Accumulation of DNA breaks has been linked to several neurological disorders. However, only recently have links started to emerge between DNA damage, perturbed co-transcriptional processing, and impaired cellular clearance pathways as causal and intertwined mechanisms underpinning neurodegeneration. Walker and El-Khamisy discuss these new links with a focus on ALS.
Keywords: R-loops, autophagy, DNA repair, genomic instability, neurodegeneration
Abstract
Maintaining genomic stability constitutes a major challenge facing cells. DNA breaks can arise from direct oxidative damage to the DNA backbone, the inappropriate activities of endogenous enzymes such as DNA topoisomerases, or due to transcriptionally-derived RNA/DNA hybrids (R-loops). The progressive accumulation of DNA breaks has been linked to several neurological disorders. Recently, however, several independent studies have implicated nuclear and mitochondrial genomic instability, perturbed co-transcriptional processing, and impaired cellular clearance pathways as causal and intertwined mechanisms underpinning neurodegeneration. Here, we discuss this emerging paradigm in the context of amyotrophic lateral sclerosis and frontotemporal dementia, and outline how this knowledge paves the way to novel therapeutic interventions.
Genomic instability: a neuronal perspective
The ability to maintain genomic integrity represents a major challenge facing cells. A variety of exogenous and endogenous agents can induce DNA damage (Ciccia and Elledge, 2010) with each cell estimated to process ∼1013 lesions each day (Lindahl and Barnes, 2000). Many normal cellular processes induce DNA damage. For example, reactive oxygen species, a natural by-product of cellular respiration, oxidize nucleic acids causing DNA damage (Valko et al., 2006). The faulty action of DNA processing enzymes, such as DNA ligases and topoisomerases, induce DNA lesions (El-Khamisy et al., 2005; Ahel et al., 2006). Furthermore, the fundamental cellular process of transcription leads to additional DNA damage due to the formation of DNA/RNA hybrids (R-loops) (Skourti-Stathaki and Proudfoot, 2014). The cell is therefore under constant pressure to maintain genomic stability.
The DNA damage response comprises multiple distinct repair pathways, each taking precedence in response to different forms of damage (Ciccia and Elledge, 2010). More often, damage to the DNA duplex is restricted to one strand of the phosphodiester backbone (single-stranded breaks), but can also occur on both strands (double-stranded breaks, DSBs). Though less common, DSBs are highly mutagenic and cytotoxic (Khanna and Jackson, 2001). To address this threat, cells have evolved a high fidelity DSB repair mechanism, homologous recombination, which utilizes sister chromatids as a template for polymerase rectification (Hartlerode and Scully, 2009). During G2 and S phase, cells can utilize homologous recombination to perform error-free DSB repair. Post-mitotic neurons, however, are intrinsically homologous recombination-deficient (El-Khamisy, 2011; Rulten and Caldecott, 2013), relying on error-prone non-homologous end-joining to repair DSBs.
DNA repair defects often present themselves clinically as neurological disease (Madabhushi, et al., 2014). For example, DSB repair dysfunction leads to the neurodegenerative diseases, ataxia telangiectasia (Savitsky et al., 1995) and ataxia telangiectasia-like disorder (Stewart et al., 1999). Single-stranded break repair defects are also associated with a number of neurological diseases, including cerebellar ataxias (Moreira et al., 2001, 2004; Takashima et al., 2002; Hoch et al., 2017), and has been implicated in Cockayne syndrome (Nance and Berry, 1992) and xeroderma pigmentosum (Kraemer, et al., 1987). More recently, defective DNA repair has been characterized as an important pathological process in motor neuron disease, otherwise known as amyotrophic lateral sclerosis (ALS).
Amyotrophic lateral sclerosis
ALS is a heterogeneous disease characterized by the loss of motor neurons. While sporadic in ∼90% of patients, 10% are familial and are caused by mutations in myriad of genes. ALS-causing mutations often occur within genes associated with RNA processing or cellular degradation (Hardiman et al., 2017). Interestingly, ALS patients can develop frontotemporal dementia (FTD) in unison and the two diseases appear to co-exist in a single disease spectrum (Couratier et al., 2017). Mutations in the TARDBP gene, encoding the RNA processing factor TAR DNA-binding protein-43 (TDP-43), are known to cause ALS and FTD (Kabashi et al., 2008). In addition, mutations in other RNA processing factors, such as FUS (encoding fused in sarcoma) (Kwiatkowski et al., 2009; Vance et al., 2009), SETX (encoding senataxin) (Chen et al., 2004), and HNRNPA/B (encoding heterogeneous nuclear ribonucleoprotein A/B, hnRNPAB) (Kim et al., 2013) also cause ALS. Mutations in the SMN1 gene (encoding the core spliceosome component survival of motor neuron, SMN), lead to a severe form of juvenile motor neuron disease called spinal muscular atrophy (Melki et al., 1990). Thus, motor neuron disease is frequently associated with mutations in genes involved with RNA processing.
As well as RNA processing dysfunction, ALS is strongly associated with defects in autophagy and the ubiquitin proteasome system (UPS) (Majcher et al., 2015; Hardiman et al., 2017), which are the primary degradation pathways available to mammalian cells. Mutations in VCP (encoding valosin-containing protein, also known as p97 or Cdc48), cause ALS, FTD and a related syndrome known as inclusion body myopathy with Paget disease of bone and frontotemporal dementia (IBMPFD) (Watts et al., 2004; Johnson et al., 2010). VCP functions as a central component of the UPS (Meyer and Weihl, 2014), and is also implicated in autophagy (Majcher et al., 2015). Alike VCP, mutations in the SQSTM1 gene (encoding p62, also known as sequestosome-1) cause ALS, FTD and IBMPFD (Fecto et al., 2011; Rea et al., 2014). P62 is important for the process of ubiquitin-mediated autophagy (Pankiv et al., 2007; Katsuragi et al., 2015), and ALS-causing mutations perturb this process (Goode et al., 2016). Mutations in the UBQLN2 gene (encoding for ubiquillin-2), which acts as a dual regulator of autophagy and the ubiquitin proteasome system (Ceballos-Diaz et al., 2015; Osaka et al., 2016), also cause ALS (Deng et al., 2011). Taken together, these examples demonstrate that genetic perturbations in cellular clearance pathways are also associated with ALS.
A hexanucleotide repeat expansion contained within the first intron of the C9orf72 gene, was recently discovered to be the most common genetic cause of ALS and FTD (DeJesus-Hernandez et al., 2011; Renton et al., 2011). The C9orf72 protein is now characterized as an autophagy coactivator (Webster et al., 2016; Yang et al., 2016). C9orf72 protein levels are also reduced in patient tissue (DeJesus-Hernandez et al., 2011; Belzil et al., 2013; Waite et al., 2014), suggesting a possible role for haploinsufficiency and defective autophagy in C9orf72-ALS. In addition, RNA molecules transcribed from the C9orf72 expansion form nuclear RNA foci (Zu et al., 2013; Cooper-Knock et al., 2014). Intronic C9orf72 RNAs are also exported into the cytoplasm, and undergo repeat associated non-ATG dependent (RAN)-translation (Ash et al., 2013; Zu et al., 2013), producing five dipeptide repeat proteins. C9orf72 expansions are associated with aberrant nucleocytoplasmic transport (Freibaum et al., 2015; Jovicic et al., 2015; Zhang et al., 2015a), dysfunctional RNA processing (Lee et al., 2013; Cooper-Knock et al., 2014; Prudencio et al., 2015; Yin et al., 2017), and impaired cellular clearance mechanisms (Zhang et al., 2014; Gao et al., 2017; Gupta et al., 2017; Ramesh and Pandey, 2017). It is thought that some contribution of these factors conspires to drive neurodegeneration in C9orf72-ALS.
In line with the genetic aetiology of ALS, CNS tissues from around 97% of ALS patients, including both sporadic and familial cases, display the mislocalization and aggregation of TDP-43 (TDP-43 proteinopathy) (Neumann et al., 2006; Ling et al., 2013). Given the role of TDP-43 in nuclear RNA processing pathways (Scotter et al., 2015), TDP-43 mislocalization likely represents a loss of its nuclear function and a defect in RNA processing. Sporadic and familial ALS patient tissues also display a second disease hallmark, the accumulation and aggregation of p62. P62 binds to misfolded protein aggregates, targeting them for degradation by autophagy (Bjørkøy et al., 2005). P62 itself is degraded during autophagy (Ichimura et al., 2008) and the accumulation of p62 in ALS patients likely indicates a defect in this process. Understanding the pathological consequences of these two disease hallmarks will likely unlock the door to novel treatment options for ALS patients.
