Abstract
In photosynthetic organisms it is becoming increasingly evident that light-driven shifts in redox potential act as a sensor that initiates alterations in gene expression at both the level of transcription and translation. This report provides evidence that the expression of a cyanobacterial RNA helicase gene, crhR, is controlled at the level of transcription and mRNA stability by a complex series of interacting mechanisms that are redox regulated. Transcript accumulation correlates with reduction of the electron transport chain between QA in photosystem II and QO in cyt b6f, when Synechocystis sp. strain PCC 6803 is cultured photoautotrophically or photomixotrophically and subjected to darkness and/or electron transport inhibitors or illumination that preferentially excites photosystem II. crhR mRNA stability is also regulated by a redox responsive mechanism, which differs from that affecting accumulation and does not involve signaling initiated by photoreceptors. The data are most consistent with plastoquinol/cyt b6f interaction as the sensor initiating a signal transduction cascade resulting in accumulation of the crhR transcript. Functionally, CrhR RNA unwinding could act as a linker between redox regulated transcription and translation. The potential for translational regulation of redox-induced gene expression through RNA helicase-catalyzed modulation of RNA secondary structure is discussed.
Photosynthetic organisms must have the capacity to respond rapidly to changes in their light environment and therefore possess a sophisticated array of mechanisms to sense light quality, quantity, direction, and duration (Elich and Chory, 1997; Fankhauser and Chory, 1997, 1999). Although the regulation of gene expression as a result of light sensing by a variety of photoreceptors is well documented, it has recently become clear that expression can also be modulated by light-induced electron transport. Specifically, changes in illumination are sensed by the cells as a shift in the redox equilibrium of the plastoquinone pool (Pfannschmidt et al., 1999).
Light-induced alterations in the redox state of plastoquinone have been reported to have two main consequences, state transitions that maximize the efficiency of light harvesting and alterations in gene expression. Although the effects of the redox status of plastoquinone on state transitions are well established (Allen et al., 1989; Allen, 1992; Gal et al., 1997; Vener et al., 1998), evidence for effects on gene expression are not as well documented. In photosynthetic eukaryotes, redox signals sensed within the chloroplast have been shown to indirectly regulate nuclear gene expression. In Dunaliella tertiolecta transcription of the nuclear cab genes, which encode chlorophyll a/b-binding proteins, has been proposed to be coupled to light intensity via the redox state of plastoquinone (Escoubas, et al., 1995; Maxwell et al., 1995). In Arabidopsis the redox status of plastoquinone regulates the expression of two cytosolic ascorbate peroxidase genes during excess light stress (Karpinski et al., 1997). More recently it has been shown in mustard seedling chloroplasts that the net redox state of the plastoquinone pool directly influences the rate of transcription of chloroplast-encoded psaA and psbAB genes, resulting in rapid adjustment of the stoichiometry of the photosystem (PS) I and PSII reaction centers (Pfannschmidt et al., 1999). These and other results implicate a regulatory signal transduction cascade initiated by the activity of a redox-modulated thylakoid kinase (Allen et al., 1995; Escoubas, et al., 1995; Baginsky et al., 1997; Karpinski et al., 1997; Pfannschmidt et al., 1999). This is further supported by the demonstration that psbA expression in chloroplasts is differentially regulated by two ς-like transcription factors whose activation by phosphorylation is light dependent (Tiller and Link, 1993; Baginsky et al., 1997). Indeed it has been proposed that transcriptional regulation, and not state transitions, is the most important outcome of cellular redox sensing events mediated by the plastoquinone-plastoquinol equilibrium (Vener et al., 1998).
Evidence is also accumulating that redox signaling modulates expression of photosynthetic genes at the post-transcriptional level. In chloroplasts of the unicellular alga Chlamydomonas reinhardtii, translation of key photosynthetic mRNAs requires a multiprotein complex whose association with secondary structures in the 5′-untranslated region (UTR) of these mRNAs is modulated in a redox-responsive manner (Danon and Mayfield, 1994; Mayfield et al., 1994). In addition, mRNA stabilization mediated by polyribosome association and translation is required for the light-enhanced accumulation of Fed-1 mRNA in tobacco, whereas an inhibition of translation initiation and a repeat element in the 5′-UTR are involved in transcript instability in the dark (Dickey et al., 1998).
As a prokaryotic model of oxygenic photosynthesis, cyanobacterial transcriptional responses to light are also well documented (Mohamed and Jansson, 1989; Golden, 1995; Reyes and Florencio, 1995; Kis et al., 1998; Richter et al., 1998). In some cases the light-induced increase in transcript level has been correlated with photosynthetic electron transport (Reyes and Florencio, 1995; Kis et al., 1998). In addition, light may influence gene expression post-transcriptionally by redox-mediated effects on mRNA stability. This has been observed for the Synechocystis psbA-2 and psbA-3 transcripts whose increased stability in the dark is controlled by the cellular redox potential (Mohamed et al., 1993; Tyystjarvi et al., 1998). A similar situation has been proposed in Synechococcus, in which sequences in the 5′-UTR regions of the three psbA transcripts affect transcript turnover in response to light intensity, possibly by recruiting RNA-binding proteins required for translation or degradation (Kulkarni and Golden, 1997).
In determining the role performed by redox status in the regulation of cyanobacterial gene expression, it is significant that cyanobacteria differ from plant chloroplasts by possessing a common electron transport chain for electrons derived from photosynthetic light harvesting and respiration. In the cyanobacterial thylakoid membrane, the plastoquinone pool has an established role as the common point of electron entry from both PSII (photosynthesis) and NAD(P) H-dehydrogenases (respiration; Hirano et al., 1980; Scherer, 1990). Therefore the redox status of the plastoquinone pool depends on photosynthetic light harvesting in addition to the metabolism of endogenous respiratory substrates. In fact, respiratory electron flow can effect state transitions (Mullineaux and Allen, 1986). Whereas the majority of cyanobacteria are photoautotrophic, Synechocystis possesses the additional ability to grow at the expense of a limited number of exogenously-supplied carbon sources. We have utilized this photomixotrophic phenotype, in conjunction with various electron transport inhibitors and preferential excitation of each photosystem, to investigate the role of redox signaling in gene expression in Synechocystis.
