Summary
Cytoplasmic long non-coding RNAs have been shown to act at many different levels to control post-transcriptional gene expression, although their role in translational control is poorly understood. Here, we show that lnc-31, a non-coding RNA required for myoblast proliferation, promotes ROCK1 protein synthesis by stabilizing its translational activator, YB-1. We find that lnc-31 binds to the Rock1 mRNA as well as to the YB-1 protein and that translational activation requires physical interaction between the two RNA species. These results suggest a localized effect of YB-1 stabilization on the Rock1 mRNA. ROCK1 upregulation by lnc-31, in proliferative conditions, correlates well with the differentiation-repressing activity of ROCK1. We also show that, upon induction of differentiation, the downregulation of lnc-31, in conjunction with miR-152 targeting of Rock1, establishes a regulatory loop that reinforces ROCK1 repression and promotes myogenesis.
Keywords: long non-coding RNA, myogenesis, Rock1, YB-1, lnc-31, cell cycle, translation, proteasome, protein stability
Graphical Abstract
Highlights
-
•
lnc-31 sustains myoblast proliferation, counteracting differentiation
-
•
lnc-31 binds to Rock1 mRNA and YB-1 protein
-
•
Rock-1 translation is favored through its interaction with lnc-31 and YB-1 protein
-
•
lnc-31 counteracts YB-1 protein degradation, thus promoting Rock1 translation
Dimartino et al. demonstrate that lnc-31 is required to sustain myoblast proliferation. lnc-31 interacts with Rock1 mRNA, an inhibitor of differentiation, and promotes its translation. This activity is strengthened by binding of the translational regulator YB-1 and its lnc-31-dependent stabilization.
Introduction
Cytoplasmic long non-coding RNAs (lncRNAs) have been shown to play many roles, ranging from protein and microRNA (miRNA) sponges to antisense regulators of target transcripts. LncRNAs also can function as protein remodelers or regulators of protein modification (Rashid et al., 2016, Ballarino et al., 2016). Most of these activities rely on the ability of lncRNAs to act as scaffolds where both RNA and protein components can be assembled in a composite and combinatorial fashion, thus allowing the tethering of different molecules and their concerted and localized action.
In this study, we have examined the interactors and mode of action of lnc-31, which was identified initially as a murine lncRNA expressed in proliferating myoblasts and downregulated upon muscle differentiation. Notably, lnc-31 was shown to play a significant role in sustaining cell proliferation and in counteracting differentiation (Ballarino et al., 2015), even though the molecular mechanism at the basis of this control was not clarified. An interesting feature of lnc-31 is that it harbors in its third exon the precursor sequence of miR-31. Several experiments have indicated that while the primary lnc-31 transcript is exclusively nuclear and, if processed by Drosha, can give rise to miR-31, the mature lnc-31 has cytoplasmic localization and does not contribute to the miR-31 pool. Therefore, lnc-31 and miR-31 originate from the same nuclear precursor through two mutually exclusive pathways (Ballarino et al., 2015). Because miR-31 activity also has been linked to cell proliferation (Laurila and Kallioniemi, 2013, Zhang et al., 2011, Liu et al., 2010, Cacchiarelli et al., 2011), it can be inferred that the activities of both the long and small RNAs, deriving from the same primary transcript, converge on common regulatory pathways.
In this study, we show that lnc-31 modulates the expression of important key factors regulating the maintenance of the myoblast proliferation state. By investigating the molecular mechanism of lnc-31 action, we discovered that it associates with several mRNAs and specific proteins, among them, the Rock1 mRNA, a known inhibitor of myogenesis (Zhang et al., 2012, Charrasse et al., 2006), and the translational regulator YB-1, which we demonstrate is necessary for the activation of Rock1 translation. We show that lnc-31 stabilizes the YB-1 factor, thus allowing its positive effect on Rock1 mRNA translation.
Results and Discussion
lnc-31 Knockdown Affects the Expression of Genes Coordinating Cell-Cycle Exit and Differentiation
To address the molecular mechanism of lnc-31 action in the cytoplasmic compartment, we performed transcriptome analysis on total RNA of C2C12 murine myoblasts treated either with scramble small interfering RNA (siRNA) (SCR) or with siRNA against lnc-31 (si1-lnc-31). Under these conditions, efficient downregulation of the mature lnc-31 was obtained, and no change in miR-31 levels was detected (Figure 1A), ensuring that the effects observed are not caused by miR-31. Next-generation RNA sequencing (NGS) was applied to control (SCR) and si1-lnc-31-treated samples (Table S1). Hierarchical clustering of the samples based on their gene expression profile proved that the transcriptome of C2C12 cells depleted of lnc-31 is altered significantly in comparison with that of SCR-treated cells (Figure S1A). Differential expression analysis led to the identification of a set of 353 genes (Table S2) affected by lnc-31 depletion (adjusted p value < 0.05) (Figure 1B).
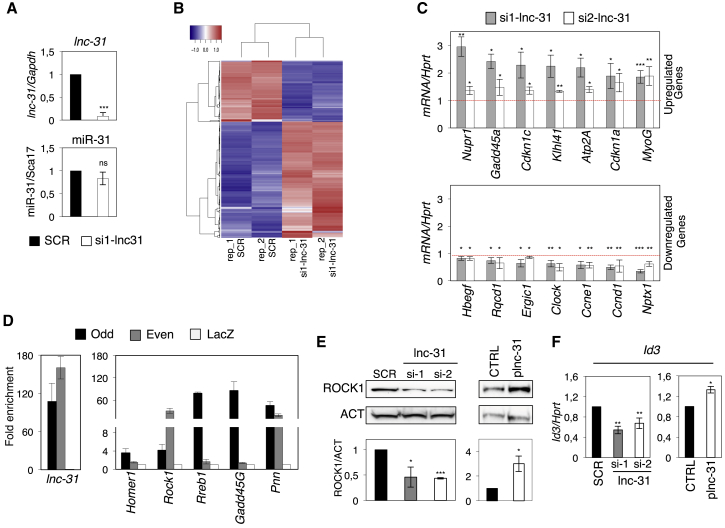
Figure 1.
lnc-31 Knockdown Affects Gene Expression of Proliferating Murine Myoblasts
(A) Graphs showing the level of lnc-31 and miR-31 in C2C12 cells treated with SCR or si1-lnc-31. lnc-31 and miR-31 levels were normalized against Gapdh and Sca17, respectively, and expressed as relative quantity with respect to the SCR sample set to a value of 1.
