See Fratta and Isaacs (doi:10.1093/brain/awy091) for a scientific commentary on this article.
Mutations in the RNA binding proteins TDP-43 and hnRNP A1 are associated with ALS. Deshaies et al. report that TDP-43 modulates hnRNP A1 splicing so as to elevate levels of an isoform containing an elongated prion-like domain, hnRNP A1B. Patients with documented TDP-43 pathology show neuronal hnRNP A1B cytoplasmic accumulation.
Keywords: amyotrophic lateral sclerosis, TDP-43, hnRNP A1, alternative splicing, protein aggregation
Abstract
See Fratta and Isaacs (doi:10.1093/brain/awy091) for a scientific commentary on this article.
The RNA binding proteins TDP-43 (encoded by TARDBP) and hnRNP A1 (HNRNPA1) are each mutated in certain amyotrophic lateral sclerosis cases and are often mislocalized in cytoplasmic aggregates within motor neurons of affected patients. Cytoplasmic inclusions of TDP-43, which are accompanied by a depletion of nuclear TDP-43, are observed in most amyotrophic lateral sclerosis cases and nearly half of frontotemporal dementia cases. Here, we report that TDP-43 binds HNRNPA1 pre-mRNA and modulates its splicing, and that depletion of nuclear TDP-43 results in increased inclusion of a cassette exon in the HNRNPA1 transcript, and consequently elevated protein levels of an isoform containing an elongated prion-like domain, referred to as hnRNP A1B. Combined in vivo and in vitro approaches demonstrated greater fibrillization propensity for hnRNP A1B, which drives protein aggregation and is toxic to cells. Moreover, amyotrophic lateral sclerosis patients with documented TDP-43 pathology showed neuronal hnRNP A1B cytoplasmic accumulation, indicating that TDP-43 mislocalization may contribute to neuronal vulnerability and loss via altered HNRNPA1 pre-mRNA splicing and function. Given that TDP-43 and hnRNP A1 each bind, and thus modulate, a third of the transcriptome, our data suggest a much broader disruption in RNA metabolism than previously considered.
Introduction
Amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease, is a neurodegenerative disease affecting upper and lower motor neurons causing progressive muscle wasting, denervation and eventual respiratory failure (Ling et al., 2013). ALS can occur in isolation, or more commonly in association with frontotemporal dysfunction, which can include frontotemporal dementia (FTD) (Strong et al., 2017). FTD is characterized clinically by neuropsychological, speech and language deficits typical of frontotemporal dysfunction, and neuropathologically by either pathological neuronal and glial deposits of either TDP-43 or tau. In the former instance, there is a striking similarity to those observed in ALS, suggesting the existence of a continuum of disease (Ling et al., 2013; Weishaupt et al., 2016). While RNA binding proteins are a major focus of current ALS/FTD research, it is not well understood how these proteins interact and/or coordinate RNA processing and metabolism. Disease-causing mutations in several RNA binding proteins, including TDP-43 (encoded by TARDBP) (Neumann et al., 2006; Scotter et al., 2015), heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1, HNRNPA1) and A2 (hnRNP A2, HNRNPA2B1) (Kim et al., 2013; Scotter et al., 2015; Kanekura et al., 2016), are reported to promote aggregate formation in familial and sporadic ALS/FTD cases. Mislocalization of TDP-43 into cytoplasmic aggregates is a major disease feature as it is observed in 97% of all ALS cases and 45% of FTD cases (Ling et al., 2013). Neurons featuring TDP-43 cytoplasmic inclusions typically also demonstrate nuclear TDP-43 depletion, suggesting a probable disruption in normal nuclear functions of TDP-43 such as mRNA stabilization, transcription, splicing or mRNA nucleocytoplasmic transport (Neumann et al., 2006; Ling et al., 2013). It remains debated whether these histological observations translate to a loss of nuclear TDP-43 function, whether aggregated TDP-43 acquires a cytoplasmic-oriented toxic gain of function (Lee et al., 2012), or both.
HnRNP A1 is a 34 kDa protein featuring two RNA-recognition motifs (RRM) and a prion-like/glycine-rich domain (prion-like domain) that is also broadly implicated in RNA metabolism (Mayeda et al., 1994; Bekenstein and Soreq, 2013; Jean-Philippe et al., 2013). It is highly abundant in motor neurons and is implicated in the disease mechanism(s) relevant to spinal muscular atrophy (Kashima and Manley, 2003; Kashima et al., 2007; Chen et al., 2017). Alternative splicing of HNRNPA1 transcripts results in the inclusion of a cassette exon, giving rise to a longer 38 kDa isoform of relatively low abundance and unknown function. This longer version, referred to as hnRNP A1B, contains an additional 52 amino acids in the prion-like domain encoded by exon 7B (Buvoli et al., 1990; Bekenstein and Soreq, 2013). The biological significance of hnRNP A1B remains to be fully elucidated.
Here, we demonstrate that TDP-43 can bind to HNRNPA1 pre-mRNA to modulate its alternative splicing. Furthermore, nuclear depletion of TDP-43 drives exon 7B inclusion. The resulting longer isoform hnRNP A1B is aggregation-prone and negatively impacts cell survival. Finally, in ALS patients with documented TDP-43 cytoplasmic inclusions/nuclear depletion, hnRNP A1B accumulation is observed as pronounced cytoplasmic aggregates that are distinct from TDP-43 inclusions. Thus, our data indicate cross-regulation between two ALS/FTD relevant RNA binding proteins and predict disruptions in RNA metabolism that are more extensive than previously considered.
Materials and methods
Constructs
Flag-TDP-43ΔNLSsiRes was generated using the QuickChange® II Site-Directed mutagenesis kit (Agilent Technologies) on pCS2-Flag-TDP-43WT with the forward primer 5′-CAACTATCCAAAAGATAACGCAGCAGCAATGGATGAGACAGATGC-3′ and its complementary reverse to mutate the nuclear localization signal (NLS), as previously published (Winton et al., 2008). Modifications to render TDP-43 plasmids siRNA-resistant were done with the forward primer 5′-CTTCCTAATTCTAAGCAGAGGCAGGACGAGCCTTTGAGAAGC-3′ and its complementary reverse. pCMV-Myc-A1 and pCMV-Myc-A1B were subcloned from pIND-A1-Myc and pIND-A1B-Myc, respectively (Yang et al., 1994), and inserted between EcoRI and HindIII sites in pCMV(SV40), which was previously modified to include the Myc tag. The D262V/D314V mutation was introduced via the QuickChange® II Site-Directed mutagenesis kit (Agilent Technologies) with the following primers: Forward, 5′-GGTGGTGGAAGCTACAATGTTTTTGGGAATTACAACAATC-3′ and Reverse, 5′-GATTGTTGTAATTCCCAAAAACATTGTAGCTTCCACCACC-3′.
Cell culture and transfection
HeLa and HEK293FT cells were cultured in Dulbecco’s high glucose modified Eagle medium (DMEM, GE Healthcare) supplemented with 10% foetal bovine serum (FBS, Wisent) and 2 mM l-glutamine (Sigma). CB3 cells (parental and stable transfectants) were cultured in Minimum Essential Media (MEM) Alpha Modification (GE Healthcare) supplemented with 10% FBS (Wisent) and 1% Penicillin-Streptomycin (Thermo Fisher Scientific). Cells were collected 72 h after transfection with 125 pmol Stealth siRNA using Lipofectamine® 2000 (Invitrogen), according to the manufacturer’s instructions. The sequences used were: Control siRNA: #12935-200, (Invitrogen), TDP-43 (TARDBP) siRNA #1: 5′-AAGAUGAGAACGAUGAGCCCAUUGA-3′, and TDP-43 siRNA #2: 5′-AAGCAAAGCCAAGAUGAGCCUUUGA-3′ (used for all experiments). For filter trap and intracellular aggregate experiments, HEK293FT and HeLa cells, respectively, were transfected with Myc-tagged cDNA for 24 h using Lipofectamine® LTX and Plus reagent (Invitrogen).