We show that the light-responsive accumulation of an RNA helicase-encoding transcript is mediated by the redox state of the electron transport chain, most probably as a result of signaling of the redox state of plastoquinone through plastoquinol interaction with the cyt b6f complex, in the cyanobacterium Synechocystis. We have therefore designated this gene crhR (cyanobacterial RNA helicase redox). Treatments predicted to increase electron flow between QA in PSII and QO in cyt b6f enhance crhR mRNA and CrhR protein accumulation. crhR transcript accumulation is also regulated at the post-transcriptional level since crhR mRNA stability responds to the overall cellular redox state. CrhR protein is constitutively present regardless of the redox state. The deduced amino acid sequence of crhR is characteristic of the DEAD-box family of RNA helicases, proteins that unwind duplex regions of RNA, thereby altering the availability of the substrate RNA for participation in subsequent metabolic events (Linder et al., 1989; Schmid and Linder, 1992; Pause and Sonenberg, 1993; Fuller-Pace, 1994). We discuss the potential physiological significance of redox-regulated RNA helicase gene expression and the possible effects the resulting RNA unwinding activity may have on the translational regulation of redox-responsive mRNAs.
RESULTS
The Light-Responsive Pattern of crhR Expression Is Altered in Response to Glc Metabolism
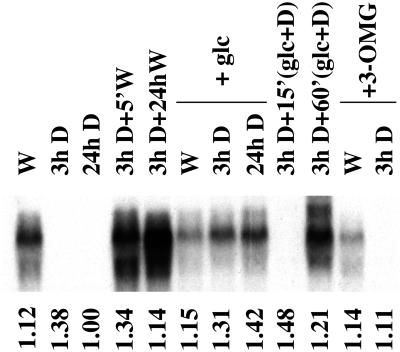
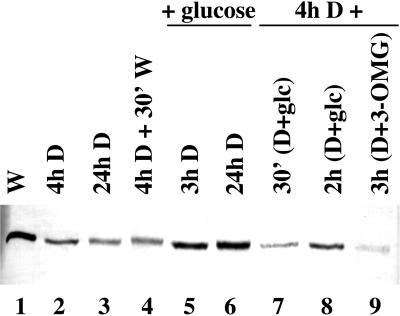
Northern-blot analysis of total RNA isolated from photoautotrophic Synechocystis cultures indicated that crhR transcript is constitutively detected during growth in the light, but is not detected within 3 h after onset of dark treatment and remains undetectable thereafter (Fig. 1, W, 3h D, and 24h D). Following a 2-h dark incubation, transcript accumulation is rapidly recovered by illumination, with detectable levels of crhR transcript observed within 5 min (Fig. 1, 3h D + 5′W) and remains constitutively expressed in the light thereafter (Fig. 1, 3h D + 24hW). The dramatic response of crhR transcript accumulation to alterations in the light regime is not a result of differential RNA loading as the levels of a control RNA, RNase P, are not altered significantly by these treatments (Fig. 1, relative RNase P levels shown below each lane). This demonstration that crhR expression is responsive to light, both to initiate and to maintain transcript accumulation, prompted further investigation into the specific mediator of this light signal.
Figure 1.
crhR transcript accumulation in response to illumination and/or exogenous carbon sources. A DNA probe internal to the crhR coding region was used to detect the crhR transcript in total RNA (6 μg) extracted from mid-log phase Synechocystis cultures grown photoautotrophically, or with addition of Glc (5 mm) or 3-OMG (5 mm), and treated as indicated. The 1.6-kb crhR transcript is shown. W, White light; D, darkness. Relative RNase P transcript levels are shown below each lane as a control for RNA loading.
Although the majority of cyanobacteria are obligate photoautotrophs, Synechocystis possesses the ability to import and respire appropriate exogenously-supplied organic carbon compounds. Since the presence of exogenous Glc stimulates respiration and has effects on photosynthetic activity, crhR expression was determined after light and dark treatment of cultures grown in the presence of Glc (5 mm). Although Glc does not alter crhR transcript accumulation in photoautotrophically grown cells (Fig. 1, compare lanes W and W+glc), levels observed in the dark are significantly affected. crhR transcript levels similar to those observed during continuous growth in white light are constitutively detected in cells after transfer into darkness in the presence of Glc (Fig. 1, W+glc, 3h D+glc, and 24h D+glc). Glc induction of transcript accumulation in the dark is in stark contrast to the deficiency of transcript in cultures grown in the absence of Glc (Fig. 1, compare 3h D and 24h D with 3h D+glc and 24h D+glc). Since these cultures were grown in the presence of light and Glc, it was necessary to determine whether the addition of Glc alone is sufficient to induce crhR expression, or whether a synergistic interaction with light is required. For this purpose, a photoautotrophic culture was incubated in the dark for 3 h, at which point crhR transcript was no longer detectable. Subsequent Glc addition was performed in a darkroom under a red safelight, a condition that did not induce crhR expression (data not shown). crhR transcript was not detected 15 min after Glc addition during continued incubation in the dark, but was detected within 60 min (Fig. 1, 3h D+15′glc+D and 3h D+60′glc+D), and was maintained in the absence of light for a minimum of 48 h (data not shown). From these results it is apparent that Glc induces crhR transcript accumulation in dark-treated cultures, but accumulation is slower in comparison to the induction observed by illumination. These results suggest that the Glc-induced dark accumulation of crhR transcript requires metabolism of the exogenous Glc.
To confirm that respiration of the exogenous Glc is a prerequisite for induction of crhR transcript accumulation, cells were grown in the presence of O-methyl-d-gluco-pyranose (3-OMG; 5 mm), a Glc analog that is transported into, but not metabolized by, Synechocystis (Flores and Schmetterer, 1986). In the presence of 3-OMG, crhR transcript declines to undetectable levels after a 3-h dark treatment (Fig. 1, W+3-OMG and 3h D+3-OMG), results identical to those observed in dark-treated photoautotrophic cells. This decline is, however, in contrast to the continued accumulation of crhR transcripts in dark-treated cultures grown in the presence of Glc. Thus metabolism of the exogenous Glc is required to induce crhR transcript accumulation in the dark.
