG4:LEU2:HIS1 allele, the A-YFG1 interval provided 419 bp of homology. For construction of the UME6:BRG1:HIS1 allele, the A-YFG1 interval provided 403 bp of homology. In our experiments, we avoided extending the homology between the YFG1 and YFG2 cassettes to 160 bp in length by adding 80 bp of homology at sequence B. We were concerned that the presence of sequence B in the YFG2 cassette could allow integration of only YFG2 and M1 downstream of the yfg1Δ deletion scar in the B-F interval.
The first (leftmost) segment includes one of the coding regions, YFG1, that corresponds to one of the deletions in the yfg1Δ/Δ yfg2Δ/Δ double mutant (Fig. 1). It also includes YFG1 5′ and 3′ flanking regions. The overall assembly will be directed to integrate at the YFG1 genomic locus to replace a yfg1Δ coding region deletion allele. Therefore, the YFG1 5′ flanking region does not need to include all sequences necessary for proper expression; integration by recombination at the A region will join the YFG1 DNA to the complete genomic 5′ flanking region that extends to the left in the diagram. The segment needs to include sufficient 3′ flanking sequences for YFG1 expression, extending to the B region. This requirement arises because the 3′ flanking end of the YFG1 segment will be fused to novel sequences from the YFG2 locus, the C region, in the integrated assembly.
The second (middle) segment includes the second coding region, YFG2, that corresponds to a deletion in the yfg1Δ/Δ yfg2Δ/Δ double mutant (Fig. 1). This segment includes YFG2 5′ and 3′ flanking regions that extend to regions labeled C and D. These flanking regions must include all sequences necessary for proper expression because both regions are joined to novel neighboring sequences, the B and E regions, specified by the PCR primers.
The third (rightmost) segment includes a marker for selection, M1, along with all necessary 5′ and 3′ flanking sequences for expression (Fig. 1). It has homology to the YFG2 segment (D-E region) at one end and to the genome neighboring the yfg1Δ allele (F region) at the other end. The 5′-to-3′ orientation of each gene (M1, YFG1, and YFG2) is arbitrary; the diagram illustrates the actual relative orientations of the genes in the experiments we present below.
Homologous recombination should not allow integration of M1 on its own. However, the YFG1 and YFG2 segments may serve as adaptors to enable homologous recombination events to insert a YFG1:YFG2:M1 concatenated assembly in place of the yfg1Δ allele (Fig. 1). Genomic integration of the concatemer may be stimulated by double-strand cleavage of the yfg1Δ allele directed by the CRISPR-Cas9 system (Fig. 1).
Genetic rescue of arg4, leu2, and his1 mutations in strain SN152.
We chose the popular laboratory strain SN152, which has arg4Δ/Δ, leu2Δ/Δ, and his1Δ/Δ mutations, for an initial test of the multigene rescue strategy. The strategy is expected to yield Arg+ Leu+ His+ transformants through recombinational concatemerization and integration of ARG4, LEU2, and HIS1 cassettes. We designed three cassettes with concatenating homology as depicted in Fig. 1, using the C. albicans ARG4 gene in the position of YFG1, the Candida albicans LEU2 gene in the position of YFG2, and the Candida dubliniensis HIS1 gene in the position of M1. Integration of the cassettes was targeted by homology to the arg4Δ::dpl200 mutant locus in the SN152 genome, and transformants were selected through acquisition of only a His+ phenotype. If the other two genes were assembled into an integrating concatemer, then His+ transformants should be Arg+ and Leu+ as well. Transformation of approximately 3 µg of each cassette yielded seven His+ transformants from three separate transformations (Table 1). One transformant could not be propagated on media selective for His+ strains and was not studied further. Five of the remaining six transformants grew when replica plated to minimal SD medium and thus were Arg+ Leu+ His+. This observation indicates that the ARG4, LEU2, and HIS1 genes were all maintained. Thus, genetic rescue with three cassettes is possible, though the transformant recovery frequency is low (13, 14).
TABLE 1 .
SN152 His+ Leu+ Arg+ transformation outcomes
| Expt or parameter | No. of His+ colonies with no sgRNA |
No. of His+ colonies with sgRNA |
|---|---|---|
| 1 | 4 | 831 |
| 2 | 3 | 716 |
| 3 | 0 | 402 |
| Avg ± SD | 2.33 ± 2.08 | 650 ± 222 |
| % prototrophic | 83 | 94 ± 2.7 |
The CRISPR-Cas9 system has been used to increase the rate of DNA integration through generation of a genomic double-strand break that stimulates homology-directed repair. To test the effect of CRISPR-Cas9 on concatemer integration, we included DNA cassettes specifying Cas9 and a single guide RNA (sgRNA) that targets the arg4Δ::dpl200 locus in our transformation mixture. These components increased the recovery of His+ transformants by roughly 300-fold (Table 1). Approximately 94% of His+ transformants grew when replica plated to SD medium, thus indicating once again that the ARG4, LEU2, and HIS1 genes were all maintained. This result indicates that multiple cassettes with concatenating homology may successfully integrate with selection for only a single cassette and that integration is greatly enhanced by the introduction of a genomic double-strand break at the integration site.
Genetic rescue of a ume6Δ/Δ brg1Δ/Δ mutant strain.
