CONSPECTUS
The ability to sense and manipulate the state of biological systems has been extensively advanced during the last decade with the help of recent developments in physical tools. Unlike standard genetic and pharmacological perturbation techniques – knockdown, overexpression, small molecule inhibition – that provide a basic on/off switching capability, these physical tools provide the capacity to control the spatial, temporal, and mechanical properties of the biological targets.
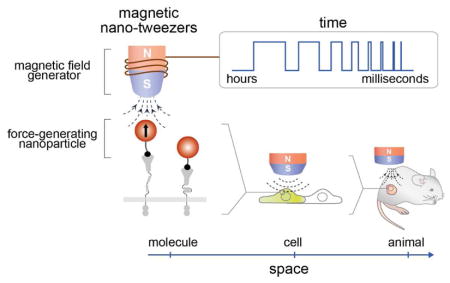
Among the various physical cues, magnetism offers distinct advantages over light or electricity. Magnetic fields freely penetrate biological tissues and are already used for clinical applications. As one of the unique features, magnetic fields can be transformed into mechanical stimuli which can serve as a cue in regulating biological processes. However, their biological applications have been limited due to a lack of high-performance magnetism-to-mechanical force transducers with advanced spatiotemporal capabilities. In this account, we present recent developments in magnetic nano-tweezers (MNTs) as a high-performance tool for interrogating the spatiotemporal control of cells in living tissue.
MNTs are comprised of force-generating magnetic nanoparticles and field generators. Through proper design and the integration of individual components, MNTs deliver controlled mechanical stimulation to targeted biomolecules at any desired space and time. We first discuss about MNT configuration with different force-stimulation modes. By modulating geometry of the magnetic field generator, MNTs exert pulling, dipole-dipole attraction, and rotational forces to the target specifically and quantitatively. We discuss the key physical parameters determining force magnitude, which include magnetic field strength, magnetic field gradient, magnetic moment of the magnetic particle, as well as distance between the field generator and the particle. MNTs also can be used over a wide range of biological time scales — By simply adjusting the amplitude and phase of the applied current, MNTs based on electromagnets allow for dynamic control of the magnetic field from microseconds to hours. Chemical design and the nanoscale effects of magnetic particles are also essential for optimizing MNT performance. We discuss key strategies to develop magnetic nanoparticles with improved force-generation capabilities with a particular focus on the effects of size, shape, and composition of the nanoparticles. We then introduce various strategies and design considerations for target-specific biomechanical stimulations with MNTs. One-to-one particle-receptor engagement for delivering a defined force to the targeted receptor and the small size of the nanoparticles are important. Finally, we demonstrate the utility of MNTs for manipulating biological functions and activities with various spatial (single molecule/cell to organisms) and temporal resolution (microseconds to days).
MNTs have the potential to be utilized in many exciting applications across diverse biological systems spanning from fundamental biology investigations of spatial and mechanical signaling dynamics at the single-cell and systems levels to in vivo therapeutic applications.
Graphical Abstract
1. Introduction
Mechanical transducers, devices that convert one form of energy to another via mechanical elements, are commonly employed in many current areas of science and technology1. Cells utilize miniaturized single-molecule mechanical transducers that convert extracellular physical cues into intracellular biochemical or neuroelectrical signals, (i.e. mechanoreceptors).2,3 Surprisingly, these tiny molecular transducers can detect spatial and temporal information related to the environment of the cell, such as the mechanical stiffness, and elicit corresponding cellular responses.2–4 For example, the spatial distribution of integrin-binding ligands within the extracellular matrix differentially regulates downstream cell signaling processes, determining spreading, motility, polarization, and differentiation of the cell.5–9 Auditory hair cells expressing mechanosensitive ion channels selectively detect oscillatory mechanical stimulation (i.e., sound-induced mechanical motion of the inner ear basal membranes) of specific sound waves.4,10,11
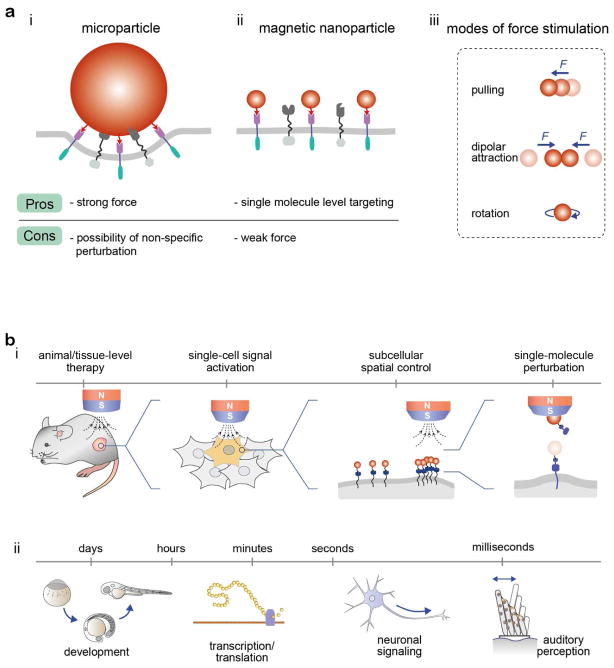
Spatiotemporal tools for applying controlled stimulation to mechanoreceptors allow us to interrogate and understand the operating mechanisms underlying these complex mechanotransduction pathways. By measuring the cellular responses to mechanical perturbations in space and time, mechanoreceptors transducing cell signals can be systematically investigated. For several decades, various microscale tweezers, including atomic force microscopy, magnetic tweezers, and optical tweezers, have played prominent roles in the study of biological systems.12 These tweezers have provided many exciting new discoveries owing to their strong force (~nanonewton, nN) generating capabilities. However, many of these techniques suffer from disadvantages associated with having rather large dimensions (~μm) and their mode of force stimulation is mostly limited to pulling mode (Figure 1a(i), Table 1).13
Figure 1.
