Abstract
Background:
Harvey-Ras (H-Ras) is an important guanosine triphosphatase protein for the regulation of cellular growth and survival. Altered Ras signaling has been observed in different types of cancer either by gene amplification and/or mutation. The H-Ras oncogene mutations are well reported, but expression of the H-Ras gene is still unknown.
Objective:
This study aimed to examine both protein and messenger-RNA (mRNA) expressions of H-Ras in oral squamous cell carcinoma (OSCC) and analyzed the association with risk habits and the clinicopathological profile of cases.
Methodology:
A total of 65 tissue specimens of OSCC (case group) and equal number of normal tissues (control group) were included in this study. H-Ras protein and mRNA expressions were analyzed using immunohistochemical and quantitative real time-polymerase chain reaction techniques, respectively.
Results:
The H-Ras protein was significantly overexpressed in the oral carcinoma group compared to the normal group (P = 0.03). Most of the OSCC cases showed positive staining with moderate expression, while negative and moderate staining was high in the control group. The majority of H-Ras positive cases were found in individuals with multiple risk habits including tobacco chewing. The risk of H-Ras positivity was 1.46 times higher in smokers than non-smokers. H-Ras positivity increased in cases affected with buccal mucosa site and higher grade of carcinoma. Relative mRNA level of H-Ras was significantly elevated in oral carcinoma as compared with the control group (P ≤ 0.001). Protein and mRNA levels of H-Ras in case group was poorly correlated.
Conclusion:
H-Ras oncogene expression was markedly higher in oral carcinoma, and it can be a prognostic marker and target for an effective molecular therapy.
Keywords: Gene expression, H-Ras, immunohistochemistry, oral squamous cell carcinoma (OSCC)
INTRODUCTION
Oral cancer is the sixth and third most common cancer globally and in India, respectively.[1] The incidence rate of oral cancer in India is over 83,000 new cases and 46,000 deaths annually.[2] Approximately 90% of oral cancer cases are diagnosed for squamous cell type carcinoma. Therefore, oral cancer is also called oral squamous cell carcinoma (OSCC).[3] Tobacco-related practices such as chewing or smoking, alcohol consumption and HPV are well-established risk factors for oral cancer. Moreover, approximately 10% of OSCC cases were identified for HPV infection in the North Indian population.[4]
A number of transforming cellular oncogenes have been identified and isolated from different types of human tumor. Categorization of these oncogenes provides an understanding of cancer at the molecular level. In this context, attention has focused on the Harvey-Ras (H-Ras) gene in oral cancer. The RAS gene family consists of three functional genes, H-Ras, Kristen Ras (K-Ras: isoform A and isoform B) and Neuroblastoma Ras (N-Ras) encoding four highly similar, small and conserved Ras proteins (or p21 proteins), which located on the inner surface of the plasma membrane.[5]
The Ras proteins have intrinsic guanosine triphosphatase (GTPase) activity that transduces the growth signal from the cell surface to intracellular effectors through mitogenic activating protein kinase (MAPK), c-Jun N-terminal kinase (JNK) and p38-kinase pathways, which regulate normal cell proliferation function.[6,7] The RAS GTPases activation regulates through cycle between GDP bound inactive and GTP bound active state with the help of guanine nucleotide exchange factors and GTPase-activating proteins (GAPs).[8]
For the last two decades, it has been shown that Ras genes are frequently mutated in different types of tumor and participate in their proliferation and maintenance.[9] G12D, G12V, G13D, G13R, Q61H and Q61R are possible missense mutations occurring at codons G12, G13 and Q61; but their frequencies are not consistent. A remarkable variation in incidence of activational mutations of H-Ras gene in different ethnic groups with oral cancer has been recognized to be very different (0-55%).[10,11,12,13,14] H-Ras gene is most commonly mutated gene than the K-Ras and N-Ras in oral cancer. Moreover, G12V, Q61 R and G13R are reported as predominant mutations in H-Ras gene.[15] A study defined that K-Ras protein was highly expressed in advance stages of OSCC with higher grade carcinogenesis.[16] However, most of the reports detailed the frequency of all three isoforms of Ras gene mutations, but none of them investigated the exact pattern of H-Ras protein expression in cancers. Hence, in this perspective, we analyzed the H-Ras protein and messenger RNA (mRNA) expression in OSCC and normal tissues of the oral cavity. In addition, an attempt was also made to identify an association of H-Ras expression with clinicopathological profiles and risk habits in the North Indian population.