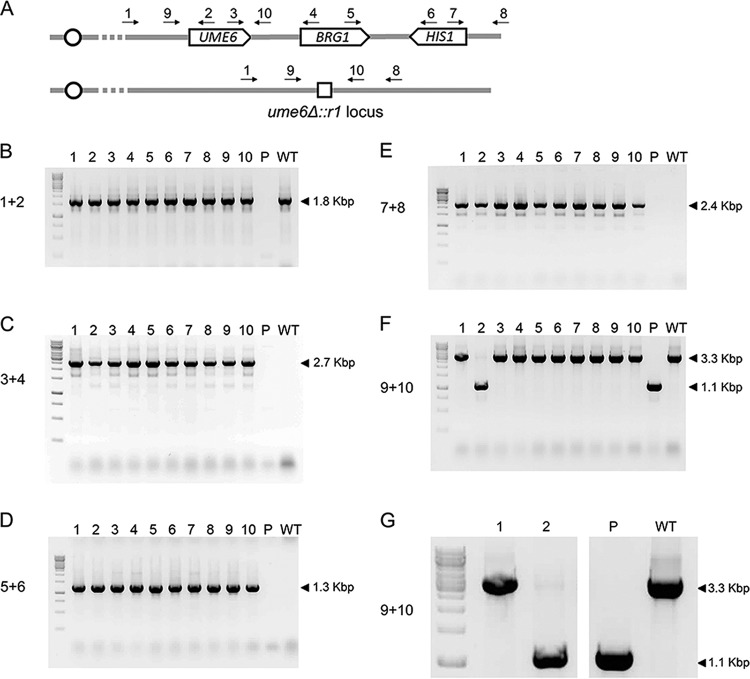
We also tested our genetic rescue strategy with a ume6Δ/Δ brg1Δ/Δ double mutant strain. UME6 and BRG1 encode transcription factors that promote C. albicans filamentation and biofilm formation (15–18). These two genes have large promoter and regulatory regions, and cloning of the two large regions for a conventional complementing construct could be laborious. We designed three PCR products with concatenating homology (Fig. 1): a UME6 segment (YFG1), a BRG1 segment (YFG2), and a C. dubliniensis HIS1 segment (M1). Integration of the concatenated cassettes was targeted to the ume6Δ::r1 locus. The ume6Δ::r1 locus has a deletion of the UME6 coding region; it contains the UME6 upstream and downstream regions flanking a 360-bp segment (designated r1) of vector pRS424 (19). The strain was transformed with approximately 3 µg of each PCR product along with cassettes specifying Cas9 and a single guide RNA that targets the ume6Δ::r1 allele. We recovered more than 200 His+ transformants from a single transformation. Ten transformants were selected for PCR genotyping. All 10 transformants yielded the PCR product (primers 1 plus 2 [Fig. 2A and B]) expected from joining of the UME6 coding region with the UME6 5′ flanking region. All 10 transformants also yielded the PCR product (primers 3 plus 4 [Fig. 2A and C]) expected from the presence of the UME6 coding region adjacent to the BRG1 coding region. In addition, all 10 transformants yielded the PCR product (primers 5 plus 6 [Fig. 2A and D]) expected from the presence of the BRG1 coding region adjacent to the HIS1 coding region. Finally, all 10 transformants yielded the PCR product (primers 7 plus 8 [Fig. 2A and E]) expected from the presence of the HIS1 coding region adjacent to the UME6 3′ region. These results are consistent with the structure expected for integration of a concatenated UME6:BRG1:HIS1 DNA segment at the ume6Δ::r1 locus in the genome. Therefore, these genotyping results indicate that concatemer integration occurred in the majority of transformants.
FIG 2 .
Genotyping UME6:BRG1:HIS1 candidates. (A) Primer pairs for detection of UME6, BRG1, and HIS1. The UME6:BRG1 locus is depicted in the first line with the expected location and integration of UME6, BRG1, and HIS1 depicted. Primers 1 and 2 are used for detection of UME6 at the ume6Δ::r1 locus. Primer 1 anneals upstream of any predicted recombinational junction. Primers 3 and 4 are used to detect BRG1 integration downstream of UME6. Primers 5 and 6 are used to detect HIS1 integration downstream of BRG1. Primers 7 and 8 are used to detect HIS1 at the ume6Δ::r1 locus. Primer 8 sits downstream of any predicted recombinational junction. The ume6Δ::r1 locus is depicted on the second line, with the genetic scar consisting of a single repeat from the r1HIS1r1 cassette indicated by a small white box. Primers 9 and 10 bind upstream and downstream of the ume6Δ::r1 locus and can be used for detection of the ume6Δ::r1 allele. (B) PCR products for detection of UME6 at its native locus using primer pair 1 plus 2. Amplification of a 1.8-kbp band is consistent with the presence of UME6 at its native locus. (C) PCR products for detection of the UME6:BRG1 junction using primer pair 3 plus 4. Amplification of a 2.7-kbp band is consistent with the presence of BRG1 downstream of UME6. (D) PCR products for detection of the BRG1:HIS1 junction using primer pair 5 plus 6. Amplification of a 1.3-kbp band is consistent with the presence of HIS1 downstream of BRG1. (E) PCR products for detection of integration of HIS1 at the ume6Δ::r1 locus using primer pair 7 plus 8. Amplification of a 2.4-kbp band is consistent with the presence of HIS1 at the ume6Δ::r1 locus. (F) PCR products for detection of homozygous integration using primer pair 9 plus 10. Amplification of a 1.1-kbp band suggests the presence of a ume6Δ::r1 allele (see the bottom line in panel A). Amplification of a 3.3-kbp band is consistent with the presence of either a wild-type (WT) UME6 or concatenated UME6:BRG1 allele (see the top line in panel A). A 1.1-kbp band is observed in the parental strain and in the strain in lane 2, indicating the presence of at least one copy of the ume6Δ::r1 allele. The absence of a 1.1-kbp band in the strains in lanes 1 and 3 to 10 suggests homozygous integration of the concatenated allele. (G) Expanded insert of panel F for detection of homozygous integration. Lane 2 contains a 3.3-kbp band which is absent in the parental strain. The 3.3-kbp band in lane 2 is consistent with integration of at least one UME6 allele, which suggests that this strain underwent heterozygous integration of the UME6:BRG1 concatemer.