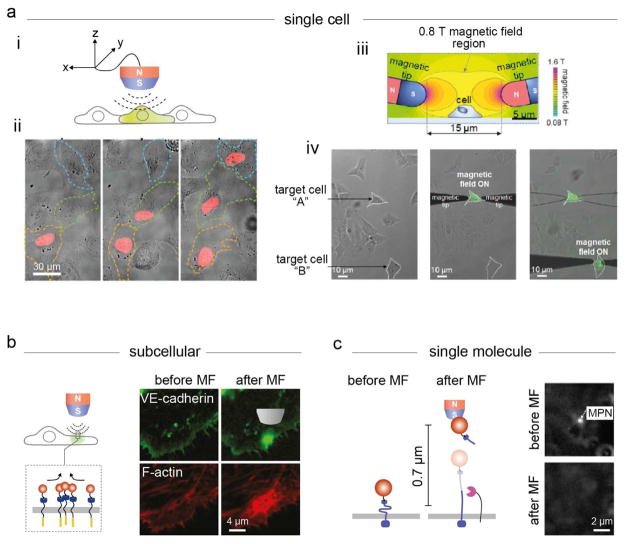
(a) Comparison between micro- and nanoparticles as force generation components with microscale (i) and nanoscale (ii) tweezers, respectively. (iii) Modes of force stimulation. (b) Applications of magnetic nano-tweezers (MNTs) in the regulation of biological process on various spatial and temporal scales. (i) Regulation of cell signaling on various biological spatial scales from organisms/tissues, to single cells and to single molecules is possible by manipulating the nanoparticles with an external magnetic field. (ii) Extremely fast or slow mechanotransduction processes can be precisely recapitulated using MNTs.
Table 1.
Comparison of magnetic nano-tweezers with conventional tweezers*.
| Nanoscale tweezers | Conventional tweezers | ||||
|---|---|---|---|---|---|
| Magnetic nano-tweezers (MNTs) | Magnetic tweezers | Optical tweezers | AFM | ||
| Probe type | Single magnetic nanoparticle | Clusters of magnetic nanoparticles | Magnetic microparticles | Polystyrene or silica microparticles | AFM Cantilever |
| Primary mode of force stimulation | - Pulling - Dipolar-attraction - Rotation |
- Pulling | - Pulling - Rotation |
- Pulling | - Pulling - Push |
| Force range | Weak (~ hundreds of fN) | Strong (~ sub-nN) | Strong (10−2 – 102 pN) | Strong (10−1 – 102 pN) | Very strong (101 – 104 pN) |
| Target dimension | Single molecule to cell and tissue | Single cell to tissue | Single molecule to cell and tissue | Single molecule to single cell | Single molecule to single cell |
| Target labeling density (probes/cell) | High (101–105 probes/cell#) | Low (one or a few probes/cell) | Low (one or a few probes/cell) | Low (one probe/cell) | |
general observation, but can be varied depending on the experimental condition, instrument, and setup
depends on the target receptor density
Recently, as a new enabling nanoscale tool, magnetic nano-tweezers (MNTs) have been developed for the controlled stimulation of targeted receptors.13–22 MNTs use magnetic nanoparticles with sizes comparable to those of conventional proteins as a force-generating component (Figure 1a(ii)). Therefore, MNTs can target specific cells, deliver a defined mechanical force at a desired location and time by tuning the configuration of the external magnets, and hence regulate cellular responses. They can create multiple modes of force stimulation, including pulling, dipolar attraction, and rotation (Figure 1a(iii)). This capacity of the tool has opened exciting opportunities for interrogating cell signaling that are not available with conventional microscale tweezers. Regulation of cell signaling on various biological spatial scales from macroscopic organisms/tissues, to single cells, and even to single molecules is possible by manipulating the nanoparticles with an external magnetic field (Figure 1b(i)).13–20 This ability has enabled the remote control of animal development and behaviors,14,15 single-cell and multi-cell signaling control,13,15–22 subcellular localized aggregation of receptors,13,20 and single receptor activation.13,20 MNTs also allow for receptor manipulation over wide ranges of temporal scales that cover the dynamics of most biological events. Extremely fast (e.g., sound perception; μs-ms) or slow (e.g., tissue pressure changes during development; hours to days) mechanotransduction processes can be precisely recapitulated using MNTs (Figure 1b(ii)).21,22
To summarize the features of MNTs presented here, we provide a comparison with the features of conventional tweezers in Table 1.12,23,24
2. Modes of force stimulation in MNTs
MNTs are comprised of two components: force-generating magnetic nanoparticles and an external magnetic field generator. By using a gradient or uniform magnetic field, several different modes of force stimulation can be achieved to steer the magnetic nanoparticles. Here, we discuss basic principles of force-generation using different field application modes. Construction of external magnetic field generators required for these modes is described in previous publications by our lab and other researchers.20,22,25
2.1. Pulling mode under gradient magnetic field
The forces involved in cellular events span a broad range from a few femtonewtons (fN) to sub-nanonewtons.12,26 Single-membrane protein diffusion based on Brownian motion is typically on the order of tens of femtonewtons.14,27 In contrast, the rupture force of a single protein domain-domain interaction is roughly estimated to be 1–100 piconewtons (pN),28 and forces exerted by single cells (e.g., hair cell deflection) can reach sub-nanonewtons.29,30
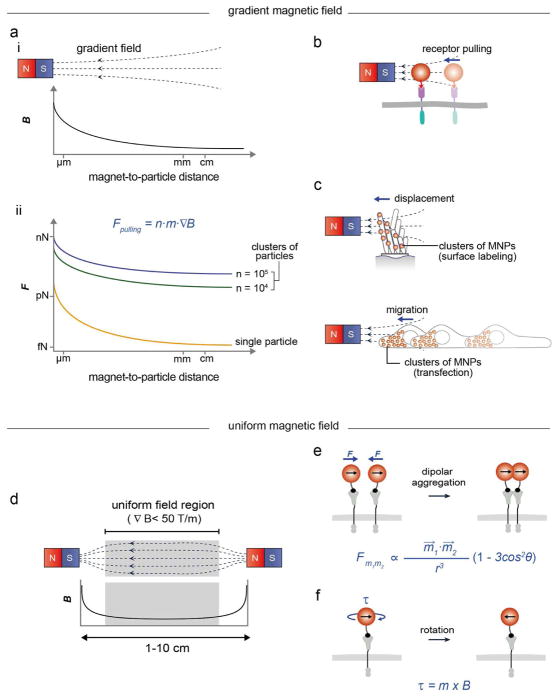
Magnetic nanoparticles in a gradient magnetic field (Figure 2a(i)) experience a pulling force (Figure 2a(ii)) proportional to the number of magnetic nanoparticles (n), the magnetic moment of the nanoparticles (m), and the magnetic field gradient (∇B), where Fpulling = n·m·∇B, B ≫ kBT/m (kB, Boltzmann constant; T, temperature).31–33 Typically, a single magnetic nanoparticle can have a pulling force in the femtonewton range, which is sufficient for the translocation of membrane receptors requiring ca. 0.1 femtonewton (Figure 2a(ii), orange and Figure 2b).13,14,20
Figure 2.