Immunoblotting
Lysates were prepared with RIPA buffer (150 mM NaCl, 50 mM Tris pH 7.4, 1% Triton™ X-100, 0.1% SDS, 1% sodium deoxycholate) with protease inhibitors, subjected to SDS-PAGE, transferred to nitrocellulose and blocked with 5% powdered milk in PBS-T. Membranes were incubated with mouse anti-hnRNP A1 (9H10; Abcam), mouse anti-Actin (MP Biomedicals), rabbit anti-c-Myc (Sigma), mouse anti-alpha tubulin (DM1A; Abcam), rabbit anti-TDP-43 (Proteintech), rabbit anti-GAPDH (Cell Signaling) and rabbit anti-HuR (Millipore) in blocking buffer. Membranes were subsequently incubated with HRP-conjugated secondary antibodies (Jackson Immunoresearch) and revealed with ECL chemoluminescence (Thermo Fisher Scientific). Films were quantified by densitometry using Photoshop. Subcellular fractions were prepared using the NE-PER™ Nuclear and Cytoplasmic Extraction kit (Thermo Scientific), according to the manufacturer’s instructions. Sprague-Dawley rat tissues were homogenized with a manual tissue grinder in five volumes of homogenization buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, with protease inhibitors). Homogenates were subsequently adjusted to 1% SDS and 1% NP-40, incubated 10 min on ice and 10 min at room temperature. Some tissues were further processed with a fine-tip probe sonicator (three pulses 10 s on, 30 s off, 40% amplitude) to reduce viscosity of the homogenates. Homogenates were cleared by centrifugation at 16 000g for 20 min at 4°C, supernatants recovered and the protein concentration quantified via BCA (Pierce). Tissue lysates were separated by SDS-PAGE and subsequently immunoblotted.
Immunoprecipitation
Lysates were prepared in immunoprecipitation buffer (50 mM Tris pH 8, 150 mM NaCl, 0.1 mM EDTA, 0.5% NP-40 and 10% glycerol) with protease inhibitors and subsequently triturated with a 25G needle. Protein G Dynabeads® (Thermo Fisher) were prepared according to the manufacturer’s instructions. Three hundred micrograms of cell lysate was immunoprecipitated overnight with rabbit anti-hnRNP A1B (custom) at 4°C. Immunoprecipitates were eluted with 2.5× Laemmli buffer and immediately analysed by immunoblotting with rabbit anti-hnRNP A1B (custom), mouse anti-hnRNP A1 (9H10; Abcam).
Immunofluorescence
Coverslips were fixed in 4% formaldehyde/PBS, washed with PBS, permeabilized in 0.1% Triton™ X-100/PBS and blocked in 0.1% bovine serum albumin/PBS. Coverslips were incubated with primary antibodies: mouse anti-Flag (Sigma), rabbit anti-c-Myc (Sigma), goat anti-c-Myc (Sigma), rabbit anti-TDP-43 (Proteintech), mouse anti-TDP-43 (Millipore), rabbit anti-hnRNP A1B (custom), diluted in 0.1% BSA/PBS; washed once with 0.1% Triton™ X-100/PBS and then twice with 0.1% BSA/PBS. Coverslips were then incubated with secondary antibodies: donkey anti-rabbit Alexa 488 (Thermo Fisher Scientific), donkey anti-mouse Alexa 488 (Jackson Immunoresearch), donkey anti-rabbit Alexa 594 (Jackson Immunoresearch) and donkey anti-mouse Alexa 594 (Jackson Immunoresearch) diluted in 0.1% BSA/PBS, washed, labelled with TO-PRO-3 iodide (Thermo Fisher Scientific), and mounted using ProLong® Antifade (Thermo Fisher Scientific). Images were collected on a Leica TCS SP5 confocal microscope equipped with 40× (1.25 N.A.) oil objective and the Leica Application Suite imaging software. To quantify the levels of hnRNP A1B, 35–70 cells per condition were analysed per experiment. To quantify aggregates, a minimum of 100 cells per genotype were analysed per experiment.
RNA immunoprecipitation
HeLa cells were lysed in 10 mM Tris, pH 7.4, 50 mM NaCl, 0.5% Triton™ X-100, with protease inhibitors, triturated subsequently through 25G and 18G needle syringes, incubated for 5 min at room temperature, centrifuged at 13 000g and the supernatant collected. Aliquots of 3–7 mg of pre-cleared lysate were immunoprecipitated at 4°C overnight with mouse anti-TDP-43 (Abnova) or mouse anti-Flag (Sigma) pre-bound to Protein G Dynabeads® (Thermo Fisher Scientific). Immuno-precipitates were treated with DNase (Qiagen) and RNA was recovered with TRIzol® (Invitrogen). Equal amounts of RNA were reverse transcribed using the QuantiTect® Reverse Transcription kit (Qiagen). cDNA was amplified by standard PCR with the following primers: TARDBP exon 2 forward: 5′-ACCGAAGACCTGAAAGAG-3′; TARDBP exon 3 reverse: 5′-GGAAGTTTGCAGTCACACCAT-3′; HNRNPA1 exon 4 forward: 5′-GGGCTTTGCCTTTGTAACCT-3′; HNRNPA1 exon 6 reverse: 5′-ACGACCGAAGTTGTCATTCC-3′; CAMK2A forward: 5′-CCACAGGGGCTTTAGGAGA-3′; CAMK2A reverse: 5′-GCTGCTGCCGCTTTTGTA-3′; HSPA4 forward: 5′-TGAAGGAGACAGCCGAAAG-3′; HSPA4 reverse: 5′-TGCATCAGTATAGAAACAAGGAA-3′.
Quantitative PCR, reverse transcription-PCR and RNA stability
RNA was extracted with the RNeasy® mini kit (Qiagen) and equal amounts were reverse transcribed via the QuantiTect® Reverse Transcription kit (Qiagen), according to the manufacturer’s instructions. PrimeTime Standard qPCR assays (IDT) for HNRNPA1 exons 1–2 (NM_031157, Hs.PT.58.38919354), HNRNPA1 exons 8–9 (NM_031157, Hs.PT.58.458382, detects exon 7B) and ACTB exon 6-6 (NM_001101, Hs.PT.56a.40703009.g) were used to quantify mRNA levels. To determine mRNA stability, HeLa cells were treated with 5 µg/ml of actinomycin D (Sigma) for 2, 4 or 8 h prior to RNA extraction and analysed as above. The QuantStudio™ 7 Flex Real-Time PCR System (Life Technologies) was used for qPCR. To assess exon 7B inclusion, cDNA was amplified with HNRNPA1 exon 7 forward: 5′-CGGTGGGAATGACAACTTCGGT-3′, HNRNPA1 exon 8 reverse: 5′-TTGAGGACTGATTGTTGTAATTCCC-3′, ACTB exon 5 forward: 5′-CTGTGGCATCCACGAAACTA-3′, ACTB exon 6 reverse: 5′-AGTACTTGCGCTCAGGAGGA-3′, TARDBP exon 4 forward: 5′-GTCTCTTTGTGGAGAGGACTTGATC-3′ and TARDBP exon 5 reverse: 5′-GGTTTGGCTCCCTCTGCATG-3′. Amplicons were visualized on a 4% agarose gel.
Luciferase assays
HeLa cells were transfected with siRNA for 48 h, and then subsequently transfected for 24 h with reporter plasmids including the 5′ and 3′ untranslated region (UTR) of human HNRNPA1 (SwitchGear Genomics) and relevant controls (for promoter assay: GAPDH and a vector containing random sequence, R01; for 3′ UTR: GAPDH, GRN and vector containing random sequence, R03) using FuGENE® (Promega). The LightSwitch™ Assay (Promega) reagents were added according to the manufacturer’s instructions and luciferase activity was assessed with a Synergy H4 Hybrid Multi-Mode Microplate reader (Biotek).
In vitro splicing assay
HeLa cells were depleted or not of TDP-43 by shRNA shTDP-43 forward 5′-GATCCCCACTACAATTGATATCAAATTCAAGAGATTTGATATCAATTGTAGTGTTTTTGGAAA-3′ and shTDP-43 reverse 5′-AGCTTTTCCAAAAACACTACAATTGATATCAAATCTCTTGAATTTGATATCAATTGTAGTGGG-3′. Nuclear extracts were prepared (Dignam et al., 1983) and incubated with hnRNP A1 7B pre-mRNA (Blanchette, 1999) and a competing oligo (TG)13 or a non-targeting control oligo 5′-TCGCAGCAAACTCCGGCACTT-3′; or recombinant TDP-43 (TP710010, Origene) for 2 h at 30°C. Distally and proximally spliced products were amplified by RT-PCR using the following primers: T3–5′ forward 5′-GGGAACAAAAGCTGGGTACCG-3′ and E-Ad reverse 5′-GAGTTTGTCCTCAACCGCGA-3′ and separated on native PAGE gels.