METHODOLOGY
The present study was carried out in one of the best tertiary care hospital of North India, King George's Medical University (K.G.M.U.) which is located in Lucknow, district of Uttar Pradesh (U. P.) state, India. This medical university facilitates to the health needs of people from the North India and the Eastern India as well. However, in this study, all the participants were belonged from the North India.
A total of 65 biopsies and surgically removed tumor tissues of the oral cavity were collected from suspected and confirmed oral carcinoma cases from January 2013 to June 2016 at the Department of Surgical Oncology and Department of Oral and Maxillofacial Surgery, K.G.M.U., Lucknow, U.P., India. Only histopathologically confirmed OSCC tissue samples were included in the case group. Equal number of age and sex-matched oral tissue samples from nonmalignant lesions of the oral cavity were also collected as a control group. Only those nonmalignant tissue sections which showed no evidence of dysplasia, i.e., normal cell morphology in histopathological examination were considered as controls. The study protocol was approved by the Institutional Ethics Committee, K.G.M.U., Lucknow, U.P., India. Demographic details and history of risk habits were recorded using a predesigned standard questionnaire. Details of clinical and pathological characteristics of each patient were noted down from their case history sheet. Patients who had completed or were undergoing therapy or diagnosed for secondary or any other malignancy, were excluded from the study.
All tissue samples (cases and controls) were collected in 4% formaldehyde solution for histological diagnosis and immunohistochemical analysis of the H-Ras protein. Tissues collected in RNA later solution (Invitrogen, USA) were used for relative mRNA expression analysis. Hematoxylin and Eosin (HE) staining was performed to diagnose the type, grade and stage of tumor in tissue sections according to WHO guidelines and the AJCC cancer staging manual for the histological and clinical classification of oral lesions.[17,18]
The immunohistochemistry (IHC) of the H-Ras protein was performed on paraffin-embedded 4 μm tissue sections. Detection of the H-Ras protein was done using polyclonal primary antibody against H-Ras (sc-68743, Santa Cruz, USA; 1:100 dilution) and horseradish peroxidase (HRP) enzyme labeled-secondary antibody detection kit (TL-015-QHD, Thermo Fisher Scientific, USA) as per the manufacturer's protocol. The sections were incubated overnight with the primary antibody at 4°C after blocking endogenous peroxidase activity with 3% hydrogen peroxide and protein block reagent. Antigen retrieval of tissue sections was done in Tris-EDTA buffer (pH 9.0) using Pascal retrieval system (Dako, Denmark). The HRP-labeled secondary antibody was applied on tissue sections with primary antibody and incubated for 30 min in a dark chamber at room temperature. Positive staining was visualized using diaminobenzidine (DAB) and counterstained with Mayer's hematoxylin. Normal skin tissue was used as positive control for H-Ras protein.
The interpretation of H-Ras protein expression in IHC was carried out in the tumor hotspot, including invasion fronts. Five hundred tumor cells were viewed at high-magnification power (×40) in tumor hotspots and scored by assessing the cytoplasmic staining, using a semi-quantitative method in at least five fields. The staining score was ranged from 0 to +3; 0 negative expression (<5% positive-stained cells), +1 low expression (5%–20% positive-stained cells), +2 moderate expression (21%–50% positive-stained cells) and +3 strong expression (>50% positive-stained cells). Scores of 2 and 3 were defined as the H-Ras positive expression.[19]
Relative mRNA expression of the H-Ras gene was analyzed by quantitative real time-polymerase chain reaction (qRT-PCR). Total RNA was isolated from OSCC and control oral tissue samples by RNeasy Mini Kit (Qiagen, Germany). cDNA was synthesized from isolated RNA samples by reverse transcription, followed by PCR amplification with high capacity cDNA reverse transcription kit (Applied Biosystem, USA). qRT-PCR was performed in Lightcycler 96 system (Roche, USA) using Power SYBR Green PCR Master Mix (Applied Biosystem, USA). The specific primers H-Ras (NM001130442.1) forward 5’-TTTGAGGACATCCACCAGTACA-3’, reverse 5’-GCCGAGATTCCACAGTGC-3’ (product size 118 bp)[20] and β-actin (XM006715764.1) forward 5’-ATCGTGCGTGACATTAAGGAGAAG-3’, reverse 5’-AGGAAGGAAGGCTGGAAGAGTG-3’ were used as internal control (product size 179 bp). The annealing temperature was standardized by Gradient PCR at 57°C–62°C for both gene, and 58°C was considered as final annealing temperature [Figure 1]. qRT-PCR reactions for each sample were performed in triplicates. The qRT-PCR thermal profile for H-Ras and β-actin gene was 94°C: 10 min (94°C: 30 s, 58°C: 35 s, 72°C: 1 min) ×45 cycles and final extension at 72°C for 5 min. Fold change was calculated by relative gene quantification method (2-ΔΔCT method).[21]
Figure 1.