We anticipated that we may recover homozygous integrants as a result of Cas9-sgRNA cleavage of both ume6Δ::r1 alleles (12). PCR analysis indicated that 9 of the 10 genotyped transformants lacked a detected ume6Δ::r1 allele and thus were likely homozygous integrants (primers 9 plus 10 [Fig. 2A and F]). One genotyped transformant was apparently heterozygous (primers 9 plus 10 [Fig. 2A, F, and G]). These results argue that the majority of our transformants were homozygous for the recombinational concatemer, integrated to replace the ume6Δ::r1 locus in the genome.
For phenotypic rescue assays, we focused on one transformant, strain MH281, that carried the integrated UME6:BRG1:HIS1 concatenation product and was presumably homozygous due to the absence of a ume6Δ::r1 allele. We refer to this strain as the validation strain. Three assays were used to assess phenotypic rescue: filamentation, biofilm formation, and expression of UME6- and BRG1-responsive genes (11, 15–18). We compared these phenotypes in the wild-type strain, a ume6Δ/Δ single mutant, a brg1Δ/Δ single mutant, a ume6Δ/Δ brg1Δ/Δ double mutant, and the UME6:BRG1 validation strain. We sought to use strains with matched nutritional requirements; therefore, for example, the ume6Δ/Δ brg1Δ/Δ strain was a His+ derivative of the His− parental ume6Δ/Δ brg1Δ/Δ double mutant that had been transformed to create the validation strain. Similarly, the other strains had auxotrophies complemented.
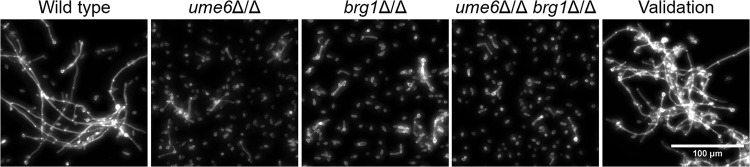
Filamentation was assayed by microscopic observation after growth in yeast extract-peptone-dextrose (YPD) plus serum at 37°C (Fig. 3). The wild-type strain formed long hyphae with few detached yeast-form cells. The ume6Δ/Δ brg1Δ/Δ double mutant failed to form filaments under these conditions. The ume6Δ/Δ and brg1Δ/Δ mutants each formed short hyphae intermixed with a large proportion of yeast-form cells. The UME6:BRG1 validation strain, like the wild type, formed long hyphae and few yeast-form cells. These observations indicate that the inability of the double mutant strain to undergo filamentation is due to the ume6Δ/Δ brg1Δ/Δ genotype, because (i) each single mutant has a filamentation defect and (ii) the UME6:BRG1 concatemer restores filamentation when introduced into the ume6Δ/Δ brg1Δ/Δ background.
FIG 3 .
Filamentation phenotypes of mutant and validation strains. Fluorescence images of cells with Calcofluor white staining after 4 h of growth in YPD plus serum. The wild-type (WT) SN250, ume6Δ/Δ mutant, brg1Δ/Δ mutant, ume6Δ/Δ brg1Δ/Δ double mutant, and UME6:BRG1 validation strains were assayed for phenotypic rescue of filamentation under planktonic growth conditions. The wild-type strain forms long filamentous hyphae, whereas the ume6Δ/Δ mutant, brg1Δ/Δ mutant, and ume6Δ/Δ brg1Δ/Δ double mutant grow primarily as yeast-form cells. The UME6:BRG1 validation strain forms long filamentous hyphae comparable to those of WT cells.