ALS-linked RNA processing factors are guardians of the genome
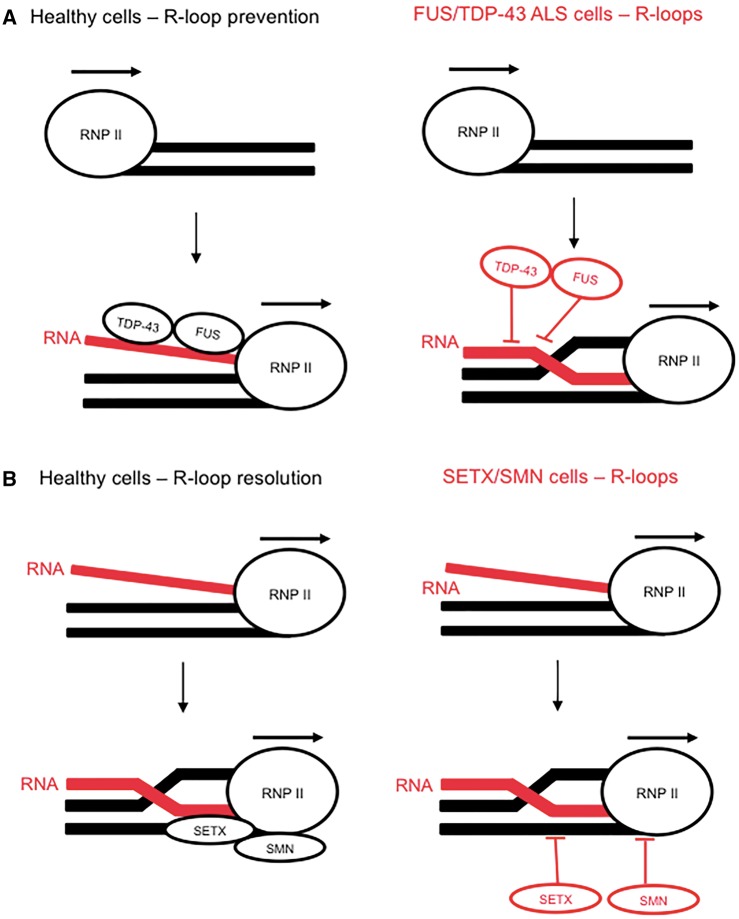
In the past decade, it has become apparent that transcription is a primary source of DNA damage, and that RNA processing factors are important guardians of the genome (Dutertre et al., 2016). RNA/DNA hybrids form during transcription when the nascent RNA transcript anneals to the template DNA strand, displacing the complementary DNA strand, and yielding a three-stranded nucleic structure known as an R-loop (Aguilera and Garcia-Muse, 2012). R-loop accumulation leads to genomic instability (Huertas and Aguilera, 2003; Li et al., 2005; Tuduri et al., 2009; Sollier et al., 2014), and thereby couples RNA processing to the regulation of genomic stability. This was first observed when yeast mutant strains lacking members of the THO complex (mammalian equivalent of the transcription and export, TREX, complex) displayed increased numbers of R-loops and DNA breaks (Huertas and Aguilera, 2003). Similar results were observed after serine/arginine-rich splicing factor 1 (SRSF1) depletion in vertebrates (Li et al., 2005), demonstrating that RNA processing factors are important regulators of R-loop homeostasis. Mechanistically, it is speculated that RNA processing factors prevent R-loop formation in a preventative fashion (Fig. 1A), by binding to the nascent RNA transcript and blocking R-loop formation (Chan et al., 2014b). In follow-up studies, a separate class of transcription and export proteins, known as hnRNPs, were linked to the regulation of R-loops (Santos-Pereira et al., 2013). HNRNP mutations cause ALS and FTD (Kim et al., 2013), but whether these mutations induce R-loop-mediated genomic instability has not yet been reported.
Figure 1.
ALS-linked RNA processing factors are guardians of the genome. (A) Several factors associated with RNA processing, including FUS and TDP-43, are required to maintain R-loop homeostasis. Their depletion leads to R-loop-mediated genomic instability. (B) The resolution of R-loops involves the recruitment of the RNA/DNA helicase, SETX, which is recruited to R-loops by SMN - defects in either cause motor neuron cell death.
A similar phenomenon has since been observed for the ALS-linked RNA processing factors, FUS and TDP-43 (Hill et al., 2016). Depletion of either leads to an increase in DSBs, which are reduced by overexpression of the R-loop-specific nuclease, RNase H1. TDP-43 and FUS form damage-induced nuclear foci, which co-localize with RNA polymerase 2 and phosphorylated histone H2AX (γH2AX) (Hill et al., 2016). These data suggest that TDP-43 and FUS function at the interface between transcription and DNA repair, likely by controlling R-loop levels. Based on this work, another interesting hypothesis is that TDP-43 proteinopathy may function similar to TDP-43 depletion, and induce R-loop-associated genomic instability. This would be of significance as 97% of ALS cases display TDP-43 proteinopathy (Neumann et al., 2006; Ling et al., 2013). Other ALS-mutant RNA processing factors exhibit similar cytoplasmic mislocalization (Kapeli et al., 2017), suggesting that the nuclear-cytoplasmic shuttling of RNA binding proteins is an important disease mechanism. Indeed, defects in nucleocytoplasmic transport have emerged as an important pathological mechanism in C9orf72-ALS (Freibaum et al., 2015; Jovicic et al., 2015; Zhang et al., 2015a). As well as the cytoplasmic accumulation of RNA processing proteins, the nuclear accumulation of mRNA has also been observed in C9orf72-ALS disease models (Rossi et al., 2015), suggesting that improper RNA export is a consequence of dysfunctional RNA processing. An interesting avenue for future research would be to assess the association between nucleocytoplasmic transport defects, dysfunctional RNA processing, and the levels of R-loop-associated genomic instability.
In addition to R-loop protection via RNA processing factors, R-loops can be resolved by RNase H1, which degrades the RNA strand of RNA/DNA hybrids (Cerritelli and Crouch, 2009). A number of RNA/DNA helicases have also been implicated in the resolution of R-loops (Skourti-Stathaki et al., 2011; Yuce and West, 2013; Hodroj et al., 2017; Song et al., 2017). For instance, the RNA/DNA helicase, SETX, has been linked to the resolution of R-loops (Skourti-Stathaki et al., 2011; Yuce and West, 2013). Notably, autosomal dominant mutations in the SETX gene are known to cause a juvenile form of ALS, referred to as ALS4 (Chen et al., 2004). In addition, autosomal recessive mutations in SETX can cause ataxia with oculomotor apraxia 2 (AOA2) (Moreira et al., 2004). Disease mutations appear to occur within the helicase domain or within its N-terminal protein–protein interaction domain, suggesting either loss of helicase activity or loss of functional interactions is pathological.
Nonsense mutations in the SMN1 gene lead to a juvenile form of motor neuron disease called spinal muscular atrophy (SMA) (Melki et al., 1990). Recent evidence suggests that SMN is also important for the resolution of R-loops (Zhao et al., 2016). Mechanistically, the tudor domain of SMN was shown to be a requisite for the recruitment of SETX to R-loops, and spinal muscular atrophy patient fibroblasts display increased levels of R-loops and DSBs (Zhao et al., 2016). In addition, a mouse model of spinal muscular atrophy recapitulated these two hallmarks of genomic instability (Jangi et al., 2017), suggesting that SMN is an important factor in the prevention of R-loop-driven genomic instability. These reports suggest that motor neuron cells may be particularly sensitive to perturbations in R-loop homeostasis, caused by defects in RNA binding proteins (Fig. 1B).
R-loops are enriched at GC-rich sites of the genome (Chan et al., 2014a), which may occur because guanine-rich RNA: cytosine-rich DNA duplexes are thermodynamically more stable than the respective DNA: DNA duplex (Roy and Lieber, 2009). Susceptibility to R-loops is also increased by the presence of G-quadruplex structures (Duquette et al., 2004; Aguilera and Garcia-Muse, 2012), which assemble in G-rich DNA sequences. The C9orf72 repeat expansion has a high GC content and is known to form G-quadruplex structures (Haeusler et al., 2014; Zhou et al., 2015; Brcic and Plavec, 2017). Unsurprisingly, R-loops have been identified in vitro following the transcription of C9orf72 expansions (Haeusler et al., 2014). C9orf72 RNA also forms G-quadruplex structures, which sequester a number of RNA binding proteins (Cooper-Knock et al., 2014). The sequestration of certain RNA processing factors may impede their function in R-loop prevention. In-line with this idea, cells expressing C9orf72 expansion constructs, as well as C9orf72-ALS post-mortem tissues, display increased R-loop levels (Walker et al., 2017). The accumulation of R-loops in C9orf72 expansion expressing cells is a source of genomic instability, since overexpression of SETX reduces levels of DSBs. SETX overexpression also reduced cellular toxicity in C9orf72 expansion-expressing cells (Walker et al., 2017), indicating that excessive R-loop accumulation is able to promote cell death in cellular models of ALS. Taken together, recent evidence suggests that RNA misprocessing, in the context of ALS, results in R-loop accumulation and the formation of toxic DSBs.