Modes of force stimulation in MNTs. (a–c) Pulling force modes. (a) Gradient magnetic field generated by a single magnet (i) and the corresponding force magnitude exerted by magnetic nanoparticles (MNPs) as a function of the magnet-to-particle distance and the number of MNPs (ii). (b) The pulling force exerted by a single MNP allows for the translocation of membrane receptors. (c) The displacement (top) and migration (bottom) of cells can induced with clusters of MNPs. (d–f) Dipolar attraction and rotation modes. (c) Uniform magnetic field generated between two magnets. (e, f) Dipolar attraction between magnetic nanoparticles induces the receptor clustering (e) and the rotation of particles for torque generation (f). n, number of nanoparticles; m⃗, magnetic moment; B, magnetic field strength; ∇B, magnetic field gradient, r, distance between nanoparticles θ, angle between the direction of external magnetic field and the centerline of two magnetic nanoparticles; τ, torque.
The pulling force generated by magnetic nanoparticles can be easily increased to the piconewton range by i) shortening the magnet-to-particle distance or ii) producing clusters of magnetic nanoparticles. When magnetic nanoparticles are positioned within hundreds of nanometers of the magnet, individual nanoparticles can exert piconewton forces due to the steep magnetic field gradient.13,20 A conformational change can be triggered in proteins with such a force magnitude.13,20 Labeling a cell with multiple nanoparticles (i.e., clusters of 103 – 105 magnetic nanoparticles) through either the binding of surface receptors or intracellular transfection can exert stronger forces (Figure 2a(ii), green and blue).21,22 The advantage is that MNTs can apply piconewton-range forces to targets located a few centimeters (~ 3 cm) away from the magnet. For example, these clusters of magnetic nanoparticles allow for the remote in vivo manipulation of cell displacement (Figure 2c, top) and migration in animals (Figure 2c, bottom).21,22
2.2. Dipolar attraction and rotation modes under uniform magnetic field
A magnetic field generator design that exerts uniform magnetic fields provides another way to steer magnetic nanoparticles. In this case, the magnetic field induces magnetization of the nanoparticles; however, due to the field uniformity (∇B < 50 T/m, Figure 2d), the nanoparticles do not experience a pulling force arising from the field gradient. Instead, when the nanoparticles are in close proximity (< 100 nm) to one another, they experience magnetic dipole-dipole attraction force, which can induce aggregation (Figure 2e).34 When two magnetic particles are placed under the uniform magnetic field, force (Fm1m2)is defined as follows34:
where, r is interparticle distance, m⃗1, m⃗2 are magnetic dipole moments of nanoparticles, and θ is angle between the direction of external magnetic field and the centerline of two magnetic nanoparticles. This magnetic dipolar attraction force can be in the range of tens of femtonewtons at a relatively weak external magnetic field strength with a long working distance (1–10 cm).14,15
Distinct from other force microscopy tools, magnetic particles can rotate under a rotating external magnetic field.35 A magnetic particle having magnetic moment (m) can rotate to align with the direction of external magnetic field (B), and hence generate torque (τ), where τ = m × B (Figure 2f).36 This rotating mode has been extensively used for investigating the dynamics of single molecules such as supercoiled DNA with magnetic particles.37,38 Also, magnetic twisting cytometry on cells has been demonstrated for studying force transmission across the cell membrane and assessing cell stiffness.39,40
3. Temporal resolution of MNTs
MNTs based on permanent magnets can provide only a static magnetic field. In this case, the temporal resolution (i.e., magnetic field on/off) for the fast mode can be on the order of a few seconds. On the other hand, MNTs based on electromagnets allow for dynamic control of the magnetic field and hence the magnetic nanoparticles. By simply adjusting the amplitude and phase of the applied current, the magnetic field strength and orientation can be manipulated. This unique property of electromagnets provides programmed and rapid manipulation of magnetic particles and hence the targeted biomolecules with high temporal precision. Recently, a micro-electromagnet comprising a permalloy needle (tip diameter: 10 μm) housed inside a water cooling jacket was developed.21,22 It can selectively generate a magnetic field within a single cell with a 50 μs temporal resolution without noticeable heat generation. This ability of the micro-electromagnets is particularly useful for investigating rapid mechanotransduction pathways such as hearing at the single-cell level.21,22
4. Design of force-generating magnetic nanoparticles
Magnetic nanoparticles are conjugated to targeting moieties for the selective stimulation of desired targets. Several critical considerations should be taken into account when designing magnetic nanoparticles and conjugating them with targeting moieties to produce a force-generating component.
4.1. Magnetic nanoparticles
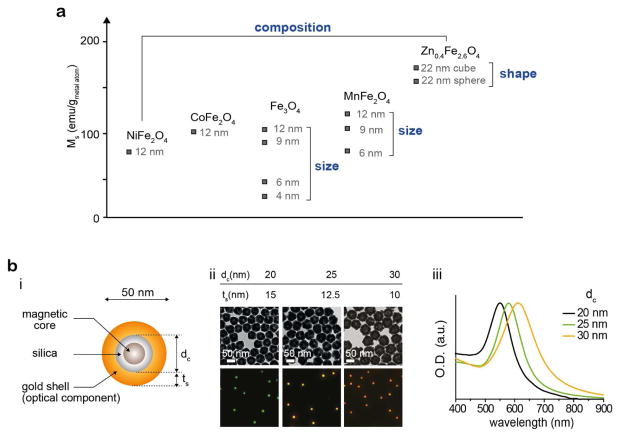
Among the various types of magnetic nanoparticles, ferrite-based nanoparticles are most commonly utilized for MNTs due to their excellent biocompatibility. Typical ferrite (that is, iron oxide) nanoparticles can usually exert a few tens of femtonewtons force under magnetic field.16 The force exertion capability of these magnetic nanoparticles can be improved by engineering ferrite nanoparticles (Figure 3a).41–44 By tuning the size (4–100 nm)41,44, composition (Ni, Mn, Co and Zn doping)41,42 and shape (spheres and cubes),43 single ferrite nanoparticles can exert forces up to sub-piconewtons.13–15,17,20–22
Figure 3.