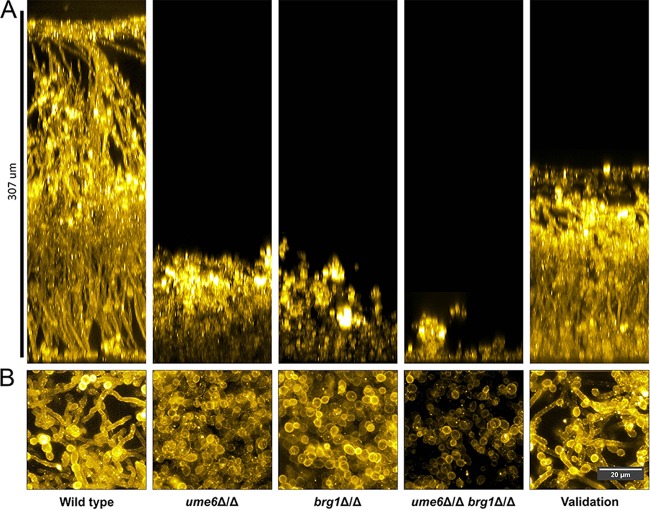
Biofilm formation was assayed by confocal microscopy after growth on silicone squares in YPD alone at 37°C for 24 h (Fig. 4). The wild-type strain formed a robust biofilm of 300 µm in depth with abundant hyphae evident in both side-view and apical-view projections. The ume6Δ/Δ brg1Δ/Δ mutant was severely defective in biofilm formation: few cells adhered to the silicone surface, and no hyphae were apparent (Fig. 4A and B). The ume6Δ/Δ and brg1Δ/Δ single mutants were also defective, yielding biofilms composed exclusively of yeast-form cells and with a depth of ~50 to 70 µm, as reported previously for 48-h biofilms (11). The UME6:BRG1 validation strain produced a biofilm with an intermediate depth, a depth between that of the wild-type strain and those of the single mutant strains. Abundant hyphae were evident in side-view and apical-view projections (Fig. 4A and B). These observations indicate that the inability of the double mutant strain to produce a biofilm is due to the ume6Δ/Δ brg1Δ/Δ genotype, because (i) each single mutant has a biofilm defect, and (ii) the UME6:BRG1 concatemer restores biofilm when introduced into the ume6Δ/Δ brg1Δ/Δ background. However, the UME6:BRG1 concatemer enables only partial rescue of the biofilm defect, because the validation strain does not produce as extensive a biofilm as the wild-type strain does.
FIG 4 .
Side-view and apical-view projections of mutant and validation strain biofilms. The WT SN250 strain, ume6Δ/Δ mutant, brg1Δ/Δ mutant, ume6Δ/Δ brg1Δ/Δ double mutant, and UME6:BRG1 validation strains were assayed for phenotypic rescue of biofilm formation. (A) Lateral projections of C. albicans WT, mutant, and validation strain biofilms. A wild-type strain forms a large robust biofilm, while the ume6Δ/Δ, brg1Δ/Δ, and ume6Δ/Δ brg1Δ/Δ mutant strains form defective biofilms. The UME6:BRG1 validation strain forms a biofilm with more than half the thickness of the WT biofilm. (B) Apical-view projections of representative sections of each biofilm. A wild-type strain forms a robustly filamentous biofilm, whereas ume6Δ/Δ, brg1Δ/Δ, and ume6Δ/Δ brg1Δ/Δ mutant strains form biofilms with largely reduced filamentation. The UME6:BRG1 strain forms a biofilm with visibly filamentous cells.
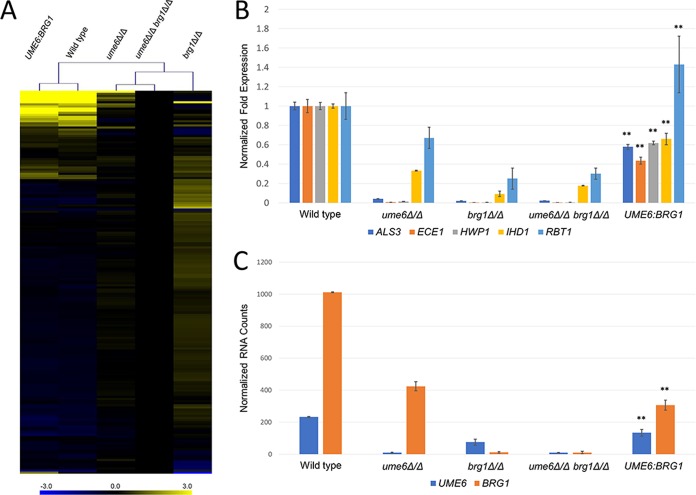
Gene expression was measured by Nanostring on RNA samples from cells grown for 4 h in YPD at 37°C. The probe set assayed RNA levels for a panel of 182 genes, including 166 environmentally responsive genes (11). The wild-type strain had significantly different RNA levels (>2-fold change; P < 0.05) than the ume6Δ/Δ brg1Δ/Δ double mutant for 18 genes in addition to the deleted genes UME6 and BRG1 (Fig. 5A). Almost all of these RNA levels were also significantly different in the wild-type strain compared to the ume6Δ/Δ and brg1Δ/Δ single mutant strains. The affected genes included the core hyphal-associated genes ALS3, ECE1, HWP1, IHD1, and RBT1 (Fig. 5B), as expected (15–18). The validation strain expressed the affected RNAs at levels similar to those of the wild-type strain (Fig. 5A). Most of the core hyphal-associated genes had RNA levels of roughly 50% of the level in the wild-type strain. These observations confirm that the concatenated UME6:BRG1 construct partially rescues the biofilm defect of the ume6Δ/Δ brg1Δ/Δ double mutant.
FIG 5 .