(B) Heatmap showing the hierarchical clustering of differentially expressed genes in C2C12 cells treated as they were in (A).
(C) Graphs showing the level of the selected mRNAs in C2C12 cells treated with SCR or si1- and si2-lnc-31. The expression levels were normalized against Hprt and expressed as relative quantity with respect to the SCR sample set to a value of 1 (red dashed line).
(D) Graphs showing the levels of lnc-31 (left) and 5 selected transcripts (right) recovered upon lnc-31 RNA pull-down. Values are expressed as “fold enrichment” with respect to the LacZ sample set to a value of 1.
(E) Western blot analysis using ROCK1 antibodies on protein extracts from C2C12 cells treated as in (C) or transfected with either pcDNA3.1 (CTRL) or with plnc-31 plasmid. ACTININ (ACT) was used as loading control. The results of the densitometric analyses are shown below.
(F) Graph showing the levels of Id3 mRNA in samples treated as in (E). The expression levels were normalized against Hprt mRNA and expressed as relative quantity with respect to SCR or CTRL samples set to a value of 1.
Error bars represent SDs of at least 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ns (not significant; p > 0.05) correspond to paired two-tailed Student’s t tests.
A Gene Ontology term enrichment analysis showed that the downregulated genes are related to cell-cycle processes, while the upregulated ones mostly cluster in the muscle system process and muscle contraction categories (Figure S1B). This correlates well with a previous study that showed that lnc-31 plays an important role in sustaining myoblast proliferation counteracting differentiation (Ballarino et al., 2015). Validation of 14 selected genes was performed by quantitative real-time PCR (qRT-PCR) on samples treated with siRNAs targeting two different lnc-31 regions (Figures 1C and S1C). Figure 1C shows that Nupr1 appears in the list of genes that are upregulated upon lnc-31 knockdown. This gene is induced in the G1 phase and acts as a brake on the cell cycle, regulating the transition to differentiation. In particular, NUPR1 is able to interact and positively modulate MYOD function, favoring its acetylation by p300 (Sambasivan et al., 2009). Indeed, upregulation of Nupr1 upon lnc-31 knockdown correlated with the activation of several MYOD target genes, such as the muscle-specific transcription factor Myogenin, the Ca2+ pump Serca1 (Atp2a1; Jin et al., 2014), and the cell-cycle inhibitors p21CIP1 and p57KIP2 (Cdkn1a and Cdkn1c; Figure 1C). Among the downregulated genes, we found Cyclin D (Ccnd1) and Cyclin E (Ccne1) (Ballarino et al., 2015), which prevent differentiation by phosphorylation of MYOD and MEF2, respectively (Figure 1C; Lazaro et al., 2002, Tintignac et al., 2000). Notably, these targets were altered in an opposite manner when lnc-31 was overexpressed by using a plasmid (plnc-31) carrying the lnc-31 sequence depleted of the Drosha processing region to prevent miR-31 production (Figure S1D). On the contrary, the rescue of lnc-31 expression upon its depletion recovered the levels of only some of these genes (data not shown). This likely can be explained by the fact that in the absence of lnc-31, the upregulation of genes, such as Myogenin and Atp2a1, causes an anticipation of myogenic differentiation that is difficult to retrieve.
Taken together, these data suggest that the activity of lnc-31 in proliferating myoblasts is to sustain the expression of cell-cycle-promoting genes while repressing pro-myogenic ones, therefore indicating a potential important role in controlling the switch between cell-cycle exit and progression into terminal differentiation.
lnc-31 Interacts with Specific mRNAs
To identify the mechanism of lnc-31 action, we started with the identification of its RNA interactors. A set of five biotinylated antisense DNA oligonucleotides (Odd) was used to pull down lnc-31 using cytoplasmic extracts from proliferating murine C2C12 myoblasts. A set of probes against LacZ mRNA (LacZ) was used as a negative control. Samples retrieved upon RNA pull-down through Odd and LacZ oligonucleotides were analyzed by NGS to identify mRNAs associated with lnc-31 (Table S1). The NGS analysis resulted in a list of 92 transcripts that may be considered lnc-31 interactors (Table S3).
The NGS data were validated by RNA pull-down experiments using an additional set (Even) of five biotinylated antisense DNA oligonucleotides. qRT-PCR analysis revealed that both sets of oligonucleotides allowed specific pull-down of lnc-31, whereas the LacZ oligonucleotides did not show any enrichment (Figure 1D).
From the list obtained from the NGS analysis (Table S3), we selected five mRNAs for further validation. Only two of them, Rho-associated coiled-coil containing protein kinase 1 (Rock1) and Pinin (Pnn), were significantly enriched with both sets of oligonucleotides (Figure 1D). To test the effect of lnc-31 on these targets, we measured the Pnn and Rock1 RNA and protein levels upon lnc-31 knockdown. In these conditions neither the protein nor the mRNA of Pnn showed any significant variation (Figure S1E); instead, in the case of Rock1, while the mRNA was unaffected (Figure S1F), the protein was significantly downregulated (Figure 1E). Moreover, with respect to lnc-31 knockdown, the ectopic expression of lnc-31 caused an increase in the ROCK1 protein (Figure 1E) without affecting its mRNA levels (Figure S1F). All of these data indicated a possible positive effect of lnc-31 on Rock1 mRNA translation. Notably, the expression of Id3, known to be an indirect target of ROCK1 (Iwasaki et al., 2008), also decreased as a consequence of ROCK1 downregulation and increased upon lnc-31 overexpression (Figure 1F). All of these data led us to select ROCK1 for further investigation.