Hexapeptide assembly and filter trap
Wild-type hnRNP A1 (NDGSNF, DGSNFG), wild-type hnRNP A1B (GSGSNF, SGSNFG) and mutant hnRNP A1D262V (SYNVFG) hexapeptides were synthesized by GenScript. Peptides (5 mM) were dissolved in fibrillization buffer (40 mM HEPES-KOH, 75 mM KCl, 1 mM dithiothreitol). Dissolved peptides were used immediately for fibril-assembly reactions via addition of 5 µM Thioflavin T (Sigma). The reaction was monitored at λex/em 440/482 nm, 25°C, with agitation, for 2 h on a Synergy H4 Hybrid Multi-Mode Microplate reader (Biotek). Samples were applied for 2 min to Formvar®-coated (polyvinyl formate) 200-mesh nickel grids evaporated with carbon, negatively stained with 2% aqueous uranyl acetate for 2 min, and examined at 80 kV with a FEI Tecnai 12 (FEI) transmission electron microscope. Filter trap assays were performed with HEK293FT and CB3 lysates prepared by freeze-thaw extraction in PBS with protease inhibitors. Thirty to sixty micrograms of whole cell lysate was diluted in 150 µl PBS with protease inhibitors and passively filtered through a non-protein binding cellulose acetate membrane (0.2 µm, GE Healthcare) for 1 h using a Bio-Dot® microfiltration apparatus (Bio-Rad). The membrane was blocked with 5% powdered milk in PBS-T and incubated with rabbit anti-c-Myc (Sigma) or mouse anti-hnRNP A1 (9H10; Abcam).
Cell viability
Equivalent numbers of CB3 cells were incubated at 37°C for 72 h then stained with trypan blue (Multicell) and counted manually with a haemocytometer.
HnRNP A1B antibody
Polyclonal antibodies against hnRNP A1B were generated in rabbit using the synthetic peptide CYGGSGSYDSYNNGG as immunogen (Genscript). This peptide corresponds to amino acids 281–294 in the hnRNP A1B variant (exon 7B, NP_112420.1) with the addition of an initial N-terminal cysteine for increased antigenicity. The amino acid stretch was chosen using the OptimumAntigen™ design tool (Genscript) to ensure uniqueness and solubility.
Patient material
Individuals were diagnosed at the ALS Clinic at London Health Sciences Center, using the revised El Escorial Criteria. Informed consent was obtained from all participants with approval from the local ethics review committee. Details of the cases used in this study are provided in Supplementary Table 1 and include six sporadic ALS cases and three familial cases (one FUS 3′ UTR*41 G>A, one C9ORF72, one C9ORF72/RGNEF), all with documented TDP-43 pathology (Keller et al., 2012) and five non-neurological disease controls. (Note, pathogenecity of FUS 3′ UTR*G>A is not known.) Paraffin-embedded spinal cord sections (6 µm) from five control cases and five ALS cases were labelled with mouse anti-hnRNP A1 (4B10; 1:100) or rabbit anti-hnRNP A1B (1:200). Antigen retrieval was performed for 15 min using sodium citrate (10 mM sodium citrate, 0.05% Tween-20, pH 6.0) and a pressure cooker (2100 Retriever; Aptum Biologics). Endogenous peroxidase was quenched with 3% hydrogen peroxide (VWR). Primary antibody incubation was performed at 4°C overnight in blocking buffer (5% BSA, 0.3% Triton™ X-100 in PBS). After washing in PBS, secondary antibody (1:200 biotinylated IgG) incubation was performed for 1 h at room temperature in blocking buffer. Antigen:antibody complexes were visualized using the Vectastain ABC Kit (Vector Laboratories) with DAB substrate. To verify the specificity of the custom hnRNP A1B antibody, peptide competition was performed in one sporadic ALS case that had previously shown hnRNP A1B pathology. The diluted antibody was incubated with the immunogenic peptide (1:20 dilution) for 1 h at room temperature prior to application to the tissue. Staining proceeded as described above. A positive control incubated at room temperature without blocking peptide was run in parallel. Harris’ haematoxylin was used to counterstain tissues for visualization. Images were collected with an Olympus BX45 light microscope at 20× and 100×.
Immunofluorescence and confocal microscopy was subsequently conducted on six ALS cases (three originally included in the DAB labelling experiments, plus three additional ALS cases previously established to have TDP-43 pathology). After sodium citrate pressure cooker antigen retrieval, tissue was probed with rabbit anti-hnRNP-A1B (1:200) and mouse anti-TDP-43 2E2-D3 (1:500, Abcam ab57105) overnight at 4°C. The following day, tissue was incubated with goat anti-rabbit Alexa Fluor® 488 (1:200, Life Technologies) and donkey anti-mouse Alexa Fluor® 568 (1:200, Life Technologies) for 1 h at room temperature. After washing and mounting, images were acquired using a Zeiss LSM 510 Meta NLO multiphoton confocal microscope.
Statistical analysis
Statistical significance of experiments was determined using Student’s t-test with GraphPad Prism software. Error bars represent the standard error of the mean (SEM).
Results
Loss of nuclear TDP-43 increases hnRNP A1B protein levels
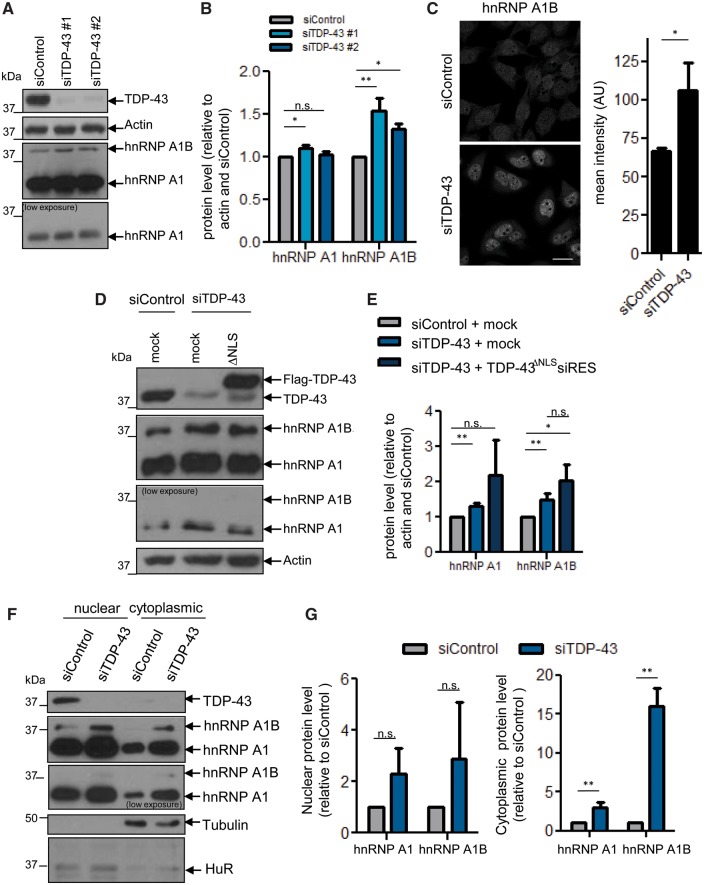
To investigate the impact of TDP-43 on hnRNP A1, we depleted endogenous TDP-43 from HeLa cells via siRNA. Immunoblotting demonstrated that TDP-43 depletion has a minimal impact on the steady state level of a 34 kDa band reactive with hnRNP A1 antibodies 4B10 (data not shown) and 9H10 (Fig. 1A and B). However, a second more-slowly migrating band at 38 kDa was upregulated in cells treated with two independent siRNAs specific for TDP-43. This 38 kDa protein is consistent with a previously described isoform, hnRNP A1B, which arises from inclusion of an alternate exon (Buvoli et al., 1990; Yang et al., 1994; Blanchette and Chabot, 1997; Chabot et al., 1997). To confirm this possibility, we generated an antibody that demonstrates a preference for hnRNP A1B (Supplementary Fig. 1A). Using this antibody, which we confirm is specific for hnRNP A1B in native conditions (Supplementary Fig. 1B and C) and quantitative immunofluorescence, we confirmed that TDP-43 depletion causes a ∼1.5-fold increase in hnRNP A1B (Fig. 1C). As ALS/FTD cases typically feature a nuclear depletion of TDP-43 rather than an outright loss of TDP-43 expression, we next examined hnRNP A1 levels in cells in which we repleted endogenous cytoplasmic TDP-43 via exogenous expression of cDNA encoding a siRNA-resistant version of TDP-43 with an inactivated NLS (FLAG-TDP-43ΔNLSsiRES). Immunofluorescence of transfectants confirmed the loss of the typical nuclear localization of TDP-43 (data not shown), as previously reported (Winton et al., 2008; Scotter et al., 2015; Xiao et al., 2015; Prpar Mihevc et al., 2016). Importantly, the 38 kDa band was also more abundant in these conditions (Fig. 1D and E). Subcellular fractionation indicated that the cytoplasmic protein levels of hnRNP A1 and hnRNP A1B were increased in TDP-43-depleted cells, while nuclear levels of these proteins were not significantly changed (Fig. 1F and G). Thus, nuclear depletion and/or cytoplasmic TDP-43 accumulation increases the cytoplasmic protein levels of endogenous hnRNP A1 and hnRNP A1B.