Cellular Redox State Affects crhR Transcript Accumulation
Since it is well established that photosynthetic and respiratory electron transport share electron carriers in the cyanobacterial thylakoid membrane, the observed pattern of crhR expression suggested that redox changes could be the signal that regulates crhR transcript accumulation. To confirm the regulation by redox potential and to differentiate between the involvement of either a particular electron carrier or the overall cellular redox state, crhR transcript accumulation was determined following treatment of cultures with electron transport inhibitors that interrupt either respiratory or photosynthetic electron transport or both. Specifically, 3-(3, 4-dichlorophenyl)-1,1-dimethylurea (DCMU) inhibits electron transfer from PSII to plastoquinone, whereas 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) inhibits electron exit from plastoquinone to the cytochrome b6f complex (Trebst, 1980) and thus the two inhibitors have opposite effects on the net redox state of the plastoquinone pool: oxidation by DCMU and enhanced reduction by DBMIB.
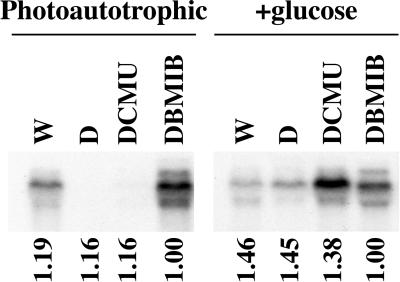
DCMU and DBMIB differentially affected crhR transcript accumulation and the effects are altered by the metabolism of exogenous Glc (Fig. 2). DCMU treatment of photoautotrophic cultures results in a significant decline in crhR transcript levels, in contrast to DBMIB treatment, which enhances accumulation. These inhibitors, however, enhance crhR transcript levels in illuminated and dark-treated cultures grown in the presence of Glc. Although DCMU and DBMIB interrupt light-generated (photosynthetic) linear electron flow between the two photosystems, only the block imposed by DCMU can be compensated for by Glc respiration. This is accomplished by the enhanced respiration of Glc, which occurs in the light in the presence of DCMU, with the resulting electrons entering the electron transport chain downstream of the blockage, at plastoquinone. The enhanced level of transcript accumulation observed in the presence of DCMU and Glc differs, however, as electrons generated by respiration are supplied to plastoquinone at a level that would not be expected to increase accumulation above that observed in cells grown in the dark plus Glc (Fig. 2, D+glc). DBMIB enhancement of transcript accumulation occurs irrespective of the presence of exogenous Glc since electrons are being supplied by PSII. These results indicate transcript accumulation is dependent on the reduction of electron carriers between QA in PSII and QO in cyt b6f.
Figure 2.
crhR transcript accumulation in response to electron transport inhibitors. Total RNA was extracted from illuminated mid-log phase Synechocystis cultures grown photoautotrophically or photoautotrophically with Glc, then treated with darkness for 2 h or with continued illumination in the presence of the indicated electron transport inhibitors for 1 h. crhR transcript abundance was determined as described in Figure 1. W, White light; D, darkness. Relative RNase P transcript levels are shown below each lane as a control for RNA loading.
Light Quality Modulates crhR Transcript Accumulation
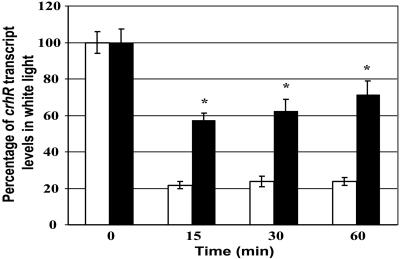
Additional confirmation that the redox status of the electron transport chain between QA in PSII and QO in cyt b6f mediates crhR transcript accumulation was obtained by determining crhR transcript accumulation under conditions designed to preferentially excite each photosystem independently. Cultures were grown in white light, followed by the indicated filtered illumination for 15, 30, and 60 min. The light 1 (L1) filter, with a transmittance spectrum that gives 50% transmittance at 650 nm and 0% below 580 nm, provides light that preferentially excites PSI and shifts the redox equilibrium to the oxidized state. The light 2 (L2) filter, giving 50% transmittance at 560 nm, preferentially excites PSII shifting the redox equilibrium to the reduced form. Figure 3 shows that at all time points crhR transcript levels are significantly elevated by illumination with L2 light compared with L1 light, namely when reducing conditions predominate. These results also indicate that alterations in the redox state of the electron transport chain between QA in PSII and QO in cyt b6f are the signals that initiate the light-induced accumulation of crhR transcripts.
Figure 3.
crhR transcript accumulation during preferential stimulation of PSI (L1, 650 nm) or PSII (L2, 560 nm). Photoautotrophic Synechocystis cells cultured in white light (0 time) and transferred to either L1 (white bars) or L2 (black bars) light for the indicated times. crhR transcript levels, determined as described in Figure 1, are expressed as a percentage of transcript detected in white light (0 time). L1 preferentially excites PSI and oxidizes the plastoquinone pool, whereas L2 preferentially excites PSII and reduces the plastoquinone pool. Statistical analysis was performed by ANOVA followed by Fisher's lsd test. Pooled results from three independent cultures are presented as the mean ± se. CrhR transcript levels were not significantly different within either the L1 or L2 time series. Asterisk indicates L2 transcript levels that are significantly different from the corresponding L1 level.
crhR mRNA Stability Is Regulated Differentially from Accumulation
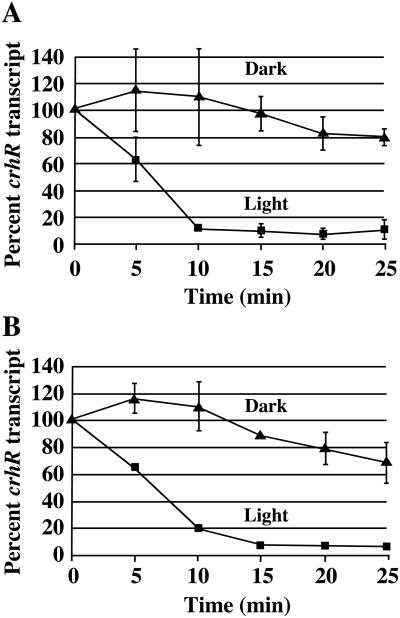
Since differential effects of light on mRNA stability may affect transcript levels in cyanobacteria, it was important to determine the crhR transcript half-life. Rifampicin was added simultaneously with light or dark treatment to cultures grown in the presence or absence of Glc, followed by determination of crhR transcript levels. Under photoautotrophic conditions the crhR transcript half-life is significantly shorter in the light than in the dark, with values of 6 and 36 min, respectively (Fig. 4A). Essentially identical half-lives were observed in Glc grown cultures, with the crhR transcript half-life again being shorter in the light than in the dark, with values of 6 and 38 min, respectively (Fig. 4B). Evidently, mRNA stability contributes significantly to the regulation of crhR transcript accumulation, with crhR transcript being more stable in the dark. Furthermore, although crhR transcript stability is affected by light versus darkness, Glc metabolism did not affect the half-life. This indicates that the post-transcriptional control of crhR mRNA stability occurs via a mechanism that responds to light, but is separate from the transcriptional regulation mediated by the redox state of plastoquinone.