A computational prediction (see Experimental Procedures) indicated the presence of a 22nt-long region of potential pairing between the third exon of lnc-31 and the 5′ UTR of Rock1 (Figure 2A). To test the relevance of this interaction in translational control, we measured the luciferase activity of a construct carrying the Rock1 5′ UTR fused to the Renilla luciferase cDNA (Luc/5′Rock) in conditions of lnc-31 downregulation or overexpression. Downregulation of lnc-31 was performed in proliferating myoblasts, while the overexpression was tested in HeLa cells (Figure S2A) to enable the dissection of the specific activity of lnc-31/YB-1 on the Rock1 mRNA independently of the effects on myogenic differentiation. Figure 2B shows that lnc-31 downregulation reduced RENILLA activity (upper panels), whereas its overexpression had the opposite effect (lower panels). Notably, Renilla mRNA and miR-31 levels were the same in both conditions (Figure S2A), suggesting that lnc-31 acts at the translational level and miR-31 does not contribute to the observed phenotype.
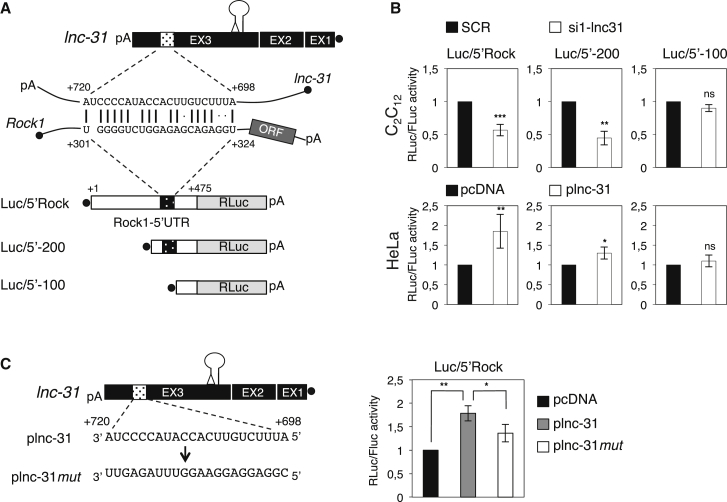
Figure 2.
A Specific Pairing Region Is Required for Rock1 Regulation Mediated by lnc-31
(A) Schematic representation of lnc-31 and Rock1 transcripts. The sequence and the nucleotide position of the pairing region are indicated together with the different constructs used for the luciferase assays (Luc/5′Rock; Luc/5′-200, and Luc/5′-100). The stem loop depicts the pre-miR-31, while the triangle represents the deletion of the Drosha processing site.
(B) Luciferase activities derived from cells transfected with the constructs described in (A) and with SCR/si1-lnc-31 (C2C12, upper panels) and pcDNA/plnc-31 (HeLa, lower panels). Values are expressed as ratio of RENILLA (RLuc) versus FIREFLY (FLuc) activities. The values are expressed as relative quantity with respect to the SCR or pcDNA samples set to a value of 1.
(C) Left: sequence of the pairing region of lnc-31 (plnc-31) and of its mutant derivative (plnc-31mut). Right: Luciferase activities derived from HeLa cells transfected with Luc/5′Rock construct together with pcDNA, plnc-31, or plnc-31mut. Values are expressed as the ratio of RLuc versus FLuc activities. The values are expressed as relative quantity with respect to the pcDNA sample set to a value of 1.
Error bars represent SDs of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ns (not significant; p > 0.05) correspond to paired two-tailed Student’s t tests.
To understand whether the predicted pairing region in the 5′ UTR of Rock1 is relevant for lnc-31-mediated regulation, we derived luciferase constructs with progressive deletions of the 5′ UTR (Figure 2A). Figure 2B shows that the Luc/5′-200 construct, depleted of 200nt but still containing the predicted pairing region, responds to lnc-31 depletion and overexpression; instead, the Luc/5′-100 construct, lacking the pairing region, fails to respond to lnc-31 modulation.
A mutant version of lnc-31 with nucleotide substitutions in the pairing region also was derived (plnc-31mut; Figure 2C) and tested for the ability to regulate Luc/5′Rock expression. Compared to the wild-type lnc-31, the mutant shows a lesser ability to upregulate the Renilla luciferase, even if with a subtle, but significant, effect (Figure 2C). This could be because the identified pairing region may act in conjunction with other neighboring sequence elements. Also, in this case, the levels of the Renilla mRNA were the same in both conditions (Figure S2B). These data indicated that the pairing region is involved in the regulation operated by lnc-31 on the 5′ UTR of Rock1.
The positive control exerted by lnc-31 on Rock1 translation has important implications for the control of myogenesis. In proliferating myoblasts, elevated levels of ROCK1 are required to sustain the phosphorylation of the transcription factor FKHR and its cytoplasmic retention (Nishiyama et al., 2004), thus determining the lack of activation of pro-myogenic genes (Bois and Grosveld, 2003) and the induction of Id3, a potent inhibitor of myogenic differentiation (Iwasaki et al., 2008). On the contrary, after the induction of differentiation, the decrease of lnc-31, and therefore of ROCK1 levels, allows the nuclear translocation of FKHR and the activation of the myogenic program.