Double-strand break repair signalling
Following DSB induction, the MRN (Mre11, Rad50, Nbs1) complex form foci at the break site (Williams et al., 2007), which promotes ataxia-telangiectasia mutated (ATM) autophosphorylation at serine 1981 (Uziel et al., 2003). ATM then sets into motion the DNA damage response: histone H2AX is phosphorylated on serine 139 by ATM, ATR and DNA-PK (Rogakou et al., 1998; Blackford and Jackson, 2017). Phosphorylated H2AX recruits the mediator of DNA damage checkpoint 1 (MDC1) (Hartlerode and Scully, 2009), inducing the recruitment of the E3 ubiquitin ligase, RNF8 (Kolas et al., 2007), which associates with phosphorylated TQ sites of MDC1 and attaches mono-K63-linked ubiquitin chains to H2A(X) (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007). RNF168 associates with these mono-ubiquitylated chains, and further catalyses the mono- and poly-ubiquitylation of H2A(X) (Stewart et al., 2009). The ubiquitylation of H2A(X) by RNF168 is mandatory for the recruitment of 53BP1 and BRCA1 to sites of DSBs (Wang and Elledge, 2007), whose recruitment dictates DSB repair choice towards non-homologous end-joining or homologous recombination, respectively (Ciccia and Elledge, 2010). Interestingly, 53BP1 recruitment to sites of DSBs is also required for concentrating ATM into nuclear foci at DSBs (Noon et al., 2010), suggesting a reciprocal relationship between ATM and 53BP1 in mediating DSB repair.
Defective autophagy leads to defective DNA repair signalling
P62 plays a central role in selective autophagy and the UPS (Katsuragi et al., 2015), which require its LC3-binding (LCB) and ubiquitin-associated (UBA) domains. While p62 resides predominantly in the cytoplasm, it is also found in the nucleus (Wang et al., 2016). P62 binds to misfolded protein aggregates and targets them for degradation (Bjørkøy et al., 2005). During p62-mediated autophagy, p62 is degraded along with its substrate, therefore autophagy self-regulates the cellular levels of p62 (Ichimura et al., 2008). In turn, the accumulation of protein aggregates or defects in autophagy leads to p62 accumulation (Korolchuk et al., 2009; Wang et al., 2016). Recently, a seminal discovery demonstrated that excessive p62 acts as a negative regulator of DSB repair signalling (Wang et al., 2016). Initially, it was observed that p62 depletion leads to increased levels of nuclear ubiquitylated protein foci, while p62 overexpression induced the opposing effect and similarly decreased levels of H2A ubiquitylation. Overexpression of p62 also prevented damage-induced 53BP1 foci formation, which could also be recapitulated by treating wild-type cells with the autophagy inhibitor, 3-methyladenine (3-MA). Importantly, 3-MA did not induce DSB repair defects in p62 depleted cells, demonstrating that autophagy inhibition leads to DSB repair impairment in a p62-dependant manner.
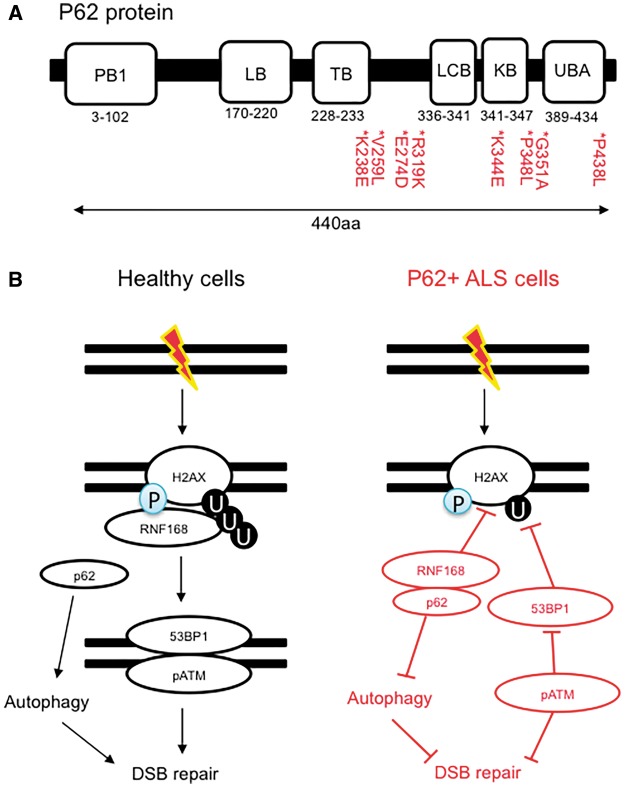
Biochemical assays demonstrated that p62-mediated DSB repair inhibition was a consequence of RNF168 sequestration, an E3 ubiquitin ligase that functions to recruit 53BP1 to DNA breaks (Stewart, 2009). P62 binds to the MIU (motif interacting with ubiquitin) domain of RNF168, which is important for its E3 ligase activity (Wang et al., 2016). On the other hand, the LIM-binding (LB) domain of p62 (residues 170–220) mediates RNF168 sequestration and DSB repair inhibition. These data demonstrate that the accumulation of p62 impairs the DNA damage response. Interestingly, ALS-causing p62 mutations can occur in or around the LC3 domain of p62 (Fig. 2A), causing autophagy defects and the accumulation of mutant p62 (Rubino et al., 2012; Goode et al., 2016). An interesting future experiment would be to determine whether ALS-causing mutations in p62 cause DSB repair defects.
Figure 2.
Defective autophagy leads to defective DNA repair signalling. (A) A schematic depicting the structure of p62 highlighting ALS, FTD, and IBMPFD missense mutations. (B) Autophagy maintains functional DNA repair by preventing p62 accumulation. In the context of ALS, where p62 accumulates, RNF168-mediated H2A ubiquitylation (U) is perturbed, leading to impaired DSB repair and genomic instability.
C9orf72 repeat expansions lead to genomic instability
Excitingly to the field of ALS, p62 accumulation is a hallmark feature of patient CNS tissues (Blokhuis et al., 2013), including C9orf72-ALS (Troakes et al., 2012), suggesting that this hallmark pathology may be associated with defects in DNA repair. Recently, it was demonstrated that C9orf72 repeat expansions cause defective DNA repair, as indicated by decreased H2A ubiquitylation, impaired 53BP1 and phosphorylated ATM (pATM) repair foci, and increased levels of DSBs (Walker et al., 2017). RNF168-mediated H2A ubiquitination is important for regulating 53BP1 and ATM mediated repair (Noon et al., 2010). In turn, RNF168 overexpression or p62 depletion restored DSB repair and reduced levels of DSBs in C9orf72 expansion expressing cells (Fig. 2B). These data demonstrate that defective autophagy, indicated by p62 accumulation, is a driver of genome instability in C9orf72-ALS disease models (Walker et al., 2017). It is likely that this phenomenon also occurs in non-C9orf72 ALS subtypes that exhibit p62 accumulation.
Increased DNA damage response activation in C9orf72-ALS patients has been corroborated by two separate reports (Farg et al., 2017; Walker et al., 2017), which demonstrated that motor neurons from C9orf72-ALS patients display increased levels of ϒH2AX phosphorylation. It was demonstrated by immunohistochemistry that phosphorylated levels of ATM were increased in C9orf72-ALS motor neurons (Farg et al., 2017), though the authors did not observe or quantify pATM repair foci. Importantly, defective ATM signalling in C9orf72-ALS was found to be the result of RNF168 dysfunction, a process that promotes the focal concentration of ATM but not its overall activation (Noon et al., 2010). Using western blotting from unfractionated spinal cord homogenates, it was reported that C9orf72-ALS patient tissues have increased levels of 53BP1 protein (Farg et al., 2017). Importantly, the extraction protocol used by the authors does not appear to extract the chromatin-bound fraction of tissue lysates, where 53BP1 protein engaged in DNA repair is enriched. Thus, neither pATM immunohistochemistry nor 53BP1 western blotting are indicative of DNA repair capacity. A crucial future experiment is to assess levels of 53BP1 repair foci in C9orf72-ALS tissues, particularly in those cells that are enriched with the hallmark accumulation of p62, and to repeat the immunoblotting experiments to specifically examine the levels of chromatin-bound 53BP1.