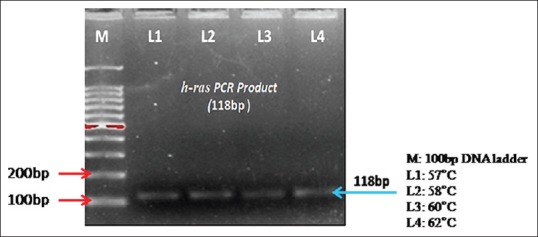
Two percent Agarose gel showing amplified product of H-Ras at different temperatures by gradient PCR
Statistical analysis
Statistical analysis was performed using SPSS software (version 16; IBM corp., IL, USA). Continuous data were summarized as mean ± standard error (SE), while categorical data in number and percentage. Two-tailed Chi-square test and unpaired t-test were used to assess the associations between two parameters. Correlation between two continuous data was calculated using Pearson's correlation coefficient (r) method. Associations of demographical characteristics, risk habits and clinicopathologic variables with H-Ras expression status were calculated with binary logistic regression method. Odd ratio (OR), with 95% confidence interval, was used to assess the risk value of biomarker. P < 0.05 was considered statistically significant.
RESULTS
HE-stained histopathological sections showed proliferated atypical squamous epithelial cell with high nuclear-cytoplasmic ratio, enlarged hyperchromatic nuclei few having prominent nucleoli and variable amount of scanty cytoplasm presenting low-grade dysplasia arranged in tubercular and cluster patterns. At places epithelial, keratin pearl formation was seen. Moderate (+2 score) and strong (+3 score) immunostaining of the H-Ras protein was observed in most cases. However, tissue sections of oral nonmalignant lesions depicted normal squamous epithelial lining based on an intact basement membrane. Several cells of normal tissue section exhibited the negative and moderate cytoplasmic immuno-reactivity of H-Ras [Figure 2a–d].
Figure 2.
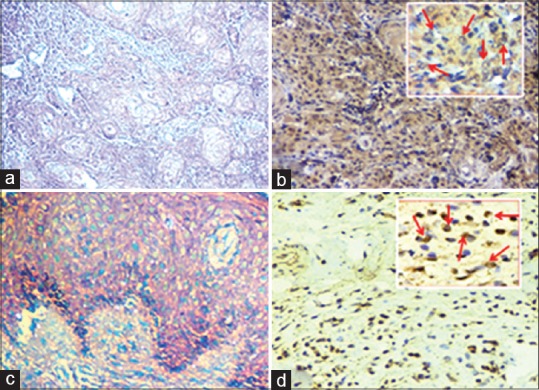
Photomicrograph showing HE section and immunohistochemistry images of diagnosis and H-Ras protein expression: (a) HE-stained tissue section of oral squamous cell carcinoma (OSCC) case (digital magnification, ×100), (b) IHC image showing strong cytoplasmic immunoexpression in oral squamous cell carcinoma (DAB, digital magnification, ×100), Inset shows the brown color-stained H-Ras positive cells (OSCC) (DAB, digital magnification, ×400), (c) HE of normal tissue section of nonmalignant lesions of oral cavity (digital magnification, ×100), (d) Weak immunostaining in normal tissue (DAB, digital magnification, ×100), Inset shows brown colored H-Ras positive cells (DAB, digital magnification, ×400)
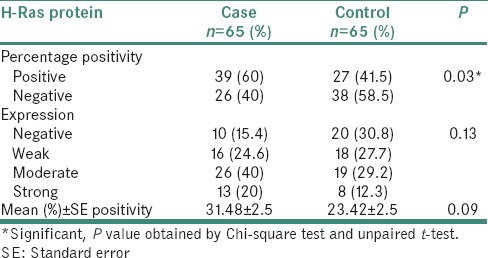
The percentages positivity and expression patterns of the H-Ras protein in case and control groups are shown in Table 1. Out of a total of 65 cases, 39 cases (60%) showed H-Ras positive expression whereas, 27/65 (41.5%) normal tissues expressed positive immunostaining. This difference was significantly associated with oral carcinoma (P = 0.03). The mean (±SE) percent positivity of H-Ras protein in the cases versus controls was 31.48 ± 2.5 versus 23.42 ± 2.5, respectively. The subcategorization of percentage positive staining of H-Ras illustrated that 10/65 OSCC cases showed negative expression, 16/65 cases weak expression, 26/65 cases moderate expression and 13/65 cases showed strong expression. However, in the control group, 20/65 samples presented negative expression followed by moderate, weak and strong expression.