Notably, the decrease of ROCK1 upon differentiation, caused by reduced levels of lnc-31, is further reinforced by the parallel upregulation of miR-152 (Figure S2C), which we demonstrated as targeting the 3′ UTR of Rock1 (Figure S2D).
lnc-31 Interacts with YB-1 and Regulates Rock1 Translation
The pull-down experiment described above also was used to recover protein factors associated with lnc-31. The protein composition of the pull-down samples was analyzed by mass spectrometry. From a list of proteins with a probability score >70, identified by at least three peptides and with a low background in the LacZ control, a few candidates were selected for further validation (Table S4). The interactions were confirmed by western blot analysis of RNA pull-down samples obtained by using both Odd and Even oligonucleotides and the LacZ pool. Among the selected protein factors, Y-box protein 1 (YB-1) showed a specific interaction with lnc-31 in both Odd and Even samples, while EEF2, TUBA6, and HNRPM did not confirm the mass spectrometry data (Figure 3A). The interaction between lnc-31 and YB-1 also was proved by UV-crosslinking immunoprecipitation (CLIP) assay using YB-1 antibodies (Figure 3B). The 3′ UTR region of Yb-1 mRNA and Akt1 mRNA were used as positive controls because they already have been shown to interact with YB-1 (Dong et al., 2009, Skabkina et al., 2005). Snhg12, an lncRNA expressed at comparable levels to those of lnc-31, was instead used as a control reference. Because of the intrinsic high affinity of YB-1 for RNA molecules, some binding also was detected for the control reference; however, lnc-31 interaction with YB-1 had much higher results. Indeed, it has been shown that YB-1 has a slight but reliable preference for certain sequences and that other RNA-binding proteins can compete with YB-1 for RNA binding, thus directing YB-1 toward more specific sequences (Dong et al., 2009, Skabkina et al., 2005, Evdokimova et al., 2006). Notably, CLIP experiments using human myoblasts indicated that the YB-1/lnc-31 interaction also is conserved in humans (Figure S3A).
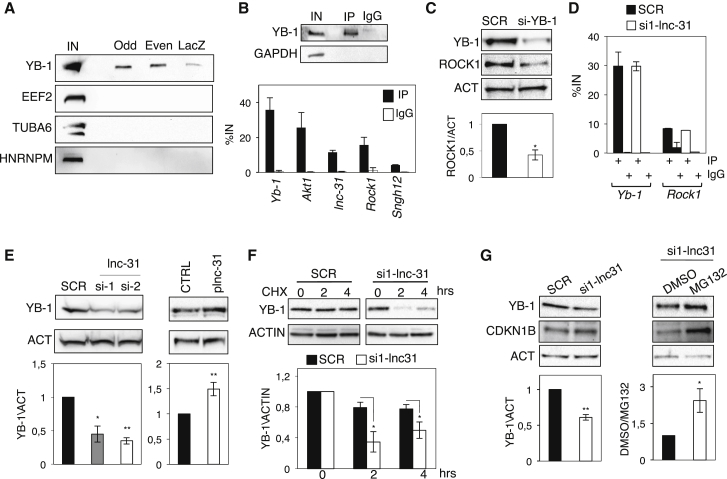
Figure 3.
lnc-31 Interacts and Stabilizes YB-1 Protein that Controls Rock1 Translation
(A) Western blot analysis of proteins retrieved from lnc-31 RNA pull-down.
(B) Upper: western blot with YB-1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (negative control) antibodies of protein extracts from YB-1 CLIP experiment. Input sample (IN) accounts for 2% of the extract. Lower: graph showing the results of two CLIP experiments using YB-1 antibodies. The graph shows the enrichment (%IN) in IP and IgG samples for Yb-1, Akt1, lnc-31, and Rock1 transcripts and for the control reference Sngh12 RNA.
(C) Western blot of proteins from C2C12 cells treated with SCR and si-YB-1 using YB-1 and ROCK1 antibodies. ACT is used as loading control. Quantification of ROCK1 protein levels from three independent experiments is shown below. ROCK1 levels were normalized against ACT levels and expressed with respect to the SCR sample set to a value of 1.
(D) Graph showing the results of two CLIP experiments using YB-1 antibodies and C2C12 cells treated with SCR and si1-lnc-31. The graph shows the enrichment (%IN) in IP and IgG samples for the Yb-1 and Rock1 mRNAs.
(E) Western blot analysis using YB-1 antibodies on proteins from C2C12 cells treated with SCR or si1-/si2-lnc-31 or transfected with either pcDNA3.1 (CTRL) or plnc-31. ACT was used as loading control. Densitometric analyses of three independent experiments are shown below.
(F) Upper: western blot of proteins from C2C12 cells treated with CHX in the presence of SCR or si1-lnc-31 for the indicated time. Lower: densitrometric analyses of YB-1 protein levels from three independent experiments. YB-1 protein levels were normalized against ACTIN levels and expressed with respect to the SCR and si-lnc-31 time “0” set to a value of 1.
(G) Western blot analysis, using YB-1 and CDKN1B antibodies, of proteins from C2C12 cells treated as in (D) (left) or with si1-lnc-31 in the presence of DMSO or MG132 (right). ACT was used as loading control. Densitometric analyses of three independent experiments are shown below.
Error bars represent SD of three independent experiments (unless differently specified). ∗p < 0.05 and ∗∗p < 0.01 correspond to paired two-tailed Student’s t tests.
YB-1 has been shown to control many DNA- and RNA-dependent processes (Lyabin et al., 2014), among which are the control of stability and translation of several classes of mRNAs (El-Naggar et al., 2015, Ohashi et al., 2011, Tanaka et al., 2012, Evdokimova et al., 2009). In particular, it has been suggested that the activity of YB-1 in regulating translation resides in its ability to induce mRNA structural arrangements by promoting non-specific strand displacement and annealing (Nekrasov et al., 2003, Skabkin et al., 2001).
Indeed, the knockdown of Yb-1 in proliferating myoblasts resulted in an approximately 2-fold decrease in ROCK1 protein (Figure 3C). It is worth noting that in the same conditions, the amounts of lnc-31 and Rock1 mRNA were unaffected (Figure S3B), indicating that the effect of YB-1 on Rock1 was exerted at the translational level.