Figure 1.
Loss of nuclear TDP-43 increases hnRNP A1B protein levels. (A–C) siRNA-treated cells were (A) immunoblotted for TDP-43, Actin and hnRNP A1/A1B (9H10) and (B) quantified by densitometry. (C) The level of hnRNP A1B was also assessed by immunofluorescence using a custom hnRNP A1B specific antibody and the mean intensity was quantified. (D and E) Cells were depleted of endogenous TDP-43 and transfected with mock or Flag-TDP-43ΔNLSsiRES cDNA (ΔNLS). Levels of hnRNP A1 and hnRNP A1B were assessed by western blot and quantified by densitometry. (F and G) Nuclear and cytoplasmic fractions from siRNA-treated cells were immunoblotted for TDP-43, hnRNP A1/A1B (9H10), Tubulin and HuR and quantified by densitometry. For all, the mean ± SEM of three to five independent experiments is plotted. *P < 0.05, **P < 0.02. Scale bar = 25 µm.
Endogenous TDP-43 binds and regulates HNRNPA1 mRNA
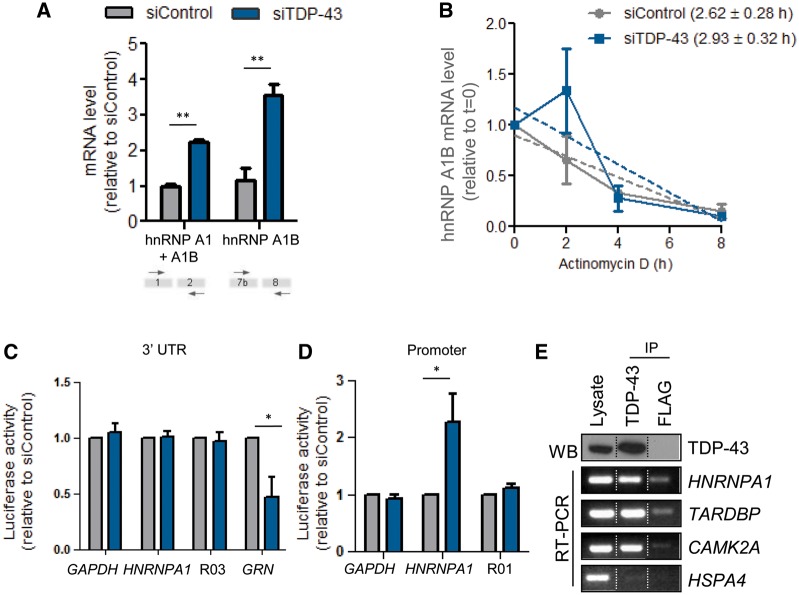
Since TDP-43 has multiple functions related to RNA metabolism (Lagier-Tourenne et al., 2010), we hypothesized that TDP-43 could regulate HNRNPA1 mRNA expression. Indeed, TDP-43 siRNA-mediated depletion induced ∼2-fold and 3.5-fold increases of total hnRNP A1/A1B and hnRNP A1B mRNA, respectively, as detected by qPCR probe sets, which detected either both transcripts or uniquely hnRNP A1B (Fig. 2A). This increase was not due to increased mRNA stability since the half-life of hnRNP A1B transcripts was comparable between control and TDP-43 siRNA treated cells (2.62 ± 0.28 h versus 2.93 ± 0.32 h, P = 0.06; Fig. 2B). The half-life of HNRNPA1 transcripts was also comparable in both conditions (data not shown). Additional data indicated that TDP-43 does not influence HNRNPA1 mRNA via the 3′ UTR, as demonstrated using a luciferase-reporter assay (Fig. 2C). However, it was also noted that the HNRNPA1 promoter is more active in TDP-43 depleted conditions (Fig. 2D). We next questioned whether TDP-43 could interact with HNRNPA1 transcripts using immunoprecipitation of endogenous TDP-43 followed by RT-PCR. Indeed, HNRNPA1 transcripts were recovered from TDP-43 immunocomplexes but not from those prepared with an antibody for a non-endogenous target (FLAG, Fig. 2E). CAMK2A and TARDBP were used as positive controls, and HSPA4 was used as a negative control (Wang et al., 2008).
Figure 2.
TDP-43 binds and modulates HNRNPA1 mRNA. (A) Quantitative PCR with a probe set recognizing both hnRNP A1 and A1B (exon 1–2) or only hnRNP A1B (exon 7b-8) transcripts. Data was normalized to ACTN. (B) Quantitative PCR for hnRNP A1B transcripts, normalized to 18S, following actinomycin D treatment. (C) Luciferase assay using HeLa cells first treated with siRNA and then co-transfected with the reporter vectors containing the luciferase gene located downstream of the HNRNPA1 3′ UTR. GAPDH 3′ UTR and a random sequence (R03) serve as negative controls. GRN is included as positive control. (D) Luciferase assay using HeLa cells first treated with siRNA and then co-transfected with the reporter vectors containing the luciferase gene upstream of the promoter. The GAPDH promoter and random sequence (R01) serve as negative controls. (E) TDP-43 protein and its associated transcripts were co-immunoprecipitated from HeLa cells. TDP-43 bound mRNAs were extracted from the precipitates, reverse transcribed and amplified for HNRNPA1, TARDBP, CAMK2A and HSPA4. FLAG IP serves as a control. Dotted lines indicate that the samples are not contiguous lanes. For all, the mean ± SEM of three to four independent experiments is plotted. *P < 0.05, **P < 0.02.
TDP-43 influences HNRNPA1 pre-mRNA splice site selection
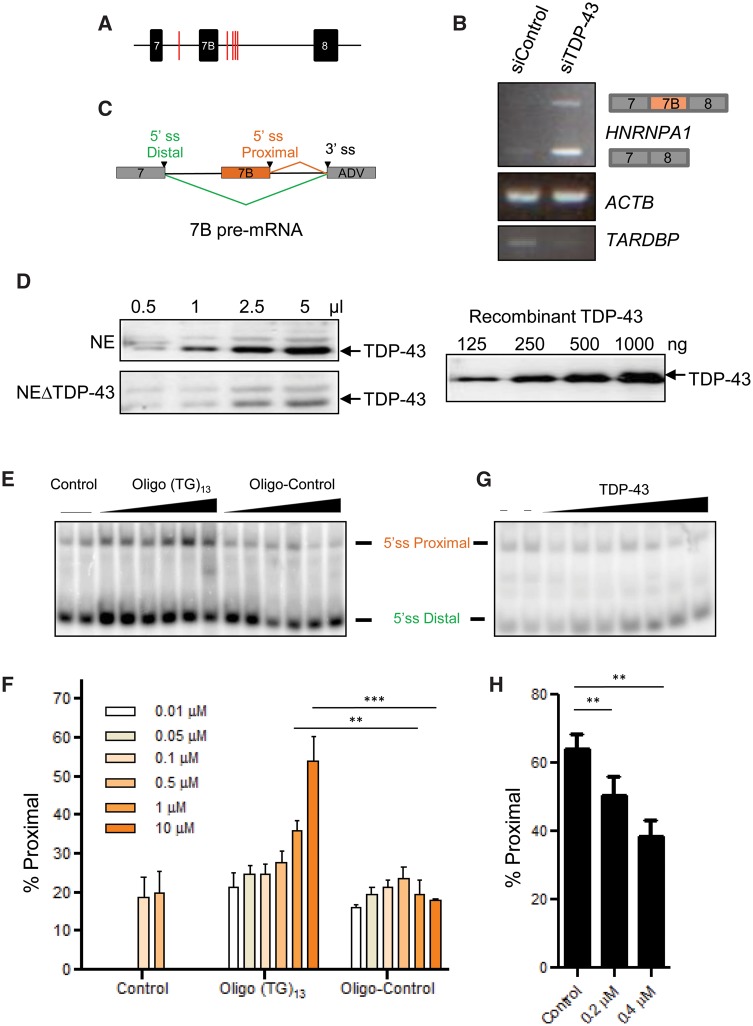
Previously, it has been demonstrated that TDP-43 depletion results in the differential splicing of more than 230 genes, favours UG/TG-rich sequences and is implicated in the alternative splicing of several genes implicated in neuronal development and neurological disease (Sephton et al., 2011; Tollervey et al., 2011; Colombrita et al., 2015; Mohagheghi et al., 2016). Given that HNRNPA1 is previously reported to be alternatively spliced (Chabot et al., 1997), and that our data indicated that TDP-43 can bind HNRNPA1 transcripts and influence their levels, we hypothesized that TDP-43 may influence HNRNPA1 alternative splicing. HnRNP A1B arises from inclusion of exon 7B, which encodes an additional 52 amino acids. Using RBPmap, a computational tool that predicts RNA binding protein binding sites, we identified potential TDP-43 binding sites in intron 7 (between exon 7 and 7B) as well as in intron 8 (between exon 7B and 8) (Paz et al., 2014) (Fig. 3A). Indeed, siRNA-mediated depletion of TDP-43 followed by semi-quantitative RT-PCR using primers flanking exon 7B show increased amounts of HNRNPA1 transcripts with exon 7B included (Fig. 3B, top), which is nearly undetectable in siControl lysates. In addition, and consistent with Fig. 2A, the band corresponding to HNRNPA1 with exon 7B excluded was also increased (Fig. 3B, bottom).