In other research, the expression of C9orf72-related poly-glycine-arginine dipeptide repeats (poly-GR DPRs) was shown to cause increased levels of genomic instability (Lopez-Gonzalez et al., 2016). Unlike previous work, poly-GA DPRs did not induce γH2AX foci formation, though they were not quantified. A similar increase in DSBs was observed in C9orf72-ALS induced pluripotent stem cell-derived motor neurons, when compared to non-ALS controls (Lopez-Gonzalez et al., 2016). Treatment with antioxidants partially decreased levels of nuclear DNA damage, suggesting that elevated levels of reactive oxygen species (or defective repair of reactive oxygen species-induced lesions) contributes to genome instability in C9orf72-ALS. In summary, there is now a wealth of evidence showing that C9orf72 expansions cause genomic instability. This is likely driven by multiple sources: (i) RNA processing dysfunction and R-loops accumulation; (ii) defects in autophagy and p62-mediated inhibition of DNA repair; and (iii) mitochondrial perturbations and the generation of reactive oxygen species.
Familial ALS proteins are double-strand break repair players
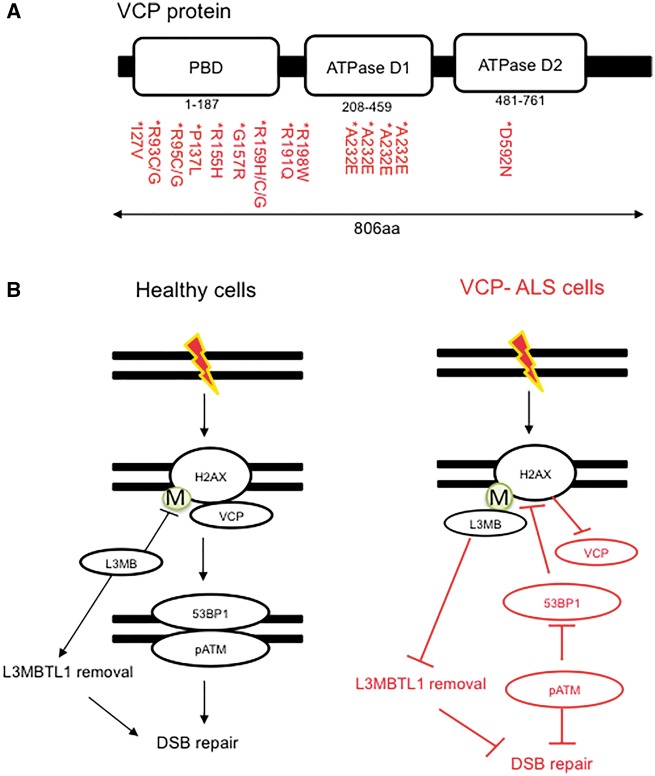
ALS-linked genomic instability can therefore arise, indirectly, due to deficits in RNA processing and/or cellular clearance pathways. In addition to these mechanisms, proteins commonly mutated in ALS have been shown to play a more direct role in DSB repair. For example, VCP is a novel DSB repair player and its depletion leads to elevated levels of DSBs (Acs et al., 2011; Meerang et al., 2011). Mechanistically, VCP functions to catalyse the removal of lysine 48 ubiquitin conjugates at DNA damage sites, which is obligatory for the recruitment of DNA repair factors. As in C9orf72-ALS, VCP depletion impairs the recruitment of the DNA repair factor, 53BP1, to sites of DNA damage (Acs et al., 2011; Meerang et al., 2011), suggesting a common theme by which 53BP1-mediated DNA repair is defective in ALS (Fig. 3A). In the absence of VCP, L3MBTL1 is constitutively bound to H4K20me2, a histone residue that is recognized by the tudor domain of 53BP1, thereby blocking 53BP1 recruitment (Acs et al., 2011). VCP is therefore required to remove L3MBTL1 and unmask H4K20me2 residues, permitting 53BP1 recruitment (Fig. 3B).
Figure 3.
The familial ALS protein VCP promotes DNA repair. (A) A schematic depicting the structure of VCP protein highlighting ALS, FTD, and IBMPFD missense mutations. (B) VCP functions to catalyse the removal of ubiquitylated proteins from chromatin, including L3MBTL1. The removal of L3MBTL1 from methylated (M) histone H2A(X) permits 53BP1 recruitment to DSBs, enabling DNA repair by non-homologous end-joining. Loss of VCP leads to impaired removal of L3MBTL1, leading to impaired non-homologous end-joining and genomic instability.
VCP has been further implicated in DSB repair, by removing lysine 48 conjugated Ku70/80 rings from DNA (van den Boom et al., 2016). Failure to remove Ku70/Ku80 rings perturbs DSB repair, leading to genomic instability. How ALS causing mutations in VCP affect DSB repair, however, is not yet clear. Some ALS-causing VCP mutations affect the ATP binding domain of VCP (Fig. 3A) (Schuetz and Kay, 2016). Since the ATPase activity is required for its function in regulating DSB repair (Meerang et al., 2011; van den Boom et al., 2016), it is likely that these mutations result in an impairment of DNA repair signalling. Furthermore, due to its additional role in cellular degradation pathways, VCP mutations cause p62 accumulation in patient samples (Ayaki et al., 2014), and may therefore cause additional defects in DNA repair through p62-dependant processes. Taken together, these data indicate that VCP has a direct role in repairing DNA breaks.
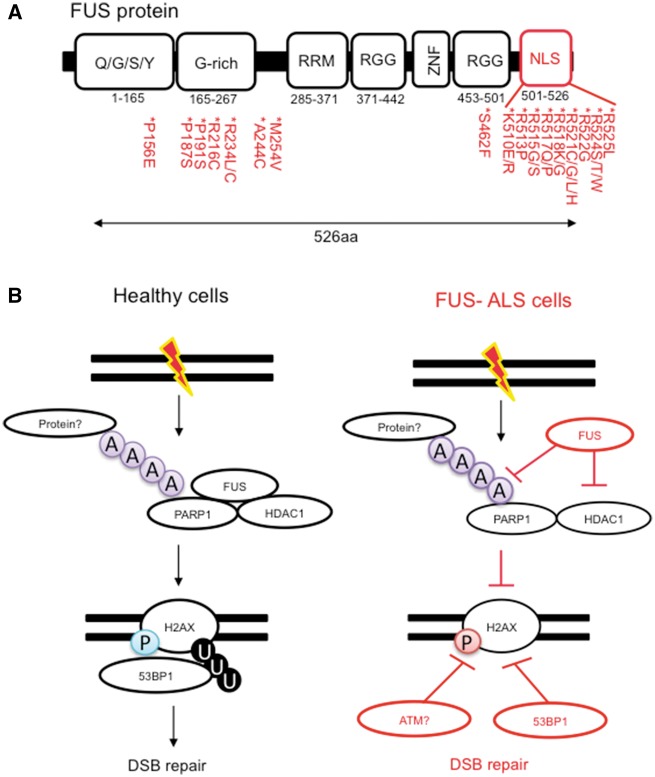
In addition to VCP, the ALS-linked protein FUS has also been implicated in repairing DNA breaks (Fig. 4A) (Wang et al., 2013; Rulten et al., 2014). FUS depletion leads to impaired 53BP1 foci and increased DNA damage (Wang et al., 2013), bearing striking similarities to what has been observed following the expression of C9orf72 expansions and the depletion of VCP (Acs et al., 2011; Meerang et al., 2011; Walker et al., 2017). FUS recruitment to DSBs is facilitated by interactions with HDAC1, and ALS-linked FUS mutations impede this interaction and perturb DSB repair (Fig. 4A and B) (Wang et al., 2013). Finally, FUS-ALS patient tissues display increased DNA damage, as measured by ϒH2AX levels in neuronal cells. In a separate study, FUS recruitment to DNA damage was dependent on PARP-1 catalytic activity (Rulten et al., 2014). It was also shown that FUS binds directly to PAR chains and that ALS-causing mutations decrease this interaction (Rulten et al., 2014). Taken together, these data indicate that FUS is a member of the DNA damage response, and indicate that ALS-associated defects in FUS lead to defects in DNA repair processes.
Figure 4.
The familial ALS protein FUS promotes DNA repair. (A) A schematic depicting the structure of the FUS protein with ALS missense mutations highlighted. (B) FUS functions to regulate the DNA damage response through interactions with HDAC1 and PARP1. These interactions are important for H2AX phosphorylation (P) and 53BP1-mediated DSB repair. Loss of FUS or ALS-linked mutations leads to defective DSB repair and genomic instability.