Figure 4.
crhR transcript stability is differentially regulated by light and Glc. crhR transcript levels are expressed as a percentage of crhR mRNA in illuminated cultures. Statistical analysis was performed as described in Figure 3. A, Photoautotrophic Synechocystis cultures were treated with rifampicin and white light or darkness for the times indicated. Statistical analysis indicates all time points except 0 and 5 min are significantly different between light and dark treatments. B, Cultures grown in the presence of Glc were treated with rifampicin and white light or darkness for the times indicated. Statistical analysis indicates all time points except 0 are significantly different between light and dark treatments.
We then asked whether the light-induced decrease in crhR transcript stability involved light sensing by photoreceptors or light-induced effects on a cellular redox component other than between QA and QO. When rifampicin was added simultaneously with DCMU or DBMIB to illuminated cultures grown in the presence or absence of Glc, crhR transcript half-lives increased significantly, becoming stable over the 30-min time course of the experiment (data not shown). Inhibition of linear electron flow therefore abolishes the light-dependent regulation of crhR transcript stability, suggesting that crhR transcript stability responds to cellular redox conditions and not specifically to either the redox state of the electron transport chain between QA and QO or to photoreceptor-mediated light sensing.
CrhR Protein Levels Vary from Transcript Accumulation
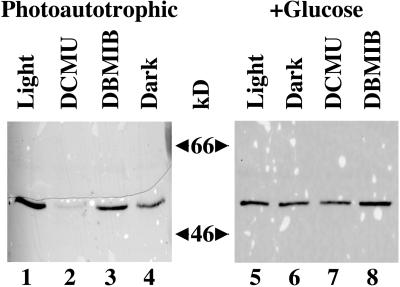
In contrast to crhR transcript levels, CrhR protein is observed constitutively in Synechocystis regardless of the transcript level (Fig. 5). Comparison of the relative levels of CrhR protein in response to shifts in the redox state of the electron transport chain indicates, however, that they respond similarly to the observed alterations in crhR mRNA accumulation (Fig. 5). CrhR protein levels respond to light-dark cycles as the level detected in photoautotrophic cultures grown in the light declines after 4 and 24 h in the dark (Fig. 5, lanes 1–3) and then increase upon subsequent illumination (Fig. 5, lane 4). Dark treatment of photomixotrophic cultures had no affect as CrhR protein levels remain at the level seen in photoautotrophic cultures (Fig. 5, compare lanes 5 and 6 with lane 1). The decline in CrhR levels observed in photoautotrophic cultures incubated in the dark for 4 h is also reversed, in a time-dependent and light-independent manner, by Glc addition and continued incubation in the dark (Fig. 5, lanes 7 and 8). The rate at which Glc induces the recovery of protein levels in the dark is slower than that observed in the light, as observed for transcript accumulation (compare lanes 4 and 7). Finally, as expected from transcript analysis and in contrast to exogenous Glc, 3-OMG does not induce CrhR protein accumulation in the dark (Fig. 5, lane 9). The detection of CrhR protein under all growth conditions indicates that a basal level of CrhR protein is present in the cells regardless of the cellular redox state.
Figure 5.
Redox regulation of CrhR protein accumulation. Synechocystis cell-free extracts were obtained from cultures grown in the presence or absence of light and/or Glc or 3-OMG, as indicated. Total protein (15 μg) was separated on a 10% (w/v) SDS-PAGE gel, transferred to a nitrocellulose membrane, and immunostained with polyclonal antiserum generated against a CrhR His-tagged fusion protein. The 55-kD immunoreactive polypeptide corresponding to CrhR is shown. W, White light; D, darkness.
CrhR protein accumulation in the presence of the electron transport inhibitors is shown in Figure 6. As expected from transcript analysis, DCMU inhibition of electron flow reduces CrhR protein levels compared with those observed in photoautotrophic cultures (Fig. 6, lanes 1 and 2), whereas CrhR levels are not significantly altered by DBMIB, which permits electron flow to plastoquinone, but not past cyt b6f (Fig. 6, lane 3). In contrast to transcript accumulation, CrhR protein levels are not affected significantly by a 4-h dark treatment (Fig. 6, lane 4). In agreement with transcript accumulation, Glc does not alter protein levels in light, dark, or DBMIB-treated cells (Fig. 6, lanes 5, 6, and 8), whereas Glc does restore protein accumulation in the presence of DCMU (Fig. 6, lane 7). These results reveal a correlation between CrhR protein and crhR mRNA transcript accumulation in response to conditions that result in reduction of the electron transport chain between QA and QO, except in the dark. The correlation does not hold for conditions that result in oxidation of the electron transport components as CrhR protein is constitutively observed in dark treated cells, suggesting that the mechanisms regulating protein and transcript accumulation differ in the dark.
Figure 6.
Electron transport inhibitor effect on CrhR protein levels. CrhR protein was detected in total protein (15 μg) isolated from illuminated Synechocystis cultures grown in the absence (photoautotrophic) or presence of Glc and treated for 4 h in darkness or 3 h with the indicated electron transport inhibitors, as described in Figure 5.
DISCUSSION
The importance of environmental sensing by light-induced shifts in cellular redox status is becoming increasingly evident in photosynthetic organisms. To be specific, the expression of a minimum of seven genes has been reported to be regulated by the redox state of plastoquinone at either the transcriptional or post-transcriptional levels. A mechanism by which the regulation of gene expression at the two levels can be linked, however, has not been proposed. The results presented here indicate that expression of a cyanobacterial RNA helicase, crhR, is controlled by complex interactions between a series of redox-mediated mechanisms that regulate both transcription and mRNA stability. It is important to note that since RNA helicases have the potential to alter translational efficiency by modulation of RNA secondary structure, a redox-responsive RNA helicase may provide photosynthetic organisms with the ability to regulate expression of redox-responsive genes at the translational level.