The UV-CLIP experiment shown in Figure 3B indicates that Rock1 mRNA also is found in association with YB-1. Because after UV-crosslinking the samples were treated with high salt buffers, we suggest that YB-1 is able to bind lnc-31 and Rock1 mRNA independently. In contrast to YB-1/lnc-31 interaction, the binding between YB-1 and the Rock1 mRNA was not conserved in humans because the levels of this mRNA, recovered upon immunoprecipitation, were comparable to those of the RPS6KB1 RNA, which was used as control reference (Figure S3A; Dong et al., 2009). In addition, the control exerted by hsa-lnc-31 on ROCK1 expression is not conserved because the hsa-lnc-31 depletion did not have any effect on ROCK1 protein and mRNA levels (Figure S3C)
To test whether lnc-31, through its interaction with Rock1 mRNA, was able to promote the formation of the YB-1/Rock1 mRNA complex, we performed UV-CLIP assays with YB-1 antibodies in myoblasts treated with siRNAs against lnc-31. Figure 3D shows that in the absence of lnc-31, YB-1 was still able to bind Rock1 mRNA, indicating that lnc-31 was not required for the download of YB-1 onto the Rock1 mRNA. However, the absence of lnc-31 affected greatly the overall levels of the YB-1 protein, despite the unaltered accumulation of its mRNA levels (Figures 3E and S3D). The YB-1 protein but not the mRNA became upregulated upon lnc-31 overexpression (Figures 3E and S3D). These data suggested that lnc-31 may control Yb-1 translation, protein stability, or both. To address this point, we treated C2C12 cells with the protein synthesis inhibitor cycloheximide (CHX) in the presence of SCR or si1-lnc-31 siRNAs. Figure 3F shows that, in si1-lnc-31-treated cells, the half-life of YB-1 is markedly reduced, suggesting that lnc-31 affects mainly YB-1 stability.
Because it has been reported that the 20S proteasome is able to degrade YB-1 in a ubiquitin- and an ATP-independent manner and that this event is inhibited by the association of YB-1 with target RNAs (Sorokin et al., 2005, Wei et al., 2016), we tested whether the decrease in the YB-1 protein, upon lnc-31 depletion, was caused by protein degradation. The treatment of C2C12 cells with the proteasome inhibitor MG132, in association with the depletion of lnc-31, gave rise to the consistent stabilization of YB-1 (Figure 3G). As control, the levels of the CDKN1B protein, a known target of the 20S proteasome, increased upon MG132 treatment (Besson et al., 2006). Moreover, the knock down of lnc-31 in the presence of MG132 did not cause any increase in YB-1 ubiquitination (data not shown). In conclusion, all of these experiments indicate that lnc-31 controls the stability of YB-1 protein by limiting its degradation in a ubiquitin-independent/proteasome-dependent manner.
Notably, the lnc-31-dependent stabilization of YB-1 did not have any effect on the translation of c-Myc mRNA, known to be controlled by YB-1 but not to be bound by lnc-31 (Figures S4A and 4B; Cobbold et al., 2010). Moreover, lnc-31 depletion caused the increase in cMYC-nick (Figure S4B), the product of calpain-mediated cleavage of the full-length protein, previously described as occurring in differentiating myoblasts (Conacci-Sorrell et al., 2010), which indicates again the pro-differentiation effects of lnc-31 depletion.
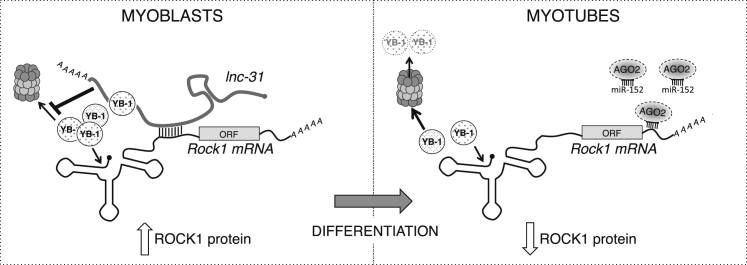
Figure 4.
lnc-31 Interacts with Rock1 mRNA and Mediates Its YB-1-Dependent Translation
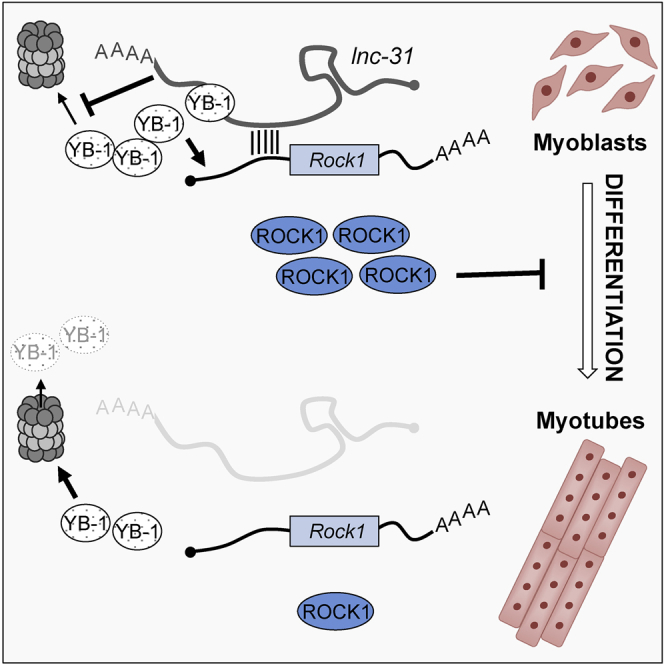
Schematic representation of lnc-31 mode of action. In proliferating myoblasts (left), lnc-31 promotes the translation of Rock1 through the binding of its mRNA and the inhibition of proteasome-mediated YB-1 degradation. In myotubes (right), the poor levels of lnc-31 are not sufficient to sustain such activities leading to the downregulation of Rock1 expression. This loop would be further reinforced by the upregulation during myogenesis of miR-152, making it possible to establish robust negative control of Rock1 expression.
Linking this effect to the specificity of lnc-31/Rock1 mRNA interaction and to the requirement of lnc-31 for efficient Rock1 translation, we suggest that in proliferating myoblasts, lnc-31, when bound to Rock1 mRNA, can locally control the stability of YB-1 on Rock1 mRNA (Figure 4). Upon differentiation, the low levels of lnc-31 would contribute to ROCK1 downregulation through the reduced stabilization of YB-1. This loop would be reinforced further by the upregulation during myogenesis of miR-152, making it possible to establish robust negative control of Rock1 expression (Figure 4).