More recently, it was shown that mutation variants of the NIMA-related kinase 1 (NEK1) gene confer susceptibility to ALS (Kenna et al., 2016). NEK1 has been implicated in the repair of DSBs by homologous recombination (Spies et al., 2016), functioning to phosphorylate Rad54 and ensuring the adequate removal of chromatin-associated Rad51 filaments, which is required for homologous recombination completion. Mutations in C21orf2, an interactor of NEK1, are also associated with ALS (van Rheenen et al., 2016). Moreover, its depletion similarly leads to defects in homologous recombination (Fang et al., 2015). Many canonical homologous recombination factors, such as BRCA1 and BRCA2, have been linked to the processing of R-loops (Bhatia et al., 2014; Hatchi et al., 2015; Tan et al., 2017; Zhang et al., 2017). We therefore would like to propose that NEK1 and C21orf2 play a role in controlling R-loop homeostasis, which may explain at least in part their association with ALS.
Heterochromatin: protecting from R-loops but perturbing DNA repair
Eukaryotic DNA is highly organized, typically the cell houses active genes in open euchromatic DNA, while non-transcribed or repressed genetic material is housed in tightly bound heterochromatin. In addition to promoting genomic instability, R-loops have been shown to alter the chromatin landscape by increasing levels of heterochromatic DNA. Of note, R-loop accumulation increases the levels of H3S10P (Castellano-Pozo et al., 2013), a heterochromatic histone mark usually associated with mitosis. More recently, the same group demonstrated that R-loop-driven H3S10P formation is a requisite for subsequent genomic instability (Garcia-Pichardo et al., 2017). Whether levels of H3S10P are therefore perturbed in in ALS samples is yet to be tested.
It was also demonstrated that H3K9 methylation is enriched in response to R-loop accumulation (Castellano-Pozo et al., 2013). Increased H3K9 methylation has been reported in C9orf72-ALS (Belzil et al., 2013; Walker et al., 2017), which might be driven by the aberrant accumulation of R-loops in ALS (Haeusler et al., 2014; Walker et al., 2017). Interestingly, this mark has also been associated with Friedreich’s ataxia and fragile X syndrome, whereby trinucleotide repeat expansions were shown to cause R-loop-dependent H3K9me2 deposition (Groh et al., 2014). H3K9 methylation marks and R-loops are also enriched at GC-rich transcriptional pause sites (Skourti-Stathaki et al., 2014). It is postulated that H3K9 methylation may play a protective role, since abolishment of all H3K9 methylation induces massive R-loop accumulation and genomic instability in Caenorhabditis elegans (Zeller et al., 2016). These data couple R-loop accumulation to the presence of heterochromatic DNA. An attractive model for this association is that the formation of repressive heterochromatin serves to reduce transcriptional activity and protect from dangerous R-loop-mediated instability.
While heterochromatin may protect the cell from the toxic effects of R-loops, the accumulation of heterochromatin represents an obstacle for DNA repair. Work in the Jeggo laboratory demonstrated that heterochromatic DSBs display slower repair kinetics than euchromatic breaks (Goodarzi et al., 2008; Noon et al., 2010). Interestingly, ATM and 53BP1 are particularly required for the repair of heterochromatic DSBs (Dimitrova et al., 2008) and the inhibition of either impairs the resolution of heterochromatic DNA breaks (Goodarzi et al., 2008; Noon et al., 2010). Mechanistically, ATM-mediated phosphorylation of KRAB-associated protein 1 (KAP1), a bona fide heterochromatic protein, was shown to release KAP1 from heterochromatin and enable DSB repair (Goodarzi et al., 2008). 53BP1 was subsequently shown to be upstream of ATM-mediated phosphorylation of KAP1 (Noon et al., 2010), acting to concentrate active ATM at sites of DSBs. It is conceptualized that the phosphorylation and mobility of KAP1 promotes local chromatin decondensation that enables DNA repair factor access to the break site. In line with this, the genetic inhibition of KAP1 improves heterochromatic DSB repair kinetics in ATM inhibited cells (Goodarzi et al., 2008). As such, heterochromatic DSBs are particularly challenging to cells because they require additional chromatin remodelling prior to repair.
Paradoxically, recent evidence suggests that the presence of local heterochromatic DNA is also a requirement for DSB repair signalling (Sun et al., 2009; Kaidi and Jackson, 2013; Burgess et al., 2014). The heterochromatic histone mark, H3K9me3, is required for the damage-dependent activation of the acetyltransferase KAT5 (Sun et al., 2009). KAT5 activation, in turn, is required for the activation of ATM (Kaidi and Jackson, 2013). More recently, it was shown that chromatin compaction activates the DNA damage response, specifically proteins that occur upstream in DSB repair signalling, such as γH2AX (Burgess et al., 2014). Importantly, excessive chromatin compaction impaired the completion of DNA repair and caused reduced cellular viability (Burgess et al., 2014). These data suggest that, while heterochromatin is an important aspect of initial DSB repair signalling, it is ultimately a barrier to the proper completion of DNA repair.
As such, the careful organization of DNA into euchromatin and heterochromatin is critical to cellular health. Evidently, perturbations in these processes have been linked to human disease, such as premature ageing (Zhang et al., 2015b) and ataxia telangiectasia (Li et al., 2012, 2013). Chromatin relaxation, via HDAC inhibition or enhancer of zest homologue 2 (EZH2) depletion, was able to reduce the cellular and neurological phenotype of ataxia telangiectasia mouse models (Li et al., 2012, 2013), suggesting a pathological role for excessive heterochromatin in the neurological aspect of the disease. Elevated levels of the heterochromatin have also been reported in C9orf72-ALS samples (Belzil et al., 2013; Walker et al., 2017). Interestingly, HDAC inhibition was able to reduce DSBs and cellular toxicity in models of C9orf72-ALS (Walker et al., 2017). These data implicate excessive heterochromatin formation as a pathological feature of ataxia telangiectasia and ALS, highlighting the potential of HDAC inhibitors as a means to prevent neuronal cell death. Whether excessive heterochromatin is the cause or consequence of defective ATM, a feature in both disorders, is yet to be determined. Nevertheless, these data offer a plausible therapeutic approach and a new insight into the role of ATM in controlling chromatin compaction.
Defective ATM signalling provides an unexpected link between amyotrophic lateral sclerosis and ataxia telangiectasia
Defective ATM signalling provides a molecular link between ALS and ataxia telangiectasia. Curiously, a recently characterized murine model of ataxia telangiectasia displays motor neuron cell loss (Quek et al., 2017), indicating that motor neurons may be particularly sensitive to perturbations in ATM signalling. As well as its canonical role in DSB repair, ATM signalling is important for the repair of topoisomerase I (TOP1) cleavage complexes (Das et al., 2009). In non-replicating cells, TOP1 cleavage complexes are primarily repaired by TDP1 (El-Khamisy et al., 2005), and TDP1 mutations cause the neurodegenerative disease spinocerebellar ataxia with axonal neuropathy (SCAN-1) (Takashima et al., 2002). ATM phosphorylates TDP1 (Das et al., 2009) to enhance TOP1 cleavage complex resolution. In addition to TDP1 and ATM, a scaffold protein, XRCC1, further facilitates the repair of TOP1 cleavage complexes (Das et al., 2009). Recently, XRCC1 mutations were identified in a patient with cerebellar ataxia (Hoch et al., 2017). As well as promoting neurodegeneration in man, Atm, Tdp1 and Xrcc1 deletion leads to the accumulation of TOP1 cleavage complexes in mice (Katyal et al., 2007, 2014; Alagoz et al., 2013). Exposure to camptothecin, which specifically induces TOP1 cleavage complexes, also leads to neurodegeneration in mice (Katyal et al., 2014), further highlighting the pathogenic consequences of TOP1 cleavage complex in neurons.
Interestingly, TOP1 cleavage complexes are also increased in cells expressing C9orf72 repeat expansions (Walker et al., 2017). This is in-line with the observation that C9orf72 expansions impair ATM signalling. Since defective ATM signalling was a consequence of p62 accumulation, p62 accumulation in other ALS subtypes likely drives the formation of TOP1ccs. Recently, it was shown that ATM is activated by R-loops, independently from the formation of DSBs (Tresini et al., 2015). ATM activation by R-loops enhances the mobility of the spliceosome, ultimately leading to mis-splicing and the retention of introns within mRNA transcripts (Tresini et al., 2015). The function of ATM kinase activity in mediating the cellular response to R-loops is yet to be elucidated, though it is tempting to speculate that these data represent yet another ATM-mediated repair pathway. In line with this idea, restoring ATM signalling in cells expressing C9orf72 expansion expressing cells was able to reduce R-loop levels (Walker et al., 2017). The function of ATM signalling in the regulation of R-loop homeostasis, in both health and disease, represents a fruitful avenue for future research.
Why does defective ATM signalling caused by C9orf72 expansions not predispose to cancer or immunodeficiency?