RNA expression levels in mutant and validation strains. RNA was extracted from WT SN250, ume6Δ/Δ mutant, ume6Δ/Δ brg1Δ/Δ double mutant, and UME6:BRG1 validation strains grown for 4 h in YPD at 37°C and then analyzed with a NanoString nCounter. Three biological replicates were assayed, and the average of all replicates was used for further analysis. brg1Δ/Δ mutant gene expression was drawn from Woolford et al., assayed under the same conditions as this experiment (4 h YPD, 37°C) and analyzed using the same NanoString code set (11). (A) A heat map shows log2 fold expression of environmentally responsive genes between strains normalized against expression in a ume6Δ/Δ brg1Δ/Δ double mutant. Log2 fold expression is depicted on a color gradient ranging between −3.0 (blue) and +3.0 (yellow). The tree depicts clustering of strains according to an algorithm for minimum Manhattan distance (MeV software). The ume6Δ/Δ mutant, brg1Δ/Δ mutant, and ume6Δ/Δ brg1Δ/Δ double mutant cluster, showing similar profiles with reduced expression of environmentally responsive genes. The UME6:BRG1 validation strain clusters with the wild-type strain, showing similar expression profiles. (B) Mean fold change in the expression of hypha-associated genes ALS3, ECE1, HWP1, IHD1, and RBT1 in mutant strains and validation strain normalized against expression in the wild-type strain. Values are means ± standard deviations (SDs) (error bars). Mean fold change values that are significantly different (P < 0.01 by Tukey-Kramer test) from the values for the mutant strains are indicated by two asterisks. (C) Mean normalized counts of UME6 and BRG1 mRNA between WT, mutant, and validation strains. Values are means ± SDs. Mean RNA counts that are significantly different (P < 0.01 by Tukey-Kramer test) from the mean RNA counts of all other strains are indicated by two asterisks.
Partial rescue by UME6:BRG1 may result from incompletely restored expression of the UME6 or BRG1 gene. We were concerned in particular about BRG1 expression for two reasons. First, its genomic 5′ flanking region extends for over 10 kbp to the next open reading frame, but only 1.6 kbp had been included in our concatenation cassette. Second, Brg1 is a positive regulator of UME6 expression, so a reduction in BRG1 expression may cause a reduction in UME6 expression as well (18). Our Nanostring measurements indicated that the validation strain expressed BRG1 at ~30% of the wild-type level and expressed UME6 at ~60% of the wild-type level (Fig. 5C). These results are consistent with the model that the UME6:BRG1 concatenated construct allows partial phenotypic rescue because expression of the BRG1 and UME6 genes is only partially restored.
DISCUSSION
Genetic interaction analysis is a vital tool to help decipher functional relationships in pathways, networks, and protein complexes (2). Central to this analysis is the ability to construct and validate multigene mutants. In this report, we have described a simple approach to introduce multiple DNA segments as an integrated genomic concatemer to generate a validation strain. The validation strain is then used in assays for rescue of multigene mutant phenotypes. We refer to the approach as concatemer assembly for rescue of mutant abilities (CARMA) in order to convey features of both the technical procedure and experimental objective.
The CARMA approach builds upon well-founded principles. Homologous recombination can seal a break in a target DNA, as demonstrated by Ma et al. with plasmid DNA in S. cerevisiae (20). Multiple DNA fragments with overlapping homology can be assembled through in vivo recombination in S. cerevisiae, as used for example in high-throughput construction of gene deletion cassettes (21) and in construction of a synthetic bacterial genome (22). A genomic CRISPR cut in a C. albicans mutant allele can enable integration of the wild-type allele, as shown by Nguyen et al. (23). We view CARMA as an amalgam of the principles demonstrated in these prior studies.
CARMA requires some attention to the extent of functional regulatory regions. Integration of the multigene concatemer at one of the mutant loci reconstitutes that gene with its native genomic 5′ flanking region. We chose to integrate specifically at the ume6Δ::r1 locus because UME6 is known to have a large 5′ regulatory region that includes 7 kbp or more upstream of the open reading frame (24, 25). Long 5′ regulatory regions are also found among other genes connected to filamentation, including NRG1, HWP1, and ALS3 (24, 26, 27). The BRG1 5′ regulatory region has not been subjected to detailed analysis, and we included 1.6 kbp of 5′ flanking sequence in our genetic rescue segment. For many genes, we have found that ~1.6 kbp is sufficient for full rescue. However, we note that the BRG1 5′ region extends for more than 10 kbp before the next open reading frame, so it seems possible that it is yet another filamentation-related gene with a long 5′ regulatory region. We infer that 1.6 kbp of 5′ flanking sequence is not sufficient for full BRG1 expression, because BRG1 RNA levels were lower in the validation strain than in the wild-type strain. The reduced BRG1 expression was nonetheless sufficient for substantial rescue of the double mutant phenotypes. Perhaps the best quantitative measures of function were the levels of expression of UME6- and BRG1-dependent target genes, which were significantly greater than those observed for either single mutant or the double mutant.