Table 1.
H-Ras protein expression and percentage positivity in cases and controls
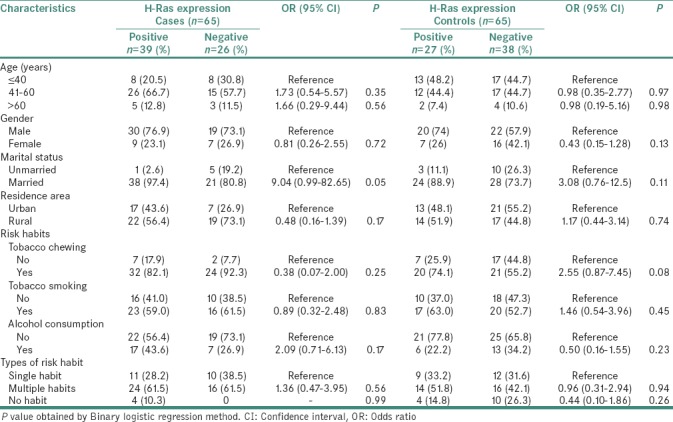
Among H-Ras positive cases, most cases (26/39; 66.7%) were from the age group 41–60 years. In controls, H-Ras positivity was more frequent in the age group ≤40 years compared to 41–60 and >60 years age groups. In this study, H-Ras positivity was high in males in both case and control groups. All the demographic variables of both groups did not show any significant association with H-Ras positivity. Nearly 82.1% of H-Ras-positive cases were associated with habit of tobacco chewing, and 74.1% of controls with H-Ras positive expression had similar habits. Moreover, a higher risk (1.46) of H-Ras positivity was estimated in smokers than nonsmokers of control group. Furthermore, we subcategorized group of all cases and controls according to type of risk habits; single, multiple (habituated with any two or more type of tobacco-related and alcohol consumption habits) and no risk habit. It was shown that majority (61.5%) of H-Ras-positive cases were attributed with multiple risk habits (OR = 1.36, P = 0.56) [Table 2].
Table 2.
Association of H-Ras protein expression with demographic characteristics and risk-habits in cases and controls
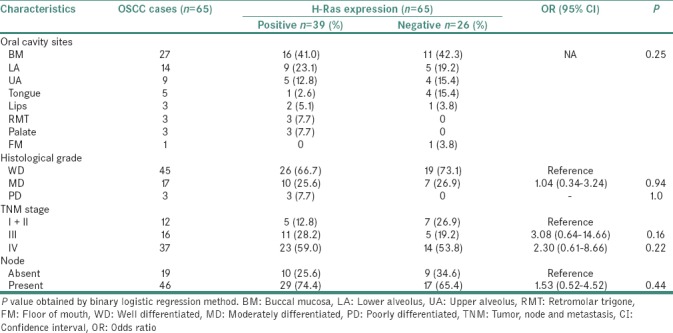
The association of clinicopathological profile of OSCC cases and H-Ras-protein expression is presented in Table 3. Buccal mucosa was the most common affected site in both H-Ras positive as well as negative cases. H-Ras protein expression was detected in most of Stage III (28.2%) and Stage IV (59%) oral cancer patients. In histopathological investigations, H-Ras positivity was enhanced in higher grade carcinoma. Most cases with lymph node involvement showed H-Ras positive expression.
Table 3.
Association of H-Ras protein expression with clinicopathological variables in oral squamous cell carcinoma cases
In the present study, out of the total tissue samples of both groups, nine tissue samples of OSCC cases and thirteen tissue samples of control samples were dropped out due to either insufficient samples or poor yields of isolated total RNA. The H-Ras mRNA fold change level in cases (n = 56) range (min-max) 0.46–6.38, median 1.28 and mean ± standard deviation [SD] 1.62 ± 1.02 was analyzed. However, in the control group, mRNA fold change range was 0.34–2.47 with a median value of 0.97 and mean ± SD 1.10 ± 0.50. mRNA level of H-Ras was significantly elevated in oral carcinoma compared to control group (P ≤ 0.001) [Figure 3].