One of the primary features of ataxia telangiectasia is cancer predisposition, namely lymphomas (Taylor et al. 2004). The ATM deficiency in C9orf72-ALS patients, however, does not appear to predispose to cancer. While ataxia telangiectasia and C9orf72-ALS share defective ATM signalling, the nature and origin of this defect are fundamentally different. The ATM defect in C9orf72-ALS originates from the impaired ability to degrade misfolded protein aggregates, which leads to p62 accumulation and RNF168 inhibition (Walker et al., 2017). RNF168 deficiency impairs 53BP1-mediated sustained signalling of ATM, a non-canonical pathway complementing the canonical ATM activation cascade initiated by the MRN complex (Noon et al., 2010). Notably, RNF168 dysfunction causes another human disease typified by radiosensitivity, immunodeficiency and learning difficulties called RIDDLE syndrome, which similarly does not appear to be linked to cancer predisposition (Stewart et al., 2007). In ataxia telangiectasia, oncogenesis is thought to arise as a consequence of defective DSB repair by homologous recombination and defective cell-cycle checkpoint activation (Cremona and Behrens, 2014). RIDDLE cells, however, display normal levels of homologous recombination and functional ATM-mediated cell-cycle checkpoint, potentially protecting patients from cancer (Stewart et al., 2007). Thus, one possible explanation for the absence of cancer in C9orf72-ALS is the retention of functional homologous recombination and ATM-mediated cell cycle checkpoint, akin to RIDDLE. While this provides a plausible explanation for the absence of cancer in C9orf72-ALS, it does not explain the apparent absence of immunodeficiency, a hallmark of defective non-homologous end-joining in RIDDLE cells. In dividing cells, misfolded protein aggregates are asymmetrically distributed into daughter cells, leaving one of the daughter cells aggregate-free (Rujano et al., 2006). Therefore, even if p62 accumulation occurs in ALS lymphocytes, non-homologous end-joining dysfunction would only persist for a single cell cycle and thus the toxic effect of protein aggregation is inherently ‘diluted’ in cycling cells. Indeed, protein aggregates typically impact non-dividing neuronal cells (Ross and Poirier, 2004; Taylor et al., 2004; Bjørkøy et al., 2005; Rujano et al., 2006; Stewart et al., 2007; Cremona and Behrens, 2014). We favour the latter explanation in which the magnitude of toxic effects caused by protein aggregation is higher in non-cycling compared to cycling cells, providing an explanation for the apparent absence of immunodeficiency in C9orf72-ALS. The extent of protein aggregation and the preset threshold at which non-cycling cells can tolerate may also explain the selective vulnerability of specific neuronal populations to the toxic effects of protein aggregations.
The vulnerability of neurons to oxidative DNA damage
Despite representing only 2% of total body mass, one-fifth of all oxygen is consumed by the brain (Nicholls and Budd, 2000). Because they metabolize a high proportion of inhaled oxygen, neurons are likely to be highly susceptible to damage from reactive oxygen species. The primary free radical that is formed during cellular respiration is O2•− (superoxide) (Murphy, 2009), which is dismutated into hydrogen peroxide (H2O2) by superoxide dismutase 1 (SOD1). SOD1 mutations cause ALS (Rosen et al., 1993) and result in high levels of oxidative stress (Tsang et al., 2014). Reactive oxygen species generated by mutant SOD1 expression induce breaks in the mitochondria and nucleus of human cells (Tsang et al., 2014; Chiang et al., 2017). These breaks also require processing by TDP1 (Ben Hassine and Arcangioli, 2009; Chiang et al., 2017), and mutant SOD1 expression in a TDP1 knock-out background was toxic (Chiang et al., 2017). Thus, ALS-causing SOD1 mutations lead to the over-accumulation of reactive oxygen species, causing oxidative DNA damage requiring TDP1-mediated repair. Elevated levels of reactive oxygen species have also been reported in other ALS subtypes, including C9orf72-, FUS-, TDP-43- and sporadic-ALS disease models (Smith et al., 2017).
Therapeutic potential of PARP inhibition and NAD(+) replacement strategies
Several lines of independent evidence have linked ALS to genomic instability. Perhaps the most important question, though, is how can we utilize this knowledge to modulate the disease? Cerebellar ataxias are commonly caused by mutations in DNA repair proteins (Madabhushi et al., 2014). It has become apparent that cells defective in DNA repair processes typically display PARP1 hyperactivation (Lee et al., 2009; Krenzlin et al., 2012; Fang et al., 2014; Scheibye-Knudsen et al., 2014; Hoch et al., 2017), as greater demand becomes bestowed upon PARP1-mediated repair pathways. In-line with this idea, XRCC1 knock-out mouse models display PARP1 hyperactivation (Lee et al., 2009; Hoch et al., 2017). Deletion of the Parp1 gene in Xrcc1 knock-out mice is able to prevent neurodegeneration, indicating that the overactivation of PARP1 is a neurotoxic consequence of defective DNA repair (Hoch et al., 2017).
PARP enzymes catalyse the addition of poly-ADP-ribose (PAR) chains onto glutamate or serine residues of target proteins (Bai, 2015). Cellular pools of NAD+ are the donor molecule, which PARP enzymes catalyse during this process. It is postulated that PARP1 hyperactivation leads to the overconsumption of NAD+, underpinning the neurodegeneration observed in XRCC1-null mice (Hoch et al., 2017). Indeed, several neurological diseases that are characterized by defective DNA repair, including ataxia telangiectasia, are associated with PARP1 hyperactivation and low levels of cellular NAD+ (Krenzlin et al., 2012; Fang et al., 2014, 2016; Scheibye-Knudsen et al., 2014). These data suggest that pharmacological inhibition of PARP1 may be protective. However, current PARP1 inhibitors have been shown to induce the trapping of PARP1 onto chromatin (Pommier et al., 2016). PARP trapping would likely cause neurotoxicity by inducing DNA lesions and blocking transcriptional elongation, making current PARP inhibitors unsuitable as disease-modifying agents for treating neurodegeneration. Next generation PARP1 inhibitors, which block the enzymatic action of PARP1 while preventing its trapping onto chromatin, may offer a promising therapeutic potential for ALS and related diseases.
As well as PARPs, cellular NAD+ is further consumed by another family of enzymes called sirtuins (SIRTs) (Vassilopoulos et al., 2011). SIRTs function as NAD+ dependent lysine deacetylases, which regulate a number of biological processes, including metabolism and DNA repair (Michan and Sinclair, 2007). SIRT1, for instance, promotes DSB repair by deacetylating and in turn activating HDAC1 (Dobbin et al., 2013). SIRT1 activity appears to be impeded by PARP1 hyperactivation (Cantó et al., 2015). In addition, NAD+ replenishment has been shown to reactivate SIRT1 and prevent neurodegeneration (Scheibye-Knudsen et al., 2014), thereby compensating for PARP1 hyperactivation. An attractive model for these data is that compromised DNA repair leads to PARP1 hyperactivation, resulting in the depletion of NAD+ levels, limiting NAD+ dependent SIRTs. The repletion of NAD+ therefore restores SIRT1 function, reduces genomic instability, and ultimately prevents neurodegeneration. As such, NAD+ replenishment strategies represent a promising new strategy for neurological diseases characterized by DNA repair dysfunction. Indeed, the repletion of NAD+ has been shown to positively modulate the disease course in models of ataxia telangiectasia, Cockayne syndrome and xeroderma pigmentosium (Krenzlin et al., 2012; Fang et al., 2014, 2016; Scheibye-Knudsen et al., 2014).
Given the emerging role of dysfunctional DNA repair, particularly defective ATM-mediated repair, in ALS it may be possible that similar strategies would confer therapeutic benefits. Indeed, motor neurons are particularly sensitive to perturbations in cellular NAD+ levels. Deletion of intracellular nicotinamide phosphoribosyltransferase (iNAMPT), the rate-limiting enzyme involved in NAD+ biosynthesis, leads to a motor neuron disease phenotype in mice (Wang et al., 2017). It is striking that the specific targeting of NAD+ levels causes a motor neuron disease phenotype. Moreover, iNAMPT protein levels were significantly reduced in ALS patient spinal cord tissues (Wang et al., 2017), reinforcing the notion that NAD+ replenishment could offer new therapeutic benefit in ALS.
The selective sensitivity of motor neurons to genomic instability
Motor neurons, like Purkinje cells that degenerate in ataxia telangiectasia and related disorders, are among the largest cells of the human body. Their large size is probably linked to high levels of cellular respiration, elevated levels of transcription and in turn, a high dependency upon functional cellular clearance pathways to maintain cellular homeostasias. Unsurprisingly, defective RNA processing, impaired cellular clearance pathways, and dysfunctional mitochondrial respiration are linked to the pathogenesis of ALS—all of which drive genomic instability. Defective RNA processing leads to genomic instability due to the formation of R-loops, while dysfunctional mitochondrial respiration leads to oxidative damage. Autophagy defects lead to genomic instability due to the over accumulation of p62 and the suppression of DSB repair signalling. The high metabolic, transcriptional, and autophagic activity of motor neurons may render these cells particularly vulnerable to perturbations in these, intrinsically linked, cellular processes.