Figure 3.
TDP-43 influences HNRNPA1 pre-mRNA splice site selection in vitro. (A) RBPmap results demonstrate potential TDP-43 binding sites (red lines) in the introns upstream and downstream of exon 7B. (B) RT-PCR for endogenous hnRNP A1- and A1B-encoding transcripts from HeLa cells treated with control or TDP-43 siRNA. (C) Structure of the model HNRNPA1 pre-mRNA/mini-gene. The two 5′ splice sites (ss) of exon 7 and exon 7B compete for a unique 3′ splice sites (adenovirus). (D) Western blot for TDP-43 in varying volumes of HeLa nuclear extracts depleted or not of endogenous TDP-43 (left). Different amounts of recombinant TDP-43 protein were also loaded and detected simultaneously (right). (E and F) Spliced products were amplified by RT-PCR and separated on native PAGE gels. Oligo (TG)13 competes away the activity of endogenous TDP-43. The percentage of proximal splicing was quantified and expressed as mean ± SEM of three independent experiments, **P < 0.02, ***P < 0.005. (G and H) Addition of recombinant TDP-43 protein restores splicing activity in extracts prepared from cells depleted of TDP-43 by stable shRNA expression. The percentage of proximal splicing was quantified and expressed as mean ± SEM of four independent experiments. **P < 0.02.
To examine this in more detail, we used a model HNRNPA1 pre-mRNA (Blanchette, 1999) containing exon 7 and 7B and an adenoviral exon featuring a 3′ splice site (Fig. 3C). In an in vitro splicing assay using HeLa nuclear extracts with normal endogenous levels of TDP-43 (Fig. 3D, top left), the distal splice site of exon 7 is preferentially chosen and this can be competed with by increasing amounts of a TG-repeat oligonucleotide that sequesters TDP-43, but not by a non-targeting control oligonucleotide. Thus, the TG-repeat oligonucleotide shifts splice site selection towards the proximal 5′ splice site of exon 7B (Fig. 3E and F). To test if TDP-43 can repress the selection of the exon 7B 5′ splice site, we used a nuclear extract prepared from HeLa cells depleted of endogenous TDP-43 using inducible shRNA (Fig. 3D, bottom left). This nuclear extract displayed improved usage of the proximal 5′ splice site as compared to the extract containing normal levels of TDP-43 (first two lanes of Fig. 3G, quantified as ‘control’ in Fig. 3H). The addition of recombinant TDP-43 protein (Fig. 3B, right) led to a dose-dependent favouring of distal 5′ splice site (Fig. 3G and H). Thus, our in vivo and in vitro experiments indicate that TDP-43 represses the use of the 5′ splice site of exon 7B in the HNRNPA1 pre-mRNA.
Elongation of the prion-like domain confers greater fibrillization propensity in vitro
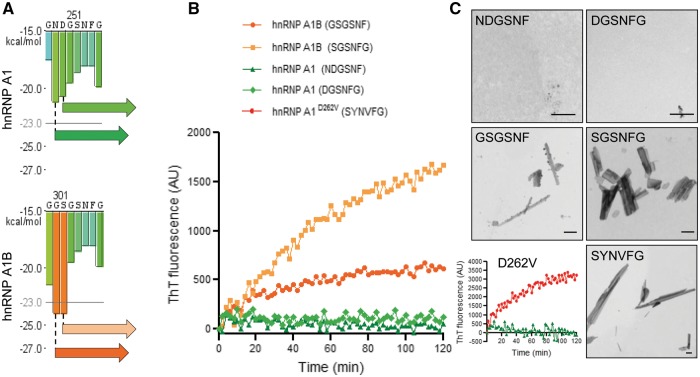
Both hnRNP A1 isoforms encode proteins with two N-terminal RRMs and a C-terminal Arg-Gly-Gly (RGG) repeat-rich domain referred to as the glycine-rich or prion-like domain (Bekenstein and Soreq, 2013). HnRNP A1B differs from hnRNP A1 uniquely by the inclusion of the cassette exon 7B, which encodes an additional 52 amino acids within the prion-like domain (Mayeda et al., 1994). The centre of this domain contains a steric zipper motif that is a critical contributor to the intrinsic tendency of hnRNP A1 to self-associate and form fibrils (Kim et al., 2013; Molliex et al., 2015). It has been previously reported that the disease-related mutation hnRNP A1D262V confers a stronger steric zipper motif, effectively accelerating fibril nucleation and polymerization (Kim et al., 2013). To determine if exon 7B inclusion and consequent elongation of the prion-like domain confers greater fibrillization propensity, we performed in silico analysis of both isoforms using ZipperDB (Goldschmidt et al., 2010), a structure-based algorithm that predicts fibril-forming segments. Specifically, hexapeptides with Rosetta energies <−23 kcal/mol are predicted to have increased fibrillization propensity (Goldschmidt et al., 2010). From this analysis, two hexapeptides (GSGSNF and SGSNFG) generated by exon 7B to 8 splicing were predicted to fibrillize (Fig. 4A and Supplementary Fig. 2). To test whether the fibrillization propensity of these peptides was increased as predicted, we performed a thioflavin T incorporation assay with these synthesized peptides. Thioflavin T, a benzothiazole dye, demonstrates increased fluorescence intensity when bound to β-sheet-rich deposits, such as the cross-β-sheet quaternary structure of amyloid fibrils (Hudson et al., 2009). As negative controls, we selected two hexapeptides, which arise from exon 7 splicing to exon 8 and have no fibrillization potential (Rosetta energies >−23 kcal/mol; NDGSNF, DGSNFG). Consistent with the in silico prediction, the two control peptides derived from hnRNP A1 did not incorporate thioflavin T. In contrast, as a positive control, the hexapeptide containing the ALS mutation D262V (SYNVFG) demonstrated robust thioflavin T incorporation, as expected (Kim et al., 2013) (Fig. 4B and C). Finally, as predicted from ZipperDB, both peptides derived from exon 7B inclusion (GSGSNF and SGSNFG) demonstrated increased thioflavin T incorporation (Fig. 4B), as well as fibrils that were readily visualized by transmission electron microscopy (Fig. 4C). These data indicate that peptides derived from the elongated hnRNP A1 variant, hnRNP A1B, have a greater in vitro fibrillization propensity compared to hnRNP A1-derived peptides.
Figure 4.
Fibrillization propensity of hnRNP A1B-derived peptides. (A) HnRNP A1 and hnRNP A1B were analysed using ZipperDB. Enlarged views of ZipperDB output illustrating the peptides generated by exon 7B inclusion and selected for further study. (B and C) Thioflavin T (ThT) incorporation assay indicates fibrillization of the peptides generated by (B) exon 7B inclusion or the ALS-causing mutation D262V. (C) Transmission electron micrographs demonstrating the fibrillization of the peptides. Data are representative of three to six experiments. Scale bar = 500 nm.