Note that, in all experiments reported here, the target integration loci are deletion alleles with fairly small relics of the initial marker; they are yfg1Δ::dpl200 or yfg1Δ::r1 alleles. Our preliminary studies suggest that a CARMA-like approach is less efficient with insertion-deletion alleles of structure yfg1Δ::ARG4. In addition, it is noteworthy that many mutant strains currently in use have different markers at the two alleles (genotype yfg1Δ::ARG4/yfg1Δ::URA3, for example). The recipient strains in the present study were all homozygous for identical mutant alleles at the target integration locus. Therefore, for existing yfg1Δ::ARG4/yfg1Δ::URA3 strains, it may be necessary to use two sgRNA genes in order to direct integration to both alleles.
A few features of CARMA seem especially useful. First, all of the DNA molecules in the transformation reaction are PCR products, including the DNA segments destined to be assembled into the integrated concatemer, and the genes specifying Cas9 and the sgRNA, which are the original constructs used by Vyas et al. (12) used with our transient CRISPR protocol (13). This feature saves some time because the investigator circumvents cloning and screening for appropriate plasmids. This feature also overcomes the potential problem that a DNA fragment may be unstable in the intermediate host used for cloning. A second useful feature is that homozygous integrants can be recovered. This feature can eliminate partial phenotypic rescue due to gene dosage effects. Third, while in CARMA, each DNA segment corresponded to a complete coding region and flanking regions, concatenating homology of PCR products could also be used for construction of fusion genes, localized mutagenesis, mapping of functional promoter elements, or introduction of allelic variants.
MATERIALS AND METHODS
Strains, media, and transformations.
C. albicans strains were all archived in 15% glycerol stocks stored at −80°C. Strains were grown on solid yeast extract-peptone-dextrose (YPD) medium for 2 days at 30°C and then in liquid YPD medium overnight at 30°C with shaking prior to all assays and transformations. Transformations were performed using the lithium acetate transformation method supplemented with transient CRISPR-Cas9 system components (13). Transformants were selected on synthetic medium lacking specified auxotrophic supplements (2% dextrose, 0.67% Difco yeast nitrogen base without amino acids, and complete supplement mixture lacking the indicated amino acids). Replica plating and patching were performed using synthetic dextrose (SD) medium (2% dextrose, 0.67% Difco yeast nitrogen base without amino acids). All strains are listed in Table S1 in the supplemental material.
Candida albicans strains used in this study. Download TABLE S1, XLSX file, 0.01 MB (9.6KB, xlsx) .
Copyright © 2018 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Preparation of DNA cassettes and genotyping.
In preparation for the amplification of wild-type alleles, genomic DNA was prepared from the SC5314 strain by a modified version of the Hoffman and Winston procedure (28).
PCR was performed for 30 cycles for all PCRs according to the manufacturer’s protocols using TaKaRa Ex Taq DNA polymerase. All primer sequences are listed in Table S2.
Oligonucleotide primer sequences. Download TABLE S2, XLSX file, 0.01 MB (10.3KB, xlsx) .
Copyright © 2018 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The C. albicans ARG4 gene was amplified from SC5314 genomic DNA using primers CaARG4 MGC/F and CaARG4 3′->CaLEU2 5′/R. The C. albicans LEU2 gene was amplified from SC5314 genomic DNA using primers CaLEU2 MGC/F and CaLEU2 3′->CdHIS1 for/R. The C. dubliniensis HIS1 cassette was amplified from plasmid pSN52 (29) using primers CdHIS1 for->CaLEU2 3′/F and CdHIS1 rev->CaARG4 3′/R (29).
The CaARG4 3′->CaLEU2 5′/R primer contains 80 bp of homology to the upstream region of the C. albicans LEU2 gene. The CaLEU2 3′->CdHIS1 for/R primer contains 80 bp of homology to the upstream region of the C. dubliniensis HIS1 cassette. The CdHIS1 for->CaLEU2 3′/F primer contains 80 bp of homology to the downstream region of the C. albicans LEU2 gene. The CdHIS1 rev->CaARG4 3′/R primer contains 80 bp of homology to the downstream region of the ARG4 locus.
The C. albicans UME6 gene was PCR amplified from SC5314 genomic DNA using primers UME6 MGC/F and UME6 3′->BRG1 5′/R. The C. albicans BRG1 gene was PCR amplified from SC5314 genomic DNA using primers BRG1 MGC/F and BRG1 3′->CdHIS1 for/R. The C. dubliniensis HIS1 cassette was amplified from plasmid pSN52 (29) using primers CdHIS1 for->BRG1 3′/F and CdHIS1 rev->UME6 3′/R (29).
The UME6 3′->BRG1 5′/R primer contains 80 bp of homology to the upstream region of the C. albicans BRG1 gene. The BRG1 3′->CdHIS1 for/R primer contains 80 bp of homology to the upstream region of the C. dubliniensis HIS1 cassette. The CdHIS1 for->BRG1 3′/F primer contains 80 bp of homology to the downstream region of the C. albicans BRG1 gene. The CdHIS1 rev->UME6 3′/R primer contains 80 bp of homology to the downstream region of the UME6 locus.
PCR genotyping.