(a) Magnetism engineering of magnetic nanoparticles for effective force generation. By tuning the size, composition, and shape of magnetic nanoparticles, a higher saturation magnetization (Ms), which is a critical factor for force generation, can be achieved. (b) Magnetoplasmonic nanoparticles (MPNs). (i) A MPN is comprised of a magnetic core and a plasmonic shell that serve as the force-generating and imaging components, respectively. (ii) Silica core size (dc)-dependent plasmonic absorption shift. ts indicates thickness of gold shells. TEM images (top) of 50 nm MPNs with 20 nm (left), 25 nm (middle) and 30 nm (right) silica cores and their corresponding scattering images (bottom). (iii) Absorption spectra of 50 nm MPNs with 20 nm, 25 nm and 30 nm silica cores. Reproduced with permission from ref 20, copyright 2017 Nature Publishing Group.
The size of magnetic nanoparticles should be carefully considered in terms of the desired cell labeling density.13 Cell membranes are densely covered by a carbohydrate matrix forming the glycocalyx.45 This sugar layer serves as a permeability barrier to foreign materials, limiting the access of large particles to the cell surface receptors. Hence, the number of nanoparticles that can bind to a cell surface is inversely correlated with particle size. For example, under identical labeling conditions, magnetic nanoparticles (50 nm) have a 104–105 times higher target labeling efficiency than magnetic microbeads (2.8 μm).13 Typically, particles that range from 10–50 nm in diameter are ideal, where relatively large particles are suitable for force microscopy applications that require strong force generation and small nanoparticles are beneficial for maximizing receptor labeling and minimizing nonspecific perturbation.13,20
In contrast to conventional microscale tweezers, to observe nanoscale particles, additional optical moieties are required since nanoparticles are too small to be detected by optical microscopy due to the diffraction limit of light. Fluorescent dyes, quantum dots, and gold nanoparticles can be used for this purpose. For example, magnetoplasmonic nanoparticles (MPNs) that have gold shells as an imaging component allow for the real-time monitoring of single nanoparticles under an optical microscope (Figure 3b(i)).13,20 The scattering colors can also be precisely tuned from 550 to 620 nm depending on the gold shell thickness (Figure 3b(ii, iii)).
4.2. Targeting moieties
Several targeting strategies have been employed for magnetic nanoparticle-receptor labeling. Among them, a common strategy is to use antibody-antigen interactions, where antibodies specific to the target receptors are covalently conjugated to the nanoparticles via chemical crosslinkers. Magnetic nanoparticles with anti-Tie2 and anti-death receptor 4 (DR 4) antibodies were synthesized via carbodiimide crosslinker chemistry and used for targeting and controlling Tie2 and DR 4 receptors, respectively.14,17 The other method is to decorate magnetic nanoparticles with native ligands that selectively bind to target receptors via ligand-receptor interactions (e.g., IgE-FcεRI16 or delta-like ligand (Dll)-Notch13). The genetically encoded targeting of receptors with magnetic nanoparticles is also possible via recombinant DNA technology. In this case, various polypeptide protein tags such as Myc-, His- and SNAP-tags can be inserted into a target receptor sequence and labelled with the corresponding antibody-conjugated or small molecule labeled magnetic nanoparticles.13,20
While various targeting strategies are available, several important design considerations for the targeting domain should be taken into account. To deliver a defined force to the targeted receptor, a one-to-one particle-receptor engagement with minimized nonspecific receptor perturbation is important. The presence of multiple ligands per nanoparticle is not desirable because the force generated by a single particle is distributed across multiple receptors.13,20 Also, the presence of multiple ligands can induce receptor crosslinking and change the receptor dynamics, including diffusion, translocation, and signal transduction.13,14,20 The simplest way to achieve monovalent conjugation is through the stoichiometric control of the targeting moieties and magnetic nanoparticles.13,14,20 For example, nanoparticles bearing a single oligonucleotide can be obtained via either steric exclusion or charge-based valency discrimination. Targeting biomolecules can be further conjugated via complementary DNA or RNA hybridization or end-functionalization of the oligonucleotide strand.
5. Spatiotemporal manipulation of cell signaling with MNTs
5.1. Manipulation of biological activities on different spatial scales
Early attempts to use magnetic nano-tweezers focused on stimulating a group of cells located in the proximity of a magnetic needle in a cell culture. Aggregation and activation of FcεRI receptors in cells was reported using IgE-coated magnetic nanoparticles.16 Similarly, microtubule polymerization could be regulated by magnetic nanoparticles through induced localized aggregation of Rho-GTPase and Ran GTP.18,19
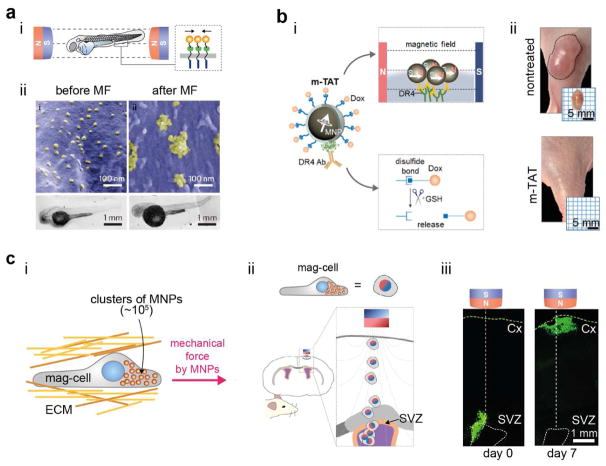
The applicability of MNTs can be further extended to in vivo systems.14,15 Magnetic nanoparticles were delivered into the yolk of a zebrafish embryo to target the ovarian TNF receptors (OTRs). Then, a uniform magnetic field was applied to induce aggregation of the OTRs by using the dipolar attraction mode to trigger apoptotic signaling (Figure 4a(i)).14 A characteristic phenotypic defect in the tail region of zebra fish was observed for the magnetically stimulated group compared with the negative control groups (Figure 4a(ii)). This approach could be further applied to a larger animal model by implementing the magnetic tandem apoptotic trigger (mTAT) concept in a system for treating multidrug resistant (MDR) cancer.15 The mTAT system could stimulate both extrinsic (i.e., DR 4 signaling) and intrinsic (i.e., DNA damage) apoptotic signals through magnetically controlled receptor aggregation and intracellular anti-cancer drug release (Figure 4b(i)). This ability led to the complete removal of MDR cancer from a mouse xenograft model, which was not possible with chemical drug treatment alone (Figure 4b(ii)).