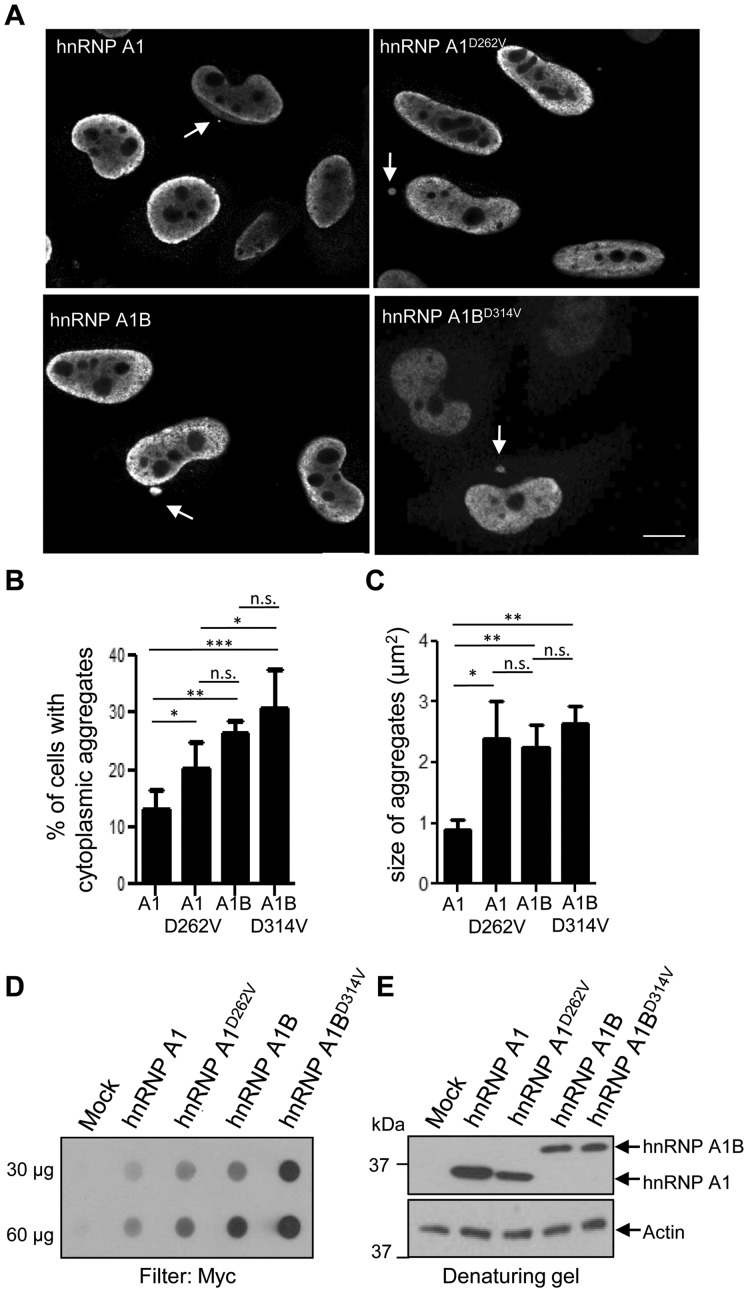
HnRNP A1B generates cytoplasmic aggregates comparable to an ALS-causing mutation
To determine if the increased fibrillization propensity observed in vitro relates to formation of cytoplasmic aggregates in vivo, we performed immunofluorescence on HeLa cells transfected with Myc-tagged hnRNP A1, hnRNP A1D262V, hnRNP A1B or the same mutation in hnRNP A1B (encoded as hnRNP A1BD314V). Cytoplasmic aggregates were detected in all transfected cells (Fig. 5A). As expected, cells expressing the ALS-linked mutation hnRNP A1D262V formed aggregates more often than hnRNP A1-expressing cells. However, quantification revealed that hnRNP A1B and hnRNP A1BD314V transfectants form cytoplasmic aggregates approximately twice as often as hnRNP A1 expressing cells (Fig. 5B). Moreover, the proportion of hnRNP A1B transfectants forming aggregates was comparable to cells expressing the rare mutation hnRNP A1D262V. We also noted that the cytoplasmic aggregates formed following hnRNP A1B expression (wild-type or D314V mutant) are approximately twice as large as those formed by exogenous hnRNP A1, and are of comparable size as those generated by cells expressing hnRNP A1D262V (Fig. 5C), consistent with our in vitro fibrillization data.
Figure 5.
The inclusion of exon 7B promotes aggregation in vivo. (A and B) HeLa cells were transfected with the indicated Myc-tagged cDNA and immunostained for Myc. Scale bar = 10 µm. Arrows indicate cytoplasmic aggregates. (B) Quantification of the percentage of cells with cytoplasmic aggregates and (C) quantification of the size of the aggregates. The mean ± SEM of 3–4 independent experiments is plotted, a minimum of 100 cells per genotype were analysed per experiment. *P < 0.05, ***P < 0.015. (D) HEK293FT cells were transfected with the indicated cDNA. Aggregation was assayed by filter retardation using non-binding cellulose acetate membranes. (E) Whole cell lysates were immunoblotted for Myc and actin. Data are representative of three independent experiments.
To extend these results biochemically, we used a filter retardation assay (filter trap) in which protein aggregates >0.2 µm are retained on a non-binding cellulose acetate membrane. Compared to HEK293FT cells transfected with an empty vector, lysates from Myc-hnRNP A1-expressing cells demonstrated a low basal level of retained material, which was exacerbated by the D262V mutation. Interestingly, expression of hnRNP A1B yielded levels of aggregated material comparable to those of hnRNP A1D262V, whereas inclusion of the same mutation in hnRNP A1B (encoded as hnRNP A1BD314V) greatly exacerbated protein aggregation (Fig. 5D). Immunoblotting of whole cell lysates indicated near equivalent expression of the various constructs (Fig. 5E). These results demonstrate that elongation of the prion-like domain in hnRNP A1 drives protein aggregation in vivo.
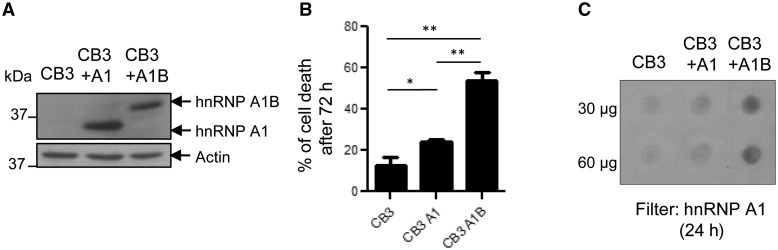
HnRNP A1B expression is cytotoxic
The cellular consequence(s) of hnRNP A1B expression remains unknown. To evaluate the effect of hnRNP A1B on cell survival, we took advantage of the CB3 cell line which lacks endogenous hnRNP A1 expression and CB3 subclones stably expressing exogenous hnRNP A1 or hnRNP A1B at comparable levels (Fig. 6A) (Ben-David et al., 1992; Yang et al., 1994). CB3 cells with stable hnRNP A1 expression demonstrate a modest, but significant increase in cell death compared to the parental CB3 cell line (P < 0.05). However, stable hnRNP A1B expression resulted in a 4.3-fold increase in cell death (Fig. 6B; P < 0.002). To determine if hnRNP A1B aggregation positively correlates with toxicity, we performed a filter trap assay 24 h post-seeding. We observed that CB3 cells expressing hnRNP A1B demonstrate more aggregated material than those expressing hnRNP A1 (Fig. 6C), consistent with our results in HEK293FT cells (Fig. 5D). Thus, exogenous expression of hnRNP A1B induces aggregate formation and is detrimental to cellular survival.
Figure 6.
HnRNP A1B expression causes cell death. (A) Whole lysates of CB3 cells, which lack endogenous hnRNP A1 or were modified to re-express one of the isoforms, were immunoblotted for hnRNP A1/A1B (9H10) and actin. (B) Viability of CB3 cell lines was assayed by trypan blue dye exclusion. The mean ± SEM of three independent experiments is plotted. *P < 0.05 **P < 0.002. (C) Aggregation was assayed by filter retardation using non-binding cellulose acetate membranes and immunoblotted for hnRNP A1/A1B (9H10). Data are representative of three independent experiments.