Because Rock1 mRNA has a highly structured GC-rich (≈70%) 5′ UTR (Figure S3C), which also has the potential to form a G-quadruplex (Figure S3D; Kikin et al., 2006), and in consideration of the ability of YB-1 to melt secondary structures, induce structural rearrangements, or both (Evdokimova et al., 2006, Evdokimova et al., 2009, El-Naggar et al., 2015, Skabkin et al., 2001), the hypothesis that YB-1 could positively affect Rock1 translation by favoring the structural remodeling of its 5′ UTR is attractive.
Experimental Procedures
Cell Culture and Treatments
Mouse myoblasts (C2C12, American Type Culture Collection [ATCC]) were cultured in growth medium (GM; DMEM with 20% fetal bovine serum [FBS], 2 mM l-glutamine, and penicillin/streptomycin) and differentiated using a differentiation medium (DM) containing 0.5% FBS. HeLa cells were cultured in DMEM with 10% FBS, 2 mM l-glutamine, and penicillin/streptomycin. Human myoblasts were cultured in GM (DMEM with 10% FBS, 2 mM l-glutamine, 50 mg/mL insulin, 25 ng/mL basic fibroblast growth factor (FGFb), 1 ng/mL epidermal growth factor (EGF), and penicillin/streptomycin.
C2C12, human myoblasts, and HeLa cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific), according to the manufacturer’s instructions. For plasmid transfection, 2 μg DNA was used for plnc-31 and plnc-31mut constructs, while 40 ng DNA was used for all of the psiCHECK- constructs; for siRNA transfection, a final concentration of 80 nM was used for SCR (AllStars Negative Control siRNA, QIAGEN) and si-lnc31, and a final concentration of 50 nM was used for SCR (siGENOME Non-Targeting siRNA, Dharmacon) and si-YB-1 (siGENOME Mouse Ybx1 [22608]SMARTpool, Dharmacon). Luciferase assays were performed using the Dual Glo luciferase assay (Promega), according to the manufacturer’s protocol.
For all of the treatments, cells were harvested for protein and RNA analyses after 48 hr. For the CHX chase, CHX at a final concentration of 100 μg/mL was added to the medium. To inhibit proteasome, MG132 at a final concentration of 50 μM, was used, while DMSO was added to the medium of the control samples.
RNA Preparation and Analysis
Total RNA was extracted using the Direct-zol RNA MiniPrep kit (Zymo Research) with on-column DNase treatment, according to the manufacturer’s instructions. For the UV-CLIP and RNA pull-down experiments, the RNA was extracted using QIAzol reagent and miRNEasy spin columns (QIAGEN), according to the manufacturer’s specifications.
Reverse transcription was carried out with SuperScript VILO cDNA Synthesis Kit (Life Technologies) and the cDNA samples were analyzed by qRT-PCR using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific). For miRNA detection, the cDNA synthesis was carried out using miScript II RT Kit (QIAGEN), and the qRT-PCR was performed using miScript SYBR Green PCR Kit (QIAGEN).
Native Pull-Down
C2C12 myoblasts cultured in GM were washed twice with complete PBS and collected in 1 mL of buffer A (Tris-HCl, pH 8, 20 mM; NaCl, 10 mM; MgCl2, 3 mM; NP-40, 0.1%; glycerol, 10%; DTT, 1 mM; protease and RNase inhibitor), incubated on ice for 10 min, and then centrifuged at 2500 rpm for 5 min. The supernatant representing cytoplasmic extracts was recovered while pellets were discarded. The protein concentration of the cytoplasmic extracts was assessed by Bradford assay. Cytoplasmic extract, 1 mg, was used for each sample (Odd, Even, and LacZ) and was precleared with 50 μL Promega Streptavidin MagneSphere Paramagnetic Particles, equilibrated in hybridization buffer (Tris-HCl, pH 7.4, 50 mM; NaCl, 150 mM; MgCl2, 1 mM; NP-40, 0.05%; EDTA, 10 mM; DTT, 1 mM), and supplemented with protease and RNase inhibitors, for 30 min on a rotating wheel at room temperature. A total of 1 μL biotinylated oligonucleotides mix (20 μM each) was added to the precleared extracts and incubated for 2 hr at room temperature on a rotating wheel. Streptavidin MagneSphere Paramagnetic Particles, 50 μL, were added to each sample and incubated for 30 min at room temperature on a rotating wheel. After the incubation, the paramagnetic particles were collected through a magnetic support and washed three times with 1 mL hybridization buffer. Finally, the paramagnetic particles were divided in two aliquots for RNA and protein analyses.
The sequences of the biotinylated oligonucleotides belonging to Odd, Even, and LacZ sets are listed in the Supplemental Experimental Procedures.
Protein Analysis
Cells were harvested with protein extraction buffer (Tris, pH 7.5, 100 mM; EDTA, 1 mM; SDS, 2%; protease inhibitor cocktail [PIC] 1x (cOmplete, EDTA-free, Roche), incubated for 10 min on ice, and centrifuged at 13,000 rpm for 10 min at 4°C. Proteins (15–30 μg) were loaded on 4%–12% bis-Tris-acrylamide gel (Life Technologies) and transferred to a nitrocellulose membrane. The membrane was blocked in 5% milk and hybridized with the following antibodies: YB-1 (ab76149, Abcam), PNN (A301-022A-M, Bethyl), ROCK1 (c8f7, Cell Signaling), eEF2 (2332S, Cell Signaling), TUBA6 (ab-191299, Abcam), HNRPM (FL-218 sc-20975, Santa Cruz Biotechnology), anti-cMyc (D84C12, Cell Signaling), anti-CDKN1B (sc-1641, Santa Cruz Biotechnology), anti-ACTIN (A3854, Sigma), and anti-ACTININ (H-300, sc-15335, Santa Cruz Biotechnology); with VeriBlot for secondary antibody (ab131366, Abcam). All of the images were captured using the Molecular Imager ChemiDoc XRS+ (Bio-Rad), and the densitometric analyses were performed using the associated Image Lab software (Bio-Rad).