Figure 4.
In vivo manipulation of biological activities with MNTs at the tissue level. (a) Magnetic activation of apoptotic signaling in zebra fish. (i) The aggregation of ovarian TNF receptors (OTRs) triggered by the dipolar attraction force induce apoptosis. (ii) SEM images of aggregated magnetic nanoparticles on the cell surface and the corresponding tail region bending. (b) Schematic illustration of magnetic tandem apoptosis trigger (m-TAT) and its dual modes of stimulation (i), and photographs of xenograft tumor-bearing mice obtained after treatment (day 14) (ii). (c) Guiding of cell migration by using a magnetic pulling force. Magnetically labelled cells (mag-cell) can remotely control cell migration (i) and thus deliver cells from the subventricular zone (SVZ) to the cortex (Cx) within a rat brain (ii). (iii) Immunohistochemistry images showing magnetism-guided cell migration within a rat brain. The green color indicates magnetic nanoparticle-loaded cells. Reproduced with permission from ref 14, copyright 2012 Nature Publishing Group; Reproduced with permission from refs 15, 46, Copyright 2016, 2018 American Chemical Society.
In contrast to the dipolar attraction mode, the usage of the pulling mode in in vivo applications is often limited due to exponential decay of the magnetic field strength. However, when loaded with clusters of magnetic nanoparticles (~105), cells can experience a piconewton-level pulling force even when located a few centimeters away from the magnet. Such magnetic nanoparticles loaded cells, termed magnetically-labeled cells (Mag-Cells), can be pulled by external magnet and thus can be delivered to a target location within in vivo tissue environments such as a rat brain (Figure 4c(i–iii).46
By implementing suitable magnetic nanoparticles and improved magnet designs, MNTs can be used for single-cell and subcellular signaling control. Specifically, under a gradient magnetic field with a magnet-to-particle distance of sub-micrometers, magnetic nanoparticles can exert force on targeted Notch receptors, leading to mechanical activation of Notch receptor signaling (Figure 5a(i)). In a model Gal4/upstream activation sequence (UAS) reporter system, the transcriptional profiles of targeted cells could be systematically tuned at any desired location with single-cell resolution (Figure 5a(ii)). Single-cell apoptosis has also been demonstrated with a m-TAT system (Figure 5a(iii, iv)).15 A pair of micro-magnets applied a uniform magnetic field only to the targeted cell region, induced DR 4 aggregation and initiated apoptosis signaling. MNTs further allowed for interrogation of the subcellular receptor signaling dynamics under a gradient magnetic field (Figure 5b).13,20 Magnetic nanoparticles could induce the localized aggregation of cadherin and hence promote stable adherens junction formation at a desired subcellular location.
Figure 5.
Manipulation of biological activities with MNTs from the single-cell to the single-molecule level. (a) Single-cell signaling control. (i) Mechanical activation of Notch receptor signaling. (ii) The mCherry fluorescence increases under sequential magnetic field application to 3 target cells. (iii) Magnetic field simulation for single-cell-level apoptosis induction by m-TAT. (iv) Sequential activation of apoptosis signaling. The green fluorescence signal (FITC-annexin V) indicates the induction of apoptosis. (b) Interrogation of subcellular VE-cadherin-mediated adherens junction formation: (top) MPN-induced VE-cadherin clustering; (bottom) F-actin assembly recruitment. (c) Single-molecule activation of mechanoreceptors in live cells. Under the application of a strong pulling force (9 pN, top), the immediate detachment of the MPN is detected under dark-field microscopy (bottom). Reproduced from ref 13, Copyright 2016, with permission from Elsevier; Reproduced with permission from ref 15, Copyright 2016 American Chemical Society; Reproduced with permission from ref 20, copyright 2017 Nature Publishing Group.
MNTs can also be used for the single-molecule activation of mechanoreceptors in live cells (Figure 5c).13,20 The key advantages of MNT-based single-molecule studies in live cells compared with conventional single-molecule force microscopy are that monovalent nanoparticles ensure one-to-one engagement of the force-generating particle with the targeted receptor and do not perturb the dynamic properties and mechanical environment of the receptors.
5.2. Toward the interrogation of rapid mechanotransduction dynamics
The time scale of biological events varies from microseconds to days.47 Among them, sound perception is one of the most rapid mechanotransduction process.4,10,11 Human hair cells sense the mechanical vibration of sound in the frequency range of 20–20,000 Hz.11 While MNTs that cover biological time scales of minute to hours (Notch receptor signaling-based DNA transcription/translation13,20) and days (apoptosis14 and angiogenesis17) have been successfully demonstrated, those with high-speed capabilities have not been possible until recently.