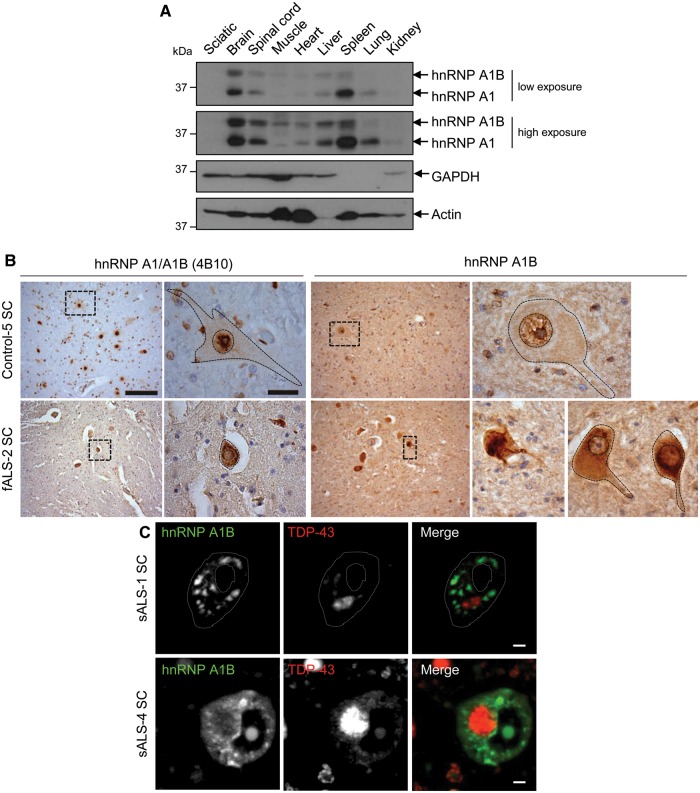
HnRNP A1B accumulates in the cytoplasm of ALS spinal motor neurons
Little is known about the physiological expression of hnRNP A1 and A1B. By immunoblotting of Sprague-Dawley rat tissue lysates, we found that hnRNP A1 is readily detected in brain and spleen, and is less abundant in all other examined tissues. HnRNP A1B shares a similar pattern but with highest levels observed in the brain (Fig. 7A). Assuming that the antibody (9H10) recognizes the two forms of hnRNP A1 with similar affinities, we estimate hnRNP A1B steady state protein levels to be ∼70% that of hnRNP A1 in CNS tissues. This is significantly higher than the distribution previously reported in HeLa cells where hnRNP A1B was reported as 5% of hnRNP A1 (Buvoli et al., 1990), but is consistent with other reports on rodent tissues (Hanamura et al., 1998; Liu et al., 2016). Neither isoform was detected in sciatic nerve.
Figure 7.
HnRNP A1B is expressed in the CNS and forms cytoplasmic aggregates in ALS patient neurons. (A) Rat tissue lysates were immunoblotted for hnRNP A1/A1B (9H10), GAPDH and actin. (B) Low magnification images of hnRNPA1 and hnRNP A1B, detected with antibody 4B10 (left) or a custom anti-hnRNP A1B antibody (right). Low magnification scale bar = 100 µm. Higher magnifications are from boxed regions in lower magnification images in B, scale bar = 10 µm. (C) Confocal images of spinal motor neurons from two ALS cases stained for hnRNP A1B and TDP-43. Scale bar = 5 μm.
To determine whether this same relationship between the presence of TDP-43 mislocalization and misregulation of HNRNPA1 splicing is relevant in ALS, we evaluated spinal motor neurons of five ALS cases in which TDP-43 pathology had been previously documented (Keller et al., 2012). Using an antibody that detects both hnRNP A1 and A1B (antibody 4B10), immunoreactivity was primarily in the nuclei of control patients. In contrast, antibody 4B10 demonstrated robust cytoplasmic labelling in ALS patient neurons (Fig. 7B). Using the antibody raised against a peptide unique to hnRNP A1B, we observed weaker nuclear labelling in control patient neurons coupled with a robust and intense cytoplasmic staining in all five ALS cases. The specificity of this labelling was confirmed by successful competition with the immunogenic peptide (Supplementary Fig. 1D). Immunofluorescence analysis of six ALS cases (three of which overlap with the immunohistochemistry study) revealed that hnRNP A1B was accumulated in multiple cytoplasmic inclusions in motor neurons depleted of nuclear TDP-43 and featuring round cytoplasmic TDP-43 inclusions in five of the six cases examined (Fig. 7C). Moreover, in all cases, TDP-43 and hnRNP A1B cytoplasmic inclusions did not co-localize. It is noteworthy that two of these cases are familial, thus implying that hnRNP A1B inclusions are broadly relevant to ALS. Interestingly, in one case where TDP-43 presented as a cytoplasmic skein-like inclusion, hnRNP A1B cytoplasmic labelling was absent. In conclusion, hnRNP A1B is enriched in CNS tissues and its accumulation in ALS motor neurons positively correlates with, but does not co-localize with, TDP-43 cytoplasmic inclusions.
Discussion
TDP-43 plays multiple roles in RNA metabolism including transcription, miRNA processing, stress granule dynamics, RNA transport, nucleocytoplasmic shuttling and splicing (Sephton et al., 2011; Aulas et al., 2012; Scotter et al., 2015; Mohagheghi et al., 2016). However, it remains unclear whether the cytoplasmic mislocalization/aggregation of TDP-43 observed in most ALS and nearly half of FTD cases results in a gain and/or loss of function (Wang et al., 2008; Lee et al., 2012; Conicella et al., 2016). Here, we demonstrate that the loss of nuclear TDP-43, either via cellular depletion or by cytoplasmic mislocalization, impacts the closely related RNA binding protein, hnRNP A1. Specifically, we found that TDP-43 binds HNRNPA1 transcripts but does not influence mRNA stability. Instead, we found that TDP-43 modulated 5′ splice site selection of HNRNPA1 pre-mRNA such that TDP-43 depletion favoured exon 7B inclusion, resulting in elevated levels of the longer splice variant hnRNP A1B. That this change in the ratio between hnRNP A1/A1B isoforms was not detected in previously reported RNA-Seq experiments could be due to the inherent limitation of RNA-Seq approaches to detect low abundance transcripts such as hnRNP A1B (Labaj et al., 2011). The similarity in the change of hnRNP A1B levels between total TDP-43 depletion and cytoplasmic mislocalization/nuclear depletion are consistent with recent data demonstrating that reducing cellular TDP-43 levels and TDP-43 cytoplasmic localization have comparable effects (Prpar Mihevc et al., 2016). Interestingly, we also noted that TDP-43 may function as a transcriptional repressor (or sequester an enhancer) of HNRNPA1 via the promoter; this is worthy of future exploration.
The presence of the low abundance hnRNP A1B protein produced by an alternative splicing event was reported 25 years ago, but little is understood about its biological significance in healthy or pathological conditions (Buvoli et al., 1990). Here we demonstrated that exon 7B inclusion increases fibrillization propensity in vitro and drives protein aggregation in vivo. In fact, hnRNP A1B forms cytoplasmic aggregates similar in abundance to those formed by the ALS-causing mutation hnRNP A1D262V, although they were noted to be 2-fold larger. Liquid-liquid phase separation, mediated by the low complexity domains of RNA binding proteins, is an emerging molecular mechanism proposed to explain the formation of histological inclusions (Lin et al., 2015; Molliex et al., 2015; Patel et al., 2015; Conicella et al., 2016). Specifically, it is proposed that high local concentration of prion-like domain/low complexity domain-containing proteins initiates phase separation to give rise to ribonucleoprotein (RNP) granules, the maturation of which is limited by fibril formation. Furthermore, it is proposed that these structures can convert to pathological RNP granules if fibril formation is excessive (Lin et al., 2015). It has been demonstrated that hnRNP A1 undergoes liquid-liquid phase separation and can assemble into hydrogels composed of cross-β fibrils (Molliex et al., 2015). Although it remains to be demonstrated, we would predict that hnRNP A1B can similarly undergo phase separation. Our data indicate that loss of nuclear TDP-43 shifts HNRNPA1 alternative splicing. This increases the levels of hnRNP A1B, a protein that is inherently prone to fibril/aggregate formation, and thus may be a contributing factor in the evolution of RNP granules into pathological inclusions.
Cytoplasmic hnRNP A1 inclusions have previously been reported in post-mortem ALS cases (Kim et al., 2013; Kanekura et al., 2016). However, the antibody used in these previous studies is unable to differentiate between the shorter, more abundant hnRNP A1 protein and the longer, less abundant hnRNP A1B protein. Here, using an antibody that uniquely detects hnRNP A1B, we demonstrate that within ALS motor neurons bearing round TDP-43 cytoplasmic inclusions, hnRNP A1B is mislocalized to the cytoplasm and accumulates as aggregates that do not contain TDP-43. Intriguingly, novel disease-related mutations in exon 7B have recently been reported in ALS patients with flail arm syndrome (Paz et al., 2014). That these disease-causing mutations are confined to the alternatively spliced exon 7B is significant and implies that hnRNP A1B dysfunction is relevant. While the physiological function of hnRNP A1B remains to be demonstrated, we observed that increasing hnRNP A1B protein levels positively correlated with protein aggregation. Indeed, in cells where the endogenous HNRNPA1 gene was inactivated, hnRNP A1B was much more toxic than hnRNP A1.