UV-CLIP Assay
C2C12 and human myoblasts cultured in GM were UV-crosslinked at 4,000 × 100 μJ cm−2 energy. Cells were resuspended in buffer A (as described above), incubated for 10 min on ice, and then centrifuged at 2500 rpm for 5 min. Cytoplasmic extracts (supernatant) were recovered while pellets were discarded. The protein concentration was assessed by Bradford assay. A total of 1 mg cytoplasmic extract was used for each sample (immunoprecipitation [IP] and immunoglobulin G [IgG]) and precleared with 40 μL protein G agarose beads (Millipore) for 1 hr on a rotating wheel in 1 mL NT2 buffer final volume (NT2: Tris-HCl, pH 7.5, 50 mM; NaCl, 150 mM; MgCl2, 1 mM; NP-40, 0.25%; protease and RNase inhibitor). A total of 10% of the final volume was recovered as input (IN), while the remaining 900 μL was incubated with 10 μg of YB-1 or IgG (rabbit sc-2027) antibodies for 1 hr at 4°C. Protein G agarose beads, 50 μL, equilibrated in NT2 buffer, were added to each sample and incubated for 2 hr at 4°C. The beads were then collected through centrifugation at 2000 rpm and washed three times with 1 mL NT2 buffer and three times with NT2-HS buffer (Tris-HCl, pH 7.5, 50 mM; NaCl, 500 mM; MgCl2, 1 mM; NP-40, 0.25%; protease and RNase inhibitor). The beads were finally resuspended in 200 μL radioimmunoprecipitation assay (RIPA) buffer (Tris-HCl, pH 7, 10 mM; NaCl, 100 mM; EDTA, 1 mM; SDS, 0.5%), while 100 μL of the same buffer was added to the IN samples. A total of 50 μL of each sample was used for protein analysis. For the RNA extraction, IP, IgG, and IN samples were incubated for 1 hr at 70°C in the presence of 7.5 μL proteinase K (Ambion) and RNase inhibitor. Five volumes of TRIzol were added to each sample, and the RNA extraction and the cDNA generation were performed as described above.
Acknowledgments
This work was partially supported by grants from the European Research Council 2013 (AdG 340172–MUNCODD), the Italian Foundation for Research on Amyotrophic Lateral Sclerosis (AriSLA) full grant 2014 (ARCI), Téléthon (GGP16213), the Epigen-Epigenomics Flagship Project, the Human Frontiers Science Program Award RGP0009/2014, AFM-Téléthon (17835), and Fondazione Roma (call 2013).
Author Contributions
D.D., M.M., and I.B. conceptualized the project and designed the experiments. D.D., M.B., J.M., and D.M. performed the experiments in C2C12, human myoblasts and HeLa cells. A.C. performed the data analysis of all of the RNA sequencing data. J.D. and A.B. performed the mass spectrometry and data analysis. M.M., G.M., and I.B. supervised the experiments. M.M. and I.B. wrote the manuscript. I.B. acquired the funding for the study. All of the authors approved the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: April 17, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.101.
Contributor Information
Mariangela Morlando, Email: mariangela.morlando@uniroma1.it.
Irene Bozzoni, Email: irene.bozzoni@uniroma1.it.
Data and Software Availability
The accession number for all RNA-seq data reported in this paper is GEO: GSE102158.
Supplemental Information
The genomic coordinates and the p value of the overlapping peaks identified by Piranha software are reported.
References
- Ballarino M., Cazzella V., D’Andrea D., Grassi L., Bisceglie L., Cipriano A., Santini T., Pinnarò C., Morlando M., Tramontano A., Bozzoni I. Novel long noncoding RNAs (lncRNAs) in myogenesis: a miR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol. Cell. Biol. 2015;35:728–736. doi: 10.1128/MCB.01394-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballarino M., Morlando M., Fatica A., Bozzoni I. Non-coding RNAs in muscle differentiation and musculoskeletal disease. J. Clin. Invest. 2016;126:2021–2030. doi: 10.1172/JCI84419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson A., Gurian-West M., Chen X., Kelly-Spratt K.S., Kemp C.J., Roberts J.M. A pathway in quiescent cells that controls p27Kip1 stability, subcellular localization, and tumor suppression. Genes Dev. 2006;20:47–64. doi: 10.1101/gad.1384406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bois P.R., Grosveld G.C. FKHR (FOXO1a) is required for myotube fusion of primary mouse myoblasts. EMBO J. 2003;22:1147–1157. doi: 10.1093/emboj/cdg116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacchiarelli D., Incitti T., Martone J., Cesana M., Cazzella V., Santini T., Sthandier O., Bozzoni I. miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep. 2011;12:136–141. doi: 10.1038/embor.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S., Comunale F., Grumbach Y., Poulat F., Blangy A., Gauthier-Rouvière C. RhoA GTPase regulates M-cadherin activity and myoblast fusion. Mol. Biol. Cell. 2006;17:749–759. doi: 10.1091/mbc.E05-04-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold L.C., Wilson L.A., Sawicka K., King H.A., Kondrashov A.V., Spriggs K.A., Bushell M., Willis A.E. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene. 2010;29:2884–2891. doi: 10.1038/onc.2010.31. [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell M., Ngouenet C., Eisenman R.N. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480–493. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Akcakanat A., Stivers D.N., Zhang J., Kim D., Meric-Bernstam F. RNA-binding specificity of Y-box protein 1. RNA Biol. 2009;6:59–64. doi: 10.4161/rna.6.1.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Naggar A.M., Veinotte C.J., Cheng H., Grunewald T.G., Negri G.L., Somasekharan S.P., Corkery D.P., Tirode F., Mathers J., Khan D. Translational activation of HIF1α by YB-1 promotes sarcoma metastasis. Cancer Cell. 2015;27:682–697. doi: 10.1016/j.ccell.