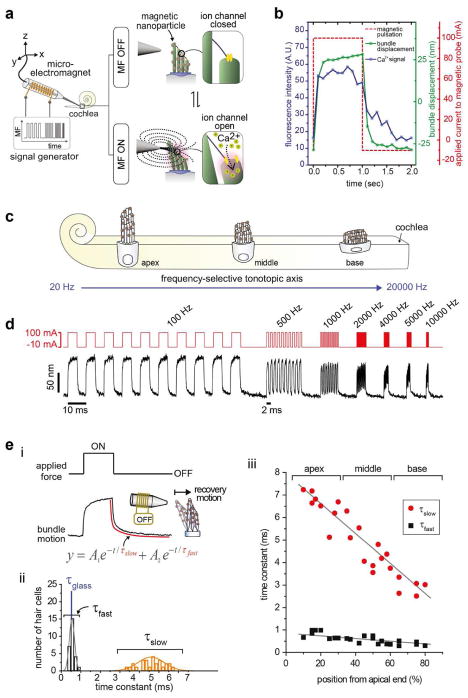
To interrogate the fast dynamic mechanotransduction process, ultra-fast MNTs consisting of 40 nm Zn-doped ferrite nanocubes and a micro-electromagnet were developed.21,22 Individual hair bundles labeled with magnetic nanoparticles can be deflected with an applied magnetic field (Figure 6a). Approximately 1,400 magnetic nanoparticles are bound to each hair bundle, and they provide a pulling force of up to 280 pN. Hair bundle deflection induces mechanical tension in the tip links and opens the ion channels, allowing for an influx of Ca2+. The gating of the mechanosensitive ion channels coincides with the applied magnetic pulses from calcium imaging (Figure 6b). Indeed, ultra-fast MNTs can provide bundle oscillations of up to 50 kHz. This temporal resolution is 1,000 times faster than that of conventional MNTs (Figure 6c). When the hair bundle is stimulated with the audible frequencies of humans, its oscillations remain phase-locked over the tested frequencies (Figure 6d).
Figure 6.
MNTs for studying fast mechanotransduction dynamics with the non-contact mode. (a) Schematic illustration of the high-frequency magnetic control of hair bundle oscillation. Hair bundles labeled with 40 nm Zn-doped ferrite nanocubes are pulled with a micro-electromagnet. Mechanical tension on the tip links, induced by hair bundle deflection, opens the ion channels, creating an influx of Ca2+. (b) Gating of the mechanosensitive ion channels in a hair bundle using MNTs. The Ca2+ influx in the bundle coincides with the applied magnetic pulses. (c) Hearing range of human hair cells. Hair cells are located along the tonotopic axis of the cochlea and detect specific resonance frequencies. (d) Magnetic entrainment of hair bundle oscillations; pulse frequencies of 100, 500, 1000, 2000, 4000, 5000, and 10000 Hz are applied (top), and the hair bundle displacements are observed (bottom). (e) MNTs for interrogating the biomechanics of the hair bundle recovery process. (i) The recovery motion of the deflected hair bundle is tracked and fitted with second-order exponential functions (equation, red line) to extract two biological components (τslow and τfast). (ii) Distribution of τslow and τfast of the middle-region hair bundle. The time constant of a glass fiber (τglass, blue) is shown as a passive object for comparison. (iii) Tonotopic dependency of the time constants measured along the entire tonotopic axis from the apical to the basal region of the cochlea. Reproduced with permission from refs 21, 22, Copyright 2014, 2016 American Chemical Society.
Noncontact-mode force stimulation is another unique capacity of ultra-fast MNTs that has led to new findings in hair cell dynamics in response to mechanical perturbation. Specifically, the noncontact-mode is beneficial for the readout cellular components involved in the oscillatory dynamics of free-standing hair bundles.22 From exponential fitting of the recovery motions of hair bundles after magnetic deflection (Figure 6e(i)), two relaxation components with different time scales (τfast and τslow) were observed (Figure 6e(ii)). τfast and τslow were closely related to the passive physical properties of hair bundles and the active actomyosin interactions, respectively. Additionally, the active recovery motions of hair bundles showed unique frequency-dependent characteristics, where basal, middle, and apical hair bundles could effectively respond to their respective ranges of frequency (Figure 6e(iii)). These experiments provided new insights into the frequency selection mechanisms of hair cells along the tonotopic axis.
6. Conclusion and outlook
In this account, we present a review of novel and potentially generalizable strategies for studying biological activities with MNTs. Nano-tweezers provide key advantages over conventional force microscopy tools, particularly for interrogating and manipulating the spatiotemporal dynamics of cellular signaling and living organisms with high precision. Limitations of the systems also exist and include a relatively high magnetic field gradient for strong pulling force exertion and poor accessibility to intracellular targets. Further improvements may be achieved by developing nanoparticles that have higher magnetization values, implementing spatiotemporally optimized magnets with more precise field control (e.g., superconducting magnetic coils), and developing genetically encoded magnetic nanoparticles beyond the current ferritin system (e.g., genetic expression of magnetosomes48–50 in mammalian systems). With their unique and versatile capabilities, nano-tweezers are being developed to investigate many important biological questions during developmental, physiological, and pathological processes and to provide new therapeutic interventions.
Acknowledgments
We thank to Dr. Tae-Hyun Shin for helpful discussions. This work was supported by IBS-R026-D1 from IBS (Y.J. and J.C.), HI08C2149 from the Korea Healthcare Technology R&D Project (J.C.), 1R01GM112081 and 1R01GM126542-01 from the National Institute of General Medical Science (NIGMS) and the National Institute of Health (NIH) (Y.J.), and 1R21NS103240 from the National Institute of Neurological Disorders and Stroke (NINDS) and the NIH (Y.J.).
Biographies
Ji-wook Kim received his B.S. and Ph.D. degrees in Chemistry from Yonsei University in 2010 and 2017, respectively. He is currently a post-doctoral fellow at IBS. His research interests are in designing and constructing force generating magnetic nanoparticles and their biological applications.
Hee-Kyung Jeong graduated from Yonsei University in 2015 with a B.S. in Biochemistry and is on a track for the Ph.D. program. Her research is focused on utilizing the plasmonic nanoparticle systems to probe single molecule rotational and translation dynamics.
Kaden M. Southard received his B.S. in Chemistry from Rensselaer Polytechnic Institute in 2011. He received his Ph.D. degree in Chemistry and Chemical Biology from University of California San Francisco (UCSF) in 2017. His research interests are focused on single molecule dynamics of Notch in live cells.
Young-wook Jun is an associate professor at UCSF. He attended Yonsei University in Korea, where he received a Bachelor’s degree in Chemistry in 1999. He did his graduate studies at the Korea Advances Institute of Science. He carried out postdoctoral studies at the University of California, Berkeley, in 2007. He received several awards including the Endowed Hemming Fellowship, Sandler-UCSF New Frontier Research Award, and IUPAC Young Chemist-Honorable Mention Award. His lab focuses on the development of nanotechnology platforms to interrogate spatiotemporal dynamics of cell signaling involved in cellular development such as gene regulation, cell-cell communication, and neuron circuit formation.