Figure 3.
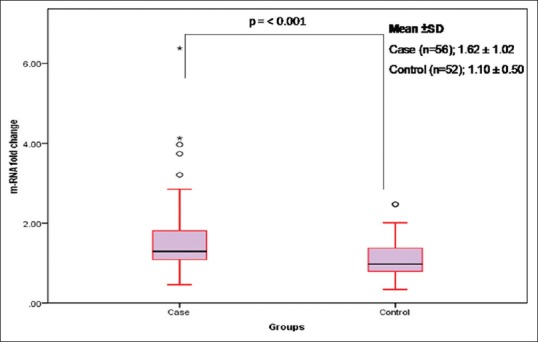
Relative messenger RNA expression of the H-Ras gene in case and control groups. Prominent horizontal line and star marks of boxes present the median and extreme values of fold change. Upper and lower bars present the distance of the 10th to 90th percentile from the median. The open circle indicates to outliers
The Pearson's correlation test showed a poor positive correlation in between protein and mRNA levels of H-Ras in the case group. However, in the control group, protein and mRNA levels were significantly correlated with each other [Figure 4].
Figure 4.
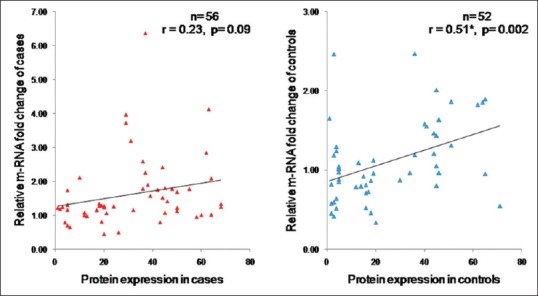
Correlation between protein and messenger RNA expression of H-Ras in case and control groups
DISCUSSION
This study shows the presence of H-Ras gene expression in oral cancer and normal control groups, examined by immunohistochemical and quantitative real-time PCR procedures. As similar to our previous study Krishna et al.,[22] the prevalence of OSCC found in this study was also high in males aged 41–60 years. Tobacco-related risk habits were more highly associated with OSCC cases in this study. The clinicopathological profile showed that buccal mucosa was the most affected site in oral cancer cases. The majority of oral cancer patients had advanced stages of tumor at the time of clinical investigations in the study.
The Ras family proteins are often deregulated in various types of human cancer. In the last three decades, various in vitro and in vivo studies confirmed that there is a significant association of specific mutated Ras family genes and their altered products with particular types of tumor.[23] The altered Ras signaling may also participate in the progression and development of other types of pathologies besides cancer and developmental syndromes such as non-obese diabetes and diabetic retinopathy.[24] Mutations in any one of three members of the Ras family are common events in human tumorigenesis. Several studies reported that most oncogenic mutations predominantly affect the K-Ras locus and express altered protein in oral carcinoma and some other cancer.[25,26,27] However, very few studies have attempted to correlate the variant feature of the H-Ras gene in different types of cancer[28] while its protein and mRNA expression patterns in oral cancer are still unexplored. Hence, in this context, we have tried to elaborate the H-Ras gene expression in oral cancer of the Northern India population. We have analyzed that the H-Ras protein was significantly overexpressed (60%) in OSCC as compared to normal tissues (41.5%) of oral mucosa. Our findings are in concordance with Cutilli et al., who carried out immunohistochemical and genetic research on the tumor suppressor p53 and H-Ras oncogene in oromaxillofacial neoplasms. They found immunohistopathological overexpression of the H-Ras protein in the majority of cases (12/15 cases; 80%) and 60% cases mutated for this gene.[29] Similarly, McDonald et al. also obtained H-Ras positive staining in 68% of cases of head and neck squamous cell carcinoma and this is in agreement with the findings of the present study.[30] We did not find any significant statistical difference in subcategories of H-Ras expression between case and control groups, whereas most tissue samples presented H-Ras positive immunostaining at moderate levels. According to the previous literature, the H-Ras protein occurs not only in malignant tumors but also in the metabolism of normal cells.[31]
H-Ras positivity in our study groups was regardless of age, gender and residential area of patients. In this investigation, the frequency of H-Ras positive expression was higher in patients who had tobacco-related habits such as chewing and smoking. Genetic alteration and increased immunoexpression level of Ras genes in tumors may reflect etiology and ethnic origins. Chang et al.[32] found that smoking, tobacco and betel quid chewing accounted for the high incidence of Ras mutations in the Indian subcontinent. In our investigation, most of the oral cancer patients had tobacco or multiple risk habits. A study conducted elsewhere reported that H-Ras mutations in codons 12, 13 and 61 related with tobacco use in oral carcinomas.[10] Hence, we can hypothesize that overexpression of H-Ras protein in the present study groups may be due to the presence of mutant-type of protein.