An extensive series of observations indicate that crhR transcript accumulation is regulated by the redox status of the electron transport chain between the QA site of PSII and the QO site of cyt b6f. In photoautotrophic cultures, crhR transcript accumulation is not observed in the dark, but occurs constitutively in cells incubated in the light. Light sensing is not the sole regulator of transcript accumulation, since accumulation is also observed in dark-grown cells in the presence of exogenously-supplied Glc. Light harvesting and Glc metabolism donate electrons to a common electron transport chain in cyanobacteria. Interruption of this chain by electron transport inhibitors reveals that crhR transcript accumulation occurs only under conditions predicted to result in electron flow between QA and QO. Finally, altering the electron flow between the two photosystems by changing the spectral quality of illumination confirms the transcriptional effects observed as a result of the artificial interruption of electron flow. Enhancement of linear electron flow by preferential excitation of PSII results in transcript accumulation, whereas shifting the carriers to a relatively more oxidized state by preferential excitation of PSI significantly reduces crhR accumulation. The observed redox-induced effects on crhR transcript accumulation are most probably accompanied by corresponding changes in the rate of crhR transcription. In fact, similar effects on the rate of transcription mediated by light-induced changes in electron flow have been interpreted to result from corresponding alterations in the redox state of plastoquinone for the accumulation of the chloroplast-encoded psaA and psbAB transcripts in mustard (Pfannschmidt et al., 1999). The data reported here are consistent with this interpretation. In fact, activation of the signal transduction pathway leading to altered transcription rates may be initiated by plastoquinol binding to QO in cyt b6f. This interaction is known to activate the thylakoid protein kinase leading to state transitions (Allen et al., 1989; Allen, 1992; Vener et al., 1998; Gal et al., 1997).
Activation of the thylakoid protein kinase could also initiate a signal cascade leading to crhR transcription. Thus although the redox status of plastoquinone itself may not be the direct regulator of the signal cascade, it is still the key sensor of electron flow through the electron transport chain. This conclusion is supported by the observation that direct reduction of cyt b6f by cyclic electron flow is not sufficient to induce crhR transcript accumulation. If this were the case then transcript accumulation should be observed in cells exposed to DCMU in the light, which is not the case. The DBMIB results are also compatible with this interpretation as DBMIB is known to occupy the QO site in cyt b6f resulting in constitutive activation of the thylakoid protein kinase (Gal et al., 1997). Furthermore, the DBMIB data indicate that the redox status of electron carriers downstream of cyt b6f, such as ferredoxin, are not involved in transcript accumulation. Although DBMIB is known to be leaky, the consistent results obtained with a range of inhibitor concentrations indicate that DBMIB is not acting as an electron shuttle bypassing cyt b6f.
We further show that crhR transcript stability contributes significantly to transcript accumulation through a mechanism that is also regulated by a redox-responsive mechanism. crhR transcripts are less stable in the light than in the dark, similar to the pattern of light-responsive stability described for the Synechocystis psbA transcripts (Mohamed et al., 1993; Tyystjarvi et al., 1998), but in contrast to the identical half-lives observed for the Synechocystis glnA transcript (Reyes and Florencio, 1995). It is significant that enhanced electron flow in the presence of exogenous Glc does not influence crhR transcript stability. These observations lead to the following conclusions. An enhanced rate of crhR mRNA turnover in the light confirms that transcription must also be occurring at an elevated rate to maintain detectable levels of the transcript. Furthermore, the observation that crhR mRNA stability differs only in response to light versus darkness, independent of the metabolism of exogenous Glc, implies that stability is regulated separately from transcription. This conclusion is further supported by the observation that crhR stability increases significantly in the presence of electron transport inhibitors, independent of the metabolism of exogenous Glc. Elimination of mRNA stability regulation by the interruption of linear electron flow between QA and QO indicates that the mechanism regulating mRNA stability responds to redox-mediated system that differs from that regulating accumulation and not light-sensing by photoreceptors. Although the mechanism regulating crhR mRNA stability remains to be determined, it follows that continued translation of crhR transcripts couldcontribute to and result from the increased mRNA stability observed in the dark, such that the cells could maintain CrhR protein levels in the absence of transcription. The constitutive presence of CrhR protein in the cells supports these predictions.
The constitutive presence of CrhR protein irrespective of the redox status implies that either a low, but undetectable, level of crhR transcription and/or enhanced CrhR protein stability occurs in the dark. In photoautotrophic cells, crhR transcription in the dark could be initiated by electron flow into plastoquinone from the respiration of endogenous Glc, producing a low level of CrhR synthesis. Respiration declines in the dark, however, as endogenous glycogen stores are depleted (Hirano et al., 1980) with the resulting decrease in electron flow correlating with the observed decline in detectable transcript, but not protein levels. The inability to detect crhR transcript in dark-treated cells combined with the constitutive presence of CrhR therefore suggests CrhR protein stability may increase in the dark. In this scenario the dark-regulated increases in both transcript and protein stability would combine to maintain CrhR protein levels in the dark, in the absence of crhR transcription. If this is indeed the case, then the observed Glc-induced accumulation of crhR transcript and CrhR protein in the dark is a reflection of the artificial enhancement in the rate of electron flow generated by respiration of exogenously supplied Glc. This would imply that a threshold limit of electron transport is required to activate the redox-mediated increase in crhR transcript accumulation—a level that is not achieved by dark respiration in the absence of exogenous Glc. This scenario best fits our observations and suggests that the physiological role of this redox-responsive pathway is to sense and respond to light-generated and not respiratory-generated electron flow.
Although further analysis is required to discriminate between these two possibilities, the lack of a correlation between transcript and protein levels has also been observed for the redox-regulated gene, glnA, in Synechocystis (Reyes and Florencio, 1995). Irrespective of the dark level of crhR transcription, the constitutive presence of CrhR protein provides the potential for RNA unwinding in the dark or if inactive, its rapid activation upon resumption of electron flow. Thus CrhR catalyzed RNA unwinding may be required for the efficient expression of genes whose products are required for and/or are a consequence of electron flow.