Acknowledgement
The authors would like to thank Matt Shaw for critically reading the manuscript.
Funding
This work is funded by a Wellcome Trust Investigator Award (103844) and a Lister Institute of Preventative Medicine fellowship to Sherif F. El-Khamisy.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- DSB
double-stranded break
- FTD
frontotemporal dementia
References
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol 2011; 18: 1345–50. [DOI] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell 2012; 46: 115–24. [DOI] [PubMed] [Google Scholar]
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 2006; 443: 713–16. [DOI] [PubMed] [Google Scholar]
- Alagoz M, Chiang SC, Sharma A, El-Khamisy SF. ATM deficiency results in accumulation of DNA-topoisomerase I covalent intermediates in neural cells. PLoS One 2013; 8: e58239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 2013; 77: 639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaki T, Ito H, Fukushima H, Inoue T, Kondo T, Ikemoto A, et al. Immunoreactivity of valosin-containing protein in sporadic amyotrophic lateral sclerosis and in a case of its novel mutant. Acta Neuropathol Commun 2014; 2: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P. Biology of Poly(ADP-Ribose) polymerases: the factotums of cell maintenance. Mol Cell 2015; 58: 947–58. [DOI] [PubMed] [Google Scholar]
- Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol 2013; 126: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Hassine S, Arcangioli B. Tdp1 protects against oxidative DNA damage in non-dividing fission yeast. EMBO J 2009; 28: 632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 2014; 511: 362–5. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171: 603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell 2017; 66: 801–17. [DOI] [PubMed] [Google Scholar]
- Blokhuis AM, Groen EJ, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol 2013; 125: 777–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brcic J, Plavec J. ALS and FTD linked GGGGCC-repeat containing DNA oligonucleotide folds into two distinct G-quadruplexes. Biochim Biophys Acta 2017; 1861 (5 Pt B): 1237–45. [DOI] [PubMed] [Google Scholar]
- Burgess RC, Burman B, Kruhlak MJ, Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell Rep 2014; 9: 1703–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Menzies K, Auwerx J. NAD(+) metabolism and the control of energy homeostasis—a balancing act between mitochondria and the nucleus. Cell Metab 2015; 22: 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez-Alegre M, et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 2013; 52: 583–90. [DOI] [PubMed] [Google Scholar]
- Ceballos-Diaz C, Rosario AM, Park HJ, Chakrabarty P, Sacino A, Cruz PE, et al. Viral expression of ALS-linked ubiquilin-2 mutants causes inclusion pathology and behavioral deficits in mice. Mol Neurodegener 2015; 10: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J 2009; 276: 1494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet 2014a; 10: e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Hieter P, Stirling PC. Mechanisms of genome instability induced by RNA-processing defects. Trends Genet 2014b; 30: 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am J Hum Genet 2004; 74: 1128–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SC, Meagher M, Kassouf N, Hafezparast M, McKinnon PJ, Haywood R, et al. Mitochondrial protein-linked DNA breaks perturb mitochondrial gene transcription and trigger free radical–induced DNA damage. Sci Adv 2017; 3: e1602506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40: 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 2014; 137 (Pt 7): 2040–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couratier P, Corcia P, Lautrette G, Nicol M, Marin B. ALS and frontotemporal dementia belong to a common disease spectrum. Rev Neurol 2017; 173: 273–9. [DOI] [PubMed] [Google Scholar]
- Cremona CA, Behrens A. ATM signalling and cancer. Oncogene 2014; 33: 3351–60. [DOI] [PubMed] [Google Scholar]
- Das BB, Antony S, Gupta S, Dexheimer TS, Redon CE, Garfield S, et al. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J 2009; 28: 3667–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72: 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 2011; 477: 211–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 2008; 456: 524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci 2013; 16: 1008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 2004; 18: 1618–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre M, Lambert S, Carreira A, Amor-Guéret M, Vagner S. DNA damage: RNA-binding proteins protect from near and far. Trends Biochem Sci 2016; 39: 141–9. [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF. To live or to die: a matter of processing damaged DNA termini in neurons. EMBO Mol Med 2011; 3: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 2005; 434: 108–13. [DOI] [PubMed] [Google Scholar]
- Fang EF, Kassahun H, Croteau DL, Scheibye-Knudsen M, Marosi K, Lu H, et al. NAD(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab 2016; 24: 566–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 2014; 157: 882–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Lin H, Wang X, Zuo Q, Qin J, Zhang P. The NEK1 interactor, C21ORF2, is required for efficient DNA damage repair. Acta Biochim Biophys Sin 2015; 47: 834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Konopka A, Soo KY, Ito D, Atkin JD. The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis. Hum Mol Genet 2017; 26: 2882–96. [DOI] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula S, Liu E, Yang Y, Chen W, et al. Sqstm1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol 2011; 68: 1440–6. [DOI] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 2015; 525: 129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Almeida S, Lopez-Gonzalez R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J 2017; 36: 2931–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pichardo D, Canas JC, Garcia-Rubio ML, Gomez-Gonzalez B, Rondon AG, Aguilera A. Histone mutants separate R loop formation from genome instability induction. Mol Cell 2017; 66: 597–609. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 2008; 31: 167–77. [DOI] [PubMed] [Google Scholar]
- Goode A, Butler K, Long J, Cavey J, Scott D, Shaw B, et al. Defective recognition of LC3B by mutant SQSTM1/p62 implicates impairment of autophagy as a pathogenic mechanism in ALS-FTLD. Autophagy 2016; 12: 1094–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh M, Lufino MM, Wade-Martins R, Gromak N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet 2014; 10: e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Lan M, Mojsilovic-Petrovic J, Choi WH, Safren N, Barmada S, et al. The proline/arginine dipeptide from hexanucleotide repeat expanded C9ORF72 inhibits the proteasome. eNeuro 2017; 4: 0249–16. doi: 10.1523/ENEURO.0249-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 2014; 507: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardiman O, Al-Chalabi A, Chio A, Corr EM, Logroscino G, Robberecht W, van den Berg LH. Amyotrophic lateral sclerosis. Nat Rev Dis Primers 2017; 3: 17071. [DOI] [PubMed] [Google Scholar]
- Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J 2009; 423: 157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Livingston DM. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 2015; 57: 636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SJ, Mordes DA, Cameron LA, Neuberg DS, Landini S, Eggan K, et al. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc Natl Acad Sci USA 2016; 113: E7701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch NC, Hanzlikova H, Rulten SL, Tetreault M, Komulainen E, Ju L, et al. XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 2017; 541: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodroj D, Recolin B, Serhal K, Martinez S, Tsanov N, Abou Merhi R, et al. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J 2017; 36: 1182–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007; 131: 901–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 2003; 12: 711–21. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kominami E, Tanaka K, Komatsu M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy 2008; 4: 1063–6. [DOI] [PubMed] [Google Scholar]
- Jangi M, Fleet C, Cullen P, Gupta SV, Mekhoubad S, Chiao E, et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc Natl Acad Sci USA 2017; 114: E2347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010; 68: 857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 2015; 18: 1226–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 2008; 40: 572–4. [DOI] [PubMed] [Google Scholar]
- Kaidi A, Jackson SP. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature 2013; 498: 70–4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kapeli K, Martinez FJ, Yeo GW. Genetic mutations in RNA-binding proteins and their roles in ALS. Hum Genet 2017; 136: 1193–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J 2015; 282: 4672–8. [DOI] [PubMed] [Google Scholar]
- Katyal S, el-Khamisy SF, Russell HR, Li Y, Ju L, Caldecott KW, McKinnon PJ. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J 2007; 26: 4720–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katyal S, Lee Y, Nitiss KC, Downing SM, Li Y, Shimada M, et al. Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat Neurosci 2014; 17: 813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna KP, van Doormaal PT, Dekker AM, Ticozzi N, Kenna BJ, Diekstra FP, et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet 2016; 48: 1037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 2001; 27: 247–54. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013; 495: 467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007; 318: 1637–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 2009; 5: 862–3. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol 1987; 123: 241–50. [DOI] [PubMed] [Google Scholar]
- Krenzlin H, Demuth I, Salewsky B, Wessendorf P, Weidele K, Burkle A, et al. DNA damage in Nijmegen Breakage Syndrome cells leads to PARP hyperactivation and increased oxidative stress. PLoS Genet 2012; 8: e1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009; 323: 1205–8. [DOI] [PubMed] [Google Scholar]
- Lee Y, Katyal S, Li Y, El-Khamisy SF, Russell HR, Caldecott KW, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci 2009; 12: 973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 2013; 5: 1178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen J, Ricupero CL, Hart RP, Schwartz MS, Kusnecov A, et al. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med 2012; 18: 783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hart RP, Mallimo EM, Swerdel MR, Kusnecov AW, Herrup K. EZH2-mediated H3K27 trimethylation mediates neurodegeneration in ataxia-telangiectasia. Nat Neurosci 2013; 16: 1745–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev 2005; 19: 2705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol 2000; 65: 127–33. [DOI] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 2013; 79: 416–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, et al. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron 2016; 92: 383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Pan L, Tsai LH. DNA damage and its links to neurodegeneration. Neuron 2014; 83: 266–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131: 887–900. [DOI] [PubMed] [Google Scholar]