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Evdokimova V., Ruzanov P., Anglesio M.S., Sorokin A.V., Ovchinnikov L.P., Buckley J., Triche T.J., Sonenberg N., Sorensen P.H. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol. Cell. Biol. 2006;26:277–292. doi: 10.1128/MCB.26.1.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V., Tognon C., Ng T., Ruzanov P., Melnyk N., Fink D., Sorokin A., Ovchinnikov L.P., Davicioni E., Triche T.J., Sorensen P.H. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Iwasaki K., Hayashi K., Fujioka T., Sobue K. Rho/Rho-associated kinase signal regulates myogenic differentiation via myocardin-related transcription factor-A/Smad-dependent transcription of the Id3 gene. J. Biol. Chem. 2008;283:21230–21241. doi: 10.1074/jbc.M710525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Kim J., Willert T., Klein-Rodewald T., Garcia-Dominguez M., Mosqueira M., Fink R., Esposito I., Hofbauer L.C., Charnay P., Kieslinger M. Ebf factors and MyoD cooperate to regulate muscle relaxation via Atp2a1. Nat. Commun. 2014;5:3793. doi: 10.1038/ncomms4793. [DOI] [PubMed] [Google Scholar]
- Kikin O., D’Antonio L., Bagga P.S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurila E.M., Kallioniemi A. The diverse role of miR-31 in regulating cancer associated phenotypes. Genes Chromosomes Cancer. 2013;52:1103–1113. doi: 10.1002/gcc.22107. [DOI] [PubMed] [Google Scholar]
- Lazaro J.-B., Bailey P.J., Lassar A.B. Cyclin D-cdk4 activity modulates the subnuclear localization and interaction of MEF2 with SRC-family coactivators during skeletal muscle differentiation. Genes Dev. 2002;16:1792–1805. doi: 10.1101/gad.U-9988R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.J., Tsai M.M., Hung P.S., Kao S.Y., Liu T.Y., Wu K.J., Chiou S.H., Lin S.C., Chang K.W. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70:1635–1644. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- Lyabin D.N., Eliseeva I.A., Ovchinnikov L.P. YB-1 protein: functions and regulation. Wiley Interdiscip. Rev. RNA. 2014;5:95–110. doi: 10.1002/wrna.1200. [DOI] [PubMed] [Google Scholar]
- Nekrasov M.P., Ivshina M.P., Chernov K.G., Kovrigina E.A., Evdokimova V.M., Thomas A.A., Hershey J.W., Ovchinnikov L.P. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J. Biol. Chem. 2003;278:13936–13943. doi: 10.1074/jbc.M209145200. [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Kii I., Kudo A. Inactivation of Rho/ROCK signaling is crucial for the nuclear accumulation of FKHR and myoblast fusion. J. Biol. Chem. 2004;279:47311–47319. doi: 10.1074/jbc.M403546200. [DOI] [PubMed] [Google Scholar]
- Ohashi S., Moue M., Tanaka T., Kobayashi S. Translational level of acetylcholine receptor α mRNA in mouse skeletal muscle is regulated by YB-1 in response to neural activity. Biochem. Biophys. Res. Commun. 2011;414:647–652. doi: 10.1016/j.bbrc.2011.09.086. [DOI] [PubMed] [Google Scholar]
- Rashid F., Shah A., Shan G. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics. 2016;14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Cheedipudi S., Pasupuleti N., Saleh A., Pavlath G.K., Dhawan J. The small chromatin-binding protein p8 coordinates the association of anti-proliferative and pro-myogenic proteins at the myogenin promoter. J. Cell Sci. 2009;122:3481–3491. doi: 10.1242/jcs.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin M.A., Evdokimova V., Thomas A.A., Ovchinnikov L.P. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J. Biol. Chem. 2001;276:44841–44847. doi: 10.1074/jbc.M107581200. [DOI] [PubMed] [Google Scholar]
- Skabkina O.V., Lyabin D.N., Skabkin M.A., Ovchinnikov L.P. YB-1 autoregulates translation of its own mRNA at or prior to the step of 40S ribosomal subunit joining. Mol. Cell. Biol. 2005;25:3317–3323. doi: 10.1128/MCB.25.8.3317-3323.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin A.V., Selyutina A.A., Skabkin M.A., Guryanov S.G., Nazimov I.V., Richard C., Th’ng J., Yau J., Sorensen P.H., Ovchinnikov L.P., Evdokimova V. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 2005;24:3602–3612. doi: 10.1038/sj.emboj.7600830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Ohashi S., Moue M., Kobayashi S. Mechanism of YB-1-mediated translational induction of GluR2 mRNA in response to neural activity through nAChR. Biochim. Biophys. Acta. 2012;1820:1035–1042. doi: 10.1016/j.bbagen.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Tintignac L.A., Leibovitch M.P., Kitzmann M., Fernandez A., Ducommun B., Meijer L., Leibovitch S.A. Cyclin E-cdk2 phosphorylation promotes late G1-phase degradation of MyoD in muscle cells. Exp. Cell Res. 2000;259:300–307. doi: 10.1006/excr.2000.4973. [DOI] [PubMed] [Google Scholar]
- Wei M.M., Zhou Y.C., Wen Z.S., Zhou B., Huang Y.C., Wang G.Z., Zhao X.C., Pan H.L., Qu L.W., Zhang J. Long non-coding RNA stabilizes the Y-box-binding protein 1 and regulates the epidermal growth factor receptor to promote lung carcinogenesis. Oncotarget. 2016;7:59556–59571. doi: 10.18632/oncotarget.10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wang Q., Zhao D., Cui Y., Cao B., Guo L., Lu S.H. The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin. Sci. (Lond.) 2011;121:437–447. doi: 10.1042/CS20110207. [DOI] [PubMed] [Google Scholar]
- Zhang J., Ying Z.Z., Tang Z.L., Long L.Q., Li K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 2012;287:21093–21101. doi: 10.1074/jbc.M111.330381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The genomic coordinates and the p value of the overlapping peaks identified by Piranha software are reported.