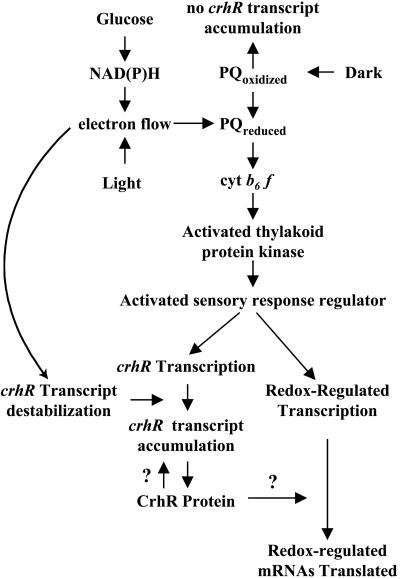
The results presented here implicate a direct regulatory coupling between the redox status of plastoquinone/cyt b6f and the transcription of a specific non-photosynthetic gene. Although various reports have described the redox regulation of gene expression in photosynthetic organisms, this is the first report that identifies a gene whose protein product provides a potential link between the changes in gene expression at the two levels. A simplified working model describing crhR expression and the potential for CrhR catalyzed RNA unwinding in the post-transcriptional regulation of expression of other redox-induced genes is shown in Figure 7. Electrons derived from either light harvesting or Glc metabolism reduce plastoquinone. Interaction of the resulting plastoquinol with cyt b6f may activate the thylakoid-associated protein kinase initiating a signal transduction pathway promoting the transcription of crhR and other redox regulated genes. Superimposed upon the redox-mediated regulation of crhR transcription is the additional control of crhR transcript accumulation by the alteration of crhR mRNA stability. Dark-induced increases in crhR stability may be beneficial during normal growth when the organisms experience transient decreases in light-induced electron flow. Under these conditions, although crhR transcription may cease, the lack of de novo synthesis of crhR mRNA would be compensated by the increase in transcript stability, allowing continued translation of CrhR and the potential to respond rapidly to subsequent increases in electron flow. In addition, CrhR RNA-unwinding activity provides the potential to self-regulate its own expression at the translational level. This model provides a testable explanation for the mechanism by which constitutively expressed mRNAs encoding photosynthetic genes in other systems are specifically translated in the light. Although an RNA unwinding requirement for translation of light- or redox-regulated mRNAs remains to be proven, the regulation of post-transcriptional processes by these signals has been reported (Danon and Mayfield, 1994; Mayfield et al., 1994; Dickey et al., 1998). The regulation of gene expression by alteration of RNA secondary structure and efficiency of translation initiation observed in these studies are hallmarks of RNA helicase activity, suggesting the potential involvement of a redox-regulated RNA helicase in these systems.
Figure 7.
A working model for the redox-mediated regulation of crhR expression. The model incorporates known elements from mustard chloroplasts and cyanobacteria. crhR expression is dynamically established by a complex interaction between redox-regulated transcription and mRNA stability. Reduction of electron carriers between QA in PSII and QO in cyt b6f in the thylakoid membrane can either be accomplished naturally by light harvesting or by the metabolism of exogenously supplied Glc. Data presented here implicate interaction of plastoquinol with cyt b6f as the environmental sensor that rapidly activates crhR expression at the transcriptional level, potentially by signaling through the signal transduction pathway known to initiate photosystem state transitions. Reduced plastoquinone has been reported to regulate directly the transcription of two chloroplast-encoded genes whose protein products are required to maintain photosynthetic electron flow. Accumulation of crhR mRNA is also regulated by a redox-regulated, but Glc metabolism-independent, mechanism. Redox regulation of transcription and mRNA stability provides a mechanism by which the cells could rapidly respond to alterations in redox conditions. The specificity of the redox regulation of crhR expression raises the intriguing possibility that CrhR-catalyzed RNA unwinding may be required for the efficient translation of its own and possibly other redox-regulated mRNAs.
RNA unwinding provided by RNA helicases appears to be an integral component of cyanobacterial response to environmental variables. We have recently shown that expression of the Anabaena sp. strain PCC 7120 RNA helicase, crhC, is regulated by temperature with expression induced specifically by cold stress (Chamot et al., 1999; Chamot and Owttrim, 2000). The data reported in this communication reflect the ability of cyanobacteria to sense and respond to changes in their environment through a complex series of regulatory pathways initiated by changes in electron flow. Redox regulation of crhR expression allows Synechocystis to respond rapidly to environmental changes, on the order of minutes, similar to the plastoquinone redox state-mediated response times observed for the photosynthetic genes, psaAB and psbA, in mustard chloroplasts (Pfannschmidt et al., 1999). In addition, RNA helicases are localized in chloroplasts (Owttrim et al., 1994) and plant mitochondria (G.W. Owttrim, unpublished observation; Gagliardi et al., 1999).
Taken together these observations indicate that the ability to respond to environmental changes by sensing shifts in redox potential has been conserved between prokaryotic and eukaryotic photosynthetic organisms and that RNA helicases are available that may be involved in the regulation of genes expressed in response to these changes. This conservation supports the proposal that the physiological requirement for the rapid regulation of gene expression by redox signaling is a factor leading to the maintenance of chloroplast and mitochondrial genetic systems (Allen, 1993; Allen and Raven, 1996). Although the plastoquinol-redox control model proposed by Allen et al. (1995) emphasizes the regulation of photosynthetic gene expression, the data presented here reveal a similar pattern of RNA helicase expression regulated by redox status. Although an RNA helicase would not be involved directly in the light-harvesting reactions per se, coordinate regulation of crhR expression implies that modulation of RNA secondary structure is required during conditions that elicit electron flow. CrhR-induced RNA unwinding activity could remove secondary structures that inhibit the efficient translation of mRNAs whose products are required under these conditions. Whether CrhR has specific RNA targets, such as light- or redox-induced mRNAs, or plays a more general role, enhancing translational efficiency, assembly of ribonucleoprotein complexes, or RNA turnover, remains to be determined.