With regard to clinicopathological parameters of OSCC cases, no significant association was observed between H-Ras positivity and tumor sites. Moreover, another striking finding of current study that out of 27 buccal carcinoma cases 16 cases (59.2%) showed overexpression of H-Ras while only one case of tongue carcinoma (1/5 cases; 20%) was positively stained for H-Ras. This finding is partially consistent with a study by Trivedi et al., in which 45% patients with buccal mucosa cancer showed higher H-Ras protein expression whereas this was 39% in tongue carcinoma patients.[33] Vairaktaris et al. examined the expression of H-Ras in an experimental model of chemically induced oral cancer in sequential stages of carcinoma; the expression was found to be progressively increased during all stages of oral carcinogenesis.[19] In agreement with this, the present study demonstrated that 26/45 well-differentiated cases and 10/17 moderately differentiated cases exhibited higher expression of H-Ras protein; however, the difference was statistically insignificant. The results of other studies are also supported by this analysis.[13,30,32] The expression pattern of H-Ras in the different stages of cancers is ambiguous. Enhanced expression of H-Ras was also defined in early stages of urinary bladder cancer.[34] However, in the current study, H-Ras expression was more pronounced in the stages of more advanced tumor and this is in accordance with previous study.[30] We also demonstrated the relative mRNA expression of the H-Ras gene. In our cases, mRNA mean fold change of H-Ras was significantly higher in comparison with normal mucosal tissue of the oral cavity (P ≤ 0.001), in harmony with K-Ras gene expression.[35] The H-Ras oncogene was also overexpressed in laryngeal and other types of head and neck cancer.[36,37] Overexpression of the H-Ras gene promoted gastric cancer aggressiveness by activating VEGFA/PI3K/AKT pathway and Ras/Raf/MAPK signaling.[38] A study also established the association of K-Ras and PI3K/AKT pathways in advance stages of oral cancer.[16] This suggested a strong association of upregulated H-Ras gene in oral cancer.
Our study also illustrated a significant positive correlation between the H-Ras protein and mRNA levels of controls, although a poor insignificant correlation was analyzed in the OSCC group. The exact interpretation of this finding remains unclear, but previous Indian studies based on etiological factors and mutational status of Ras oncogenes indicate that rare mutations or transcriptional splicing in H-Ras gene might be related to overexpression or mutant type H-Ras protein in tumoral tissues. More studies are required on the functional and mutational gene expression of H-Ras in oral carcinogenesis.
CONCLUSION
The present findings indicate that the H-Ras oncogene is markedly upregulated in oral carcinoma compared with normal oral cavity. H-Ras positivity was higher in those patients who use tobacco or have multiple risk habits. The highest score of H-Ras expression was observed in well-differentiated carcinoma and advanced stage tumor. This suggested that enhanced H-Ras expression in oral carcinoma of the North Indian population is probably due to their risk habits and genetic alteration. This study highlighted the H-Ras gene expression in oral cancer, which may extend our knowledge for better prognosis and molecular-targeted therapy by inhibiting well known MAPK and PI3K signaling pathways.
Financial support and sponsorship
The present work is partly supported by Grant-in-Aid from Council of Science and Technology, U. P., Lucknow (letter no. CST/SERPD/D-212, dated 11.05.2015).
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We would like to thank all the volunteers who participated in the study. We are grateful to Dr. Pratima Verma, Dr. Sachil Vohra and Mr. Pravin Kumar Gangwar for their constant encouragement and help in drafting of this manuscript.