Jinwoo Cheon is the Horace G. Underwood Professor at Yonsei University and the director of Institute for Basic Science (IBS) Center for Nanomedicine. He graduated from Yonsei University with a B.S. and received his Ph.D. from the University of Illinois at Urbana-Champaign. After his postdoctoral training at U.C. Berkeley and UCLA, he joined KAIST. In 2002, he moved to Yonsei University. He is the recipient of various awards, including the Ho–Am Prize, the POSCO-T. J. Park Prize, the Inchon Prize, and the Songgok Science Award. Currently, he is a senior editor of Accounts of Chemical Research and a fellow of the American Chemical Society and the Royal Society of Chemistry.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Lenk A. Electromechanical systems in microtechnology and mechatronics: electrical, mechanical and acoustic networks, their interactions and applications. Springer; Heidelberg; London: 2011. [Google Scholar]
- 2.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 3.Iskratsch T, Wolfenson H, Sheetz MP. Appreciating force and shape-the rise of mechanotransduction in cell biology. Nat Rev Mol Cell Biol. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillespie PG, Walker RG. Molecular basis of mechanosensory transduction. Nature. 2001;413:194–202. doi: 10.1038/35093011. [DOI] [PubMed] [Google Scholar]
- 5.Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagarrigue F, Anekal PV, Lee HS, Bachir AI, Ablack JN, Horwitz AF, Ginsberg MH. A RIAM/lamellipodin-talin-integrin complex forms the tip of sticky fingers that guide cell migration. Nat Commun. 2015;6:1–13. doi: 10.1038/ncomms9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Small JV, Isenberg G, Celis JE. Polarity of actin at the leading edge of cultured cells. Nature. 1978;272:638–639. doi: 10.1038/272638a0. [DOI] [PubMed] [Google Scholar]
- 9.Ingber DE. Tensegrity II. How structural networks influence cellular information processing networks. J Cell Sci. 2003;116:1397–1408. doi: 10.1242/jcs.00360. [DOI] [PubMed] [Google Scholar]
- 10.Hudspeth AJ. Integrating the active process of hair cells with cochlear function. Nat Rev Neurosci. 2014;15:600–614. doi: 10.1038/nrn3786. [DOI] [PubMed] [Google Scholar]
- 11.Mann ZF, Kelley MW. Development of tonotopy in the auditory periphery. Hear Res. 2011;276:2–15. doi: 10.1016/j.heares.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo D, Southard KM, Kim JW, Lee HJ, Farlow J, Lee JU, Litt DB, Haas T, Alivisatos AP, Cheon J, Gartner ZJ, Jun YW. A Mechanogenetic Toolkit for Interrogating Cell Signaling in Space and Time. Cell. 2016;165:1507–1518. doi: 10.1016/j.cell.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho MH, Lee EJ, Son M, Lee JH, Yoo D, Kim JW, Park SW, Shin JS, Cheon J. (Dup) A magnetic switch for the control of cell death signalling in in vitro and in vivo systems. Nat Mater. 2012;11:1038–1043. doi: 10.1038/nmat3430. [DOI] [PubMed] [Google Scholar]
- 15.Cho MH, Kim S, Lee JH, Shin TH, Yoo D, Cheon J. Magnetic Tandem Apoptosis for Overcoming Multidrug-Resistant Cancer. Nano Lett. 2016;16:7455–7460. doi: 10.1021/acs.nanolett.6b03122. [DOI] [PubMed] [Google Scholar]
- 16.Mannix RJ, Kumar S, Cassiola F, Montoya-Zavala M, Feinstein E, Prentiss M, Ingber DE. Nanomagnetic actuation of receptor-mediated signal transduction. Nat Nanotechnol. 2008;3:36–40. doi: 10.1038/nnano.2007.418. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Kim ES, Cho MH, Son M, Yeon SI, Shin JS, Cheon J. Artificial control of cell signaling and growth by magnetic nanoparticles. Angew Chem Int Ed. 2010;49:5698–5702. doi: 10.1002/anie.201001149. [DOI] [PubMed] [Google Scholar]
- 18.Etoc F, Lisse D, Bellaiche Y, Piehler J, Coppey M, Dahan M. Subcellular control of Rac-GTPase signalling by magnetogenetic manipulation inside living cells. Nat Nanotechnol. 2013;8:193–198. doi: 10.1038/nnano.2013.23. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann C, Mazari E, Lallet S, Le Borgne R, Marchi V, Gosse C, Gueroui Z. Spatiotemporal control of microtubule nucleation and assembly using magnetic nanoparticles. Nat Nanotechnol. 2013;8:199–205. doi: 10.1038/nnano.2012.246. [DOI] [PubMed] [Google Scholar]
- 20.Kim JW, Seo D, Lee JU, Southard KM, Lim Y, Kim D, Gartner ZJ, Jun YW, Cheon J. Single-cell mechanogenetics using monovalent magnetoplasmonic nanoparticles. Nat Protoc. 2017;12:1871–1889. doi: 10.1038/nprot.2017.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JH, Kim JW, Levy M, Kao A, Noh SH, Bozovic D, Cheon J. Magnetic nanoparticles for ultrafast mechanical control of inner ear hair cells. ACS Nano. 2014;8:6590–6598. doi: 10.1021/nn5020616. [DOI] [PubMed] [Google Scholar]
- 22.Kim JW, Lee JH, Ma JH, Chung E, Choi H, Bok J, Cheon J. Magnetic Force Nanoprobe for Direct Observation of Audio Frequency Tonotopy of Hair Cells. Nano Lett. 2016;16:3885–3891. doi: 10.1021/acs.nanolett.6b01392. [DOI] [PubMed] [Google Scholar]
- 23.Cordova JC, Das DK, Manning HW, Lang MJ. Combining single-molecule manipulation and single-molecule detection. Curr Opin Struct Biol. 2014;28:142–148. doi: 10.1016/j.sbi.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Lipfert J, Hao XM, Dekker NH. Quantitative Modeling and Optimization of Magnetic Tweezers. Biophys J. 2009;96:5040–5049. doi: 10.1016/j.bpj.2009.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews BD, LaVan DA, Overby DR, Karavitis J, Ingber DE. Electromagnetic needles with submicron pole tip radii for nanomanipulation of biomolecules and living cells. Appl Phys Lett. 2004;85:2968–2970. [Google Scholar]