MATERIALS AND METHODS
Bacterial Strains, Media, and Growth Conditions
Synechocystis sp. strain PCC 6803 was maintained in BG-11 liquid medium (Rippka et al., 1979) with the addition of 10 mm TES [N-tris (hydroxymethyl) methyl-2-aminoethanesulfonic acid], pH 8.2, or on BG-11 agar solidified with 1% (w/v) Bacto-Agar (Difco Laboratories, Detroit, MI). Cells were routinely cultured at 30°C with continuous illumination at an intensity of 30 μmol photons m−2 s−1. Liquid cultures were aerated by vigorous shaking (200 rpm) and bubbling with air. Glc and 3-OMG were added to a final concentration of 5 mm. Cells that had been subcultured twice with addition of Glc or 3-OMG were used as inocula for large-scale experimental cultures. Electron transport inhibitors (Sigma, St. Louis) and their final concentrations were: DCMU, 5 μm, and DBMIB, 20 μm, unless indicated). Dark conditions were obtained by wrapping growth flasks with several layers of aluminum foil. L1 was obtained using a medium red filter (Rosco no. 27) that gave 50% transmittance at 650 nm. L2 was provided by an orange filter (Rosco no. 20) that gave 50% transmittance at 560 nm (Pfannschmidt et al., 1999). The filters were inserted between the light source and the cultures with additional illumination to compensate for reductions in light intensity compared with white light. Escherichia coli DH5α, used for propagation of plasmids cs0096 and cs0096–9, and JM109, used for propagation of pRSETA plasmids, were grown in Luria-Bertani medium with addition of the appropriate antibiotics (Sambrook et al., 1989).
crhR Subcloning
The SuperCos clone cs0096 containing the crhR ORF slr0083 (Kaneko et al., 1995) was digested with EcoRI and Southern analysis, using the Anabaena sp. strain PCC 7120 RNA helicase gene crhB (Chamot et al., 1999) as probe, identified a fragment that contains the entire 1,401-bp crhR ORF plus 1.6 kb of flanking sequences, including 157 bp of the upstream ORF, slr0082. This 3.02-kb fragment was cloned into the EcoRI site in pBluescript KS+ (Stratagene, La Jolla, CA).
RNA Gel-Blot Analysis
Total RNA was extracted by mechanical lysis, aliquots (6 μg) were separated on formaldehyde gels, and were transferred to a nylon membrane (Hybond N+, Amersham, Buckinghamshire, UK) as previously described (Chamot et al., 1999). Northern blots were hybridized with the appropriate labeled DNA probe at 65°C in aqueous solution and washed at 60°C in 0.1% (w/v) SDS, 0.1× sodium chloride/sodium phosphate/EDTA (1× sodium chloride/sodium phosphate/EDTA is 0.18 m NaCl, 10 mm NaH2PO4, and 1 mm EDTA, pH 8). The crhR probe, a 325-bp internal SmaI fragment that encodes the highly conserved PTRELA, DEAD, and SAT amino acid motifs characteristic of RNA helicases was labeled with [α-32P] dCTP by the random primer method (Boehringer Mannheim, Basel). Northern blots were stripped and reprobed with the Synechocystis RNase P gene (Vioque, 1992) as a control for RNA loading. Relative RNase P levels are indicated below each autoradiogram. Treatments were performed in triplicate and each experiment repeated two to three times, using independent cultures.
Quantitation of crhR Transcript Levels in L1 and L2 and Determination of mRNA Half-Life
To determine the effect of PSI- or PSII-specific light on crhR levels, triplicate cultures were treated with L1 (650 nm) or L2 (560 nm) by inserting the indicated filters between the light source and the cultures for the indicated times, followed by RNA extraction and northern-blot analysis. mRNA stability was determined in cultures incubated in the presence or absence of light and Glc and DCMU or DBMIB, as indicated. RNA transcription was inhibited by rifampicin (400 μg/mL final concentration). Experiments were repeated in triplicate using independent cultures. For signal strength quantitation, autoradiograms were either scanned using Ofoto software (version 2, Light Source Computer Images, San Rafael, CA) and densitometric analysis performed with NIH Image software (version 1.61, Scientific Computing Resource Center, National Institutes of Health, Bethesda, MD) or detected using a PhosphorImager and analyzed using ImageQuant 4.0 software (Molecular Dynamics, Sunnyvale, CA). The results from replicate experiments were expressed as a percentage of the crhR transcript level observed in cells illuminated with white light and pooled data presented as the means ± se. Linear regression analysis was used to calculate the mRNA half-life.
Protein Analysis
Cell-free protein extracts were prepared as described for RNA extraction except the 5% (w/v) phenol-ethanol and phenol steps were omitted. After clarification, protein concentration was determined by the Bradford Assay (Bio-Rad, Hercules, CA) using bovine serum albumin as the standard. Proteins were separated on SDS-10% (w/v) polyacrylamide gels (SDS-PAGE) and transferred to nitrocellulose membranes (Bio-Rad) using a semi-dry apparatus (Tyler Research, Edmonton, Canada). Immunoreactive polypeptides were detected using rabbit anti-CrhR antibodies (1/1,000; Chamot and Owttrim, 2000). The rabbit anti-CrhR polyclonal antibodies were generated against CrhR-His tagged fusion protein purified from E. coli using the pRSETA vector (Invitrogen, Carlsbad, CA). All experiments were repeated at least twice, using independent cultures.
ACKNOWLEDGMENTS
We are grateful to Dr. Satoshi Tabata for providing cs0096, the SuperCos clone containing the Synechocystis sp. strain PCC 6803 crhR ORF (slr0083), and to Dr. Agustin Vioque for providing the Synechocystis RNase P gene. We also thank Ian Le for preliminary screening of crhR expression, Dr. Danuta Chamot for help with RNA techniques, Drs. Allen Good and Danuta Chamot for critical reading of the manuscript, and Warren Yunker for statistical analysis.
Footnotes
This work was supported in part by a grant from the Natural Sciences and Engineering Research Council of Canada (to G.W.O.).