REFERENCES
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. Lyon, France: International Agency for Research on Cancer; 2013. [Last accessed on 2017 Oct 22]. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC Cancer Base No 11. Available from: http://www.globocan.iarc.fr . [Google Scholar]
- 2.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–45. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 3.Fakhry C, Gillison ML. Clinical implications of human papillomavirus in head and neck cancers. J Clin Oncol. 2006;24:2606–11. doi: 10.1200/JCO.2006.06.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh V, Husain N, Akhtar N, Kumar V, Tewari S, Mishra S, et al. Do human papilloma viruses play any role in oral squamous cell carcinoma in North Indians? Asian Pac J Cancer Prev. 2015;16:7077–84. doi: 10.7314/apjcp.2015.16.16.7077. [DOI] [PubMed] [Google Scholar]
- 5.Repasky GA, Chenette EJ, Der CJ. Renewing the conspiracy theory debate: Does Raf function alone to mediate ras oncogenesis? Trends Cell Biol. 2004;14:639–47. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding ras: ‘it ain’t over ‘til it's over’. Trends Cell Biol. 2000;10:147–54. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 7.Krishna A, Singh S, Kumar V, Pal US. Molecular concept in human oral cancer. Natl J Maxillofac Surg. 2015;6:9–15. doi: 10.4103/0975-5950.168235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kratz CP, Niemeyer CM, Zenker M. An unexpected new role of mutant ras: Perturbation of human embryonic development. J Mol Med (Berl) 2007;85:227–35. doi: 10.1007/s00109-006-0135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young A, Lou D, McCormick F. Oncogenic and wild-type ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3:112–23. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Gimenez-Conti IB, Cunningham JE, Collet AM, Luna MA, Lanfranchi HE, et al. Alterations of p53, cyclin D1, Rb, and H-ras in human oral carcinomas related to tobacco use. Cancer. 1998;83:204–12. doi: 10.1002/(sici)1097-0142(19980715)83:2<204::aid-cncr2>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 11.Warnakulasuriya KA, Chang SE, Johnson NW. Point mutations in the Ha-ras oncogene are detectable in formalin-fixed tissues of oral squamous cell carcinomas, but are infrequent in British cases. J Oral Pathol Med. 1992;21:225–9. doi: 10.1111/j.1600-0714.1992.tb00106.x. [DOI] [PubMed] [Google Scholar]
- 12.Saranath D, Chang SE, Bhoite LT, Panchal RG, Kerr IB, Mehta AR, et al. High frequency mutation in codons 12 and 61 of H-ras oncogene in chewing tobacco-related human oral carcinoma in India. Br J Cancer. 1991;63:573–8. doi: 10.1038/bjc.1991.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakata K. Alterations of tumor suppressor genes and the H-ras oncogene in oral squamous cell carcinoma. J Oral Pathol Med. 1996;25:302–7. doi: 10.1111/j.1600-0714.1996.tb00266.x. [DOI] [PubMed] [Google Scholar]
- 14.Das N, Majumder J, DasGupta UB. Ras gene mutations in oral cancer in Eastern India. Oral Oncol. 2000;36:76–80. doi: 10.1016/s1368-8375(99)00058-5. [DOI] [PubMed] [Google Scholar]
- 15.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–51. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Rawi NH, Merza MS, Ghazi AM. PIK3CB and K-ras in oral squamous Cell carcinoma. A possible cross-talk! J Orofac Sci. 2014;6:99–103. [Google Scholar]
- 17.Barnes L, Eveson JW, Reichart P, Sidransky D. Pathology and Genetics of Head and Neck Tumours. 3rd ed. Lyon: IARC Press; 2005. WHO Classification of Tumours. [Google Scholar]
- 18.Edge SB, Byrd DR, Compton CC, Fritz AJ, Greene FL, Trotti A. AJCC Cancer Staging Manual. 7th ed. New York: Springer-Verlag; 2010. pp. 29–35. [Google Scholar]
- 19.Vairaktaris E, Papakosta V, Derka S, Vassiliou S, Nkenke E, Spyridonidou S, et al. H-ras and c-fos exhibit similar expression patterns during most stages of oral oncogenesis. In Vivo. 2008;22:621–8. [PubMed] [Google Scholar]
- 20.de Launay D, Vreijling J, Hartkamp LM, Karpus ON, Abreu JR, van Maanen MA, et al. Silencing the expression of Ras family GTPase homologues decreases inflammation and joint destruction in experimental arthritis. Am J Pathol. 2010;177:3010–24. doi: 10.2353/ajpath.2010.091053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Krishna A, Singh RK, Singh S, Verma P, Pal US, Tiwari S, et al. Demographic risk factors, affected anatomical sites and clinicopathological profile for oral squamous cell carcinoma in a North Indian population. Asian Pac J Cancer Prev. 2014;15:6755–60. doi: 10.7314/apjcp.2014.15.16.6755. [DOI] [PubMed] [Google Scholar]