- 26.Dufrene YF, Evans E, Engel A, Helenius J, Gaub HE, Muller DJ. Five challenges to bringing single-molecule force spectroscopy into living cells. Nat Methods. 2011;8:123–127. doi: 10.1038/nmeth0211-123. [DOI] [PubMed] [Google Scholar]
- 27.Alenghat FJ, Golan DE. Membrane Protein Dynamics and Functional Implications in Mammalian Cells. Curr Top Membr. 2013;72:89–120. doi: 10.1016/B978-0-12-417027-8.00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho D, Zimmermann JL, Dehmelt FA, Steinbach U, Erdmann M, Severin P, Falter K, Gaub HE. Force-driven separation of short double-stranded DNA. Biophys J. 2009;97:3158–3167. doi: 10.1016/j.bpj.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burton K, Park JH, Taylor DL. Keratocytes generate traction forces in two phases. Mol Biol Cell. 1999;10:3745–3769. doi: 10.1091/mbc.10.11.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K. Traction forces generated by locomoting keratocytes. J Cell Biol. 1994;127:1957–1964. doi: 10.1083/jcb.127.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neuman KC, Lionnet T, Allemand JF. Single-molecule micromanipulation techniques. Annu Rev Mater Res. 2007;37:33–67. [Google Scholar]
- 32.Pankhurst QA, Connolly J, Jones SK, Dobson J. Applications of magnetic nanoparticles in biomedicine. J Phys D. 2003;36:167–181. [Google Scholar]
- 33.Zhao XD, Zeng XJ, Lu C, Yan J. Studying the mechanical responses of proteins using magnetic tweezers. Nanotechnology. 2017;28:414002. doi: 10.1088/1361-6528/aa837e. [DOI] [PubMed] [Google Scholar]
- 34.Yung K, Landecker PB, Villani DD. An analytic solution for the force between two magnetic dipoles. Magn Electr Sep. 2008;9:39–52. [Google Scholar]
- 35.Jang B, Gutman E, Stucki N, Seitz BF, Wendel-Garcia PD, Newton T, Pokki J, Ergeneman O, Pane S, Or Y, Nelson BJ. Undulatory Locomotion of Magnetic Multilink Nanoswimmers. Nano Lett. 2015;15:4829–4833. doi: 10.1021/acs.nanolett.5b01981. [DOI] [PubMed] [Google Scholar]
- 36.Strick T, Allemand J, Croquette V, Bensimon D. Twisting and stretching single DNA molecules. Prog Biophys Mol Biol. 2000;74:115–140. doi: 10.1016/s0079-6107(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 37.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 38.Gore J, Bryant Z, Stone MD, Nollmann MN, Cozzarelli NR, Bustamante C. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–104. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 40.Puig-de-Morales M, Grabulosa M, Alcaraz J, Mullol J, Maksym GN, Fredberg JJ, Navajas D. Measurement of cell microrheology by magnetic twisting cytometry with frequency domain demodulation. J Appl Physiol. 2001;91:1152–1159. doi: 10.1152/jappl.2001.91.3.1152. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, Huh YM, Jun YW, Seo JW, Jang JT, Song HT, Kim S, Cho EJ, Yoon HG, Suh JS, Cheon J. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat Med. 2007;13:95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 42.Jang JT, Nah H, Lee JH, Moon SH, Kim MG, Cheon J. Critical enhancements of MRI contrast and hyperthermic effects by dopant-controlled magnetic nanoparticles. Angew Chem Int Ed. 2009;48:1234–1238. doi: 10.1002/anie.200805149. [DOI] [PubMed] [Google Scholar]
- 43.Noh SH, Na W, Jang JT, Lee JH, Lee EJ, Moon SH, Lim Y, Shin JS, Cheon J. Nanoscale magnetism control via surface and exchange anisotropy for optimized ferrimagnetic hysteresis. Nano Lett. 2012;12:3716–3721. doi: 10.1021/nl301499u. [DOI] [PubMed] [Google Scholar]
- 44.Jun YW, Huh YM, Choi JS, Lee JH, Song HT, Kim S, Yoon S, Kim KS, Shin JS, Suh JS, Cheon J. Nanoscale size effect of magnetic nanocrystals and their utilization for cancer diagnosis via magnetic resonance imaging. J Am Chem Soc. 2005;127:5732–5733. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 45.Curry FE, Adamson RH. Endothelial Glycocalyx: Permeability Barrier and Mechanosensor. Ann Biomed Eng. 2012;40:828–839. doi: 10.1007/s10439-011-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yun S, Shin TH, Lee JH, Cho MH, Kim IS, Kim J-w, Jung K, Lee IS, Cheon J, Park KI. Design of Magnetically Labeled Cells (Mag-Cells) for in Vivo Control of Stem Cell Migration and Differentiation. Nano Lett. 2018;18:838–845. doi: 10.1021/acs.nanolett.7b04089. [DOI] [PubMed] [Google Scholar]
- 47.Shamir M, Bar-On Y, Phillips R, Milo R. SnapShot: Timescales in Cell Biology. Cell. 2016;164:1302–1302e1. doi: 10.1016/j.cell.2016.02.058. [DOI] [PubMed] [Google Scholar]
- 48.Faivre D, Schuler D. Magnetotactic Bacteria and Magnetosomes. Chem Rev. 2008;108:4875–4898. doi: 10.1021/cr078258w. [DOI] [PubMed] [Google Scholar]
- 49.Araujo ACV, Abreu F, Silva KT, Bazylinski DA, Lins U. Magnetotactic Bacteria as Potential Sources of Bioproducts. Mar Drugs. 2015;13:389–430. doi: 10.3390/md13010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komeili A, Vali H, Beveridge TJ, Newman DK. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci U S A. 2004;101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]