Genotyping of the UME6:BRG1 synthetic locus was performed using four different pairs of primers. Amplification using these primer pairs yielding a DNA segment of the correct size is indicative of the expected homologous integration event. Integration of UME6 at the UME6 locus was confirmed using primers UME6 FarUP/F and UME6 Int/R. Integration of BRG1 downstream of UME6 was confirmed using primers UME6 2346 Int/F and BRG1 Int/R. Integration of the C. dubliniensis HIS1 cassette was confirmed using primers BRG1 Int/F and HIS1 Check Int/R, as well as primers HIS1 CRIME/F and UME6 FarDOWN/R.
Homozygosity was confirmed using primers UME6 MGC/F and UME6 downstream/R, which yields either a 1.1-kbp product if a ume6Δ::r1 allele is present or a 3.3-kbp product if a UME6 allele is present.
Single guide RNA cassette construction.
Fusion PCR was used to assemble sgRNA cassettes by previously described protocols (13, 19). The arg4::dpl200 sgRNA cassette was prepared using primers sgRNA/F arg4::dpl200 and SNR52/R arg4::dpl200 and targets the sequence TCAAGAATAATCCCGCTGCT. The ume6::r sgRNA cassette was prepared using primers sgRNA/F ume6::r and SNR52/R ume6::r and targets the sequence TTAATCCACTGTATAACTCG.
RNA preparation and NanoString analysis.
Overnight cultures of the relevant strains were inoculated into 25 ml of YPD to a final optical density at 600 nm (OD600) of 0.2 in 250-ml Erlenmeyer flasks. Cultures were then allowed to grow at 4 h with vigorous shaking (200 rpm) at 37°C, then collected by vacuum filtration, and stored at −80°C until ready for extraction. RNA was extracted from frozen cell cultures using the Qiagen RNeasy minikit (catalog no. 74104) with modifications as described previously (11).
Nanostring analysis of RNA samples was performed using previously described methods (11). The Nanostring code set contained 166 gene targets associated with environmental response pathways and 15 gene targets of interest for a variety of reasons (11).
Biofilm and filamentation assay imaging.
Biofilms were grown according to previously described protocols with the following modifications (11). Overnight cultures grown at 30°C were inoculated into 2 ml YPD in wells containing a silicone square (Bentec Medical Inc.) (1.5 by 1.5 cm) and allowed to adhere to the silicone square for 90 min in an incubator-shaker at 37°C with shaking at 60 rpm. The squares were then washed by brief immersion into a well containing 2 ml of phosphate-buffered saline (PBS) and then placed into a fresh well containing 2 ml YPD. After 24 h, biofilms were fixed using 2 ml of 4% formaldehyde and 2.5% glutaraldehyde in 1× PBS on an orbital shaker for 1 h at 60 rpm. The biofilms were then stained for 24 h with concanavalin A–Alexa Fluor 594 conjugate (Life Technologies) diluted to 25 µg/ml in PBS, before being imaged using a slit-scan confocal optical unit on a Zeiss Axiovert 200 microscope with a 40× 0.85-numerical-aperture (NA) oil immersion objective. The imaging processes are described in detail elsewhere (30).
The filamentation assay was performed using strains inoculated into 5 ml YPD in glass test tubes and grown for 4 h at 37°C in a roller drum at maximum speed. Following growth, the cells were fixed and stained with Calcofluor white. An aliquot of cells was then imaged using conventional fluorescence microscopy on a Zeiss Axio observer Z.1 fluorescence microscope and a 20× 0.8-NA objective.
Software.
Colony counts were collected using OpenCFU software (31). Microscopy images were compiled with ImageJ (32), Photoshop CS6, and Microsoft PowerPoint 2010. RNA counts were analyzed in Microsoft Excel 2010 and GraphPad Prism 7. Heat maps were generated using MeV software (33) and then arranged with Photoshop.
Data availability.
Nanostring transcription profiling data may be found in Table S3.
Nanostring measurements of RNA levels. Raw data, normalized probe counts, and gene expression ratios are given on three different tabs. Download TABLE S3, XLSX file, 0.2 MB (218.1KB, xlsx) .
Copyright © 2018 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We sincerely thank Frederick Lanni and Katherine Lagree for their invaluable advice and contributions and for their support and assistance. We also thank Tatyana Aleynikov and Megan Bean for their extensive technical support, as well as Surya D. Aggarwal and N. Luisa Hiller for assistance with statistical analysis.
These studies were supported by NIH/NIAID grant 1R21AI135178 (A.P.M.).