TDP-43 and hnRNP A1 are two important RNA binding proteins broadly implicated in RNA metabolism, each of which can bind thousands of transcripts (Tollervey et al., 2011; Ling et al., 2013; Bruun et al., 2016). Any alteration in steady-state protein levels, localization and/or ability to interact with protein or RNA targets would be predicted to have consequences on up to two-thirds of the transcriptome (Sephton et al., 2011; Tollervey et al., 2011; Bekenstein and Soreq, 2013; Jean-Philippe et al., 2013; Colombrita et al., 2015; Bruun et al., 2016; Geissler et al., 2016). Although the function of hnRNP A1B is unknown, it is reported to have a higher binding affinity for single stranded DNA (Buvoli et al., 1990). Thus, while it is probable that hnRNP A1B will have the same or similar functions as hnRNP A1, its mRNA targets may differ. Alternatively, or in addition, since the prion-like domain typically mediates protein–protein interactions, it is possible that shifting the ratio between hnRNP A1 and hnRNP A1B could have consequences on downstream pathways. A recent report suggests that VCP, hnRNP A1, FUS and TDP-43 constitute the core protein-protein interaction network in classical ALS (Mao et al., 2017). That these four proteins could be central in the development of ALS pathology is because of the finding that they interact with the majority of proteins implicated in the disease (Mao et al., 2017). Since TDP-43 is mislocalized in most ALS cases and half of all FTD cases, studying the targets and the function of hnRNP A1B may provide further insight towards dissecting the network of RNA binding proteins and downstream pathways that contribute to ALS pathogenesis.
Collectively, our results show that the loss of nuclear TDP-43 contributes to cellular vulnerability and loss via alterations in HNRNPA1 splicing. The resulting splice variant hnRNP A1B demonstrates an increased propensity to aggregate and enhanced toxicity. We speculate that this isoform disturbs RNA metabolism much more broadly than previously expected and is an important contributor to the cascade of events that drives ALS.
Supplementary Material
Acknowledgements
We thank the patients and families for contributing tissues to these studies. We also thank N. Arbour for access to the multi-mode plate reader, and A. Prat and the CRCHUM Cell Imaging platform for access to confocal microscopy.
Funding
This work was supported by an NSERC Discovery grant (C.V.V.) and the ALS Society of Canada – Brain Canada Arthur J. Hudson Translational Team grant (C.V.V. and M.J.S.). B.C. was supported by CIHR grant MOP-136948 and is the Pierre C. Fournier Chair in Functional Genomics. J.E.D. and H.S. were partially supported by recruitment awards from the Université de Montréal Faculty of Medicine. Q.D. was supported by A*MIDEX (ANR-11-IDEX-0001-02). HSoreq was supported by the Israeli Ministry of Science, Technology and Space, Grant No. 53140, and the Legacy Heritage Science Initiative (LHSI) of The Israel Science Foundation Grant No. 817/13; U.B. was supported by the Howard and Diana Wendy pre-doctoral fellowship. CVV is a CIHR New Investigator.
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- FTD
frontotemporal dementia
References
- Aulas A, Stabile S, Vande Velde C. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol Neurodegener 2012; 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekenstein U, Soreq H. Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles. Mol Cell Neurosci 2013; 56: 436–46. [DOI] [PubMed] [Google Scholar]
- Ben-David Y, Bani MR, Chabot B, De Koven A, Bernstein A. Retroviral insertions downstream of the heterogeneous nuclear ribonucleoprotein A1 gene in erythroleukemia cells: evidence that A1 is not essential for cell growth. Mol Cell Biol 1992; 12: 4449–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J 1999; 18: 1939–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5’ splice site region of hnRNP A1 alternative exon 7B. RNA 1997; 3: 405–19. [PMC free article] [PubMed] [Google Scholar]
- Bruun GH, Doktor TK, Borch-Jensen J, Masuda A, Krainer AR, Ohno K, et al. Global identification of hnRNP A1 binding sites for SSO-based splicing modulation. BMC Biol 2016; 14: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvoli M, Cobianchi F, Bestagno MG, Mangiarotti A, Bassi MT, Biamonti G, et al. Alternative splicing in the human gene for the core protein A1 generates another hnRNP protein. EMBO J 1990; 9: 1229–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5’ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol 1997; 17: 1776–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-C, Chang JG, Liu TY, Jong YJ, Cheng WL, Yuo CY. Securinine enhances SMN2 exon 7 inclusion in spinal muscular atrophy cells. Biomed Pharmacother 2017; 88: 708–14. [DOI] [PubMed] [Google Scholar]
- Colombrita C, Onesto E, Buratti E, de la Grange P, Gumina V, Baralle FE, et al. From transcriptomic to protein level changes in TDP-43 and FUS loss-of-function cell models. Biochim Biophys Acta 2015; 1849: 1398–410. [DOI] [PubMed] [Google Scholar]
- Conicella AE, Zerze GH, Mittal J, Fawzi NL. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 2016; 24: 1537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 1983; 11: 1475–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler R, Simkin A, Floss D, Patel R, Fogarty EA, Scheller J, et al. A widespread sequence-specific mRNA decay pathway mediated by hnRNPs A1 and A2/B1. Genes Dev 2016; 30: 1070–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA 2010; 107: 3487–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Cáceres JF, Mayeda A, Franza BR, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 1998; 4: 430–44. [PMC free article] [PubMed] [Google Scholar]
- Hudson SA, Ecroyd H, Kee TW, Carver JA. The thioflavin T fluorescence assay for amyloid fibril detection can be biased by the presence of exogenous compounds: exogenous compounds can bias thioflavin T assays. FEBS J 2009; 276: 5960–72. [DOI] [PubMed] [Google Scholar]
- Jean-Philippe J, Paz S, Caputi M. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci 2013; 14: 18999–9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekura K, Yagi T, Cammack AJ, Mahadevan J, Kuroda M, Harms MB, et al. Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum Mol Genet 2016; 25: 1803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 2003; 34: 460–63. [DOI] [PubMed] [Google Scholar]
- Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet 2007; 16: 3149–59. [DOI] [PubMed] [Google Scholar]
- Keller BA, Volkening K, Droppelmann CA, Ang LC, Rademakers R, Strong MJ. Co-aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism. Acta Neuropathol 2012; 124: 733–47. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013; 495: 467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labaj PP, Leparc GG, Linggi BE, Markillie LM, Wiley HS, Kreil DP. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics 2011; 27: i383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 2010; 19: R46–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EB, Lee VM-Y, Trojanowski JQ. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci 2012; 13: 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 2015; 60: 208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S-C, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 2013; 79: 416–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Shu S, Wang RR, Liu F, Cui B, Guo XN, et al. Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurology 2016; 87: 1763–9. [DOI] [PubMed] [Google Scholar]
- Mao Y, Kuo S-W, Chen L, Heckman CJ, Jiang MC. The essential and downstream common proteins of amyotrophic lateral sclerosis: a protein-protein interaction network analysis. PLoS One 2017; 12: e0172246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Cáceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J 1994; 13: 5483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohagheghi F, Prudencio M, Stuani C, Cook C, Jansen-West K, Dickson DW, et al. TDP-43 functions within a network of hnRNP proteins to inhibit the production of a truncated human SORT1 receptor. Hum Mol Genet 2016; 25: 534–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015; 163: 123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314: 130–3. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 2015; 162: 1066–77. [DOI] [PubMed] [Google Scholar]
- Paz I, Kosti I, Ares M, Cline M, Mandel-Gutfreund Y. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res 2014; 42: W361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prpar Mihevc S, Baralle M, Buratti E, Rogelj B. TDP-43 aggregation mirrors TDP-43 knockdown, affecting the expression levels of a common set of proteins. Sci Rep 2016; 6: 33996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotter EL, Chen H-J, Shaw CE. TDP-43 proteinopathy and ALS: insights into disease mechanisms and therapeutic targets. Neurotherapeutics 2015; 12: 352–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem 2011; 286: 1204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong MJ, Abrahams S, Goldstein LH, Woolley S, Mclaughlin P, Snowden J, et al. Amyotrophic lateral sclerosis—frontotemporal spectrum disorder (ALS-FTSD): revised diagnostic criteria. Amyotroph Lateral Scler Front Degener 2017; 18: 153–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci 2011; 14: 452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I-F, Wu L-S, Chang H-Y, Shen C-KJ. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem 2008; 105: 797–806. [DOI] [PubMed] [Google Scholar]
- Weishaupt JH, Hyman T, Dikic I. Common molecular pathways in amyotrophic lateral sclerosis and frontotemporal dementia. Trends Mol Med 2016; 22: 769–83. [DOI] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM-Y. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem 2008; 283: 13302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Sanelli T, Chiang H, Sun Y, Chakrabartty A, Keith J, et al. Low molecular weight species of TDP-43 generated by abnormal splicing form inclusions in amyotrophic lateral sclerosis and result in motor neuron death. Acta Neuropathol 2015; 130: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Bani MR, Lu SJ, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5’ splice site selection in vivo. Proc Natl Acad Sci USA 1994; 91: 6924–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.