LITERATURE CITED
- Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim Biophys Acta. 1992;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- Allen JF. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J Theor Biol. 1993;165:609–631. doi: 10.1006/jtbi.1993.1210. [DOI] [PubMed] [Google Scholar]
- Allen JF, Alexciev K, Hakansson G. Regulation by redox signaling. Curr Biol. 1995;5:869–872. doi: 10.1016/s0960-9822(95)00176-x. [DOI] [PubMed] [Google Scholar]
- Allen JF, Harrison MA, Holmes NG. Protein phosphorylation and control of excitation energy transfer in photosynthetic purple bacteria and cyanobacteria. Biochimie. 1989;71:1021–1028. doi: 10.1016/0300-9084(89)90106-5. [DOI] [PubMed] [Google Scholar]
- Allen JF, Raven JA. Free-radical-induced mutation vs redox regulation: costs and benefits of genes in organelles. J Mol Evol. 1996;42:482–492. doi: 10.1007/BF02352278. [DOI] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Link G. Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba) Plant Mol Biol. 1997;34:181–189. doi: 10.1023/a:1005802909902. [DOI] [PubMed] [Google Scholar]
- Chamot D, Magee WC, Yu E, Owttrim GW. A cold-shock induced cyanobacterial RNA helicase. J Bacteriol. 1999;181:1728–1732. doi: 10.1128/jb.181.6.1728-1732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamot D, Owttrim GW. Regulation of cold shock induced RNA helicase gene expression in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2000;182:1251–1256. doi: 10.1128/jb.182.5.1251-1256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- Dickey LF, Petracek ME, Nguyen TT, Hansen ER, Thompson WF. Light regulation of Fed-1 mRNA requires an element in the 5′-untranslated region and correlates with differential polyribosome association. Plant Cell. 1998;10:475–484. doi: 10.1105/tpc.10.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elich TD, Chory J. Phytochrome: if it looks and smells like a histidine kinase, is it a histidine kinase? Cell. 1997;91:713–716. doi: 10.1016/s0092-8674(00)80458-4. [DOI] [PubMed] [Google Scholar]
- Escoubas J-M, Lomas M, LaRoche J, Falkowsi PG. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Photomorphogenesis: light receptor kinases in plants! Curr Biol. 1999;9:R123–R126. doi: 10.1016/s0960-9822(99)80078-5. [DOI] [PubMed] [Google Scholar]
- Flores E, Schmetterer G. Interaction of fructose with the glucose permease of the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 1986;166:693–696. doi: 10.1128/jb.166.2.693-696.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace FV. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994;4:271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Gagliardi D, Kuhn J, Spadinger U, Brennicke A, Leaver CJ, Binder S. An RNA helicase (AtSUV3) is present in Arabidopsis thaliana mitochondria. FEBS Lett. 1999;458:337–342. doi: 10.1016/s0014-5793(99)01168-0. [DOI] [PubMed] [Google Scholar]
- Gal A, Zer H, Ohad I. Redox-controlled thylakoid protein phosphorylation: news and views. Physiol Plant. 1997;100:869–885. [Google Scholar]
- Golden SS. Light-responsive gene expression in cyanobacteria. J Bacteriol. 1995;177:1651–1654. doi: 10.1128/jb.177.7.1651-1654.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Satoh K, Katoh S. Plastoquinone as a common link between photosynthesis and respiration in a blue-green alga. Photosynth Res. 1980;1:149–162. doi: 10.1007/BF00020594. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803: I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 1995;2:153–166. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell. 1997;9:627–640. doi: 10.1105/tpc.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis M, Zsiros O, Farkas T, Wada H, Nagy F, Gombos Z. Light-induced expression of fatty acid desaturase genes. Proc Natl Acad Sci USA. 1998;95:4209–4214. doi: 10.1073/pnas.95.8.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RD, Golden SS. mRNA stability is regulated by a coding-region element and the unique 5′-untranslated leader sequences of the three Synechococcus psbA transcripts. Mol Microbiol. 1997;24:1131–1142. doi: 10.1046/j.1365-2958.1997.4201768.x. [DOI] [PubMed] [Google Scholar]
- Linder P, Lasko PF, Ashburner M, Leroy P, Nielen PJ, Nishi K, Schnier J, Slonimski P. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Laudenbach DE, Huner NPA. Redox regulation of light-harvesting complex II and cab mRNA abundance in Dunaliella salina. Plant Physiol. 1995;109:787–795. doi: 10.1104/pp.109.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Cohen A, Danon A, Yohn C. Translation of the psbA mRNA of Chlamydomonas reinhardtii requires a structured RNA element contained within the 5′-untranslated region. J Cell Biol. 1994;127:1537–1545. doi: 10.1083/jcb.127.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A, Eriksson J, Osiewacz H, Jansson C. Differential expression of the psbA genes in the cyanobacterium Synechocystis. Mol Gen Genet. 1993;238:161–168. doi: 10.1007/BF00279543. [DOI] [PubMed] [Google Scholar]
- Mohamed A, Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis. Plant Mol Biol. 1989;13:693–700. doi: 10.1007/BF00016024. [DOI] [PubMed] [Google Scholar]
- Mullineaux CW, Allen JF. The state 2 transition in the cyanobacterium Synechococcus 6301 can be driven by respiratory electron flow into the plastoquinone pool. FEBS Lett. 1986;205:155–160. [Google Scholar]
- Owttrim GW, Mandel T, Thomas AAM, Kuhlemeier C. Characterization of the tobacco eIF-4A gene family. Plant Mol Biol. 1994;26:1747–1757. doi: 10.1007/BF00019489. [DOI] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Helicases and RNA unwinding in translation. Curr Opin Struct Biol. 1993;3:953–959. [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Reyes JC, Florencio FJ. Electron transport controls transcription of the glutamine synthetase gene (glnA) from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol. 1995;27:789–799. doi: 10.1007/BF00020231. [DOI] [PubMed] [Google Scholar]
- Richter S, Hagemann M, Messer W. Transcription analysis and mutation of a dnaA-like gene in Synechocystis sp. strain PCC 6803. J Bacteriol. 1998;180:4946–4949. doi: 10.1128/jb.180.18.4946-4949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scherer S. Do photosynthetic and respiratory electron transport chains share redox proteins? Trends Biochem Sci. 1990;15:458–462. doi: 10.1016/0968-0004(90)90296-n. [DOI] [PubMed] [Google Scholar]
- Schmid SR, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–292. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Tiller K, Link G. Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapis alba L.) EMBO J. 1993;12:1745–1753. doi: 10.1002/j.1460-2075.1993.tb05822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebst A. Inhibitors in electron flow: tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol. 1980;69:675–715. [Google Scholar]
- Tyystjarvi T, Tyystjarvi E, Ohad I, Aro E. Exposure of Synechocystis 6803 cells to series of single turnover flashes increases the psbA transcript level by activating transcription and down-regulating psbA mRNA degradation. FEBS Lett. 1998;436:483–487. doi: 10.1016/s0014-5793(98)01181-8. [DOI] [PubMed] [Google Scholar]
- Vener AV, Ohad I, Andersson B. Protein phosphorylation and redox sensing in chloroplast thylakoids. Curr Opin Plant Biol. 1998;1:217–223. doi: 10.1016/s1369-5266(98)80107-6. [DOI] [PubMed] [Google Scholar]
- Vioque A. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 1992;20:6331–6337. doi: 10.1093/nar/20.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]