REFERENCES
- 1.Falkow S. 1988. Molecular Koch’s postulates applied to microbial pathogenicity. Rev Infect Dis 10(Suppl 2):S274–S276. http://www.jstor.org/stable/4454582. [DOI] [PubMed] [Google Scholar]
- 2.Baryshnikova A, Costanzo M, Myers CL, Andrews B, Boone C. 2013. Genetic interaction networks: toward an understanding of heritability. Annu Rev Genomics Hum Genet 14:111–133. doi: 10.1146/annurev-genom-082509-141730. [DOI] [PubMed] [Google Scholar]
- 3.Bharucha N, Chabrier-Rosello Y, Xu T, Johnson C, Sobczynski S, Song Q, Dobry CJ, Eckwahl MJ, Anderson CP, Benjamin AJ, Kumar A, Krysan DJ. 2011. A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis. PLoS Genet 7:e1002058. doi: 10.1371/journal.pgen.1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bockmühl DP, Krishnamurthy S, Gerads M, Sonneborn A, Ernst JF. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol Microbiol 42:1243–1257. doi: 10.1046/j.1365-2958.2001.02688.x. [DOI] [PubMed] [Google Scholar]
- 5.Braun BR, Johnson AD. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis D, Wilson RB, Mitchell AP. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol Cell Biol 20:971–978. doi: 10.1128/MCB.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lermann U, Morschhäuser J. 2008. Secreted aspartic proteases are not required for invasion of reconstituted human epithelia by Candida albicans. Microbiology 154:3281–3295. doi: 10.1099/mic.0.2008/022525-0. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Robbins N, O’Meara TR, Cowen LE. 2017. Extensive functional redundancy in the regulation of Candida albicans drug resistance and morphogenesis by lysine deacetylases Hos2, Hda1, Rpd3 and Rpd31. Mol Microbiol 103:635–656. doi: 10.1111/mmi.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. doi: 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- 10.Shapiro RS, Chavez A, Porter CBM, Hamblin M, Kaas CS, DiCarlo JE, Zeng G, Xu X, Revtovich AV, Kirienko NV, Wang Y, Church GM, Collins JJ. 2018. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat Microbiol 3:73–82. doi: 10.1038/s41564-017-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woolford CA, Lagree K, Xu W, Aleynikov T, Adhikari H, Sanchez H, Cullen PJ, Lanni F, Andes DR, Mitchell AP. 2016. Bypass of Candida albicans filamentation/biofilm regulators through diminished expression of protein kinase Cak1. PLoS Genet 12:e1006487. doi: 10.1371/journal.pgen.1006487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vyas VK, Barrasa MI, Fink GR. 2015. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv 1:e1500248. doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min K, Ichikawa Y, Woolford CA, Mitchell AP. 2016. Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere 1:e00130-16. doi: 10.1128/mSphere.00130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, López-Ribot JL, Kadosh D. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell 19:1354–1365. doi: 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleary IA, Lazzell AL, Monteagudo C, Thomas DP, Saville SP. 2012. BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Mol Microbiol 85:557–573. doi: 10.1111/j.1365-2958.2012.08127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du H, Guan G, Xie J, Sun Y, Tong Y, Zhang L, Huang G. 2012. Roles of Candida albicans Gat2, a GATA-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PLoS One 7:e29707. doi: 10.1371/journal.pone.0029707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang MY, Mitchell AP. 2017. Marker recycling in Candida albicans through CRISPR-Cas9-induced marker excision. mSphere 2:e00050-17. doi: 10.1128/mSphere.00050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma H, Kunes S, Schatz PJ, Botstein D. 1987. Plasmid construction by homologous recombination in yeast. Gene 58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 21.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA III, Smith HO. 2008. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen N, Quail MMF, Hernday AD. 2017. An efficient, rapid, and recyclable system for CRISPR-mediated genome editing in Candida albicans. mSphere 2:e00149-17. doi: 10.1128/mSphereDirect.00149-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Childers DS, Kadosh D. 2015. Filament condition-specific response elements control the expression of NRG1 and UME6, key transcriptional regulators of morphology and virulence in Candida albicans. PLoS One 10:e0122775. doi: 10.1371/journal.pone.0122775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Childers DS, Mundodi V, Banerjee M, Kadosh D. 2014. A 5′ UTR-mediated translational efficiency mechanism inhibits the Candida albicans morphological transition. Mol Microbiol 92:570–585. doi: 10.1111/mmi.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argimón S, Wishart JA, Leng R, Macaskill S, Mavor A, Alexandris T, Nicholls S, Knight AW, Enjalbert B, Walmsley R, Odds FC, Gow NA, Brown AJ. 2007. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot Cell 6:682–692. doi: 10.1128/EC.00340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, Nguyen QB, Wolyniak MJ, Frechette G, Lehman CR, Fox BK, Sundstrom P. 2018. Release of transcriptional repression through the HCR promoter region confers uniform expression of HWP1 on surfaces of Candida albicans germ tubes. PLoS One 13:e0192260. doi: 10.1371/journal.pone.0192260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffman CS, Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 29.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagree K, Desai JV, Finkel JS, Lanni F. 2018. Microscopy of fungal biofilms. Curr Opin Microbiol 43:100–107. doi: 10.1016/j.mib.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Geissmann Q. 2013. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS One 8:e54072. doi: 10.1371/journal.pone.0054072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Candida albicans strains used in this study. Download TABLE S1, XLSX file, 0.01 MB (9.6KB, xlsx) .
Copyright © 2018 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotide primer sequences. Download TABLE S2, XLSX file, 0.01 MB (10.3KB, xlsx) .
Copyright © 2018 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nanostring measurements of RNA levels. Raw data, normalized probe counts, and gene expression ratios are given on three different tabs. Download TABLE S3, XLSX file, 0.2 MB (218.1KB, xlsx) .
Copyright © 2018 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Nanostring transcription profiling data may be found in Table S3.
Nanostring measurements of RNA levels. Raw data, normalized probe counts, and gene expression ratios are given on three different tabs. Download TABLE S3, XLSX file, 0.2 MB (218.1KB, xlsx) .
Copyright © 2018 Huang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.