- Majcher V, Goode A, James V, Layfield R. Autophagy receptor defects and ALS-FTLD. Mol Cell Neurosci 2015; 66 (Pt A): 43–52. [DOI] [PubMed] [Google Scholar]
- Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, et al. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol 2011; 13: 1376–82. [DOI] [PubMed] [Google Scholar]
- Melki J, Sheth P, Abdelhak S, Burlet P, Bachelot MF, Frézal J, et al. Mapping of acute (type I) spinal muscular atrophy to chromosome 5q12-q14. The Lancet 1990; 336: 271–3. [DOI] [PubMed] [Google Scholar]
- Meyer H, Weihl CC. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci 2014; 127 (Pt 18): 3877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J 2007; 404: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet 2001; 29: 189–93. [DOI] [PubMed] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet 2004; 36: 225–7. [DOI] [PubMed] [Google Scholar]
- Murphy AN. In a flurry of PINK, mitochondrial bioenergetics takes a leading role in Parkinson's disease. EMBO Mol Med 2009; 1: 81–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am J Med Genet 1992; 42: 68–84. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314: 130–3. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev 2000; 80: 315–60. [DOI] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 2010; 12: 177–84. [DOI] [PubMed] [Google Scholar]
- Osaka M, Ito D, Suzuki N. Disturbance of proteasomal and autophagic protein degradation pathways by amyotrophic lateral sclerosis-linked mutations in ubiquilin 2. Biochem Biophys Res Commun 2016; 472: 324–31. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–45. [DOI] [PubMed] [Google Scholar]
- Pommier Y, O'Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med 2016; 8: 362ps317. [DOI] [PubMed] [Google Scholar]
- Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 2015; 18: 1175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek H, Luff J, Cheung K, Kozlov S, Gatei M, Lee CS, et al. A rat model of ataxia-telangiectasia: evidence for a neurodegenerative phenotype. Hum Mol Genet 2017; 26: 109–23. [DOI] [PubMed] [Google Scholar]
- Ramesh N, Pandey UB. Autophagy dysregulation in ALS: when protein aggregates get out of hand. Front Mol Neurosci 2017; 10: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Majcher V, Searle MS, Layfield R. SQSTM1 mutations–bridging Paget disease of bone and ALS/FTLD. Exp Cell Res 2014; 325: 27–37. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011; 72: 257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273: 5858–68. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993; 362: 59–62. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med 2004; 10 (Suppl): S10–17. [DOI] [PubMed] [Google Scholar]
- Rossi S, Serrano A, Gerbino V, Giorgi A, Di Francesco L, Nencini M, et al. Nuclear accumulation of mRNAs underlies G4C2-repeat-induced translational repression in a cellular model of C9orf72 ALS. J Cell Sci 2015; 128: 1787–99. [DOI] [PubMed] [Google Scholar]
- Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol 2009; 29: 3124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino E, Rainero I, Chio A, Rogaeva E, Galimberti D, Fenoglio P, et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology 2012; 79: 1556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MA, van der Want JJ, et al. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol 2006; 4: e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Caldecott KW. DNA strand break repair and neurodegeneration. DNA Repair 2013; 12: 558–67. [DOI] [PubMed] [Google Scholar]
- Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gomez-Herreros F, et al. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res 2014; 42: 307–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira JM, Herrero AB, Garcia-Rubio ML, Marin A, Moreno S, Aguilera A. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev 2013; 27: 2445–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995; 268: 1749–53. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Mitchell SJ, Fang EF, Iyama T, Ward T, Wang J, et al. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab 2014; 20: 840–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz AK, Kay LE. A dynamic molecular basis for malfunction in disease mutants of p97/VCP. Elife 2016; 5: e20143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotter EL, Chen HJ, Shaw CE. TDP-43 proteinopathy and ALS: insights into disease mechanisms and therapeutic targets. Neurotherapeutics 2015; 12: 352–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 2014; 516: 436–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 2014; 28: 1384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 2011; 42: 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Shaw PJ, De Vos KJ. The role of mitochondria in amyotrophic lateral sclerosis. Neurosci Lett 2017, in press. doi: 10.1016/j.neulet.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 2014; 56: 777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Hotz-Wagenblatt A, Voit R, Grummt I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev 2017, in press. doi: 10.1101/gad.300624.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies J, Waizenegger A, Barton O, Surder M, Wright WD, Heyer WD, et al. Nek1 regulates Rad54 to orchestrate homologous recombination and replication fork stability. Mol Cell 2016; 62: 903–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Stankovic T, Byrd PJ, Wechsler T, Miller ES, Huissoon A, et al. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci USA 2007; 104: 16910–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle 2009; 8: 1532–8. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 1999; 99: 577–87. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 2009; 136: 420–34. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 2009; 11: 1376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet 2002; 32: 267–72. [DOI] [PubMed] [Google Scholar]
- Tan SLW, Chadha S, Liu Y, Gabasova E, Perera D, Ahmed K, et al. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell 2017; 169: 1105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair 2004; 3: 1219–25. [DOI] [PubMed] [Google Scholar]
- Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature 2015; 523: 53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troakes C, Maekawa S, Wijesekera L, Rogelj B, Siklos L, Bell C, et al. An MND/ALS phenotype associated with C9orf72 repeat expansion: abundant p62-positive, TDP-43-negative inclusions in cerebral cortex, hippocampus and cerebellum but without associated cognitive decline. Neuropathology 2012; 32: 505–14. [DOI] [PubMed] [Google Scholar]
- Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XF. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun 2014; 5: 3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 2009; 11: 1315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 2003; 22: 5612–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact 2006; 160: 1–40. [DOI] [PubMed] [Google Scholar]
- van den Boom J, Wolf M, Weimann L, Schulze N, Li F, Kaschani F, et al. VCP/p97 extracts sterically trapped Ku70/80 rings from DNA in double-strand break repair. Mol Cell 2016; 64: 189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen W, Shatunov A, Dekker AM, McLaughlin RL, Diekstra FP, Pulit SL, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet 2016; 48: 1043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009; 323: 1208–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilopoulos A, Fritz KS, Petersen DR, Gius D. The human sirtuin family: evolutionary divergences and functions. Hum Genomics 2011; 5: 485–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite AJ, Baumer D, East S, Neal J, Morris HR, Ansorge O, et al. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging 2014; 35: 1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C, Herranz-Martin S, Karyka E, Liao C, Lewis K, Elsayed W, et al. C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat Neurosci 2017; 20: 1225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA 2007; 104: 20759–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, et al. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 2013; 16: 1383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang Q, Bao R, Zhang N, Wang Y, Polo-Parada L, et al. Deletion of nampt in projection neurons of adult mice leads to motor dysfunction, neurodegeneration, and death. Cell Rep 2017; 20: 2184–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma K, et al. Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol Cell 2016; 63: 34–48. [DOI] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 2004; 36: 377–81. [DOI] [PubMed] [Google Scholar]
- Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, et al. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J 2016; 35: 1656–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 2007; 85: 509–20. [DOI] [PubMed] [Google Scholar]
- Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, et al. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv 2016; 2: e1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Lopez-Gonzalez R, Kunz RC, Gangopadhyay J, Borufka C, Gygi SP, et al. Evidence that C9ORF72 dipeptide repeat proteins associate with U2 snRNP to cause mis-splicing in ALS/FTD patients. Cell Rep 2017; 19: 2244–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuce O, West SC. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol 2013; 33: 406–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P, Padeken J, van Schendel R, Kalck V, Tijsterman M, Gasser SM. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat Genet 2016; 48: 1385–95. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015a; 525: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, et al. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 2015b; 348: 1160–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chiang HC, Wang Y, Zhang C, Smith S, Zhao X, et al. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat Commun 2017; 8: 15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol 2014; 128: 505–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 2016; 529: 48–53. [DOI] [PubMed] [Google Scholar]
- Zhou B, Liu C, Geng Y, Zhu G. Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Sci Rep 2015; 5: 16673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA 2013; 110: E4968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]