- 23.Shimizu N, Ohtsubo M, Minoshima S. MutationView/KMcancerDB: A database for cancer gene mutations. Cancer Sci. 2007;98:259–67. doi: 10.1111/j.1349-7006.2007.00405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanwar M, Kowluru RA. Diabetes regulates small molecular weight G-protein, H-ras, in the microvasculature of the retina: Implication in the development of retinopathy. Microvasc Res. 2008;76:189–93. doi: 10.1016/j.mvr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitale-Cross L, Amornphimoltham P, Fisher G, Molinolo AA, Gutkind JS. Conditional expression of K-ras in an epithelial compartment that includes the stem cells is sufficient to promote squamous cell carcinogenesis. Cancer Res. 2004;64:8804–7. doi: 10.1158/0008-5472.CAN-04-2623. [DOI] [PubMed] [Google Scholar]
- 26.Camps C, Sirera R, Bremnes R, Blasco A, Sancho E, Bayo P, et al. Is there a prognostic role of K-ras point mutations in the serum of patients with advanced non-small cell lung cancer? Lung Cancer. 2005;50:339–46. doi: 10.1016/j.lungcan.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer. 2011;50:307–12. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 28.Johne A, Roots I, Brockmöller J. A single nucleotide polymorphism in the human H-ras proto-oncogene determines the risk of urinary bladder cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:68–70. [PubMed] [Google Scholar]
- 29.Cutilli T, Papola F, Di Emidio P, Corbacelli A. P53 tumor suppressor protein and H-RAS oncogene in maxillofacial tumors: Immunohistochemical and genetic investigation, induction chemotherapy response and prognosis evaluation. J Chemother. 1998;10:411–7. doi: 10.1179/joc.1998.10.5.411. [DOI] [PubMed] [Google Scholar]
- 30.McDonald JS, Jones H, Pavelic ZP, Pavelic LJ, Stambrook PJ, Gluckman JL, et al. Immunohistochemical detection of the H-ras, K-ras, and N-ras oncogenes in squamous cell carcinoma of the head and neck. J Oral Pathol Med. 1994;23:342–6. doi: 10.1111/j.1600-0714.1994.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 31.Paterson IC, Eveson JW, Prime SS. Molecular changes in oral cancer may reflect aetiology and ethnic origin. Eur J Cancer B Oral Oncol. 1996;32B:150–3. doi: 10.1016/0964-1955(95)00065-8. [DOI] [PubMed] [Google Scholar]
- 32.Chang KW, Sarraj S, Lin SC, Tsai PI, Solt D. P53 expression, p53 and Ha-ras mutation and telomerase activation during nitrosamine-mediated hamster pouch carcinogenesis. Carcinogenesis. 2000;21:1441–51. [PubMed] [Google Scholar]
- 33.Trivedi TI, Tankshali RA, Goswami JV, Shukla SN, Shah PM, Shah NG, et al. Identification of site-specific prognostic biomarkers in patients with oral squamous cell carcinoma. Neoplasma. 2011;58:217–26. doi: 10.4149/neo_2011_03_217. [DOI] [PubMed] [Google Scholar]
- 34.Goebell PJ, Knowles MA. Bladder cancer or bladder cancers? Genetically distinct malignant conditions of the urothelium. Urol Oncol. 2010;28:409–28. doi: 10.1016/j.urolonc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Han J, Wang L, Wang X, Li K. Downregulation of microrna-126 contributes to tumorigenesis of squamous tongue cell carcinoma via targeting KRAS. Med Sci Monit. 2016;22:522–9. doi: 10.12659/MSM.895306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiaris H, Spandidos DA. Analysis of H-ras, K-ras and N-ras genes for expression, mutation and amplification in laryngeal tumours. Int J Oncol. 1995;7:75–80. doi: 10.3892/ijo.7.1.75. [DOI] [PubMed] [Google Scholar]
- 37.Kiaris H, Spandidos DA, Jones AS, Vaughan ED, Field JK. Mutations, expression and genomic instability of the H-ras proto-oncogene in squamous cell carcinomas of the head and neck. Br J Cancer. 1995;72:123–8. doi: 10.1038/bjc.1995.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu XY, Liu WT, Wu ZF, Chen C, Liu JY, Wu GN, et al. Identification of HRAS as cancer-promoting gene in gastric carcinoma cell aggressiveness. Am J Cancer Res. 2016;6:1935–48. [PMC free article] [PubMed] [Google Scholar]


